Abstract
Introduction
Cattle temperament is an important factor that affects the profitability of beef cattle enterprises, due to its relationship with productivity traits, animal welfare and labor safety. Temperament is a complex phenotype often assessed by measuring a series of behavioral traits, which result from the effects of multiple environmental and genetic factors, and their interactions. The aims of this study were to perform a genome-wide association study and detect genomic regions, potential candidate genes and their biological mechanisms underlying temperament, measured by flight speed (FS) test in Nellore cattle.
Materials and Methods
The genome-wide association study (GWAS) was performed using a single-step procedure (ssGBLUP) which combined simultaneously all 16,600 phenotypes from genotyped and non-genotyped animals, full pedigree information of 162,645 animals and 1,384 genotyped animals in one step. The animals were genotyped with High Density Bovine SNP BeadChip which contains 777,962 SNP markers. After quality control (QC) a total of 455,374 SNPs remained.
Results
Heritability estimated for FS was 0.21 ± 0.02. Consecutive SNPs explaining 1% or more of the total additive genetic variance were considered as windows associated with FS. Nine candidate regions located on eight different Bos taurus chromosomes (BTA) (1 at 73 Mb, 2 at 65 Mb, 5 at 22 Mb and 119 Mb, 9 at 98 Mb, 11 at 67 Mb, 15 at 16 Mb, 17 at 63 Kb, and 26 at 47 Mb) were identified. The candidate genes identified in these regions were NCKAP5 (BTA2), PARK2 (BTA9), ANTXR1 (BTA11), GUCY1A2 (BTA15), CPE (BTA17) and DOCK1 (BTA26). Among these genes PARK2, GUCY1A2, CPE and DOCK1 are related to dopaminergic system, memory formation, biosynthesis of peptide hormone and neurotransmitter and brain development, respectively.
Conclusions
Our findings allowed us to identify nine genomic regions (SNP windows) associated with beef cattle temperament, measured by FS test. Within these windows, six promising candidate genes and their biological functions were identified. These results may contribute to a better comprehension into the genetic control of temperament expression in Nellore cattle.
Introduction
Cattle temperament is operationally defined as the set of an animal’s behaviors in response to handling by humans, attributing these reactions to general aspects of fearfulness [1, 2]. This concept is closely related to individual differences of reactivity in stressful or challenging situations [3, 4]. There is a growing interest on cattle temperament due to its association with productivity traits, animal welfare and labor safety [5, 6], stimulating cattle producers and researchers to look for a better understanding of genetic and environmental factors that influence the expression of temperament traits, and its impacts on cattle performance and corporate profitability.
Flight speed (FS) test is among the most used behavioral methods to assess beef cattle temperament, reflecting aspects of general fear and agitation [2, 7]. This test was validated in many studies, some of them based on physiological stress indicators [8–10], besides being proved that it is reliable and repeatable in both, the short and long term [11, 12]. Additionally, this method has been described as objective, safe and simple to implement routinely on farm, besides the possibility of integrating this measurement in animal breeding programs requiring minimal additional costs [13–15].
Heritability estimates for FS are moderate to high, ranging from 0.17 ± 0.07 to 0.54 ± 0.16 for Bos taurus [14, 15] and Bos indicus breeds [16–18]. Additionally, a series of studies have shown favorable genetic correlations of FS with important economic traits, e.g. better temperament (lower FS or higher flight time) was associated with higher average daily gain (ADG) [14, 19], better reproductive performance [20, 21], higher carcass weight and yield [22], and improved meat tenderness [23]. Based on these one can assume that the selection for temperament using FS is feasible and will not produce unfavorable responses on overall performance traits.
Given its phenotypic complexity and, probably, polygenic nature, temperament could potentially be studied using genomic approaches, such as genome-wide association studies (GWAS), in order to identify genomic regions or QTL that affect the underlying genetic architecture related to individual variability [24]. In fact, there were some efforts to identify QTL that account for the variation on beef and dairy cattle temperament. These findings led to several candidate genes distributed throughout the chromosomes, which were associated with a variety of temperament indicators in Bos taurus cattle and their crosses, such as: milking temperament [25, 26], isolation temperament and habituation [27], docility [28], flight from feeder and social separation [29], tethering test, weighting test, separation and restraint test [30, 31].
Despite of these initiatives, the use of GWAS to identify candidate regions of the genome associated with temperament of Zebu cattle (and others tropically adapted beef breeds) is still scarce in the literature. We found only two published papers using GWAS for beef cattle temperament, one of them aimed to identify molecular markers associated with temperament at weaning, measured by subjective social separation score in Nellore-Angus crossbred cattle [32]. In the second, a preliminary GWAS study was conducted for FS using BovineSNP50 genotypes in several European crosses (Angus, Hereford, Simmental, Limousin, Charolais, Gelbvieh and Red Angus) [33].
Zebu breeds and their crosses are commonly used for extensive cattle production in warm climates, usually with few contact with humans. Under these conditions, cattle becomes more excitable, impoverishing the animal welfare and increasing risks of labor accidents [5]. Given the importance of assessing cattle temperament and the rapid development of genomic tools, we decided to perform a genome-wide association study aiming to detect genomic regions, potential candidate genes and their biological mechanisms underlying temperament expression (measured by FS test) in Nellore cattle.
Materials and Methods
Animals and management
The Committee of Ethical Use of Animals from the Faculty of Agricultural and Veterinary Sciences, São Paulo State University, Jaboticabal—SP, Brazil (Certified n. 0016/14), approved this research.
Data collection was conducted in a private Nellore herd belonging to Agropecuária Jacarezinho Ltda®, which has two production units located in the southeast and the northeast regions of Brazil. The field study in both production units was permitted by Dr. Ian Hill, the Chief Executive Officer of Agropecuária Jacarezinho Ltda®. The handling procedures were similar in both units. Soon after birth, calves were assigned in handling groups according to the sex, keeping in the same group until weaning (artificially done when the calves were, approximately, 210 days old).
The first performance evaluation is conducted at weaning age, considering weight and visual scores for conformation, finishing precocity and muscling. These data is combined in an index, which is used to define the animals that would stay in the herd after weaning or not (remaining 50% of males and 90% of females). The selected animals are relocated in new handling groups, remaining in the same group from weaning to yearling (approximately 550 days of age), when a second performance evaluation is conducted. This is done by repeating the previous measurements (weaning and scoring each animal for conformation, finishing precocity and muscling), besides measuring scrotal circumference, scoring temperament and assessing breed characteristics. Based on these measurements obtained at weaning and yearling age a new selection index is calculated, and the results are used to define the animals to be culled (50% of males and 10% of females). Breed characteristics and temperament score are not included in the index, however they are assumed as independent criteria, discarding animals with the worst grade for each trait. The pedigree data set included 162,645 animals with 49,597 dams and 1,306 sires. A summary of the data set is shown in Table 1.
Table 1. Summary of data set structure and descriptive statistics for flight speed (FS) phenotypes, genotyped animals and pedigree using single-step genomic-BLUP methodology in Nellore cattle.
| Number of animals in the relationship matrix | 162,645 |
| Number of sires | 1,306 |
| Number of dams | 49,597 |
| Number of phenotyped animals | 16,660 |
| Number of CGa | 735 |
| Number of SNPs after quality control | 455,374 |
| Number of genotyped animals remaining after quality control | 1,384 |
aCG, contemporary group.
Assessment of cattle temperament
Flight speed (FS) test was “devised to measure the time taken for an animal to cover a set distance after leaving a confined area” [17]. In this study the measurements were carried out when each animal left the cattle crush after weighing. The time taken by each animal to cover a known distance (ranging from 1.25 to 3.0 m, according to the facilities design) was recorded with an electronic device, comprising two pairs of photoelectric cells, a stopwatch and a processor. Time and distance were used to calculate FS phenotype, in m/s. The assumption of this test is that higher speed animals have more excitable temperament. The FS records were taken from 16,660 animals that were born between 2008 and 2012. This test was carried out simultaneously with the performance evaluation at yearling, trying to minimize the interference in the farms handling routines. In the present dataset, FS ranged from 0.08 to 6.67 m/s, with an average (± SD) of 2.35 ± 1.04 m/s.
Genotype information and quality control
A total of 1,405 phenotyped animals were genotyped with high density bead array technology: Illumina Infinium HD Assay® and Illumina HiScan System®. The High Density Bovine SNP BeadChip contains 777,962 SNP markers spread across the genome at a mean distance of 3.43 kb between markers. Genotyped animals, born between 2008 and 2009, were randomly selected following the criteria of at least 5 animals in each contemporary group. As part of quality control (QC), the following were excluded: SNP markers with unknown genomic position, located on sex chromosomes, monomorphic and markers with minor allele frequency (MAF) bellow 0.05, call rate bellow 90%, with excess markers heterozygous genotypes and those who had a mean intensity of the low cluster. Individuals with call rate bellow 90% were also excluded. After QC, a total of 455,374 SNPs and 1,384 genotyped animals remained.
