Abstract
Hedgehog (Hh) signalling is a potent regulator of cell fate and function. While much is known about the events within a Hh-stimulated cell, far less is known about the regulation of Hh-ligand production. Drosophila Hyperplastic Discs (Hyd), a ubiquitin-protein ligase, represents one of the few non-transcription factors that independently regulates both hh mRNA expression and pathway activity. Using a murine embryonic stem cell system, we revealed that shRNAi of the mammalian homologue of hyd, Ubr5, effectively prevented retinoic-acid-induced Sonic hedgehog (Shh) expression. We next investigated the UBR5:Hh signalling relationship in vivo by generating and validating a mouse bearing a conditional Ubr5 loss-of-function allele. Conditionally deleting Ubr5 in the early embryonic limb-bud mesenchyme resulted in a transient decrease in Indian hedgehog ligand expression and decreased Hh pathway activity, around E13.5. Although Ubr5-deficient limbs and digits were, on average, shorter than control limbs, the effects were not statistically significant. Hence, while loss of UBR5 perturbed Hedgehog signalling in the developing limb, there were no obvious morphological defects. In summary, we report the first conditional Ubr5 mutant mouse and provide evidence for a role for UBR5 in influencing Hh signalling, but are uncertain to whether the effects on Hedgehog signaling were direct (cell autonomous) or indirect (non-cell-autonomous). Elaboration of the cellular/molecular mechanism(s) involved may help our understanding on diseases and developmental disorders associated with aberrant Hh signalling.
Introduction
In multicellular organisms Hedgehog (Hh) morphogens play an essential role in tissue/organ development [1] and their subsequent maintenance [2]. Acting as an extracellular signalling molecule, Hh ligands signal in a predominantly paracrine manner to convey information to neighbouring cells. Tight regulation of Hh ligand expression ensures temporal-spatial generation of morphogen gradients, which in turn ensure a well co-ordinated and appropriate cellular response [3]. The importance of correct Hh expression patterns is clear from the numerous human diseases (e.g., gastrointestinal, pancreatic and skin cancers) [4] and developmental disorders (e.g., cyclopia, cleft lip and limb abnormalities) [5] that result from its misexpression.
Mammals express three Hh ligands (Sonic-, Indian and Desert-Hedgehog) that exhibit distinct expression patterns throughout the body (EMBL Expression Atlas). Engagement of Hh ligands with one of their receptors Patched (Ptch1) [6–8] results in derepression of the Hedgehog signal transduction pathway, activation of the GLI family zinc finger (GLI) family of transcription factors and the subsequent transcription of GLI target genes. Activation of the Hh pathway (HhP) influences a wide range of cellular responses that include promotion of proliferation, differentiation and suppression of apoptosis[1]. Hedgehog signalling affects cell behaviour in multiple tissues and is heavily implicated in the communication between cells, including adult stem cells and their niches[2].
A large body of work has focused on investigating Sonic hedgehog (Shh) expression and function during animal development, with a particular focus on the developing limb[9]. Within the embryonic limb bud, Shh expression is spatially restricted to a posteriorly located zone of polarising activity (ZPA)[10], where its expression is regulated by a long-range enhancer element called the zone of polarizing activity regulatory sequence (ZRS) [11]. SHH expression within the ZPA, and its subsequent diffusion/transport across the tissue, sets up a posterior-anterior morphogen gradient that acts to instruct distinct cell fates and govern digit formation [9]. A number of human disease-associated point mutations within, or chromosomal translocations affecting, Shh’s regulatory regions underlie deregulated SHH/Shh expression and digit abnormalities [12].
While the molecular details concerning Shh DNA regulatory elements are well-described [13], far less is known about the proteins and upstream signalling pathway(s) that regulate Shh expression. In mammals Ras-associated signalling appears to promote Shh expression [14], while in Drosophila, Hyperplastic Disc (Hyd) suppresses hh ligand expression [15, 16]. Mechanistically, Hyd appears to regulate hh expression through influencing Shaggy–the Drosophila ortholog of Glycogen synthase kinase β (GSK3β) [16]. The ability of Hyd and its human ortholog E3 identified by Differential Display (EDD) to bind GLI2 [16], one of the HhP’s major transcriptional effectors, potentially places Hyd/EDD both upstream and downstream of the Hedgehog ligand activity. Here, we addressed whether Hyd’s murine homologue Ubiquitin Protein Ligase E3 Component N-recognin 5 (UBR5) could also influence Hedgehog pathway activity and ligand expression.
UBR5 contains a number of domains related to ubiquitin signalling, which include a Ubiquitin binding domain (UBA) [17], a substrate recruitment domain for N-end rule substrates called the Ubiquitin-protein ligase E3 component N-Recognin (UBR) domain [18, 19] and a catalytic Homologous to E6AP C-terminus (HECT) domain [20]–the presence of which defines UBR5 as an E3 ubiquitin-protein ligase. Functionally, UBR5 has previously been linked to DNA damage signalling [21–23], miRNA activity [24], metabolism [25] and cell cycle checkpoint control [26–30].
Our data supports a potential role for UBR5 in influencing hedgehog family member ligand expression and HhP activity, although we are unclear about the mechanism.
Materials and Methods
mES 14 cell culture and retinoic acid treatment
E14 mouse embryonic stem (mES) cells were cultured on 0.1% porcine gelatin (Sigma) coated 6-well plates at 37°C and 5% CO2 in GMEM (Invitrogen) supplemented with 10% foetal bovine serum, 103 U/ml leukaemia inhibitory factor (Millipore). Retinoic acid (Sigma) or DMSO (Sigma) vehicle was added to the media and refreshed every 24 hours.
shRNAi transfection and selection
6-well plates containing 2x106 cells in antibiotic-free GMEM were transfected with 4μg of pLKO-based, puromycin-expressing, Ubr5 shRNAi constructs (Sigma MISSION, TRCN0000003411 (Ubr5.1), TRCN0000226458 (Ubr5.2) and TRCN0000238587 (Ubr5.3) with Effectene (Qiagen) as per the manufacturer’s instruction. 24 hours later, transfected cells were selected in 0.1μg/ml puromycin (Sigma) and resistant colonies were pooled.
RNA extraction, reverse transcription and PCR
RNA was extracted from embryos, dissected limb buds or ES cells using QIAshredder homogenisers coupled with RNeasy kits (QIAGEN) as per the manufacturers instructions. cDNA was produced using a First Strand cDNA Synthesis Kit also as per manufacturer’s instructions (Roche). All semi-quantitative RT-PCR was carried out using Platinum Taq Polymerase PCR kit (Invitrogen) using a MJ Research Tetrad PTC-225 PCR machine. Primers used: Shh For GCC TAC AAG CAG TTT ATT CCC AAC and Rev CAG TGG ATG TGA GCT TTG GAT TC; Ubr5 For CTC GAG GAA AGC TAG AGC AAA AAA TAA AAA GCC CAA ATC CAG and Rev GAG CTC TAC AGC GAC ATA GGC ACC ATC TAC C; β-actin For GGC CCA GAG CAA GAG AGG TAT CC and Rev ACG CAC GAT TTC CCT CTC AGC; Nanog For ACC TGA GCT ATA AGC AGG TTA AGA C and Rev GTG CTG AGC CCT TCT GAA TCA GAC; Bmp4 For GAG GAG TTT CCA TCA CGA AGA and REV GCT CTG CCG AGG AGA TCA; Klf4 For CCA GCA AGT CAG CTT GTG AA and Rev GGG CAT GTT CAA GTT GGA TT. Quantitative RT-PCR (qRT-PCR) was carried out using the Roche Universal probe Library coupled with the Lightcycler®480 system as per manufacturer’s instructions (Roche). Gene specific assays were designed using the online Assay Design Centre Tool (www.universalprobelibrary.com): Shh For ACC CCG ACA TCA TAT TTA AGG A and Rev TTA ACT TGT CTT TGC ACC TCT GA (UPL probe 32); Ubr5 For TCA GCT CGA AGA GAG AGG ATG and Rev GCT CAG CAA TGT AGC ACG TC (UPL probe 103). The β-actin control reagent master mix was used to generate an internal reference for all reactions. Relative expression levels were determined using the ΔCt model [31].
SDS-PAGE and Western blotting
Cells and dissected limb buds were lysed in a 1%TX-100 lysis buffer and processed as described previously [32]. For the limb buds, cytoplasmic and nuclear fractions were separated using NE-PER® nuclear and cytoplasmic protein extraction kit (Thermo Scientific). Proteins (30μg) were resolved by SDS-PAGE using 3–8% Tris-acetate gradient gels with Tris-acetate Running Buffer (Invitrogen) and wet-blotted onto PVDF membrane (Millipore) overnight at 4°C using Towbin buffer. Membranes were blocked in 5% milk/PBS at room temperature (RT) for one hour and incubated with primary antibodies against: UBR5 (Santa Cruz goat EDD M-19 1:2500); β-tubulin (Sigma mouse 1:30,000) and HP-1 (Chemicon mouse 1:10,000) for one hour at RT in 0.1% Tween PBS (PBST). Membranes were then washed three times in PBST at RT, incubated with appropriate secondary antibodies: donkey αgoat-HRP and horse αmouse-HRP, both (Jackson Labs 1:10,000) and washed three times in PBST at RT. Membranes were then incubated in ECL solution and imaged using the digital ChemiDoc imaging system (Promega).
In situ hybridisation
Shh and Ptch1 vectors and probes were previously described [11]. Primers incorporating restriction enzymes sites were used to amplify the probe from full-length cDNA template temples. Products were cloned into Bluescript SK and KS for antisense and control sense probe production. Riboprobes were generated from linearised vector templates using the MEGAscript kit (Life technologies) as per the manufacturer’s instruction. Embryos were dissected and fixed in 4% paraformaldehyde (Sigma) overnight at 4°C. In situ hybridisation was carried out as previously described [11]. Expression patterns were detected by the alkaline phosphatase substrates nitro-blue tetrazolium chloride (NBT) and 5-bromo- 4-chloro-3'-indolyphosphate p-toluidine (BCIP) (Roche).
General animal work
Animal studies were approved by Medical Research Council Institute of Genetics and Molecular Medicine ‘Animal Care and Use Committee’ (applications PL21–06 and PL26–11) and carried out according to guidance issued by the Medical Research Council in Responsibility in the Use of Animals for Medical Research (July 1993), EU Directive 2010 and UK Home Office Project License no PPL 60/4424. Mice had constant access to food and water and were maintained on 12 hour light and dark cycles. Pups were weaned at three weeks old at which point ear clips were collected for genotype analysis. Timed matings were set up with E0.5 (embryonic day) taken as the morning a vaginal plug was detected. Following Schedule 1 sacrifice of the mother (CO2 asphyxiation followed by cervical dislocation), embryos were dissected out using a Leica EZ4HD dissecting microscope. Genomic DNA was extracted either from adult mouse earclips or embryo yolk sacs and analysed using the HotShot DNA extraction technique [33] using Platinum Taq (Invitrogen) according to the manufacturer’s instruction. Primers used were: Wild type Ubr5 For 5’ GTT TCT GGC AAG GTT CAG TGC; Rev 5’ CAC ACA TGC TGC ACA AAC ACA TG; Ubr5mt* For 5’ GTT TCT GGC AAG GTT CAG TGC; Ubr5mt* Rev 5’ GCC ACT ATG CGC ACA GCT GG; Ubr5WT* For 5’ CGC GAA GAG TTT GTC CTC AC; Ubr5WT* Rev 5’ GCC TCG ATC CTC CCT TTA TC; Neomycin For 5’ TGT TCC GGC TGT CAG CGC AG; Neomycin Rev 5’ GAT ATT CGG CAA GCA GGC ATC; FLP For 5’ AGG GTG AAA GCA TCT GGG AGA; FLP Rev 5’ TCA ACT CCG TTA GGC CCT TCA; Cre For 5’ GCA TTA CCG GTC GAT GCA ACG AGT GAT GAG; Cre Rev 5’ GAG TGA ACG AAC CTG GTC GAA A.