Statistical analysis
All analyzes, to estimate variance components and genetic parameters as well as the genome-wide association study (GWAS), were performed using a single-step procedure (single-step GBLUP—ssGBLUP) which combined, simultaneously, all phenotyped animals, pedigree information and genotypes in one step [34], using Bayesian inference [35] via Gibbs sampling. The animal model for FS included direct additive genetic and residual effects as random, and contemporary groups (CG: farm and year of birth, sex, farm at yearling and management groups at birth, weaning and yearling) as fixed effect. Age of animal at the time of temperament assessment was included as a covariate with linear and quadratic effects. The CG containing less than five animals and records out of the range given by the mean of the CG 3 SD were excluded from the analyses. To perform the statistical analyses, the GIBBS2F90 software was used [36] and the model can be represented in matrix form as:
where is y the vector of FS observations; is β the vector of fixed effects; a is the vector of direct additive genetic effects, assuming , where is H the relationship coefficients matrix among animals and genetic additive variance; X is the corresponding incidence matrix for the fixed effects; Z is the incidence matrix of the random additive direct genetic effects (associates a with vector y); e is the vector of the residual effect in which , where I is the identity matrix and is the residual variance. The prior distribution for genetic and residual variance components was an inverted Wishart and the posterior estimates were obtained using the POSTGSF90 program [37].
The inverse of H matrix was obtained as [38]:
where is the inverse of numerator relationship matrix for genotyped animals and G−1 is the inverse of the genomic relationship matrix. The G matrix was constructed weighting each SNP effect by its expected variance in an iterative procedure, following [39]:
In the G matrix, Z is the marker incidence matrix containing genotypes (0, 1 or 2) adjusted for allele frequency, D is a diagonal matrix with the inverse of expected SNP variance (initially D = I), and q is a weighting factor. The weighting factor was as in Vitezica et al. [40], ensuring that the average diagonal in G is close to that of A22.
Chains of 800,000 iterations were generated (S1 File) and the first 50,000 iterations were discarded. For parameter estimates, samples were kept each 100 cycles, generating chains with 7,500 cycles. Data convergence was checked through graphical analysis, sampled values versus rounds, and using the criteria proposed by Geweke [41], Heidelberger and Welch [42] and, Raftery and Lewis [43] using the R software, with Bayesian Output Analysis (BOA) package from R 2.9.0 software (The R Development Core Team 2009).
The ssGBLUP method has been used to GWAS [44–46], based on the SNP effects obtained using an iterative process, which increases the weight of SNPs with large effects and reduces the weight of SNPs with small effects as presented by Wang et al. [34]:
First step: D = I.
Calculate G matrix: G = ZDZ′q.
Calculate GEBV for all animals in the data set using ssGBLUP.
Convert GEBV to SNP effects: , where is the vector of marker effect and is the GEBV of the animals which were also genotyped.
Calculate the weight for each SNP marker: where i is the i-th SNP.
Normalized SNP weight to remain constant the total genetic variance.
Exit or Loop to step 2.
According to Habier et al. [47], the use of SNP windows captures the QTL effect better than a single SNP and is important to discriminate effects from statistical noise [48]. The iterative process was repeated three times, from step 2 to 7, and the percentage (%) of total genetic variance explained by i-th consecutive SNPs (SNP window) was calculated following Wang et al. [45]:
where ai is genetic value of the i-th region that consist of 10 consecutive SNPs, is the total genetic variance, Zj is vector of gene content of the j-th SNP for all individuals, and is marker effect of the i-th SNP within the i-th region (S2 File).
Gene prospection
Consecutive SNPs which explained 1% or more of the total genetic variance were considered as genomic window associated to FS. These regions were used to identify positional candidate genes based on the starting and ending coordinates of each window on Bos taurus genome view in the Map Viewer tool available at the National Center for Biotechnology Information (NCBI) platform (http://www.ncbi.nlm.nih.gov) UMD 3.1 version. Functional annotations were obtained using Ensembl Genome Browser (http://www.ensembl.org/index.html) and gene networks were explored by the online Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7 (http://david.abcc.ncifcrf.gov/). The identification of gene pathways and enrichment analysis (Gene Ontology and KEEG pathways) were performed in DAVID website from the Ensembl annotation. Human genes were used as background in pathway and gene network investigation. A Manhattan plot was created using the R package “ggplot2” (S3 File).
Results and Discussion
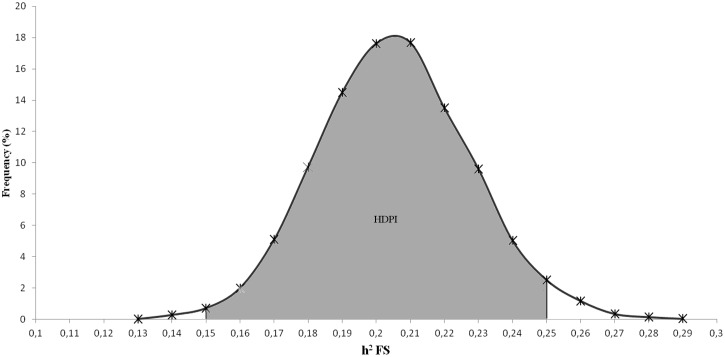
The convergence criteria used in this study, number of cycles, fixed burn-in period and number of Markov chains were sufficient for convergence of the estimated parameter. Fifty samples were discarded as a fixed burn-in period and the 7,950 interactions remained stable allowing their convergence. Additionally, the convergence was achieved with low SD and a relatively short 95% highest posterior density interval. As a result, the additive genetic () and residual variances (), and mean of posterior heritability (h2) for FS were 0.17, 0.65 and 0.21 ± 0.02, respectively. Fig 1 illustrates graphically the posterior distributions of heritability for FS, showing the highest probability density interval (HPDI) that ranged from 0.16 to 0.25.
Fig 1. Estimated posterior density of flight speed (FS) heritability in Nellore cattle.
The highest probability density interval (HPDI) is presented in gray region.
The heritability estimate (0.21 ± 0.02) is in accordance with the range described in the literature for Nellore and other beef breeds [14, 17, 18, 23] and was similar to that previously presented by our research group using a subset of these data, but not considering genomic information [15, 21]. The additive genetic variability presented by FS indicates the possibility of selecting to improve cattle temperament.
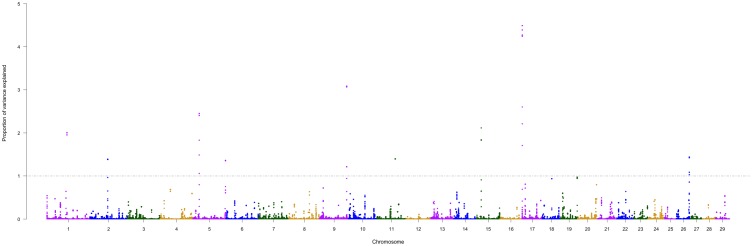
In recent years a large number of potential candidate genes have been associated with several traits of economic importance in farm animals [49, 50]. However, the selection of useful markers for specific traits, such as bovine temperament, should be based on knowledge of the relationship between physiological and/or biochemical process with variation of the phenotypic traits of interest [51]. The additive genetic variance explained by each 10-SNP moving windows is shown in a Manhattan plot (Fig 2). A summary of each SNP window that explained more than 1% of additive genetic variance and the candidate genes is shown in Table 2.
Fig 2. Manhattan plot of additive genetic variance explained by windows of 10 adjacent SNPs for flight speed (FS) in Nellore cattle.
Table 2. Summary of SNP windows that explained >1% of genetic variance for flight speed (FS) in Nellore cattle, with a list of annotated genes within each window.
| Chr | SNP windowa | Var (%)b | Genec | Gene ID | Gene Location | |
|---|---|---|---|---|---|---|
| Start, bp | Stop, bp | |||||
| 1 | 73354330 | 73406566 | 2.00 | --- | --- | --- |
| 2 | 65072013 | 65082628 | 1.39 | NCKAP5 | 786958 | 64102635–65215462 |
| 5 | 22596661 | 22604723 | 2.45 | --- | --- | --- |
| 5 | 119287445 | 119302785 | 1.36 | --- | --- | --- |
| 9 | 98759214 | 98767952 | 3.08 | PARK2 | 530858 | 98612089–99648791 |
| 11 | 67385287 | 67404876 | 1.40 | ANTXR1 | 616010 | 67334564–67590531 |
| 15 | 16598639 | 16662233 | 2.12 | GUCY1A2 | 613600 | 16604381–17100655 |
| 17 | 639678 | 671693 | 4.49 | CPE | 280753 | 546402–697925 |
| 26 | 47061401 | 47095621 | 1.44 | DOCK1 | 537203 | 46735083–47296206 |
aWindows with 10 consecutive SNPs based on UMD3.1.
bPercentage of additive genetic variance explained by each SNP window.
cCandidate genes associated to flight speed.
Using the Bos taurus genome map, six known genes were found within the significant regions (SNP windows) identified in our study. These were NCKAP5, PARK2, ANTXR1, GUCY1A2, CPE and DOCK1 located on the Bos taurus chromosomes (BTA) 2, 9, 11, 15, 17 and 26 respectively.