Creation of the conditional Ubr5mt mouse
E14 embryonic stem (ES) cells carrying the Ubr5 gene trap EUCE0171f01 were obtained from EUCOMM (European Conditional Mouse Mutagenesis Program). Chimeric mice were generated by injection of E14 mouse ES cells positive for the gene trap into C57BL/6J ES blastocysts that were transferred to pseudopregnant C57BL/6J females. Germline offspring were identified by coat colour and PCR genotyping confirmed the presence of the modified allele. Mice were then crossed to, and subsequently maintained on, a C57BL/6J background.
Conditional disruption of Ubr5 expression
For conditional studies, mice were first crossed to a mouse line expressing an enhanced form of Flippase (Flp-e) [34]. The presence of the inverted genetrap (Ubr5WT*) was confirmed by PCR. Ubr5 expression was conditionally ablated in the whole embryo or limb buds by crossing Ubr5WT* with pCAGG-Cre-ERT2[35] or Prx1-Cre [36] mice, respectively. Animal stocks were maintained as heterozygotes. Crosses were carried out using heterozygous animals to permit littermate controls and Cre was passed through the male germline. For viability studies, pregnant females underwent intraperitoneal (i.p.) injections of 4mg Tamoxifen (Sigma, stock 20mg/ml) at 11.5 days post coitum (dpc), followed by embryo collection at E13.5, E15.5 or E17.5.
X-gal staining
X-gal staining was carried out as previously described [11]. Briefly, embryos were dissected out, fixed in 4% PFA at 4°C, washed and stained overnight. Stained embryos were then imaged whole mount and then sectioned for histology.
Embryo embedding, sectioning and histological processing
Embryos were hand embedded in paraffin wax as per established in-house protocols. Wax blocks were allowed to solidify for three hours on a cold block, then mounted on wooden blocks and 4–8μm sections cut using a microtome (Leica). Sections were floated on a 45°C water bath prior to mounting on SuperFrost® slides (Fisher) which were left to dry overnight at 37°C prior to staining. Sections were de-waxed and stained using standard histological techniques for haematoxylin, alcian blue and alizarin (Sigma) staining.
Macroscopy
Colour brightfield imaging of all embryos and embryo limbs was carried out on a Nikon AZ100 macroscope attached to a QImaging Micropublisher. Images were captured using IPLab software (Scanalytics). Histological sections were scanned using a Nanozoomer slide scanner (Hamamatsu) and visualised using NDPview2 software (Hamamatsu).
Optical Projection Tomography (OPT)
OPT was carried out as previously described [37]. Briefly, embryos were fixed overnight using 4% paraformaldehyde, transferred to PBS and embedded in 1% low melting point molten agarose, dehydrated overnight and then cleared in a 2:1 mixture of benzyl alcohol to benzyl benzoate. Sample autofluorescence was analysed using a Bioptonics 3001 OPT scanner and reconstructed using Bioptonics view software.
Statistics and computer programmes
Microsoft Excel and GraphPad Prism were used for graphs and the indicated ANOVA and t-test statistical analysis. Graphs indicate p values with the following key: *** = ≤0.0001; *** = ≤0.001; ** = ≤0.01; * = ≤0.05 and ns = ≥0.05. Adobe Photoshop CS6 and Illustrator CS6 were used to produce the figures.
Results
Retinoic acid promotes Shh expression in murine ES cells
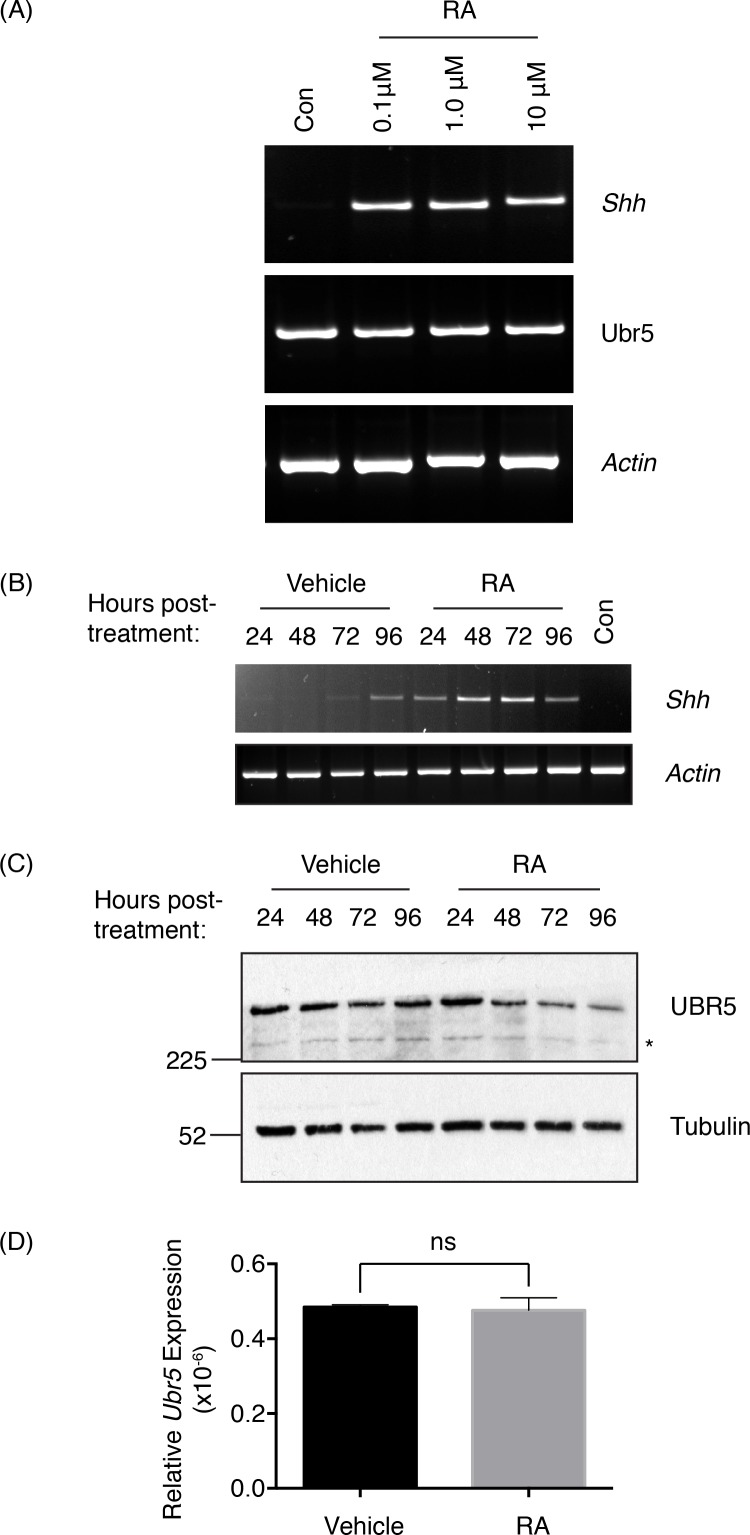
Based upon the ability of Hyd, the Drosophila orthologue of UBR5, to influence hh ligand expression we wished to examine whether murine UBR5 was capable of regulating Shh expression. We chose to use an E14 mouse embryonic stem (mES) cell system that utilises retinoic acid (RA) treatment to induce Shh expression[38]. In contrast to Okada et al, we utilised conditions to restrain embryoid body formation and ES cell differentiation (i.e., retention of LIF[39] and 10% FBS[40] in the ES cell media throughout). Initially we confirmed that RA stimulation promoted mES cells to express Shh (Fig 1A). 24 hrs post-RA-treatment, Shh levels were readily detectable by RT-PCR, with doses as low as 0.1μM RA inducing significant Shh expression [41]. More detailed analysis of the inductive response with 0.1μM RA revealed peak Shh expression 48–72hrs after treatment (Fig 1B). At 96 hrs post-treatment, an increase in Shh expression was also detected in vehicle (DMSO) treated cells, potentially reflecting a possible effect of increased cell confluence.
Fig 1. Retinoic Acid induces Shh and suppresses UBR5 expression.
Murine E14 ES cells were treated with (A) the indicated concentration of RA, or (B-D) 0.1μM RA or DMSO (Vehicle) and analysed by RT-PCR at (A) 24hrs and (B) at the indicated times post-RA-treatment. (C) SDS-PAGE and Western blotting determined UBR5 expression over the indicated RA time course. An asterisk denotes an uncharacterised, faster migrating UBR5 antibody reactive species. Tubulin was used as a loading control. (D) qRT-PCR of Ubr5 expression normalised against β-actin in mES cells 96hrs post-RA-treatment (n = 3, s.e.m indicated). Statistical analysis by Students t-test. ns = not significant.
Analysis of UBR5 protein expression in RA-treated cells revealed a significant reduction at 48hrs post-treatment (Fig 1C), which was sustained up to the end of the time course at 96hrs. To address if the loss of UBR5 expression 96hrs post-RA-treatment was due to transcriptional changes we used qRT-PCR analysis, which revealed no significant reduction (Fig 1D). These results suggested that the reduction in UBR5 expression was potentially via a post-transcriptional mechanism. Intriguingly, maximal Shh expression (48–72hrs post-RA-treatment) coincided with the marked reduction in UBR5 expression. However, at 24hrs post-RA-treatment, at a time Shh was initially induced (Fig 1B), UBR5 expression was not reduced (Fig 1C). In summary, it appeared that reduced UBR5 protein levels did not coincide with the initial induction of Shh, but did coincide with the subsequent increase in Shh expression level.
UBR5 promotes RA-mediated Shh expression
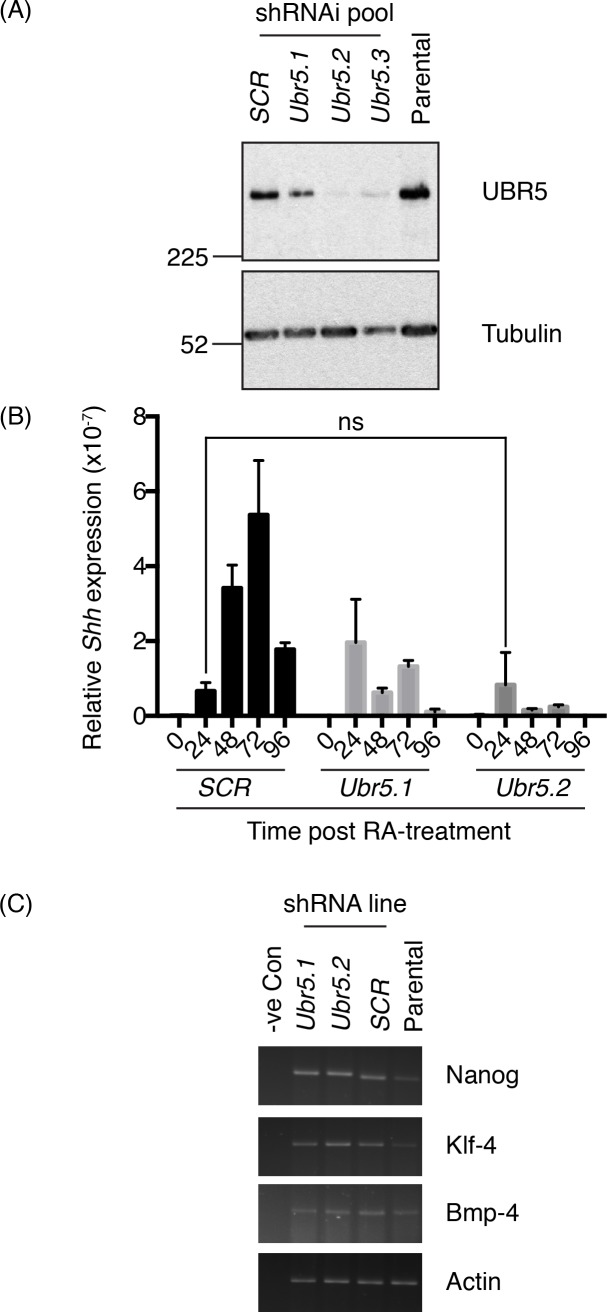
Due to the reciprocal association of UBR5 and Shh expression levels, we reasoned that, similar to Hyd’s ability to suppress hh expression [15, 16], UBR5 also functioned as a negative regulator of RA-mediated Shh expression. To address this we created stable pools of mES cells lines expressing three different Ubr5 shRNAis or a control scrambled (SCR) shRNAi. Western blot analysis revealed normal levels of UBR5 expression in the SCR pool, but moderate (Ubr5.1), intermediate (Ubr5.3) and strong (Ubr5.2) reductions in UBR5 expression in Ubr5 shRNAi pools (Fig 2A). We next challenged the SCR and Ubr5 shRNAi pools (Ubr5.1 and Ubr5.2) with RA and assessed their ability to express Shh by qRT-PCR (Fig 2B). In comparison to the SCR control line, both Ubr5 shRNAi pools exhibited dramatically impaired Shh expression responses to RA stimulation. Interestingly, the initial Shh induction at 24 hrs was either unaffected (Ubr 5.2) or enhanced (Ubr5.1) over SCR control levels. However, both Ubr5 shRNAi pools failed to significantly increase Shh expression over time. In comparison to the maximal Shh expression achieved in the SCR pool at 72hrs post RA-treatment, Ubr5 shRNAi pools exhibited an approximately three-to-five-fold decrease. We therefore concluded that in our ES cell system UBR5 promoted RA-mediated Shh expression. This finding was in contrast to the identification of Hyd as a suppressor of hh expression [15], but did support a potential role for UBR5 in regulation of hh ligand expression.
Fig 2. UBR5 is required for RA-mediated induction of Shh.
Pools of murine E14 mES cells expressing either scrambled control (SCR) or Ubr5 shRNAi were tested for (A) UBR5 expression by SDS-PAGE and Western blotting, using Tubulin as a loading control; (B) RA-mediated Shh induction by qRT-PCR (n = 3, relative expression levels to β-actin, s.e.m indicated) or (C) expression analysis of the indicated ES cell pluripotentcy markers in shRNAi pools or parental ES cells by sqRT-PCR. For (B) comparison of all matched time points post-RA-treatment between control and Ubr5 shRNAi pools revealed statistically significant differences (p = <0.01), apart from the comparison between control and Ubr5.2 at 24 hours, which was not significant (ns). Statistical analysis by one-way ANOVA and Tukey multiple comparison test.
The inability of Ubr5 shRNAi cells to maintain Shh expression could have been explained by (i) an inability of ES cells to produce Shh or (ii) differentiation of ES cells into a non-Shh-expressing cell types. To address the latter, we assessed expression of a number of ES-cell associated markers (Nanog)[42], Klf-4[43] and Bmp-4[42, 44]. sqRT-PCR analysis revealed no dramatic differences in expression levels between SCR and Ubr5shRNAi pools (Fig 2C). Therefore, based on expression analysis of a limited set of markers, Ubr5 shRNAi did not appear to affect ES pluripotentcy. While we cannot rule out a contribution of altered ES differentiation, we believe UBR5 cells plays a more direct role in influencing RA-mediated Shh expression within ES cells.
Ubr5mt/mt embryos phenocopy the Ubr5 null phenotype
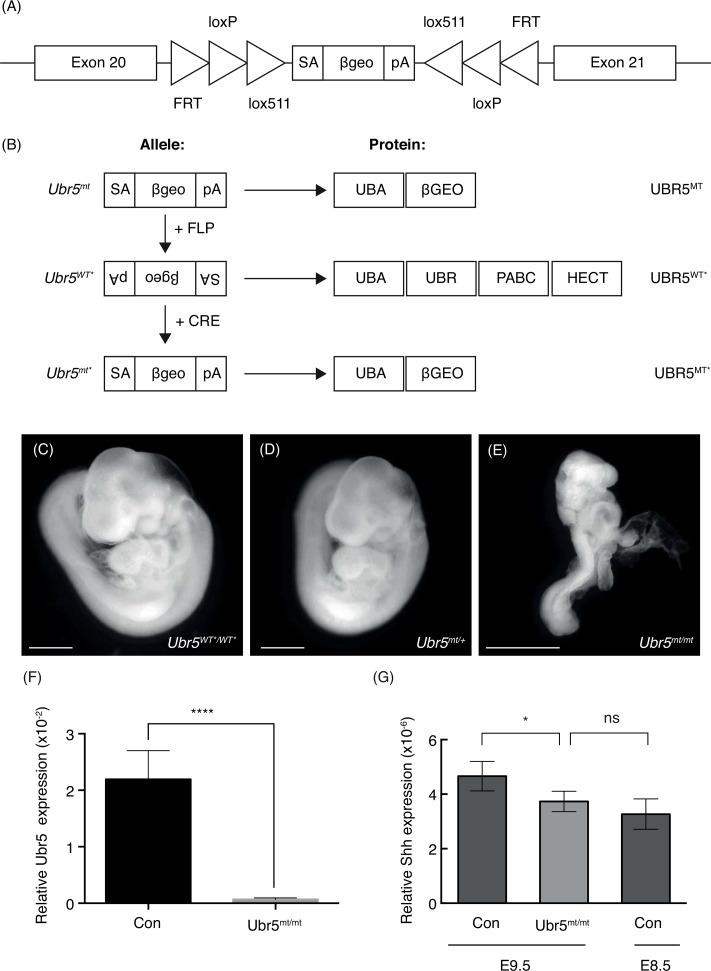
Due to its well-described roles in limb patterning and development[1], we chose to examine the effects of loss of Ubr5 function in limb bud mesenchyme. As Ubr5 null embryos are embryonic lethal [45] we developed a conditional mutant allele. We utilised a EUCOMM Ubr5 conditional gene trap (Ubr5gt) inserted between exons 20–21 (Fig 3A) to interfere with Ubr5 mRNA expression and UBR5 protein function. The gene trap was capable of existing in three distinct states [46] depending on the sequential action of FLP- and CRE-recombinases (Fig 3B): (1) in the absence of recombinase the gene trap resided in the mutagenic orientation (Ubr5mt); (2) after FLP-mediated recombination, in the non-mutagenic orientation (Ubr5WT*); and (3) after CRE-mediated recombination, in the mutagenic orientation (Ubr5mt*). When in the mutagenic orientation the predicted fusion protein consists of UBR5’s N-terminal UBA domain fused with a β-galactasidase:aminoglycoside 3' phosphotransferase fusion protein (βGEO); with the βGEO protein conferring both X-gal staining and Neomycin resistance. Such a fusion of UBR5’s UBA domain with βGEO, herein referred to as UBR5MT, was predicted to be severely functionally impaired due to lack of its important domain-associated functions: (i) E3 activity due to loss of the C-terminal catalytic HECT domain [20], (ii) N-end rule function through loss of the UBR domain [17] and (iii) miRNA regulatory function due to the absence of its Poly(A)-binding protein C-terminal (PABC) domain [47].
Fig 3. The Ubr5 gene trap phenocopies the Ubr5 null phenotype.
(A) Schematic representation of the EUCOMM EUCE0171f01 gene trap introduced in between exons 20 and 21 of the murine Ubr5 gene. FRT, loxP and lox511 recombination sites, and their orientation, are indicated as triangles and flank the gene trap encoding for a splice acceptor site (SA), a LacZ and neomycin CDS (βgeo) and a poly-adenylation signal (pA). (B) Schematic representation of the effects of FLP and Cre-mediated recombination on the gene trap and UBR5 protein/domain expression–see text for details. (C-E) Brightfield images of control (C) Ubr5WT*/WT*, (D) Ubr5mt/+ and (E) Ubr5mt/mt E9.5 embryos. Scale bar = 1mm. Whole embryo qRT-PCR analysis of (F) Ubr5 at E9.5 and (G) Shh expression in the indicated genotypes at E9.5 or E8.5. Con = Ubr5+/+. n = ≥ 6, s.e.m indicated. Statistical analysis by Students t-test. ns = not significant.
To confirm that the UBR5MT fusion protein was functionally impaired, we compared FLP-treated (Ubr5WT*/WT*) controls (Fig 3C), heterozygous (Ubr5mt/+) (Fig 3D) and homozygous (Ubr5mt/mt) Ubr5mt (Fig 3E) E9.5 embryos. Ubr5mt/mt embryos, in comparison to control or heterozygous embryos, appeared developmentally abnormal (Fig 3E), with an estimate of the apparent development stage being around E8.5. As expected these animals also exhibited a dramatic decrease in Ubr5 expression (Fig 3F). Furthermore, qRT-PCR analysis of whole embryo extracts revealed a small, but significant, decrease in Shh expression in E9.5 Ubr5mt/mt embryos (Fig 3G). This observation alone provided some support to the idea that UBR5 influenced Shh expression in vivo, even though the effects were small. However, we reasoned that the reduction in Shh expression might have simply reflected the developmental retardation of Ubr5mt/mt embryos. In agreement, Ubr5mt/mt Shh expression levels more closely resembled that of control E8.5 embryos (Fig 3G). These observations suggested that Ubr5mt/mt embryos exhibited molecular signatures (i.e., reduced Shh expression) that potentially reflected their state of embryonic retardation.
To ensure that the Ubr5mt allele functionally phenocopied the embryonic lethality of the null allele [45], we analysed the progeny of a heterozygous cross (Ubr5mt/+). The resultant litters revealed a total absence of Ubr5mt/mt animals (Table 1). Hence, we were reassured that the Ubr5 gene-trap functionally phenocopied the Ubr5 null.
Table 1. E11.5 Ubr5mt/mt embryos are not viable.
| Genotype | Expected Frequency (%) | Observed Frequency (%) |
|---|---|---|
| Ubr5+/+ | 25% | 26% (n = 7) |
| Ubr5mt/+ | 50% | 74% (n = 20) |
| Ubr5mt/mt | 25% | 0% (n = 0) |
Embryos from a Ubr5mt/+ cross were genotyped and their representation expressed as a percentage of the total litter size. n = 27. Chi squared test p = 0.0071.
Ubr5mt/mt embryos do not resemble Shh null embryos
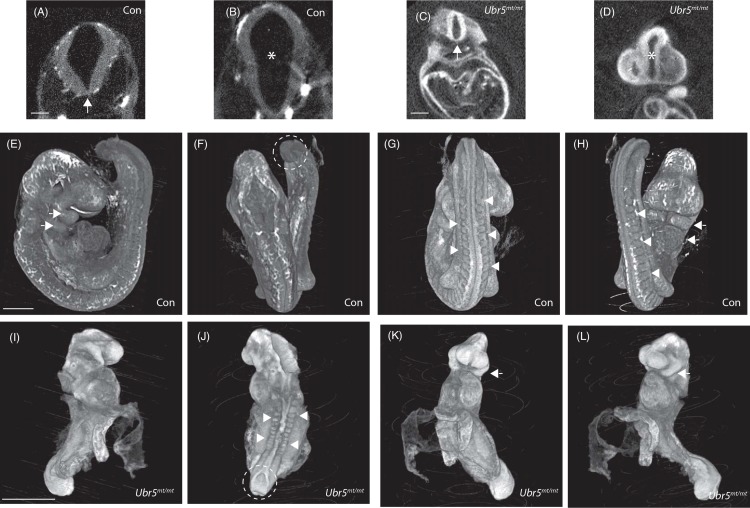
We next used optical projection tomography (OPT) to compare control (CD1) and Ubr5mt/mt E9.5 embryos in more detail (Fig 4). Specifically we wished to address whether the Ubr5mt/mt embryos bore any resemblance to Shh defective embryos. Embryos deficient in Shh die during late embryonic development and display disruption of midline structures, cyclopia and limb deformities >E10.5 [48, 49]. Unfortunately, the death and reabsorption of Ubr5mt/mt embryos by E11.5 precluded detection of any Shh-associated mid-gestation phenotypes. However, additional Shh null defects that present earlier than E10.5 were not apparent in E9.5 Ubr5mt/mt embryos: namely defects in the neural tube / floor plate (Fig 4 compare A and C, arrows) or closure of the dorsal and ventral surfaces around the dicephalic-mesencephalic junction (Fig 4 compare B and D, asterisk). Nevertheless, OPT did confirm that the morphology of these structures were clearly different to that of the age matched control embryos.
Fig 4. E9.5 Ubr5mt/mt embryos are developmentally abnormal.