The CPE (carboxypeptidase E) gene encodes a multifunctional peripheral protein that plays several non-enzymatic roles in the endocrine tissues and central nervous systems, with the highest concentration found in the brain and pituitary gland [52, 53]. Moreover, it is a prohormone-processing enzyme, which is involved in the biosynthesis of numerous peptide hormones and neurotransmitters [54, 55], including the processing of proinsulin to insulin [56]. The enzyme carboxypeptidase E is responsible for cleaving the C-terminal amino acid residues from peptide intermediates in endocrine cells and neuropeptides in peptidergic neurons [57]. This gene is conserved in different species [53] and polymorphisms in specific regions have been associated with diabetes type 2 and obesity in humans [55, 56]; obesity and infertility in mice [53, 58] and in pigs [59]; with carcass and meat quality in different cattle breeds [51, 60]. The CPE expression in the brain is directly influenced by distinct physiological or environmental stress conditions to which animals are exposed, resulting in different levels of hippocampal, cortex and amygdala neuronal degeneration and neuronal survival suggesting a role in neuroprotection [61]. Additionally, this gene was linked to deficient learning and memory [62], modulation of mood and emotional responses [57] in mice.
The functions described above for CPE support the correlation estimates presented by Hoppe et al. [14], Sant’Anna et al. [15] and Kadel et al. [22] which reported association of FS with productive traits in beef cattle, such as growth and carcass and meat quality. Additionally, these functions corroborate the relationship of temperament with learning, memory and emotional responses in human [63, 64] and cattle [65]. Thus, CPE gene is a potential candidate for future studies aiming to identify specific molecular markers associated with different levels of reactivity in cattle.
The PARK2 gene, located on BTA9 at 98 Mb, encodes the parkin protein, an ubiquitin-protein ligase (E3) which belongs to ubiquitin-proteasome system. According to Hase et al. [66] PARK2 is expressed in bovine peripheral nerves and brain. These authors showed, by immunohistochemical evidence, that parkin is located in bovine axoplasm of myelinated nerve fibers and the Schwann cell outer membrane. The parkin protein works as important machinery for intracellular quality control [67–69], marking and degrading misfolded proteins. Mutations in PARK2 gene affect mitochondrial function and apoptosis in neuronal cells [70], and have been linked with Parkinson [71–73], which is a neurodegenerative disease. Damage in the parkin protein function is associated with selective degeneration of dopaminergic neurons [66, 68]. The role of dopamine as a neurotransmitter in central nervous system is well known [74], where its action is mediated by specific receptors to control locomotion, emotional behavior, cognitive functions and memory [75]. Indeed, the dopamine receptor D4 gene (DRD4) has been extensively linked to behavior, positive affect and reactivity in mice [76], dogs [77, 78], and horses [79, 80], as well as in humans [81–83] and cattle temperament, measured by a docility test [31].
The GUCY1A2 gene, located on BTA15 at 16 Mb, encodes an alpha subunit of guanylate cyclase (GC), characterized as guanylate cyclase 1, soluble, alpha 2. The GC is activated by nitric oxide (NO) and catalyzes the conversion of intracellular guanosine-5’-triphosphate (GTP) to cyclic guanosine-3',5'-monophosphate (cGMP), a second messenger in signaling of intracellular transduction pathways [84, 85]. This enzyme has two forms: a membrane protein and a soluble form with specific kinetic properties and tissue distributions [86–88]. The membrane protein consists of a polypeptide chain with a transmembrane receptor which is activated by various endogenous peptides [89] and presents enzymatic activity [84]. The soluble GC (sGC) form is a heterodimeric protein consisting of α (α1 and α2,) and β (β1 and β2) subunits encoded by distinct genes [90]. The simultaneous expression of one α- with one β-subunit (α1β1, α2β1, α1β2 and α2β2 isoforms) is essential for enzyme catalytic activity [91, 92].
The α2 subunit, encoded by GUCY1A2 gene, was firstly identified in human fetal and bovine brain tissues [93]. Subsequently it was isolated in human brain, placenta, spleen, and uterus [94, 95] and in rat brain [96–98]. According to Gibb and Garthwaite [97] and Mergia et al. [98] the highest proportion of α2 subunit is found in the mice’s brain, hippocampus and cerebellum. The relevance of this subunit is related to the binding with NO to catalyze the conversion of GTP in cGMP. The signaling pathways NO/sCG/cGMP acts as a phosphodiesterase, ion-gated channel, and cGMP-dependent protein kinases to regulate vasodilation, smooth muscle relaxation, platelet aggregation and neurotransmission [88, 99–102].
The neuronal NO was associated with memory formation as a retrograde messenger. A disruption in exon two of the NO synthase gene increased the aggressive behavior in mice [103, 104]. Additionally, the increase of cGMP level in the brain, with greatest magnitude in the cerebellum, was associated with the occurrence of locomotor activity and depended on the duration of this behavior [105]. Thus, this candidate gene (GUCY1A2) could be related to development of the neuronal wiring of cattle temperament. Further studies are required to identify the expression of this gene in bovine brain and the molecular basis associated to highest reactivity (locomotor activity) and poor temperament (expressed by high FS) as well as with vasorelaxation and neurotransmission.
Within the SNP window of BTA26 at 47 Mb, was identified the DOCK1 gene (dedicator of cytokinesis 1), also known as DOCK180, which encodes a member of the dedicator of cytokinesis protein superfamily. In mammalian systems this family consists of eleven members named DOCK1 to DOCK11 and classified into four subfamilies, according to the sequence homology [106]. The structure of DOCK1 contains an N-terminal Src homology 3 (SH3) domain, two Dock homology regions (DHR1 and DHR2 module) and a proline-rich C-terminal region. This protein acts as a guanine nucleotide exchange factor (GEF) for the Rho GTPases (a protein family of small guanosine triphosphates), by signaling the exchange of GDP (guanosine diphosphatase) for free GTP (guanosine-5’–triphosphate). The GEF activity requires binding with engulfment and cell motility 1 protein (ELMO1) [107, 108] to form a bipartite complex, which increases stability for DOCK1 activity toward GTPase Rac1 [109] in several biological processes, such as endothelial apoptosis [108], cytoskeletal organization [110–112], cell migration and invasion [113–115], tumor cell motility and invasion [116, 117], and phagocytosis [118].
Based on the effects of Elmo1/DOCK180/Rac1 signaling cascade on cell migration and proliferation some attempts have been made to study the relationship of DOCK1 with cancer [117, 119, 120]. Additionally, the effects on migration of neurons [121–123] lead to the association of DOCK1 with brain development, playing a role in the regulation of axon guidance, dendritic spine morphogenesis in hippocampal neurons [122], axon growth [121] and axon tip motility [124]. Further investigations are important to verify the expression of DOCK1 gene on the cattle neuronal system and synaptic connectivity and neuronal behavior related to differences in temperament.
Within BTA2 window at 65 Mb the positional known gene is NCKAP5 (Nck—associated protein 5), which is a protein-coding gene interacting with an SH3 (Src homology region 3) domain of the adaptor protein Nck [125]. Initially, this gene was detected in human brain, leukocytes and fetal fibroblasts [125]. In recent years NCKAP5 has been associated with human attention deficit hyperactivity disorders [126], bipolar disorders [127], schizophrenia and bipolar disorders [128], susceptibility of primary open angle glaucoma in a Japanese population [129] and increased risk of essential hypersomnia [130] by using GWAS. However, the biological function of this gene is still unclear and future research aiming to clarify the functional analysis and validation are recommended.
Considering the expression of this gene in human brain and leukocytes cells, and the strong association with emotional expression disease, our findings lead us to consider an effect of NCKAP5 variants on temperament. Further studies are required to identify if this gene is expressed in bovine leukocyte cells. Thus, our result may contribute to elucidate the biological function of NCKAP5, considering the influence of brain system on temperament traits [131, 132] and the negative correlation between immune system and stable differences on temperament of mice [133], cattle [10, 134] and infant monkeys [135].
Within the SNP window of BTA11 at 67 Mb, is the ANTXR1 gene, also known as tumor endothelial marker 8 (TEM8) [136]. This gene encodes a type I transmembrane protein that is recognized as anthrax toxin receptor 1 [137, 138]. No research was found reporting the expression of ANTXR1 gene in cattle, however it is conserved and expressed in different tissues types of mice and humans [137, 139, 140] and dynamically expressed in chick embryogenesis [141]. The encoded transmembrane protein is a specific receptor of anthrax toxin, which is produced by opportunistic bacteria, Bacillus anthracis [138, 142, 143].
A nonsense mutation in ANTXR1 gene has been associated with human recessive GAPO syndrome [144, 145] which is characterized by a loss of function of TEM8, resulting in growth retardation, alopecia, pseudoanodontia and progressive visual impairment [146]. Moreover, some researchers reported the role of ANTXR1 in tumor vasculature formation, tumor angiogenesis [147, 148] and regulation of endothelial and fibroblastic activities [149]. ANTXR1 is within the SNP window of BTA11 and was considered as a candidate gene influencing the expression of FS, however, a possible underlying biological relationship with cattle temperament is still unclear.