Optical projection tomography images of Ubr5+/+ (Con) (A,B, E-H) and Ubr5mt/mt (C,D, I-L) E9.5 embryos. Both Ubr5mt/mt and Con embryos formed a neural floorplate (A,C arrows) and separated the plates of the diencephalic-mesencephalic junction (B,D asterisks). In comparison to control embryos, E9.5 Ubr5mt/mt embryos exhibited numerous developmental defects: an open posterior neuropore (J dashed line), irregular somites that were also reduced in number (compare G,H with J, arrowheads); lordotic curvature (L) and only one pair of pharyngeal arches (compare E,H with K,L arrows). n = 1. Scale bar (A-C) = 200μm and (E-L) = 1mm.
Further comparisons revealed that the E9.5 Ubr5mt/mt animals exhibited (i) kyphotic, rather than lordotic, curvature of the spine (Fig 4, compare E,I), (ii) an open, rather than closed, posterior neuropore (Fig 4, compare F,J dashed lines), (iii) one, rather than two, clearly defined pharyngeal arches (Fig 4, compare E,H with K,L, arrows) and (iv) ~12, rather than 24, somite pairs (Fig 4, compare G,H with J, arrowheads). Furthermore, there was some evidence of somite misalignment (Fig 4J), a defect associated with loss of Shh function [49].
Taken together these results supported the idea that Ubr5mt/mt embryos were clearly developmentally abnormal. However the E9.5 Ubr5mt/mt embryos did not resemble Shh null animals, but did phenocopy the previously reported Ubr5 null morphological phenotype [45].
UBR5 function is required for mid-gestation embryonic viability
To address the role of Ubr5 at later stages and to examine any Hh-associated defects occurring during midgestation (>E10.5) we coupled the conditional Ubr5WT* allele with the tamoxifen inducible Cre-ERT2 system [50] driven by expression from the artificial CAGG promoter [51]. Pregnant females bearing Cre-ERT2;Ubr5WT*/WT* progeny were injected daily with tamoxifen at E10.5 over four days. Genotyping the embryos at E15.5 revealed a total absence of the Cre-ERT2; Ubr5mt*/mt* genotype and a reduction in the observed/expected frequency of Cre-ERT2; Ubr5mt*/+ embryos (Table 2). In a further attempt to recover Ubr5mt*/mt* embryos for morphological analysis, we repeated the experiment and harvested the embryos at E13.5. Again, E13.5 analysis revealed the same absence of the Cre-ERT2; Ubr5mt*/mt* embryos (Table 3), as well as a detrimental effect on heterozygous embryo viability (Tables 2 and 3). These results suggested that during midgestation Ubr5 gene dosage was important for its embryonic function. Importantly, tamoxifen-mediated Cre activity alone (Cre-ERT2) had no detrimental effects on embryonic viability. Taken together this data suggested that the Ubr5WT* allele was amenable to Cre-mediated conversion into the Ubr5mt* allele and that UBR5 function was required for mid-gestation embryonic viability.
Table 2. Ubr5 is required for embryonic mid-gestational viability—E15.5 analysis.
| Genotype | Expected Frequency (%) | Observed Frequency (%) |
|---|---|---|
| Ubr5+/+ | 12.5 | 14 (n = 5) |
| Ubr5+/+; CreERT2 | 12.5 | 25 (n = 9) |
| Ubr5WT*/+ | 25 | 36 (n = 13) |
| Ubr5mt*/+; CreERT2 | 25 | 11 (n = 4) |
| Ubr5WT*/WT* | 12.5 | 11 (n = 4) |
| Ubr5mt*/mt*; CreERT2 | 12.5 | 0 (n = 0) |
Table 3. Ubr5 is required for embryonic mid-gestational viability—E13.5 analysis.
| Genotype | Expected Frequency (%) | Observed Frequency (%) |
|---|---|---|
| Ubr5+/+ | 12.5 | 20 (n = 7) |
| Ubr5+/+; CreERT2 | 12.5 | 29 (n = 10) |
| Ubr5+/WT* | 25 | 20 (n = 7) |
| Ubr5+/mt*; CreERT2 | 25 | 3 (n = 1) |
| Ubr5WT*/WT* | 12.5 | 29 (n = 10) |
| Ubr5mt*/mt*; CreERT2 | 12.5 | 0 (n = 0) |
Ubr5 function is required for mid-gestation viability. Pregnant females were injected i.p with tamoxifen at E11.5 and embryos genotyped at E13.5 (Table 3) and E15.5 (Table 2). Chi squared analysis: E15.5 = p 0.0179 and E13.5 = p = 0.0001.
Ubr5/UBR5 is expressed in the limb buds
Once the Ubr5gt construct had been functionally validated, we chose to utilise the gene trap’s lacZ reporter gene to establish UBR5 protein expression, with a particular focus on the limbs. E9.5, E10.5 and E11.5 embryos heterozygous for Ubr5mt underwent X-Gal staining (Fig 5A–5F). No significant staining was detected in control Ubr5+/+ embryos (Fig 5A, 5C and 5E). In contrast, Ubr5mt/+ embryos at E9.5, E10.5 and E11.5 (Fig 5B, 5D and 5F, respectively) exhibited widespread low-level X-Gal staining in addition to well-defined stronger signals. At all stages Ubr5mt/+ embryos revealed distinct signals associated with the dorsal edge (Fig 5B, 5D and 5F arrows) in addition to signals in the pharyngeal arches and forebrain (Fig 5B, 5D and 5F dashed lines labelled PA and FB, respectively). At E9.5, a signal within the body core potentially indicated expression within the foregut (Fig 5B, labelled FG). At E10.5, the staining along the dorso-lateral edge shifted posteriorly towards the tail (Fig 5D, posterior arrow) and by E11.5, the striated β-Gal activity was still present along the dorsal edge, but had become more medial (Fig 5F, arrows). Additionally, X-Gal staining became more prominent in the forebrain (Fig 5 dashed lines, FB) and body core (Fig 5F asterisk). Staining in both fore- and hind-limbs was detected at E10.5–11.5, with the fore-limbs exhibiting more robust staining (Fig 5D–5F dashed lines, FL and HL). In summary, the β-Gal activity exhibited dynamic expression patterns in a number of structures that included, at certain stages, both fore- and hind-limbs.
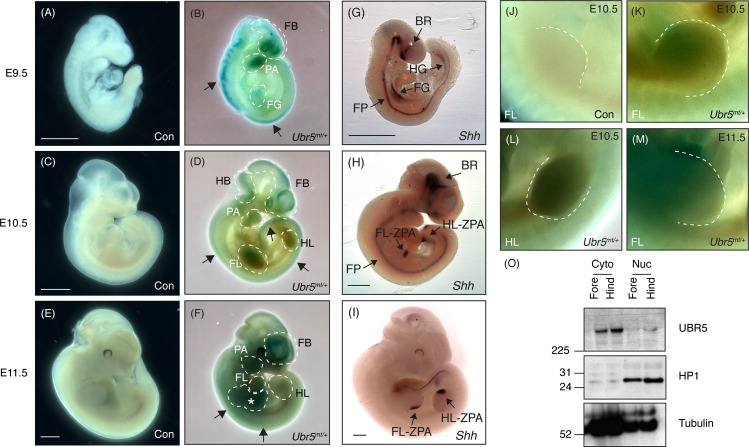
Fig 5. Expression analysis of UBR5MT, Shh and endogenous UBR5.
β-Gal activity was not detected in wild-type controls (Con) embryos (A,C,E), whereas Ubr5mt/+ embryos exhibited staining at (B) E9.5, (D) E10.5 and E11.5 (F). Dashed lines indicate areas of interest. Please see text for more details. (G-I) Shh in situ hybridisation of wild-type control embryos at the indicated embryonic stages. FB = forebrain; HB = hindbrain; PA = pharyngeal arches; FG = foregut; HG = hindgut; FL = forelimb; HL = hindlimb; BR = brain; FP = floorplate; ZPA = zone of polarising activity; an asterisk marks a potential gastrointestinal signal. Scale bars = 1mm. (J-M) Higher magnification images of E10.5 (J-L) and E11.5 (M) limbs of the indicated genotypes. A dashed line outlines the limb bud margins. Representative images shown from n = ≥3 for each stage and genotype. (O) SDS-PAGE and Western blotting of E11.5 fore- and hind-limb lysates for UBR5 expression in cytosolic (Cyto) and nuclear (Nuc) fractions. HP1 (a nuclear marker) and β-tubulin (a predominantly cytosolic marker) were used as loading and fractionation controls. n = 1.
We next wished to compare UBR5-associated β-Gal activity with Shh expression patterns. Shh in situ hybridisation at E9.5, E10.5 and E11.5 (Fig 5G–5I) revealed the expected expression in the fore- and hind-gut (labelled FG and HG, respectively), brain (labelled BR), floorplate of the neural tube (labelled FP) and the ZPAs of the fore- and hind-limbs (labelled FL-ZPA and HL-ZPA, respectively). By E11.5 Shh expression was predominantly restricted to the limb buds and the zones of polarising activity (Fig 5H and 5I labelled ZPA). In contrast to the posterioirised Shh expression in the forelimb, UBR5-associated β-Gal activity appeared to be predominantly in the anterior regions of the E10.5 and E11.5 forelimb buds (Fig 5K–5M, respectively). However, in the E10.5 hindlimb β-Gal activity was evenly expressed across the entire limb bud (Fig 5L). In general, the overall expression patterns of Shh and UBR5-associated β-Gal activity revealed no strong evidence for either a clear mutually-exclusive or -inclusive expression pattern. However, Ubr5 expression within the limb buds did provide a possibility for UBR5 to tissue autonomously regulate Hh signalling within the embryonic limb.
The β-Gal assays suggested that Ubr5/UBR5mt was expressed in E10.5 and E11.5 limb buds. To confirm that endogenous UBR5 protein was also expressed, we used SDS-PAGE and Western blotting of E11.5 limb lysates (Fig 5O). Samples were separated into crude nuclear and cytoplasmic fractions and blotted for UBR5 and cytoplasmic (tubulin) and nuclear (HP-1) markers. Analysis revealed that UBR5 was expressed in both the fore- and hind-limbs and was present in both the cytoplasmic and nuclear fractions. Due to potential cytoplasmic contamination, as indicated by the presence of β-tubulin in the nuclear fraction, we were unable to conclude that UBR5 is nuclear localised in limb bud cells. Nevertheless, techniques to identify UBR5:βGeo-associated β-Gal activity (Fig 5B, 5D, 5F and 5K–5M) and endogenous protein expression (Fig 5O) revealed that Ubr5/UBR5 was expressed in E11.5 embryonic limb buds.
Prx-Cre1-mediated recombination of the Ubr5WT* allele effectively suppresses Ubr5 expression in developing limbs
The combination of (i) our observation of UBR5’s effects on RA-mediated Shh expression, (ii) the importance of RA and SHH-mediated signalling in limb development and (iii) Ubr5/UBR5 expression within the limb prompted us to delete UBR5 expression in the developing limb. Accordingly, we chose to combine the Ubr5WT* allele with Prx1-mediated expression of Cre recombinase (Prx1-Cre). Expressed throughout the E9 limb bud mesenchyme, Prx-1-Cre mediates recombination in all mesenchymal cells by mid-bud development at E11 [36]. To confirm the efficiency of Prx1-Cre-mediated recombination, control and experimental E13.5 embryos were assayed for β-gal activity (Fig 6A–6C). Note that F1 progeny bearing both Prx1-Cre and Ubr5WT*/WT* were assumed to have undergone temporally- and spatially-restricted recombination and from herein will be indicated as Prx1-Cre;Ubr5mt*/mt*.