The identified regions and positional genes in our study, NCKAP5 (BTA2), PARK2 (BTA9), ANTXR1 (BTA11), GUCY1A2 (BTA15), CPE (BTA17) and DOCK1 (BTA26) have not been reported in Cattle QTL database [150]. Some research has described QTLs associated with several temperament traits assessed in different cattle breeds and crosses. However, each test considered distinct aspects of this complex trait and the comparison of results should be done carefully. Schmutz et al. [27] evaluated cattle behavior objectively, using an electronic movement measuring device in response to isolation during handling and habituation to the handling procedure. Considering these behavioral traits as a reflection of cattle temperament, the authors reported six associated QTLs on chromosomes 1 (14 cM), 5 (29 cM), 9 (44 cM), 11 (57 cM), 14 (19 and 35 cM) and 15 (12 cM).
On the other hand, Gutierrez-Gil et al. [29] found 29 QTLs distributed across 17 chromosomes affecting temperament traits, which were measured by flight from feeder and a social separation test (scoring to standing alert, vocalization and walking/running), in a Charolais × Holstein cattle population. While Hiendleder et al. [26] classifying dairy cows as ‘nervous/aggressive’ or ‘docile’ during milking, and reported QTLs associated with milking temperament on BTA 5 (136 cM), 18 (105 cM), 29 (20 cM) and X/Y (9 cM).
Additionally, some candidate genes have been linked to beef cattle temperament as result of GWAS using BovineSNP50 [32, 33]. Hulsman Hanna et al. [32] reported a significant association of temperament at weaning, measured in 769 Nellore-Angus crossbred cattle by subjective social separation score, with genes related to sodium ion transport and activity, particularly voltage-gate channel activity, demonstrating individual variability in nervous system responsiveness. Specifically for FS test, Lindholm-Perry et al. [33] performed a preliminary GWAS to discover genetic markers that were associated simultaneously to the variability of feed efficiency (ADG and average daily feed intake) and temperament using 1,057 beef steers of several European crosses (Angus, Hereford, Simmental, Limousin, Charolais, Gelbvieh and Red Angus). These authors reported positional candidate genes on Bos taurus chromosome 9 (quaking—QKI) and 17 (glutamate receptor, ionotropic, AMPA type subunit 2—GRIA2 and glycine receptor β –GLRB). Interestingly, the gene on BTA 9 reported by Lindholm-Perry et al. [33] is in close proximity (> 1Mb) with PARK2 gene identified in our study.
Most of the reported chromosome regions and genes, including those identified in the present study correspond to different genome locations. According to Adamczyk et al. [151] the dispersion of bovine genome regions affecting cattle temperament and behavior occurs in function of different methods to assess these traits and specific characteristics of each breed. Furthermore, the identification of regions and positional candidate genes is more accurate using high density marker panels.
An enrichment analysis was performed based on Gene Ontology and their functional pathways to identify a network among the six genes found in our study. However, no interaction among them was identified, demonstrating that each gene is associated with a different metabolic pathway. Among the genes associated with FS expression, greater attention should be given to CPE, PARK2, GUCY1A2 and DOCK1 due to their biological functions related to dopaminergic system, memory formation, biosynthesis of peptide hormone and neurotransmitter and brain development, respectively. In addition, CPE is a promising candidate gene which can influence temperament and weight gain simultaneously in cattle due to its function on proinsulin to insulin conversion and the recognized importance of insulin as hormone which modulates feeding behavior in different species [57, 152, 153].
The next step should be the identification of candidate genes associated with other methods used to assess cattle temperament, highlighting their metabolic pathways and potential interactions. This information will provide new insights about the biological mechanisms underlying the expression of this complex phenotype and better understanding about the genetic basis of cattle temperament. Moreover, further efforts are required to validate our findings in other populations in order to generate a DNA test panel for application in marked-assisted selection of Nellore temperament.
Conclusion
The genome-wide association study presented here allowed us to identify nine regions associated with beef cattle temperament, measured by FS test. Among them, six known genes were identified, contributing to a better comprehension into the genetic control of FS expression in Nellore cattle. In addition, the temperament has sufficient additive genetic variability to respond to selection, being of the same magnitude as other traits traditionally used in breeding programs. Thus, information of markers should help animal selection process in order to improve Nellore cattle temperament and, consequently, the efficiency of production systems.
Supporting Information
Contain the data used to estimate the genetic parameter for flight speed.
(TXT)
Variance explained by each 10 adjacent non-overlapping SNP window.
(TXT)
Contain the data used to generate Manhattan plot for flight speed.
(TXT)
Acknowledgments
This project was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), Brazil (Grant numbers: 2009/53609-7 and 2009/16118-5). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors thank Agropecuária Jacarezinho Ltda for allowing access to the animals and records. We would like to acknowledge Dr. Graham Plastow for the useful contributions to this paper.
Abbreviations
- SNP
Single nucleotide polymorphism
- QTL
Qualitative trait loci
- FS
Flight speed
- GWAS
Genome-wide association studies
- QC
quality control
Data Availability
Pedigree and Genotypic files belong to a third party, Agropecuária Jacarezinho, and can be obtained by contacting them directly: http://www.agrojacarezinho.com.br/. All relevant GWAS files have been included as Supporting Information.
Funding Statement
This project was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), Brazil (Grant numbers: 2009/53609-7 and 2009/16118-5). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Fordyce G, Goddard ME, Seifert GW. The measurement of temperament in cattle and the effect of experience and genotype. Anim Prod Aust. 1982;14:329–332. [Google Scholar]
- 2.Petherick JC, Holroyd RG, Doogan VJ, Venus BK. Productivity, carcass and meat quality of lot-fed Bos indicus cross steers grouped according to temperament. Aust J Exp Agric. 2002;42:389–398. [Google Scholar]
- 3.Pugh KA, Stookey JM, Buchanan FC. An evaluation of corticotropin-releasing hormone and leptin SNPs relative to cattle behaviour. Can J Anim Sci. 2011;91:567–572. [Google Scholar]
- 4.Haskell MJ, Simm G, Turner S. Genetic selection for temperament traits in dairy and beef cattle. Front Genet. 2014;5:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grandin T. Behavioral agitation during handling of cattle is persistent over time. Appl Anim Behav Sci. 1993;36:1–9. [Google Scholar]
- 6.Burrow HM. Measurements of temperament and their relationships with performance traits of beef cattle. Anim Breed Abstr. 1997;65:477–495. [Google Scholar]
- 7.Mackay JRD, Turner SP, Hyslop J, Deag JM, Haskell MJ. Short term temperament tests in beef cattle relate to long term measures of behavior recorded in the home pen. J Anim Sci. 2013;91:4917–4924. 10.2527/jas.2012-5473 [DOI] [PubMed] [Google Scholar]
- 8.Curley KO Jr., Paschal JC, Welsh TH, Randel RD. Technical note: exit velocity as a measure of cattle temperament is repeatable and associated with serum concentration of cortisol in Brahman bulls. J Anim Sci. 2006;84:3100–3103. [DOI] [PubMed] [Google Scholar]
- 9.Burdick NC, Carroll JA, Hulbert LE, Dailey JW, Willard ST, Vann RC, et al. Relationships between temperament and transportation with rectal temperature and serum concentrations of cortisol and epinephrine in bulls. Livest Sci. 2010;129:166–172. [Google Scholar]
- 10.Burdick NC, Randel RD, Carroll JA, Welsh TH Jr. Interactions between temperament, stress, and immune function in cattle. Int J Zool. 2011;2011:1–9. [Google Scholar]
- 11.Kilgour RJ, Melville GJ, Greenwood PL. Individual differences in the reaction of beef cattle to situations involving social isolation, close proximity of humans, restraint and novelty. Appl Anim Behav Sci. 2006;99:21–40. [Google Scholar]
- 12.Vetters MDD, Engle TE, Ahola JK, Grandin T. Comparison of flight speed and exit score as measurements of temperament in beef cattle. J Anim Sci. 2013;91:374–381. 10.2527/jas.2012-5122 [DOI] [PubMed] [Google Scholar]
- 13.Burrow HM, Corbet NJ. Genetic and environmental factors affecting temperament of zebu and zebu-derived beef cattle grazed at pasture in the tropics. Aust J Agric Res. 2000;51:155–162. [Google Scholar]
- 14.Hoppe S, Brandt HR, Konig S, Erhardt G, Gauly M. Temperament traits of beef calves measured under field conditions and their relationships to performance. J Anim Sci. 2010;88:1982–1989. 10.2527/jas.2008-1557 [DOI] [PubMed] [Google Scholar]
- 15.Rolfe KM, Snelling WM, Nielsen MK, Freetly HC, Ferrell CL, Jenkins TG. Genetic and phenotypic parameter estimates for feed intake and other traits in growing beef cattle, and opportunities for selection. J Anim Sci. 2011;89:3452–3459. 10.2527/jas.2011-3961 [DOI] [PubMed] [Google Scholar]
- 16.Sant’Anna AC, Paranhos da Costa MJR, Baldi F, Rueda PM, Albuquerque LG. Genetic associations between flight speed and growth traits in Nellore cattle. J Anim Sci. 2012;90:3427–3432. 10.2527/jas.2011-5044 [DOI] [PubMed] [Google Scholar]
- 17.Burrow HM, Seifert GW, Corbet NJ. A new technique for measuring temperament in cattle. Proc. Austr. Soc. Anim. Prod. 1988;17:154–157. [Google Scholar]
- 18.Prayaga KC, Corbet NJ, Johnston DJ, Wolcott ML, Fordyce G, Burrow HM. Genetics of adaptive traits in heifers and their relationship to growth, pubertal and carcass traits in two tropical beef cattle genotypes. Anim Prod Sci. 2009;49:413–425. [Google Scholar]
- 19.Sant’Anna AC, Baldi F, Valente TS, Albuquerque LG, Menezes LM, Boligon AA, et al. Genetic associations between temperament and performance traits in Nellore beef cattle. J Anim Breed Genet. 2015;132:42–50. 10.1111/jbg.12117 [DOI] [PubMed] [Google Scholar]
- 20.Burrow HM. Variances and covariances between productive and adaptive traits and temperament in a composite breed of tropical beef cattle. Livest Prod Sci. 2001;70:213–233. [Google Scholar]
- 21.Valente TS, Sant'Anna AC, Baldi F, Albuquerque LG, Paranhos da Costa MJ. Genetic association between temperament and sexual precocity indicator traits in Nellore cattle. J Appl Genet. 2015;56:349–354. 10.1007/s13353-014-0259-0 [DOI] [PubMed] [Google Scholar]
- 22.Nkrumah JD, Crews DH Jr, Basarab JA, Price MA, Okine EK, Wang Z, et al. Genetic and phenotypic relationships of feeding behavior and temperament with performance, feed efficiency, ultrasound, and carcass merit of beef cattle. J Anim Sci. 2007;85:2382–2390. [DOI] [PubMed] [Google Scholar]
- 23.Kadel MJ, Johnston DJ, Burrow HM, Graser HU, Ferguson DM. Genetics of flight time and other measures of temperament and their value as selection criteria for improving meat quality traits in tropically adapted breeds of beef cattle. Aust J Agric Res. 2006;57:1029–1035. [Google Scholar]
- 24.Jensen P, Buitenhuis B, Kjaer J, Zanella A, Mormede P, Pizzari T. Genetics and genomics of animal behaviour and welfare—challenges and possibilities. Appl Anim Behav Sci. 2008;113:383–403. [Google Scholar]
- 25.Spelman RJ, Huisma AE, Singireddy SR, Coppieters W, Arranz J, Georges M, et al. Short communication: quantitative trait loci analysis on 17 nonproduction traits in the New Zealand dairy population. J Dairy Sci. 1999;82:2514–2516. [DOI] [PubMed] [Google Scholar]
- 26.Hiendleder S, Thomsen H, Reinsch N, Bennewitz J, Leyhe-Horn B, Looft C, et al. Mapping of QTL for body conformation and behavior in cattle. J Hered. 2003;94:496–506. [DOI] [PubMed] [Google Scholar]
- 27.Schmutz SM, Stookey JM, Winkelman-Sim DC, Waltz CS, Plante Y, Buchanan FC. A QTL study of cattle behavioural traits in embryo transfer families. J Hered. 2001;92:290–292. [DOI] [PubMed] [Google Scholar]
- 28.Esmailizadeh AK, Bottema CD, Sellick GS, Verbyla AP, Morris CA, Cullen NG, et al. Effects of the myostatin F49L substitution on beef traits. J Anim Sci. 2008;86:1038–1046. 10.2527/jas.2007-0589 [DOI] [PubMed] [Google Scholar]
- 29.Gutierrez-Gil B, Ball N, Burton D, Haskell M, Williams JL, Wiener P. Identification of quantitative trait Loci affecting cattle temperament. J Hered. 2008;99:629–638. 10.1093/jhered/esn060 [DOI] [PubMed] [Google Scholar]
- 30.Glenske K, Brandt H, Prinzenberg EM, Gauly M, Erhardt G. Verification of a QTL on BTA1 for temperament in German Simmental and German Angus calves. Archiv Tierzucht. 2010;4:388–392. [Google Scholar]
- 31.Glenske K, Prinzenberg EM, Brandt H, Gauly M, Erhardt G. A chromosome-wide QTL study on BTA29 affecting temperament traits in German Angus beef cattle and mapping of DRD4. Animal. 2011;5:195–7. 10.1017/S1751731110001801 [DOI] [PubMed] [Google Scholar]
- 32.Hulsman Hanna LL, Garrick DJ, Gill CA, Herring AD, Riggs PK, Miller RK, et al. Genome-wide association study of temperament and tenderness using Bayesian approaches in a Nellore-Angus cross bred population. Livest Sci. 2014;161:17–27. [Google Scholar]
- 33.Lindholm-Perry AK, Kuehn LA, Freetly HC, Snelling WM. Genetic markers that influence feed efficiency phenotypes also affect cattle temperament as measured by flight speed. Anim Genet. 2015;46:60–64. 10.1111/age.12244 [DOI] [PubMed] [Google Scholar]
- 34.Wang H, Misztal I, Aguillar I, Legarra A, Muir WM. Genome-wide association mapping including phenotypes from relatives without genotypes. Genet Res. 2012;94:73–83. [DOI] [PubMed] [Google Scholar]
- 35.Gianola D, Fernando RL. Bayesian methods in animal breeding theory. J Anim Sci. 1986;63:217–244. [Google Scholar]
- 36.Misztal I, Tsuruta S, Strabel T, Auvray B, Druet T, Lee DH. BLUPF90 and related programs (BGF90). Proc 7th World Congress Genet Appl Livest Prod. 2002;28:21–22. [Google Scholar]
- 37.Aguilar I, Misztal I, Legarra A, Tsuruta S. Efficient computation of the genomic relationship matrix and other matrices used in single-step evaluation. J Anim Breed Genet. 2011;128:422–428. 10.1111/j.1439-0388.2010.00912.x [DOI] [PubMed] [Google Scholar]
- 38.Aguilar I, Misztal I, Johnson DL, Legarra A, Tsuruta S, Lawlor TJ. Hot topic: a unified approach to utilize phenotypic, full pedigree, and genomic information for genetic evaluation of Holstein final score. J Dairy Sci. 2010;93:743–752. 10.3168/jds.2009-2730 [DOI] [PubMed] [Google Scholar]
- 39.Van Raden PM. Efficient methods to compute genomic predictions. J Dairy Sci. 2008;91:4414–4423. 10.3168/jds.2007-0980 [DOI] [PubMed] [Google Scholar]
- 40.Vitezica ZG, Aguilar I, Misztal I, Legarra A. Bias in genomic predictions for populations under selection. Genet Res. 2011;93:357–366. [DOI] [PubMed] [Google Scholar]
- 41.Geweke J. Evaluating the accuracy of sampling-based approaches to calculating posterior moments In: Bernardo JM, Berger JO, David AP, Smith AFM, editors. Bayesian statistics. Oxford: Clarendon Press; 1992. p. 169–193. [Google Scholar]
- 42.Heidelberger P, Welch PD. Simulation run length control in the presence of an initial transient. Oper Res. 1983;31:1109–1144. [Google Scholar]
- 43.Raftery AE, Lewis SM. How many iterations in the Gibbs sampler? In: Bernardo JM, Berger JO, David AP, Smith AFM, editors. Bayesian statistics. Oxford: Clarendon Press; 1992. p. 763–773. [Google Scholar]