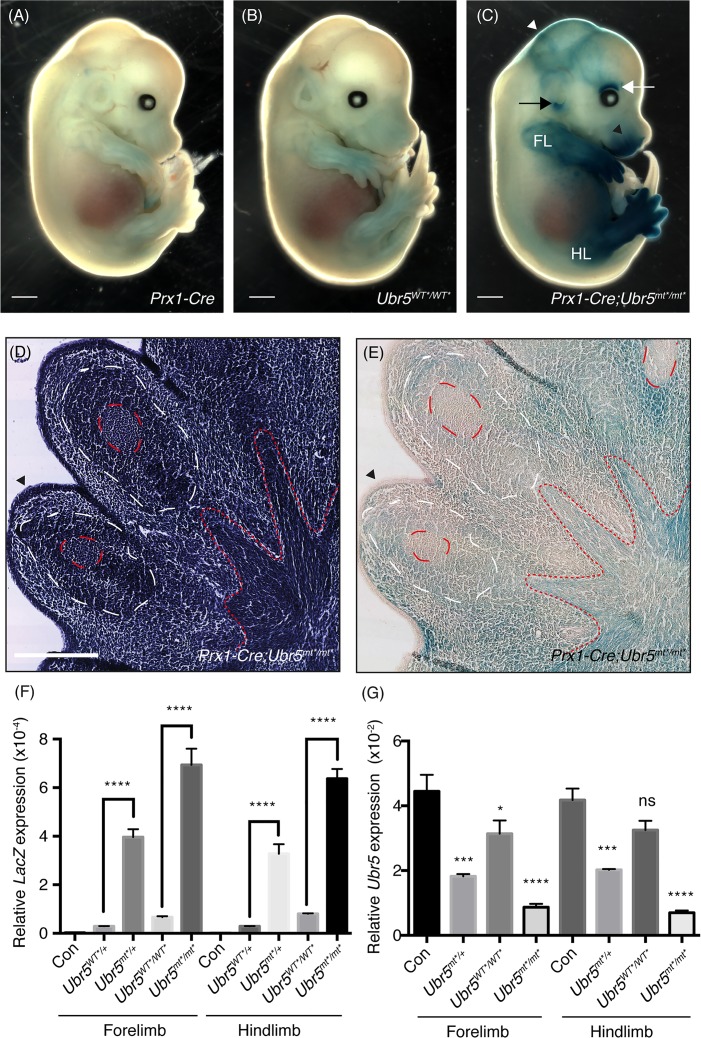
Fig 6. Prx-Cre1-mediated recombination of the Ubr5gt and loss of Ubr5 expression.
Prx1-Cre promotes UBR5MT-associated β-Gal activity (A-E), lacZ expression (F) and (G) loss of Ubr5 expression. (A-C) E13.5 embryos of the indicated genotypes stained for β-Gal activity revealed strong X-Gal staining in the embryonic limbs, in addition to other regions: above the eye, white arrow; the ear, black arrow; snout black arrowhead and head, white arrowhead. Scale bar = 1mm. (D-E) Histological sections through the footplate revealed a non-uniform UBR5MT expression pattern (red dashed lines = chondrocyte condensations; white dashed lines = dark staining mesenchyme and red dotted line = developing sinews). Scale bar = 250μm. qRT-PCR analysis of (F) LacZ expression and (G) full-length Ubr5 in dissected E13.5 fore- and hind-limb buds. Prx1-Cre in combination with heterozygosity or homozygosity for the Ubr5mt* allele exhibited (F) a dramatic increase in LacZ expression and (G) a dramatic decrease in Ubr5 expression in comparison to Prx1-Cre only (Con). The primer pairs in (G) were specific for a gene region absent in the Ubr5mt* transcript. All differences relative to the appropriate Prx1-Cre controls (Con) were statistically significant (at least p = <0.05), apart from the comparison between Con and Ubr5mt*/mt*, which was not significant (ns). n = ≥3, s.e.m indicated. Statistical analysis by one-way ANOVA and Tukey multiple comparison test.
Prx-Cre;Ubr5mt*/mt* embryos exhibited robust β-gal activity within the limbs (Fig 6C) that was absent in Ubr5+/+ and Ubr5WT*/WT* controls (Fig 6A and 6B, respectively). In Prx-Cre;Ubr5mt*/mt* embryos, significant signals were also detected outside of the limbs and included regions above the eye (Fig 6C white arrow), around the ear (Fig 6C black arrow), the snout (Fig 6C black arrowhead) and head (Fig 6C, white arrowhead). These observations indicated that both Prx1-Cre and Ubr5 were expressed within, as we well as outside of, the limb fields. In summary, these results confirmed (i) efficient Cre-mediated gene conversion and (ii) Ubr5/UBR5 expression within the fore- and hind-limbs.
Haematoxylin-based histological examination of the Prx-Cre;Ubr5mt*/mt* footplate revealed a dark staining epidermal layer (Fig 6D, arrowhead), chondrocyte condensations within the developing digits and surrounding dark staining mesenchymal cells (Fig 6D, red and white dashed lines respectively). Proximally extending structures indicative of developing sinews radiated out from the mid-region of the footplate (Fig 6D, red dotted lines). Examination of an adjacent unstained section revealed widespread β-Gal activity (UBR5MT) across the footplate (Fig 6E). Regions of high expression appeared to reside around and between the chondrocyte condensations as well as within the developing sinews. In contrast, β-Gal activity appeared to be either low or absent within the chondrocyte condensations and epidermis (Fig 6, red dashed line and arrowhead, respectively). In summary, within the Prx1-Cre;Ubr5mt*/mt* forelimb, UBR5MT exhibited a non-uniform expression pattern and appeared to be excluded from condensing chondrocytes.
We next carried out qRT-PCR analysis on Prx-Cre;Ubr5mt*/mt* limbs to quantify the extent of gene conversion through increased LacZ expression (Fig 6F) and confirm that Ubr5 expression was also reduced (Fig 6G). LacZ primers targeting expression of Ubr5mt* mRNA revealed no signal in Prx1-Cre animals, but detected low-levels in heterozygous and homozygous Ubr5WT*-bearing animals that lacked Prx1-Cre. Nevertheless, the presence of Prx1-Cre promoted a 15–40 fold increase in LacZ expression in either heterozygous or homozygous Ubr5mt* backgrounds (Fig 6F). Interestingly Prx1-Cre;Ubr5mt*/mt* exhibited only ~33% more LacZ expression than Prx1-Cre;Ubr5mt*/+, suggesting either non-linear gene dosage effects and/or limiting recombinase activity.
To quantify Ubr5 expression, we used primer pairs complementary to the 3’ end of Ubr5 mRNA to discriminate endogenous and Ubr5WT* from Ubr5mt*, alleles. In agreement with the increased β-Gal staining (Fig 6C) and LacZ expression analysis (Fig 6F), full-length Ubr5 expression levels were significantly reduced in Prx1-Cre limbs either heterozygous or homozygous for Ubr5mt* (Fig 6G); with the greatest reduction occurring in the homozygous Ubr5mt*/mt* limbs. Surprisingly, homozygosity for the non-mutagenic Ubr5WT* allele, in the absence of Prx1-Cre, caused a small but significant decrease in Ubr5 expression. This observation indicated that physical insertion of the gene-trap (Fig 6F) had a detrimental effect on Ubr5 mRNA expression.
Ubr5mt E13.5 limbs exhibit decreased Hedgehog pathway activity
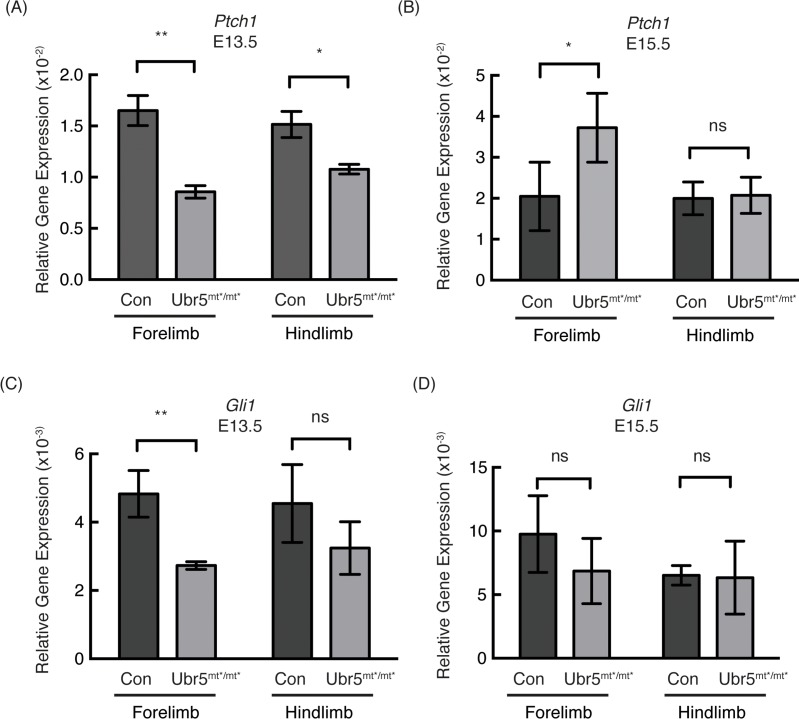
Having established that Ubr5 was expressed in the limb and that the Ubr5gt was functional, we wanted to address whether loss of Ubr5 expression in the developing limb would affect Hedgehog signalling (Fig 7). qRT-PCR analysis of the Hh pathway target gene Ptch1 revealed that at E13.5 HhP activity was repressed in both the fore- and hind-limb (Fig 7A). At E15.5, the levels of Ptch1 expression appeared to be either overcorrected (forelimb) or corrected (hindlimb) relative to control levels (Fig 7B). Similar to Ptch1 expression, Gli1 expression was also repressed in both limbs, although only the reduction in the forelimb was statistically significant (Fig 7C) and was then corrected, to some degree, by E15.5 (Fig 7D).
Fig 7. Loss of Ubr5 function resulted in decreased expression of Ptch1 and Gli1 at E13.5.
(A-D) qRT-PCR analysis of Prx1-Cre (Con) and Prx1-Cre;Ubr5mt*/mt* (Ubr5mt*/mt*) E13.5 or E15.5 embryonic fore- and hind-limbs for Ptch1 (A,B) and Gli1 (C,D) expression. qRT-PCR of target genes were normalised against β-actin (n = ≥3, s.e.m indicated). Statistical analysis by Students t-test. ns = not significant.
In summary, at E13.5 it appeared that loss of Ubr5 in the forelimb resulted in a decrease in the HhP’s transcriptional outputs. Within the hindlimb, however, the reduction in HhP transcriptional output was less pronounced, Some of the differences between fore- and hind-limbs may have reflected the difference in developmental timing between the two structures, with the forelimb initiating development prior to the hindlimb (EMouse Atlas Project). Overall, these results suggested that loss of Ubr5 function resulted in a transient repression of Hh signalling.
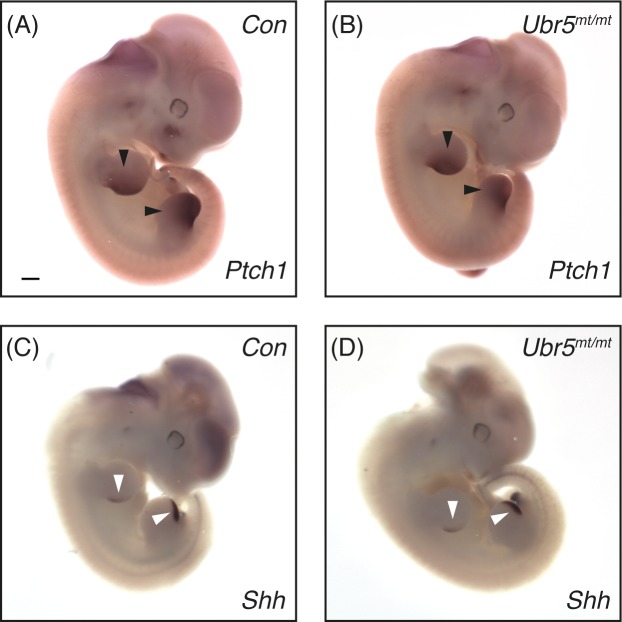
Significant quantitative changes in Ptch1 gene expression at E13.5 led us to investigate its spatial expression pattern at E11.5. Using ISH, we examined Ptch1 expression in both fore- and hind-limb buds of E11.5 embryonic fore- and hind-limbs of control Prx1-Cre (Fig 8A) and experimental Prx1-Cre; Ubr5mt/mt (Fig 8B) embryos. Comparison between the two genotypes revealed no dramatic differences in spatial expression patterns, although Ubr5mt/mt hindlimbs exhibited a small reduction in Ptch1 signal intensity (Fig 8B). Analysis of Shh expression showed no differences in its spatial expression patterns (Fig 8C and 8D). Taken together these data revealed that Prx1-Cre-mediated loss of UBR5 function did not dramatically affect Shh or Ptch1 spatial expression patterns in E11.5 embryonic limbs. In combination with the qRT-PCR analysis, we concluded that in Ubr5mt limbs the effect on Hedgehog signalling were transient and predominantly affected the magnitude of mRNA expression around E11.5-E13.5.