- 44.Dikmen S, Cole JB, Null DJ, Hansen PJ. Genome-wide association mapping for identification of quantitative trait loci for rectal temperature during heat stress in Holstein cattle. PLoS One. 2013;8:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang H, Misztal I, Aguilar I, Legarra A, Fernando RL, Vitezica Z, et al. Genome-wide association mapping including phenotypes from relatives without genotypes in a single-step (ssGWAS) for 6-week body weight in broiler chickens. Front Genet. 2014;5:134 10.3389/fgene.2014.00134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tiezzi F, Parker-Gaddis LK, Cole JB, Clay JS, Maltecca C. A genome-wide association mastitis in first parity US Holstein cows using single-step approach and genomic matrix re-weighting procedure. PLoS One. 2015;10:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Habier D, Fernando RL, Kizilkaya K, Garrick DJ. Extension of the Bayesian alphabet for genomic selection. BMC Bioinformatics. 2011;12:186 10.1186/1471-2105-12-186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun X, Fernando RL, Garrick DJ, Dekkers JCM. An iterative approach for efficient calculation of breeding values and genome-wide association analysis using weighted genomic BLUP. J Anim Sci. 2011;89:28. [Google Scholar]
- 49.Santana MHA, Utsunomiya YT, Neves HHR, Gomes RC, Garcia JF, Fukumasu H, et al. Genome-wide association study for feedlot average daily gain in Nellore cattle (Bos indicus). J Anim Breed Genet. 2014;131:210–216. 10.1111/jbg.12084 [DOI] [PubMed] [Google Scholar]
- 50.Zhang C, Wang Z, Bruce H, Kemp RA, Charagu P, Miar Y, et al. Genome-wide association studies (GWAS) indentify a QTL close to PRKAG3 affecting meat pH and colour in crossbred commercial pigs. BMC Genetics. 2015;16:33 10.1186/s12863-015-0192-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shin SC, Chung ER. Association of SNP marker in the leptin gene with carcass and meat quality traits in Korean cattle. Asian-Aust J Anim Sci. 2007;20:1–6. [Google Scholar]
- 52.Strittmatter SM, Lynh DR, Snyder SH. [3H]Guanidinoethylmercaptosuccinic acid binding to tissue homogenates. Selective labeling of enkephalin convertase. J Biol Chem. 1984;259:11812–11817. [PubMed] [Google Scholar]
- 53.Cawley NX, Wetsel WC, Murthy SR, Park JJ, Pacak K, Loh YP. New roles of carboxypeptidase E in endocrine and neuronal funciton and cancer. Endocr Rev. 2012;33:216–253. 10.1210/er.2011-1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fricker LD, Evans CJ, Esch FS, Herbert E. Cloning and sequence analysis of cDNA for bovine carboxypeptidase E. Nature. 1986;3:666–673. [DOI] [PubMed] [Google Scholar]
- 55.Chen H, Jawahar S, Qian Y, Duong Q, Chan G, Parker A, et al. Missense polymorphism in the human carboxypeptidase E gene alters enzymatic activity. Hum Mutat. 2001;18:120–131. [DOI] [PubMed] [Google Scholar]
- 56.Wang J, Zhang Y, Yang Z, Zhu T, Wang L, Chen B, et al. Association of human carboxypeptidase E exon5 gene polymorphisms with angiographical characteristics of coronary atherosclerosis in a Chinese population. Acta Pharmacol Sin 2008;29:736–744. 10.1111/j.1745-7254.2008.00798.x [DOI] [PubMed] [Google Scholar]
- 57.Manser E, Fernandez D, Loo L, Goh PY, Monfries C, Hall C, et al. Human carboxypeptidase E. Isolation and characterization of the cDNA, sequence conservation, expression and processing in vitro. Biochem J. 1990;267:517–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Naggert JK, Fricker LD, Varlamov O, Nishina PM, Rouille Y, Steiner DF, et al. Hyperproinsulinaemia in obese fat/fat mice associated with a carboxypeptidase-E mutation which reduces enzyme activity. Nat Genet. 1995;10:135–142. [DOI] [PubMed] [Google Scholar]
- 59.Cargill EJ, Baskin LC, Pomp D. Rapid communication: localization of the porcine carboxypeptidase-E gene by linkage analysis further extends region of synteny between human chromosome 4 and porcine chromosome 8. J Anim Sci. 1998;76:2211–2212. [DOI] [PubMed] [Google Scholar]
- 60.Haegeman A, Williams JL, Law A, Van Zeveren A, Peelman LJ. Mapping and SNP analysis of bovine candidate genes for meat and carcass quality. Anim Genet. 2003;34:349–353. [DOI] [PubMed] [Google Scholar]
- 61.Koshimizu H, Senatorov V, Loh YP, Gozes I. Neuroprotective protein and carboxypeptidase E. J Mol Neurosci. 2009;39:1–8 10.1007/s12031-008-9164-5 [DOI] [PubMed] [Google Scholar]
- 62.Woronowicz A, Koshimizu H, Chang SY, Cawley NX, Hill JM, Rodriguiz RM, et al. Absence of carboxypeptidase E leads to adult hippocampal neuronal degeneration and memory deficits. Hippocampus. 2008;18:1051–1063. 10.1002/hipo.20462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.LeDoux JE. Emotional memory systems in the brain. Behav Brain Res. 1993;58:69–79. [DOI] [PubMed] [Google Scholar]
- 64.Yoshino A, Kimura Y, Yoshida T, Takahashi Y, Nomura S. Relationships between temperament dimensions in personality and unconscious emotional responses. Biol Psychiat. 2005;57:1–6. [DOI] [PubMed] [Google Scholar]
- 65.Webb LE, van Reenen CG, Jensen MB, Schmitt O, Bokkers EAM. Does temperament affect learning in calves?. App Anim Behav Sci. 2015;16:33–39. [Google Scholar]
- 66.Hase A, Yamada H, Arai K, Sunada Y, Shimizu T, Matsumura K. Characterization of parkin in bovine peripheral nerve. Brain Res. 2002;15:143–149. [DOI] [PubMed] [Google Scholar]
- 67.Shimura H, Hattori N, Kubo S, Mizuno Y, Asakawa S, Minoshima S, et al. Familial Parkinson disease gene product, parkin, is a ubiquitin—protein ligase. Nat Genet. 2000;25:302–305. [DOI] [PubMed] [Google Scholar]
- 68.Mizuno Y, Hattori N, Mori H, Suzuki T, Tanaka K. Parkin and Parkinson's disease. Curr Opin Neurol. 2001;14:477–482. [DOI] [PubMed] [Google Scholar]
- 69.Fukae J, Sato S, Shiba K, Sato K, Mori H, Sharp PA, et al. Programmed cell death-2 isoform1 is ubiquitinated by parkin and increased in the substantia nigra of patients with autosomal recessive Parkinson’s disease. FEBS Lett. 2009;583:521–525. 10.1016/j.febslet.2008.12.055 [DOI] [PubMed] [Google Scholar]
- 70.Kuroda Y, Mitsui T, Kunishige M, Matsumoto T. Parkin affects mitochondrial function and apoptosis in neuronal and myogenic cells. Biochem Biophys Res Commun. 2006;348:787–793. [DOI] [PubMed] [Google Scholar]
- 71.de Mena L, Samaranch LL, Coto E, Cardo LF, Ribacoba R, Lorenzo-Betancor O, et al. Mutational screening of PARKIN identified a 3' UTR variant (rs62637702) associated with Parkinson's disease. J Mol Neurosci. 2013;50:264–269. 10.1007/s12031-012-9942-y [DOI] [PubMed] [Google Scholar]
- 72.Riley BE, Lougheed JC, Callaway K, Velasquez M, Brecht E, Nguyen L, et al. Structure and function of Parkin E3 ubiquitin ligase reveals aspects of RING and HECT ligases. Nat Commun. 2013;4:1982 10.1038/ncomms2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ambroziak W, Koziorowski D, Duszyc K, Górka-Skoczylas P, Potulska-Chromik A, Sławek J, et al. Genomic instability in the PARK2 locus is associated with Parkinson’s disease. J Appl Genet. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vallone D, Picetti R, Borrelli E. Structure and function of dopamine receptors. Neurosci Biobehav Rev. 2000;24:125–132. [DOI] [PubMed] [Google Scholar]
- 75.Jaber M, Robinson SW, Missale C, Caron MG. Dopamine Receptors and Brain Function. Neuropharmacology. 1996;35:1503–1519. [DOI] [PubMed] [Google Scholar]
- 76.Grady DL, Thanos PK, Corrada MM, Barnett JC Jr, Ciobanu V, Shustarovich D, et al. DRD4 genotype predicts longevity in mouse and human. J Neurosci. 2013;33:286–291. 10.1523/JNEUROSCI.3515-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hejjas K, Kubinyi E, Ronai Z, Szekely A, Vas J, Miklósi A, et al. Molecular and behavioral analysis of the intron 2 repeat polymorphism in the canine dopamine D4 receptor gene. Genes Brain Behav. 2009;8:330–336. 10.1111/j.1601-183X.2008.00475.x [DOI] [PubMed] [Google Scholar]
- 78.Wan M, Hejjas K, Ronai Z, Elek Z, Sasvari-Szekely M, Champagne FA, et al. DRD4 and TH gene polymorphisms are associated with activity, impulsivity and inattention in Siberian Husky dogs. Anim Genet. 2013;44:717–727. 10.1111/age.12058 [DOI] [PubMed] [Google Scholar]
- 79.Momozawa Y, Takeuchi Y, Kusunose R, Kikusui T, Mori Y. Association between equine temperament and polymorphisms in dopamine D4 receptor gene. Mamm Genome. 2005;16:538–544. [DOI] [PubMed] [Google Scholar]
- 80.Hori Y, Ozaki T, Yamada Y, Tozaki T, Kim HS, Takimoto A, et al. Breed differences in dopamine receptor D4 gene (DRD4) in horses. J Equine Sci. 2013;24:31–36. 10.1294/jes.24.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Keltikangas-Jarvinen L, Elovainio M, Kivimaki M, Lichtermann D, Ekelund J, Peltonen L. Association between the type 4 dopamine receptor gene polymorphism and novelty seeking. Psychosom Med. 2003;65:471–476. [DOI] [PubMed] [Google Scholar]
- 82.Schmidt LA, Fox NA, Perez-Edgar K, Hamer DH. Linking gene, brain, and behavior: DRD4, frontal asymmetry, and temperament. Psychol Sci. 2009;20:831–837. 10.1111/j.1467-9280.2009.02374.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ptácek R, Kuzelová H, Stefano GB. Dopamine D4 receptor gene DRD4 and its association with psychiatric disorders. Med Sci Monit. 2011;17:215–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McDonald LJ, Murad F. Nitric oxide and cGMP signaling. Adv. Pharmacol. 1995;34:263–275. [DOI] [PubMed] [Google Scholar]
- 85.Derbyshire ER, Marletta MA. Biochemistry of soluble guanylate cyclase. Handb Exp Pharmacol. 2009;191:17–31. 10.1007/978-3-540-68964-5_2 [DOI] [PubMed] [Google Scholar]
- 86.Yuen PS, Potter LR, Garbers DL. A new form of guanylyl cyclase is preferentially expressed in rat kidney. Biochemistry. 1990;29:10872–10878. [DOI] [PubMed] [Google Scholar]
- 87.Gupta G, Azam M, Yang L, Danziger RS. The β2 subunit inhibits stimulation of the α1/β1 form of soluble guanylyl cyclase by nitric oxide potential relevance to regulation of blood pressure. J Clin Invest. 1997;100:1488–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rybalkin SD, Yan C, Bornfeldt KE, Beavo JA. Cyclic GMP phosphodiesterases and regulation of smooth muscle function. Circ Res. 2003;93:280–291. [DOI] [PubMed] [Google Scholar]
- 89.Winquist RJ, Faison EP, Waldman SA, Schwartz K, Murad F, Rapoport RM. Atrial natriuretic factor elicits an endothelium-independent relaxation and activates particulate guanylate cyclase in vascular smooth muscle. Proc Natl Acad Sci. 1984;81:7661–7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Koesling D, Russwurm M, Mergia E, Mullershausen F, Friebe A. Nitric oxidesensitive guanylyl cyclase: structure and regulation. Neurochem Int. 2004;45:813–819. [DOI] [PubMed] [Google Scholar]
- 91.Harteneck C, Koesling D, Soling A, Schultz G, Bohme E. Expression of soluble guanylyl cyclase. Catalytic activity requires two enzyme subunits, FEBS Lett. 1990;272:221–223. [DOI] [PubMed] [Google Scholar]
- 92.Wedel B, Harteneck C, Foerster J, Friebe A, Schultz G, Koesling D. Functional domains of soluble guanylyl cyclase. J Biol Chem. 1995;270:24871–2485. [DOI] [PubMed] [Google Scholar]
- 93.Harteneck C, Wedel B, Koesling D, Malkewitz J, Bohme E, Schultz G. Molecular cloning and expression of a new K-subunit of soluble guanylyl cyclase. Interchangeability of the K-subunits of the enzyme. FEBS Lett. 1991;292:217–222. [DOI] [PubMed] [Google Scholar]
- 94.Russwurm M, Behrends S, Harteneck C, Koesling D. Functional properties of a naturally occurring isoform of soluble guanylyl cyclase. Biochem J. 1998;335:125–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Budworth J, Meillerais S, Charles I, Powell K. Tissue distribution of the human soluble guanylate cyclases. Biochem Biophys Res Commun. 1999;263:696–701. [DOI] [PubMed] [Google Scholar]
- 96.Koglin M, Behrends S. Cloning and functional expression of the rat alpha(2) subunit of soluble guanylyl cyclase. Biochim Biophys Acta. 2000;1494:286–289. [DOI] [PubMed] [Google Scholar]
- 97.Gibb BJ, Garthwaite J. Subunits of the nitric oxide receptor, soluble guanylyl cyclase, expressed in rat brain. Eur J Neurosci. 2001;13:539–544. [DOI] [PubMed] [Google Scholar]
- 98.Mergia E, Russwurm M, Zoidl G, Koesling D. Major occurrence of the new a2h1 isoform of NO-sensitive guanylyl cyclase in brain. Cell Signa. 2003;15:189–195. [DOI] [PubMed] [Google Scholar]
- 99.Sanders KM, Ward SM, Thornbury KD, Dalziel HH, Westfall DP, Carl A. Nitric-oxide as a nonadrenergic, noncholinergic neurotransmitter in the gastrointestinal-tract. Jpn J Pharmacol. 1992;58:220–5 [PubMed] [Google Scholar]
- 100.Warner TD, Mitchell JA, Sheng H, Murad F. Effects of cyclic GMP on smooth muscle relaxation. Adv Pharmacol. 1994;26:171–194. [DOI] [PubMed] [Google Scholar]
- 101.Munzel T, Feil R, Mulsch A, Lohmann SM, Hofmann F, Walter U. Physiology and pathophysiology of vascular signaling controlled by cyclic guanosine 3’, 5’-cyclic monophosphate-dependent protein kinase. Circulation. 2003;108:2172–2183. [DOI] [PubMed] [Google Scholar]
- 102.Potter LR. Guanylyl cyclase structure, function and regulation. Cell Signal. 2011;23:1921–1926. 10.1016/j.cellsig.2011.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Huang PL, Dawson TM, Bredt DS, Snyder SH, Fishman MC. Targeted disruption of the neuronal nitric oxide synthase gene. Cell. 1993;75:1273–1286. [DOI] [PubMed] [Google Scholar]
- 104.Nelson RJ, Demas GE, Huang PL, Fishman MC, Dawson VL, Dawson TM, et al. Behavioural abnormalities in male mice lacking neuronal nitric oxide synthase. Nature. 1995;378:383–386. [DOI] [PubMed] [Google Scholar]
- 105.Meyerhoff JL, Kant GJ, Sessios GR, Mougey EH, Pennington LL, Lenox RH. Brain and Pituitary Cyclic Nucleotide Response to Stress In: Williams R, editor. Perspectives on Behavioral Medicine: Neuroendocrine Control and Behavior. Academic Press; 1985. p. 250. [Google Scholar]
- 106.Côté JF, Vuori K. Identification of an evolutionarily conserved superfamily of DOCK180-related proteins with guanine nucleotide exchange activity. J Cell Sci. 2002;115:4901–4913. [DOI] [PubMed] [Google Scholar]
- 107.Côté JF, Vuori K. GEF what? Dock180 and related proteins help Rac to polarize cells in new ways. Trends Cell Biol. 2007;17:383–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schäker K, Bartsch S, Patry C, Stoll SJ, Hillebrands JL, Wieland T, et al. The bipartite rac1 Guanine nucleotide exchange factor engulfment and cell motility 1/dedicator of cytokinesis 180 (elmo1/dock180) protects endothelial cells from apoptosis in blood vessel development. J Biol Chem. 2015;290:6408–6418. 10.1074/jbc.M114.633701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Makino Y, Tsuda M, Ichihara S, Watanabe T, Sakai M, Sawa H, et al. Elmo1 inhibits ubiquitylation of Dock180. J Cell Sci. 2006;119:923–932. [DOI] [PubMed] [Google Scholar]
- 110.Brugnera E, Haney L, Grimsley C, Lu M, Walk SF, Tosello-Trampont AC, et al. Unconventional Rac-GEF activity is mediated through the Dock180-ELMO complex. Nat Cell Biol. 2002;4:574–582. [DOI] [PubMed] [Google Scholar]
- 111.Lu M, Ravichandran KS. Dock180-ELMO cooperation in Rac Activation. Methods Enzymol. 2006;406:388–402. [DOI] [PubMed] [Google Scholar]
- 112.Jean-Francois C, Kristiina V. GEF what? Dock180 and related proteins help Rac to polarize cells in new ways. Trends Cell Biol. 2007;17:383–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cheresh DA, Leng J, Klemke RL. Regulation of cell contraction and membrane ruffling by distinct signals in migratory cells. J Cell Biol. 1999;146:1107–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kiyokawa E, Hashimoto Y, Kobayashi S, Sugimura H, Kurata T, Matsuda M. Activation of Rac1 by a Crk SH3-binding protein, DOCK180. Genes Dev. 1998;12:3331–3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jarzynka MJ, Hu B, Hui KM, Bar-Joseph I, Gu W, Hirose T, et al. ELMO1 and Dock180, a bipartite Rac1 guanine nucleotide exchange factor, promote human glioma cell invasion. Cancer Res. 2007;67:7203–7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Smith HW, Marra P, Marshall CJ. uPAR promotes formation of the p130Cas-Crk complex to activate Rac through DOCK180. J Cell Biol. 2008;182:777–790. 10.1083/jcb.200712050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang J, Dai JM, Che YL, Gao YM, Peng HJ, Liu B, et al. Elmo1 helps dock180 to regulate Rac1 activity and cell migration of ovarian cancer. Int J Gynecol Cancer. 2014;24:844–850. 10.1097/IGC.0000000000000137 [DOI] [PubMed] [Google Scholar]
- 118.Albert ML, Kim JI, Birge RB. Alphavbeta5 integrin recruits the CrkII-Dock180-rac1 complex for phagocytosis of apoptotic cells. Nat Cell Biol. 2000;2:899–905. [DOI] [PubMed] [Google Scholar]
- 119.Zhao F, Siu MK, Jiang L, Tam KF, Ngan HY, Le XF, et al. Overexpression of dedicator of cytokinesis I (Dock180) in ovarian cancer correlated with aggressive phenotype and poor patient survival. Histopathology. 2011;59:1163–1172. 10.1111/j.1365-2559.2011.04045.x [DOI] [PubMed] [Google Scholar]