Fig 8. Loss of Ubr5 at E11.5 does not affect Shh or Ptch1 expression domains.
In situ hybridisation for expression of Hh signalling components Ptch1 (A,B) and Shh (C,D) in E11.5 embryos of Prx1-Cre (Con) and Prx1-Cre;Ubr5mt*/mt* (Ubr5mt*/mt*) embryos. Analysis revealed no significant effects on either Shh or Ptch1 expression patterns in the Ubr5mt*/mt* embryos (representative image from n = >4 of each genotype). Arrowheads indicate Shh ZPA expression domains. Scale bar = 0.5mm.
Ubr5-deficient embryonic limbs do not exhibit digit abnormalities
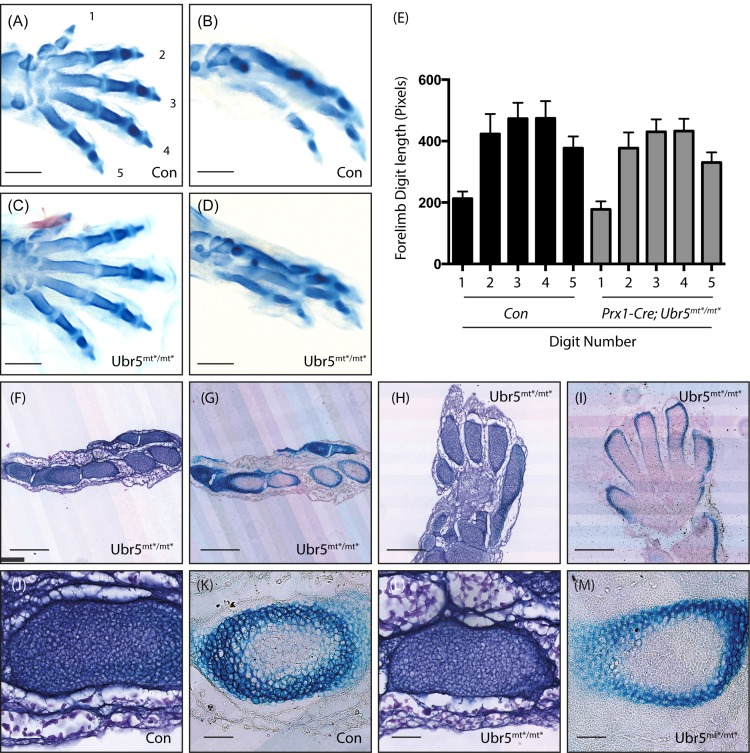
Although the E11.5 limb buds showed no obvious changes in the spatial expression of Shh, the E13.5 limbs did exhibit a reduction in HhP activity. We hypothesised that even a transient reduction might manifest a subsequent morphological/developmental defect. Accordingly, we examined the digits of E15.5 control Prx1-Cre and Prx1-Cre, Ubr5mt*/mt* fore- and hind-paws, which revealed no gross abnormalities (Fig 9A–9D). Although measurement of Ubr5mt*/mt* forelimb digits revealed a reduction in average length (Fig 9E), the effects proved not to be statistically significant.
Fig 9. Ubr5mt*/mt* limbs and digits appear morphologically normal.
Analysis of fore- (A,C, H, I) or hind-paws (B,D,F,G, J-M) of Prx1-Cre (Con) and Prx1-Cre;Ubr5mt*/mt* (Ubr5mt*/mt*) E15.5 embryos. (A-D) No obvious morphological difference were apparent between Con (A,B) and Ubr5mt*/mt* (C,D) digits. Scale bar = 1mm. (E) Measurement of forelimb digit length, numbered as in (A), revealed a reduction in the average length in Ubr5mt*/mt* animals. However, none of the comparisons between matching digits were statistically significant (p = >0.05). n = ≥6, s.e.m indicated. Statistical analysis by one-way ANOVA and Tukey multiple comparison tests. (F-M) Histological analysis of haematoxylin (F,H,J,L) and alcian blue (G,I,K,M) stained material. Histology and morphology of Ubr5mt*/mt* hind- (F,G) or fore-paws (H,I) appeared histologically and morphologically normal, (J-M) as did chondrocyte clusters within the hind-paw. Scale bar = 250μm (F-I) and 50μM (J-M).
Histological analysis of Prx1-Cre;Ubr5mt*/mt* mutant feet and paws using haematoxylin or alizarin red/alcian blue revealed no obvious defects in digit formation, or composition (Fig 9F–9I). Closer analysis of Prx1-Cre and Prx1-Cre;Ubr5mt*/mt* developing tarsals (Fig 9J–9M) also revealed no dramatic effects on chondrocyte condensations (Fig 9 compare J,L) or upon the associated deposition of Alcian-blue-reactive acidic polysaccharides (cartilage) (Fig 7 compare K,M). Therefore, it appeared that there were no obvious morphological/histological defects in Ubr5mt*/mt* digits.
Ubr5-deficient embryonic limbs exhibit reduced Ihh expression
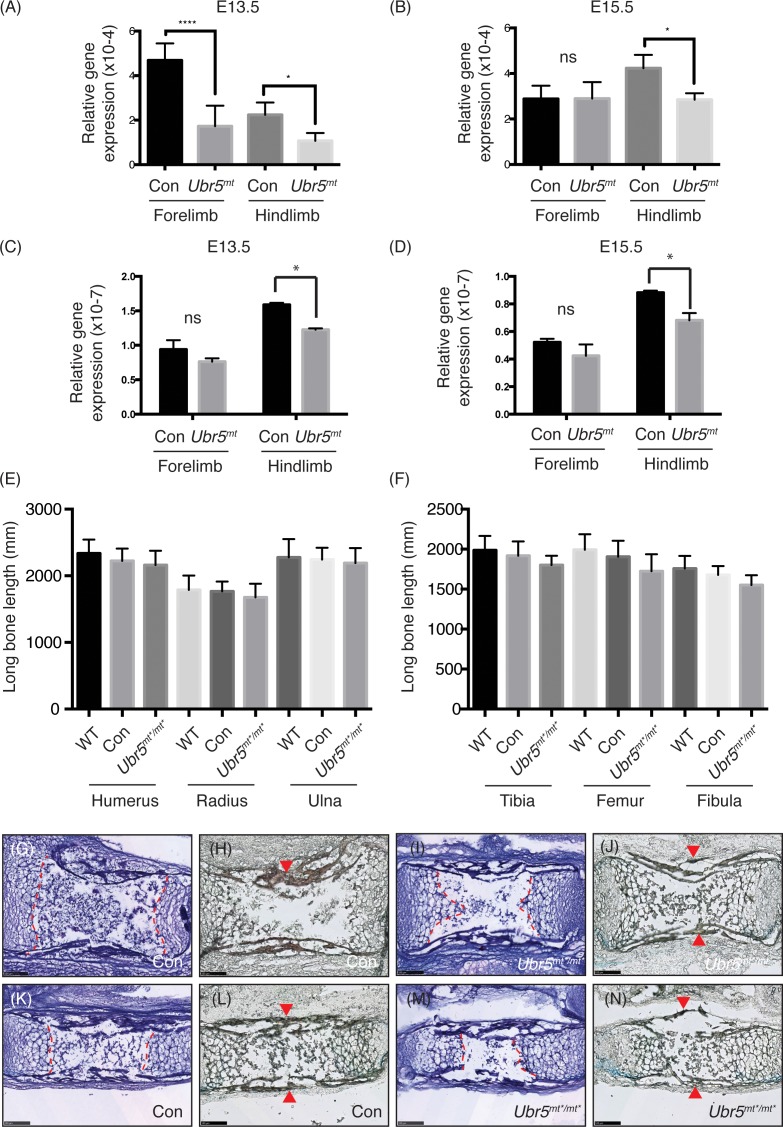
The lack of any digit phenotype suggested that the observed changes in HhP activity had no impact on digit development. In simplistic terms, Shh-mediated signalling controls limb/digit formation; whereas, Ihh regulates the subsequent growth, maturation and homeostasis of the limbs’ long bones [52, 53]. We therefore hypothesised that reduced Ihh expression could account for the decrease in HhP activity and might elicit an effect on embryonic long bone length. qRT-PCR analysis revealed a significant reduction in Ihh expression in E13.5 Prx1-Cre;Ubr5mt*/mt* limbs (Fig 10A), with only the reduction in the hind limbs persisting through to E15.5 (Fig 10B).
Fig 10. Ubr5mt*/mt* limbs exhibit reduced Ihh expression.
(A-B) Ihh and (C,D) Shh qRT-PCR analysis of control Prx1-Cre (Con) or Prx1-Cre; Ubr5mt*/mt* (Ubr5mt) fore- or hind-limbs at (A,C) E13.5 or E15.5 (B,D) revealed significant decreases in Ubr5mt limbs versus Con. qRT-PCR values were normalised against β-actin (n = 3, s.e.m indicated). Statistical analysis by Students t-test. (E,F) Measurement of the length of the indicated long bones in Ubr5+/+ (WT), Prx1-Cre (Con) or Prx1-Cre;Ubr5mt*/mt* (Ubr5mt*/mt*) animals revealed no statistically significant reductions in length. Statistical analysis by ANOVA and Tukey multiple comparison test. (G-N) Histological analysis of haematoxylin (G,I,K,M) and alcian blue + alizarin red (H,J,L,N) stained humerus (G-J) and ulna (K-N) revealed no obvious histological or morphological differences. Dashed lines = border between chondrocytes and site of primary ossification and arrowheads = alizarin red positive cortical bone. Scale bar = 100μm.
At E13.5-E15.5 Shh expression in the limb bud is dramatically reduced[54–56] and Shh analysis revealed no significant difference between control and Prx1-Cre;Ubr5mt*/mt* fore limbs at either time point (Fig 10C and 10D). However, at both developmental stages, Prx1-Cre;Ubr5mt*/mt* hindlimbs exhibited small, but significant decreases (Fig 10C and 10D). It should be noted that the Shh detected at >E13.5 was most likely in non-mesenchymal-derived developing hair follicles[57] and therefore not directly affected by Prx1-Cre-mediated loss of Ubr5 function. Overall, our data suggested that a reduction in Ihh, rather than Shh, expression potentially accounted for the observed reduction in HhP activity at E13.5 (see Fig 7).
Due to IHH’s well-established role in regulating bone growth [52, 53] we measured the length of the Prx1-Cre;Ubr5mt*/mt* long bones at E15.5 (Fig 10E and 10F). Similarly to what was observed in the digits, Prx1-Cre;Ubr5mt*/mt* long bones were on average shorter than those of Ubr5+/+ (WT) or Prx1-Cre (Con) control animals. However, the effects were not statistically significant. Finally, we chose to examine the E15.5 long bones for morphological defects. As in the digits, histological analysis of the control and Ubr5mt humerus (Fig 10E–10HJ) and ulna (Fig 10K–10N) revealed no obvious morphological differences or changes in alcian blue and alizarin red staining. In conclusion, loss of Ubr5 function correlated with a transient decrease in Ihh expression, but no obvious or statistically significant limb defects.
Discussion
In conclusion, our in vitro and in vivo findings support the initial hypothesis that UBR5 can regulate hh ligand family expression and HhP activity. However, we are uncertain as to how UBR5 influences Hedgehog signalling. One mechanism would involve an indirect route whereby UBR5 governs the production/maintenance of Hh-ligand producing / Hh-responsive cells. Such a mechanism could be extremely indirect, with defects occurring early on in development only manifesting a molecular or cellular consequence later on. Whereas a more direct role could involve UBR5 acting cell-autonomously to promote hh ligand mRNA expression and/or Hh pathway transcriptional outputs. Future efforts will attempt to resolve this uncertainty.
Our initial in vitro observations supported a cell autonomous role for UBR5 in promoting RA-mediated Shh expression. Although maximal Shh expression was impaired in UBR5-deficient ES cells, the initial RA-mediated induction was not. Therefore, these results potentially indicate two distinct phases in RA-mediated Shh expression: (i) an initial UBR5-independent induction phase and (ii) a subsequent UBR5-regulatable, amplification phase. However, we are uncertain as to how UBR5 may influence RA-mediated signalling. One possibility resides in UBR5’s ability to bind [58] the retinoic-acid responsive [59] progesterone receptor (PGR). Although no evidence exists that PGR induces Shh expression, it can promote Ihh expression [60, 61].
While our work supported a role for UBR5 in promoting Shh expression in ES cells, it was not supported by our in vivo studies. At E13.5 Prx1-Cre;UBr5mt*/mt* hindlimbs exhibited only small decreases in Shh expression that were not apparent in the forelimbs. However, significant decreases in Ihh expression and HhP activity were detected in fore- and hind-limbs at E13.5. Therefore, in Ubr5mt limbs reduced Ihh expression may have accounted for the reduction in HhP activity. The transient nature of these effects could indicate (i) a tightly restricted temporal window of UBR5 action, (ii) functional redundancy and/or (iii) existence of a compensatory/homeostatic mechanism.
Taken together our in vitro and in vivo findings imply that loss of Ubr5 function correlated with a reduction in hedgehog ligand expression and pathway activity. However, we did not establish the molecular mechanism by which UBR5 might influence transcription and/or half-life of hh-family mRNAs. A potential explanation resides in the ability of Drosophila and mammalian Hyd/UBR5 to bind Sgg/GSK3β [16, 62] and Ci/GLI [16, 63] as well as influence Sgg-mediated hh expression [16]. GSK3β’s role as a potent regulator of numerous transcription factors [64], raises the possibility that it may also regulate the activity of known hedgehog-regulatory transcription factors such as RUNX2 [65, 66], ATF4 [67], HAND2-TWIST1 [68, 69] and ETV4/5 [70].
While there is currently no evidence to suggest GSK3β regulates Shh or Ihh expression in mammals, it does bind, phosphorylate and repress GLI proteins [71, 72]. Hyd/UBR5’s ability to bind Sgg/GSK3β and Ci/GLI therefore provides it with the potential to influence Ci/GLI activity downstream of any effects on Shh/Ihh ligand expression. Finally, Hyd’s ability to bind chromatin also provides an alternative means to directly regulate gene expression [63]. Interestingly, the ability of one protein to independently regulate hh ligand expression and GLI activity is also observed with the Hh-pathway-associated kinase DYRK1B [14]. Together, these observations support the notion that initiation of Hh signalling (ligand production) and its response (GLI activity) can be independently co-ordinated by the action of a single protein.
In the Ubr5 null embryos, defective Hedgehog signalling may underlie the reported placental vascular defects [73] and embryonic death [48, 53]. Defective placental function may also explain our observed midgestation lethality of pCAGG-Cre;Ubr5mt*/mt* embryos, but we cannot exclude an essential role for UBR5 in the embryo proper. The detection of Ubr5 expression within the pharyngeal arches, a major source for artery development [74] could also provide an alternative/complementary means for UBR5 to influence embryonic vasculature. Our observation that mid-gestational loss of one copy of Ubr5 resulted in a reduction in the observed/expected progeny, suggested that correct Ubr5 gene dosage was required for mid-gestation viability. This is in contrast to a lack of any effects on progeny lacking one copy of Ubr5 from conception. We therefore hypothesise that a gene compensatory mechanism is able to cope with reduced Ubr5 gene dose from conception, but not in response to an acute decrease in UBR5 function mid-gestation.
The concepts of genetic redundancy and compensation may also explain why we observed no defect in the embryonic Prx1-Cre;Ubr5mt/mt limbs. A threshold model could also be a potential explanation, whereby only a certain magnitude of change in Hedgehog signalling would elicit a morphological defect. Such a model is strongly supported by the normal limb development of animals with a 50% reduction in Shh expression [75]. Furthermore, heterozygous mutant animals of Shh [48], Ihh [76] and Smo [77], also fail to exhibit developmental limb defects. Therefore, the morphological consequences associated with a reduction in, rather than complete loss of, a Hedgehog signalling component varies dramatically.
In summary, the generation and validation of a conditional Ubr5 mutant mouse provides a useful tool to address murine development and homeostasis. While our findings further highlight the importance of UBR5 in embryonic development and ES cell biology, they also reinforce its potential role in influencing Hedgehog signalling.
Acknowledgments
We thank Helen Caldwell for histology services.
Data Availability
All relevant data are within the paper.
Funding Statement
The work was funded by the Biotechnology and Biological Sciences Research Council (http://www.bbsrc.ac.uk) as part of a New Investigator Award BB/H012869/1 and by Edinburgh University as part of a Chancellor's Fellowship (both awarded to MD) and the Medical Research Council (http://www.mrc.ac.uk) as part of an IGMM core grant award (RH). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Briscoe J, Therond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nature reviews Molecular cell biology. 2013;14(7):416–29. Epub 2013/05/31. 10.1038/nrm3598 . [DOI] [PubMed] [Google Scholar]
- 2.Petrova R, Joyner AL. Roles for Hedgehog signaling in adult organ homeostasis and repair. Development. 2014;141(18):3445–57. 10.1242/dev.083691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Towers M, Wolpert L, Tickle C. Gradients of signalling in the developing limb. Curr Opin Cell Biol. 2012;24(2):181–7. 10.1016/j.ceb.2011.11.005 . [DOI] [PubMed] [Google Scholar]
- 4.Takebe N, Miele L, Harris PJ, Jeong W, Bando H, Kahn M, et al. Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: clinical update. Nat Rev Clin Oncol. 2015;12(8):445–64. 10.1038/nrclinonc.2015.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McMahon AP, Ingham PW, Tabin CJ. Developmental roles and clinical significance of hedgehog signaling. Curr Top Dev Biol. 2003;53:1–114. . [DOI] [PubMed] [Google Scholar]
- 6.Chen Y, Struhl G. Dual roles for patched in sequestering and transducing Hedgehog. Cell. 1996;87(3):553–63. . [DOI] [PubMed] [Google Scholar]
- 7.Marigo V, Davey RA, Zuo Y, Cunningham JM, Tabin CJ. Biochemical evidence that patched is the Hedgehog receptor. Nature. 1996;384(6605):176–9. Epub 1996/11/14. 10.1038/384176a0 . [DOI] [PubMed] [Google Scholar]
- 8.Stone DM, Hynes M, Armanini M, Swanson TA, Gu Q, Johnson RL, et al. The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature. 1996;384(6605):129–34. Epub 1996/11/14. 10.1038/384129a0 . [DOI] [PubMed] [Google Scholar]
- 9.Towers M, Tickle C. Growing models of vertebrate limb development. Development. 2009;136(2):179–90. 10.1242/dev.024158 . [DOI] [PubMed] [Google Scholar]
- 10.Riddle RD, Johnson RL, Laufer E, Tabin C. Sonic hedgehog mediates the polarizing activity of the ZPA. Cell. 1993;75(7):1401–16. . [DOI] [PubMed] [Google Scholar]
- 11.Lettice LA, Heaney SJ, Purdie LA, Li L, de Beer P, Oostra BA, et al. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet. 2003;12(14):1725–35. . [DOI] [PubMed] [Google Scholar]
- 12.Hill RE, Lettice LA. Alterations to the remote control of Shh gene expression cause congenital abnormalities. Philos Trans R Soc Lond B Biol Sci. 2013;368(1620):20120357 10.1098/rstb.2012.0357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson E, Hill RE. Long range regulation of the sonic hedgehog gene. Curr Opin Genet Dev. 2014;27:54–9. 10.1016/j.gde.2014.03.011 . [DOI] [PubMed] [Google Scholar]
- 14.Lauth M, Bergstrom A, Shimokawa T, Tostar U, Jin Q, Fendrich V, et al. DYRK1B-dependent autocrine-to-paracrine shift of Hedgehog signaling by mutant RAS. Nat Struct Mol Biol. 2010;17(6):718–25. 10.1038/nsmb.1833 . [DOI] [PubMed] [Google Scholar]
- 15.Lee JD, Amanai K, Shearn A, Treisman JE. The ubiquitin ligase Hyperplastic discs negatively regulates hedgehog and decapentaplegic expression by independent mechanisms. Development. 2002;129(24):5697–706. . [DOI] [PubMed] [Google Scholar]
- 16.Moncrieff S, Moncan M, Scialpi F, Ditzel M. Regulation of hedgehog Ligand Expression by the N-End Rule Ubiquitin-Protein Ligase Hyperplastic Discs and the Drosophila GSK3beta Homologue, Shaggy. PLoS One. 2015;10(9):e0136760 10.1371/journal.pone.0136760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kozlov G, Nguyen L, Lin T, De Crescenzo G, Park M, Gehring K. Structural basis of ubiquitin recognition by the ubiquitin-associated (UBA) domain of the ubiquitin ligase EDD. J Biol Chem. 2007;282(49):35787–95. . [DOI] [PubMed] [Google Scholar]
- 18.Matta-Camacho E, Kozlov G, Li FF, Gehring K. Structural basis of substrate recognition and specificity in the N-end rule pathway. Nat Struct Mol Biol. 2010;17(10):1182–7. Epub 2010/09/14. 10.1038/nsmb.1894 . [DOI] [PubMed] [Google Scholar]
- 19.Tasaki T, Mulder LC, Iwamatsu A, Lee MJ, Davydov IV, Varshavsky A, et al. A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons. Mol Cell Biol. 2005;25(16):7120–36. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matta-Camacho E, Kozlov G, Menade M, Gehring K. Structure of the HECT C-lobe of the UBR5 E3 ubiquitin ligase. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012;68(Pt 10):1158–63. Epub 2012/10/03. 10.1107/S1744309112036937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gudjonsson T, Altmeyer M, Savic V, Toledo L, Dinant C, Grofte M, et al. TRIP12 and UBR5 suppress spreading of chromatin ubiquitylation at damaged chromosomes. Cell. 2012;150(4):697–709. Epub 2012/08/14. 10.1016/j.cell.2012.06.039 . [DOI] [PubMed] [Google Scholar]
- 22.Okamoto K, Bartocci C, Ouzounov I, Diedrich JK, Yates JR 3rd, Denchi EL. A two-step mechanism for TRF2-mediated chromosome-end protection. Nature. 2013;494(7438):502–5. Epub 2013/02/08. 10.1038/nature11873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang T, Cronshaw J, Kanu N, Snijders AP, Behrens A. UBR5-mediated ubiquitination of ATMIN is required for ionizing radiation-induced ATM signaling and function. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(33):12091–6. Epub 2014/08/06. 10.1073/pnas.1400230111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su H, Meng S, Lu Y, Trombly MI, Chen J, Lin C, et al. Mammalian hyperplastic discs homolog EDD regulates miRNA-mediated gene silencing. Molecular cell. 2011;43(1):97–109. Epub 2011/07/06. 10.1016/j.molcel.2011.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reid MA, Wang WI, Rosales KR, Welliver MX, Pan M, Kong M. The B55alpha subunit of PP2A drives a p53-dependent metabolic adaptation to glutamine deprivation. Molecular cell. 2013;50(2):200–11. Epub 2013/03/19. 10.1016/j.molcel.2013.02.008 . [DOI] [PubMed] [Google Scholar]
- 26.Benavides M, Chow-Tsang LF, Zhang J, Zhong H. The novel interaction between microspherule protein Msp58 and ubiquitin E3 ligase EDD regulates cell cycle progression. Biochimica et biophysica acta. 2013;1833(1):21–32. Epub 2012/10/17. 10.1016/j.bbamcr.2012.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maddika S, Chen J. Protein kinase DYRK2 is a scaffold that facilitates assembly of an E3 ligase. Nat Cell Biol. 2009;11(4):409–19. 10.1038/ncb1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Munoz MA, Saunders DN, Henderson MJ, Clancy JL, Russell AJ, Lehrbach G, et al. The E3 ubiquitin ligase EDD regulates S-phase and G(2)/M DNA damage checkpoints. Cell cycle. 2007;6(24):3070–7. Epub 2007/12/13. . [DOI] [PubMed] [Google Scholar]