- 120.Pezeshkpour GH, Moatamed F, Lewis M, Hoang B, Rettig M, Mortazavi F. CRK SH3N domain diminishes cell invasiveness of non-small cell lung cancer. Genes Cancer. 2013;4:315–324. 10.1177/1947601913497573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Li X, Gao X, Liu G, Xiong W, Wu J, Rao Y. Netrin signal transduction and the guanine nucleotide exchange factor DOCK180 in attractive signaling. Nat Neurosci. 2008;11:28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kim JY, Oh MH, Bernard LP, Macara IG, Zhang H. The RhoG/ELMO1/Dock180 signaling module is required for spine morphogenesis in hippocampal neurons. J Biol Chem. 2011;286:37615–37624. 10.1074/jbc.M111.268029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Franke K, Otto W, Johannes S, Baumgart J, Nitsch R, Schumacher S. miR-124-regulated RhoG reduces neuronal process complexity via ELMO/Dock180/Rac1 and Cdc42 signalling. EMBO J. 2013;31:2908–2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Shi L. Dock protein family in brain development and neurological disease. Commun Integr Biol. 2013;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Matuoka K, Miki H, Takahashi K, Takenawa T. A novel ligand for an SH3 domain of the adaptor protein Nck bears an SH2 domain and nuclear signaling motifs. Biochem Biophys Res Commun. 1997;239:488–492. [DOI] [PubMed] [Google Scholar]
- 126.Lasky-Su J, Neale BM, Franke B, Anney RJ, Zhou K, Maller JB, et al. Genome-wide association scan of quantitative traits for attention deficit hyperactivity disorder identifies novel associations and confirms candidate gene associations. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:1345–1354. 10.1002/ajmg.b.30867 [DOI] [PubMed] [Google Scholar]
- 127.Smith EN, Bloss CS, Badner JA, Barrett T, Belmonte PL, Berrettini W, et al. Genome-wide association study of bipolar disorder in European American and African. Mol Psychiatry. 2009;14:755–763. 10.1038/mp.2009.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wang KS, Liu XF, Aragam N. A genome-wide meta-analysis identifies novel loci associated with schizophrenia and bipolar disorder. Schizophr Res. 2010;124:192–199. 10.1016/j.schres.2010.09.002 [DOI] [PubMed] [Google Scholar]
- 129.Osman W, Low SK, Takahashi A, Kubo M, Nakamura Y. A genome-wide association study in the Japanese population confirms 9p21 and 14q23 as susceptibility loci for primary open angle glaucoma. Hum Mol Genet. 2012;21:2836–2842. 10.1093/hmg/dds103 [DOI] [PubMed] [Google Scholar]
- 130.Khor SS, Miyagawa T, Toyoda H, Yamasaki M, Kawamura Y, Tanii H. et al. Genome-wide association study of HLA-DQB1*06:02 negative essential hypersomnia. Peer J. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Robinson DL. How brain arousal systems determine different temperament types and the major dimensions of personality. Pers Individ Diff. 2001;31:1233–1259. [Google Scholar]
- 132.Tuominen L, Salo J, Hirvonen J, Någren K, Laine P, Melartin T, et al. Temperament, character and serotonin activity in the human brain: a positron emission tomography study based on a general population cohort. Psychol Med. 2013;43:881–894. 10.1017/S003329171200164X [DOI] [PubMed] [Google Scholar]
- 133.Cavigelli SA, Bennett JM, Michael KC, Klein LC. Female temperament, tumor development and life span: relation to glucocorticoid and tumor necrosis factor α levels in rats. Brain Behav Immun. 2008;5:727–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bauer ME, Perks P, Lightman SL, Shanks N. Restraint stress is associated with changes in glucocorticoid immunoregulation. Physiol Behav. 2001;73:525–532. [DOI] [PubMed] [Google Scholar]
- 135.Capitanio JP, Mendoza SP, Cole SW. Nervous temperament in infant monkeys is associated with reduced sensitivity of leukocytes to cortisol's influence on trafficking. Brain Behav Immun. 2011;1:151–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.St Croix B, Rago C, Velculescu V, Traverso G, Romans KE, Montgomery E, et al. Genes expressed in human tumor endothelium. Science. 2000;289:1197–1202. [DOI] [PubMed] [Google Scholar]
- 137.Bradley KA, Mogridge J, Mourez M, Collier RJ, Young JA. Identification of the cellular receptor for anthrax toxin. Nature 2001;414:225–229. [DOI] [PubMed] [Google Scholar]
- 138.Ingram RJ, Harris A, Ascough S, Metan G, Doganay M, Ballie L, et al. Exposure to anthrax toxin alters human leucocyte expression of anthrax toxin receptor 1. Clin Exp Immunol. 2013;173:84–91. 10.1111/cei.12090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Carson-Walter EB, Watkins DN, Nanda A, Vogelstein B, Kinzler KW, St. Croix B. Cell surface tumor endothelial markers are conserved in mice and humans. Cancer Res. 2001;61:6649–6655. [PubMed] [Google Scholar]
- 140.Vargas M, Karamsetty R, Leppla SH, Chaudry J. Broad expression analysis of human ANTXR1-TEM8 transcripts reveals differential expression and novel splizce variants. Plos One. 2012;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Herrmann D, Ferrer-Vaquer A, Lahsnig C, Firnberg N, Leibbrandt A, Neubüser A. Expression and regulation of ANTXR1 in the chick embryo. Dev Dyn. 2010; 239: 680–687. 10.1002/dvdy.22194 [DOI] [PubMed] [Google Scholar]
- 142.Moayeri M, Leppla SH. The roles of anthrax toxin in pathogenesis. Curr Opin Microbiol. 2004;7:19–24. [DOI] [PubMed] [Google Scholar]
- 143.Rainey GJ, Wigelsworth DJ, Ryan PL, Scobie HM, Collier RJ, Young JA. Receptor-specific requirements for anthrax toxin delivery into cells. Proc Natl Acad Sci. 2005;102:13278–13283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Stránecký V, Hoischen A, Hartmannová H, Zaki MS, Chaudhary A, Zudaire E, et al. Mutations in ANTXR1 cause GAPO Syndrome. Am J Hum Genet. 2013;92:792–799. 10.1016/j.ajhg.2013.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bayram Y, Pehlivan D, Karaca E, Gambin T, Jhangiani SN, Serkan Erdin, et al. Whole exome sequencing identifies three novel mutations in ANTXR1 in families with GAPO Syndrome. Am J Med Genet A. 2014;9:2328–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tipton RE, Gorlin RJ. Growth retardation, alopecia, pseudoanodontia, and optic atrophy—the GAPO syndrome: report of a patient and review of the literature. Am J Med Genet. 1984;19: 209–216. [DOI] [PubMed] [Google Scholar]
- 147.Yang MY, Chaudhary A, Seaman S, Dunty J, Stevens J, Elzarrad MK, et al. The cell surface structure of tumor endothelial marker 8 (TEM8) is regulated by the actin cytoskeleton. Biochim Biophys Acta. 2011;1813:39–49. 10.1016/j.bbamcr.2010.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chen D, Bhat-Nakshatri P, Goswami C, Badve S, Nakshatri H. ANTXR1, a stem cell-enriched functional biomarker, connects collagen signaling to cancer stem-like cells and metastasis in breast cancer. Cancer Res. 2013;73:5821–5833. 10.1158/0008-5472.CAN-13-1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Besschetnova TY, Ichimura T, Katebi N, St Croix B, Bonventre JV, Olsen BR. Regulatory mechanisms of anthrax toxin receptor 1-dependent vascular and connective tissue homeostasis. Matrix Biol. 2015;42:56–73. 10.1016/j.matbio.2014.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hu ZL, Park CA, Wu XL, Reecy JM. Animal QTLdb: an improved database tool for livestock animal QTL/association data dissemination in the post-genome era. Nucleic Acids Res. 2013;41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Adamczyk K, Pokorska J, Makulska J, Earley B, Mazurek M. Genetic analysis and evaluation of behavioural traits in cattle. Livest Sci. 2013;154:1–12. [Google Scholar]
- 152.Gerozissis K. Brain insulin and feeding: a bi-directional communication. Eur J Pharmacol. 2004;490:59–70. [DOI] [PubMed] [Google Scholar]
- 153.Shiraishi J, Yanagita K, Fujita M, Bungo T. Central insulin suppresses feeding behavior via melanocortins in chicks. Domest Anim Endocrinol. 2008;34:223–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Contain the data used to estimate the genetic parameter for flight speed.
(TXT)
Variance explained by each 10 adjacent non-overlapping SNP window.
(TXT)
Contain the data used to generate Manhattan plot for flight speed.
(TXT)
Data Availability Statement
Pedigree and Genotypic files belong to a third party, Agropecuária Jacarezinho, and can be obtained by contacting them directly: http://www.agrojacarezinho.com.br/. All relevant GWAS files have been included as Supporting Information.