- 29.Scialpi F, Mellis D, Ditzel M. EDD, a ubiquitin-protein ligase of the N-end rule pathway, associates with spindle assembly checkpoint components and regulates the mitotic response to nocodazole. The Journal of biological chemistry. 2015;290(20):12585–94. Epub 2015/04/03. 10.1074/jbc.M114.625673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smits VA. EDD induces cell cycle arrest by increasing p53 levels. Cell cycle. 2012;11(4). Epub 2012/02/07. . [DOI] [PubMed] [Google Scholar]
- 31.Yuan JS, Reed A, Chen F, Stewart CN Jr. Statistical analysis of real-time PCR data. BMC Bioinformatics. 2006;7:85 10.1186/1471-2105-7-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ditzel M, Broemer M, Tenev T, Bolduc C, Lee TV, Rigbolt KT, et al. Inactivation of effector caspases through nondegradative polyubiquitylation. Mol Cell. 2008;32(4):540–53. 10.1016/j.molcel.2008.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Truett GE, Heeger P, Mynatt RL, Truett AA, Walker JA, Warman ML. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). Biotechniques. 2000;29(1):52, 4. . [DOI] [PubMed] [Google Scholar]
- 34.Buchholz F, Angrand PO, Stewart AF. Improved properties of FLP recombinase evolved by cycling mutagenesis. Nat Biotechnol. 1998;16(7):657–62. 10.1038/nbt0798-657 . [DOI] [PubMed] [Google Scholar]
- 35.Indra AK, Warot X, Brocard J, Bornert JM, Xiao JH, Chambon P, et al. Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: comparison of the recombinase activity of the tamoxifen-inducible Cre-ER(T) and Cre-ER(T2) recombinases. Nucleic Acids Res. 1999;27(22):4324–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Logan M, Martin JF, Nagy A, Lobe C, Olson EN, Tabin CJ. Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis. 2002;33(2):77–80. Epub 2002/07/12. 10.1002/gene.10092 . [DOI] [PubMed] [Google Scholar]
- 37.Sharpe J, Ahlgren U, Perry P, Hill B, Ross A, Hecksher-Sorensen J, et al. Optical projection tomography as a tool for 3D microscopy and gene expression studies. Science. 2002;296(5567):541–5. 10.1126/science.1068206 . [DOI] [PubMed] [Google Scholar]
- 38.Okada Y, Shimazaki T, Sobue G, Okano H. Retinoic-acid-concentration-dependent acquisition of neural cell identity during in vitro differentiation of mouse embryonic stem cells. Dev Biol. 2004;275(1):124–42. 10.1016/j.ydbio.2004.07.038 . [DOI] [PubMed] [Google Scholar]
- 39.Williams RL, Hilton DJ, Pease S, Willson TA, Stewart CL, Gearing DP, et al. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature. 1988;336(6200):684–7. 10.1038/336684a0 . [DOI] [PubMed] [Google Scholar]
- 40.Okabe S, Forsberg-Nilsson K, Spiro AC, Segal M, McKay RD. Development of neuronal precursor cells and functional postmitotic neurons from embryonic stem cells in vitro. Mech Dev. 1996;59(1):89–102. . [DOI] [PubMed] [Google Scholar]
- 41.Bain G, Kitchens D, Yao M, Huettner JE, Gottlieb DI. Embryonic stem cells express neuronal properties in vitro. Dev Biol. 1995;168(2):342–57. 10.1006/dbio.1995.1085 . [DOI] [PubMed] [Google Scholar]
- 42.Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113(5):643–55. . [DOI] [PubMed] [Google Scholar]
- 43.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–76. 10.1016/j.cell.2006.07.024 . [DOI] [PubMed] [Google Scholar]
- 44.Qi X, Li TG, Hao J, Hu J, Wang J, Simmons H, et al. BMP4 supports self-renewal of embryonic stem cells by inhibiting mitogen-activated protein kinase pathways. Proc Natl Acad Sci U S A. 2004;101(16):6027–32. 10.1073/pnas.0401367101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saunders DN, Hird SL, Withington SL, Dunwoodie SL, Henderson MJ, Biben C, et al. Edd, the murine hyperplastic disc gene, is essential for yolk sac vascularization and chorioallantoic fusion. Mol Cell Biol. 2004;24(16):7225–34. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schnutgen F, De-Zolt S, Van Sloun P, Hollatz M, Floss T, Hansen J, et al. Genomewide production of multipurpose alleles for the functional analysis of the mouse genome. Proc Natl Acad Sci U S A. 2005;102(20):7221–6. 10.1073/pnas.0502273102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lim NS, Kozlov G, Chang TC, Groover O, Siddiqui N, Volpon L, et al. Comparative peptide binding studies of the PABC domains from the ubiquitin-protein isopeptide ligase HYD and poly(A)-binding protein. Implications for HYD function. J Biol Chem. 2006;281(20):14376–82. . [DOI] [PubMed] [Google Scholar]
- 48.Chiang C, Litingtung Y, Lee E, Young KE, Corden JL, Westphal H, et al. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383(6599):407–13. . [DOI] [PubMed] [Google Scholar]
- 49.Tsukui T, Capdevila J, Tamura K, Ruiz-Lozano P, Rodriguez-Esteban C, Yonei-Tamura S, et al. Multiple left-right asymmetry defects in Shh(-/-) mutant mice unveil a convergence of the shh and retinoic acid pathways in the control of Lefty-1. Proc Natl Acad Sci U S A. 1999;96(20):11376–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feil R, Wagner J, Metzger D, Chambon P. Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem Biophys Res Commun. 1997;237(3):752–7. 10.1006/bbrc.1997.7124 . [DOI] [PubMed] [Google Scholar]
- 51.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108(2):193–9. . [DOI] [PubMed] [Google Scholar]
- 52.Vortkamp A, Lee K, Lanske B, Segre GV, Kronenberg HM, Tabin CJ. Regulation of rate of cartilage differentiation by Indian hedgehog and PTH-related protein. Science. 1996;273(5275):613–22. . [DOI] [PubMed] [Google Scholar]
- 53.St-Jacques B, Hammerschmidt M, McMahon AP. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 1999;13(16):2072–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Visel A, Thaller C, Eichele G. GenePaint.org: an atlas of gene expression patterns in the mouse embryo. Nucleic Acids Res. 2004;32(Database issue):D552–6. 10.1093/nar/gkh029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sears KE, Maier JA, Rivas-Astroza M, Poe R, Zhong S, Kosog K, et al. The Relationship between Gene Network Structure and Expression Variation among Individuals and Species. PLoS Genet. 2015;11(8):e1005398 10.1371/journal.pgen.1005398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Niedermaier M, Schwabe GC, Fees S, Helmrich A, Brieske N, Seemann P, et al. An inversion involving the mouse Shh locus results in brachydactyly through dysregulation of Shh expression. J Clin Invest. 2005;115(4):900–9. 10.1172/JCI23675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Petiot A, Conti FJ, Grose R, Revest JM, Hodivala-Dilke KM, Dickson C. A crucial role for Fgfr2-IIIb signalling in epidermal development and hair follicle patterning. Development. 2003;130(22):5493–501. 10.1242/dev.00788 . [DOI] [PubMed] [Google Scholar]
- 58.Henderson MJ, Russell AJ, Hird S, Munoz M, Clancy JL, Lehrbach GM, et al. EDD, the human hyperplastic discs protein, has a role in progesterone receptor coactivation and potential involvement in DNA damage response. J Biol Chem. 2002;277(29):26468–78. . [DOI] [PubMed] [Google Scholar]
- 59.Clarke CL, Roman SD, Graham J, Koga M, Sutherland RL. Progesterone receptor regulation by retinoic acid in the human breast cancer cell line T-47D. J Biol Chem. 1990;265(21):12694–700. . [PubMed] [Google Scholar]
- 60.Matsumoto H, Zhao X, Das SK, Hogan BL, Dey SK. Indian hedgehog as a progesterone-responsive factor mediating epithelial-mesenchymal interactions in the mouse uterus. Dev Biol. 2002;245(2):280–90. 10.1006/dbio.2002.0645 . [DOI] [PubMed] [Google Scholar]
- 61.Takamoto N, Zhao B, Tsai SY, DeMayo FJ. Identification of Indian hedgehog as a progesterone-responsive gene in the murine uterus. Mol Endocrinol. 2002;16(10):2338–48. 10.1210/me.2001-0154 . [DOI] [PubMed] [Google Scholar]
- 62.Hay-Koren A, Caspi M, Zilberberg A, Rosin-Arbesfeld R. The EDD E3 ubiquitin ligase ubiquitinates and up-regulates beta-catenin. Mol Biol Cell. 2011;22(3):399–411. Epub 2010/12/02. 10.1091/mbc.E10-05-0440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang G, Tang X, Chen Y, Cao J, Huang Q, Ling X, et al. Hyperplastic discs differentially regulates the transcriptional outputs of hedgehog signaling. Mech Dev. 2014;133:117–25. 10.1016/j.mod.2014.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sutherland C. What Are the bona fide GSK3 Substrates? Int J Alzheimers Dis. 2011;2011:505607 Epub 2011/06/02. 10.4061/2011/505607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Amano K, Ichida F, Sugita A, Hata K, Wada M, Takigawa Y, et al. MSX2 stimulates chondrocyte maturation by controlling Ihh expression. The Journal of biological chemistry. 2008;283(43):29513–21. Epub 2008/08/07. 10.1074/jbc.M803681200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yoshida CA, Yamamoto H, Fujita T, Furuichi T, Ito K, Inoue K, et al. Runx2 and Runx3 are essential for chondrocyte maturation, and Runx2 regulates limb growth through induction of Indian hedgehog. Genes & development. 2004;18(8):952–63. Epub 2004/04/27. 10.1101/gad.1174704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang W, Lian N, Li L, Moss HE, Perrien DS, Elefteriou F, et al. Atf4 regulates chondrocyte proliferation and differentiation during endochondral ossification by activating Ihh transcription. Development. 2009;136(24):4143–53. Epub 2009/11/13. 10.1242/dev.043281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Charite J, McFadden DG, Olson EN. The bHLH transcription factor dHAND controls Sonic hedgehog expression and establishment of the zone of polarizing activity during limb development. Development. 2000;127(11):2461–70. . [DOI] [PubMed] [Google Scholar]
- 69.Zhang Z, Sui P, Dong A, Hassell J, Cserjesi P, Chen YT, et al. Preaxial polydactyly: interactions among ETV, TWIST1 and HAND2 control anterior-posterior patterning of the limb. Development. 2010;137(20):3417–26. 10.1242/dev.051789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lettice LA, Williamson I, Wiltshire JH, Peluso S, Devenney PS, Hill AE, et al. Opposing functions of the ETS factor family define Shh spatial expression in limb buds and underlie polydactyly. Dev Cell. 2012;22(2):459–67. 10.1016/j.devcel.2011.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Price MA, Kalderon D. Proteolysis of the Hedgehog signaling effector Cubitus interruptus requires phosphorylation by Glycogen Synthase Kinase 3 and Casein Kinase 1. Cell. 2002;108(6):823–35. Epub 2002/04/17. . [DOI] [PubMed] [Google Scholar]
- 72.Jia J, Amanai K, Wang G, Tang J, Wang B, Jiang J. Shaggy/GSK3 antagonizes Hedgehog signalling by regulating Cubitus interruptus. Nature. 2002;416(6880):548–52. Epub 2002/03/26. 10.1038/nature733 . [DOI] [PubMed] [Google Scholar]
- 73.Byrd N, Becker S, Maye P, Narasimhaiah R, St-Jacques B, Zhang X, et al. Hedgehog is required for murine yolk sac angiogenesis. Development. 2002;129(2):361–72. . [DOI] [PubMed] [Google Scholar]
- 74.Frisdal A, Trainor PA. Development and evolution of the pharyngeal apparatus. Wiley Interdiscip Rev Dev Biol. 2014;3(6):403–18. 10.1002/wdev.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lettice LA, Williamson I, Devenney PS, Kilanowski F, Dorin J, Hill RE. Development of five digits is controlled by a bipartite long-range cis-regulator. Development. 2014;141(8):1715–25. 10.1242/dev.095430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gao B, Hu J, Stricker S, Cheung M, Ma G, Law KF, et al. A mutation in Ihh that causes digit abnormalities alters its signalling capacity and range. Nature. 2009;458(7242):1196–200. 10.1038/nature07862 . [DOI] [PubMed] [Google Scholar]
- 77.Zhang XM, Ramalho-Santos M, McMahon AP. Smoothened mutants reveal redundant roles for Shh and Ihh signaling including regulation of L/R asymmetry by the mouse node. Cell. 2001;105(6):781–92. . [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.