Abstract
Genetic testing for hypertrophic cardiomyopathy (HCM) can provide an important clinical marker for disease outcome and family screening. This study set out to validate our recently developed phenotype-based HCM genotype predictor score. Patients clinically diagnosed with HCM and evaluated by genetic counselors comprised the study cohort. Genotype score was derived based on clinical and echocardio-graphic variables. Total score was correlated with the yield of genetic testing. Of 564 HCM patients, 198 sought genetic testing (35 %; 55 % male; mean age at diagnosis, 50 ±20 years). Of these, 101 patients (51 %) were genotype positive for a HCM-associated genetic mutation (55 % male; mean age at diagnosis, 42 ± 18 years). Cochran-Armitage analysis showed similar, statistically significant trends of increased yields for higher genotype scores for both the original and study cohort. Validated by the current study, this scoring system provides an easy-to-use, clinical tool to aid in determining the likelihood of a positive HCM genetic test.
Keywords: Hypertrophiccardiomyopathy, Genetics, Sudden cardiac death, Genotype predictor
Introduction
Hypertrophic cardiomyopathy (HCM) is one of the most common inherited cardiac diseases affecting about 1 in 500 individuals [1]. Typically inherited in an autosomal dominant fashion, its phenotype is characterized by variable penetrance and expressivity [2]. Currently, over 24 genes associated in the pathogenesis have been identified, and clinical genetic testing has been available for over a decade. While genetic testing gene panels may vary in size depending on the company or institution providing the test, all panels at minimum include the nine established and most commonly associated myofilament HCM genes (ACTC1, MYBPC3, MYH7, MYL2, MYL3, TNNC1, TNNI3, TNNT2, and TPM1). Genotype-phenotype correlations have shown variable results among different cohorts [3–15], and while the mutant genotype is present at birth, HCM can be phenotypically silent during early childhood only to become manifest as a clinical entity during puberty, adolescence, or even later in adulthood. Following diagnosis of a patient with HCM, screening of all first degree relatives of a confirmed HCM index patient is advocated by current HCM guidelines [16, 17]. This is done using a combination of clinical history, physical examination, electrocardiography, and echocardiography, while mutation-specific confirmatory genetic testing is reserved for first degree relatives of a HCM patient after a specific HCM-associated mutation has been identified in the index case [16–18].
Currently, the yield of genetic testing for the known HCM-associated genes in a confirmed index patient ranges from 30 to 50 % [8–15]. Thus, clinical tools to determine the a priori yield of genetic testing have emerged to aid physicians and their patients in the decision whether to pursue genetic testing [5, 6, 14, 19–21]. In a recent publication from our institution [20], we reported the development of a new phenotype-based, genetic prediction score that uses six routinely assessed, clinical parameters to create a score that associates strongly with the likelihood of a positive HCM genetic test result. To establish the usability of this tool in clinical practice, we herein aimed to validate the original genotype predictor score using a new cohort of HCM patients seen by a certified genetic counselor as a routine part of their Mayo Clinic HCM evaluation.
Methods
In this IRB approved study, a retrospective review of the electronic medical record (EMR) was performed of all HCM patients seen by a cardiac genetic counselor between January 1, 2005 and June 30, 2014. The study cohort included all patients with a confirmed diagnosis of HCM using standard clinical, electrocardiographic, and echocardiographic criteria seen in the Mayo Clinic HCM Clinic. This created a distinct validation cohort to compare to our original study cohort comprised of patients evaluated at Mayo Clinic used in manuscript that first described the genotype predictor score [20]. Again, this validation cohort is distinct from the sentinel study as these patients were evaluated at Mayo Clinic in the time frame after enrollment of the research cohort had ended.
During consultation with one of our genetic counselors, all the patients were offered genetic testing for HCM using any of the commercially available genetic tests. The philosophy of the Department of Clinical Genetics at the Mayo Clinic is to inform patients, but not to sway the ultimate decision as to whether or not the patient will pursue testing. Counselors try to remain neutral in the discussion about advantages and disadvantages of testing, simply providing enough information for patients to make educated decisions. Genetic tests were chosen based on available insurance reimbursement and patient preference. Genetic analysis was performed by several commercial companies, including Transgenomic, GeneDx, Correlagen, Ambry Genetics, Harvard Partners, and the Mayo Clinic. The uptake of the genetic test was determined following review of all the patients included in this study. The genetic test result as provided on the original report was used for analysis of the genotype predictor score; variants classified as likely pathogenic, possible or probably pathogenic, or variant of uncertain significance (VUS) were considered genotype positive as reported to patient at the time of consultation. Additionally, all variants were re-analyzed at the time of this study to by determining minor allele frequency in population databases (ExAC) [22], reference in clinical databases (ClinVAR) as well as by application of various in silico mutation prediction tools (PolyPhen, SIFT, Mutation Assessor, Condel and Grantham score). Variants that were seen at MAF >0.01 % based on data not present at clinical evaluation were reclassified as VUS-likely benign after which the genotype predictor score was recalculated (Supplemental Data) [23–49].
Subsequently, the EMR of patients who opted for genetic testing was further evaluated for clinical, diagnostic, and genetic testing results, including but not limited to components of the Mayo Clinic Phenotype-Based Genotype Predictor Score. Data recorded included gender, age at diagnosis, family history of HCM (FHHCM), family history of sudden cardiac death (FHSCD), history of hypertension, symptoms at the time of diagnosis, septal shape, maximum left ventricular wall thickness (MLVWT), degree of left ventricular outflow tract obstruction (LVOTO), history of surgical myectomy or catheter-based septal ablation, presence of implantable cardioverter defibrillator (ICD), company performing testing, and specific mutation and mutation classification identified. In this study, a family history of HCM includes a first or second degree relative who has been diagnosed with HCM. Family history of sudden cardiac death includes first or second degree relatives who died suddenly and unexpectedly before the age of 40.
The Mayo genotype score, outlined in our previously published paper [20], was retrospectively calculated for each patient and correlated with their test result as reported on their genetic test report assigning one point for the presence of these established variables: age at diagnosis ≤45 years, MLVWT ≥20 mm on echocardiography, reverse curve septal shape on echocardiogram, positive FHHCM, and positive FHSCD. The presence of hypertension resulted in subtracting one point from their score. Echocardiograms of patients without documentation of their septal shape were reviewed by two independent investigators (SLM, JHA) blinded to clinical and genetic information, and disagreements on septal shape determination were unified by consensus opinion with input from a third expert reviewer (SRO).
Demographics and genotype-phenotype correlations were compared using Student’s t test and Fisher’s exact test. Cochran-Armitage trend test was utilized to establish presence of a significant trend in correlation between genotype score and yield of genetic testing for both the study cohort as well as the initial study cohort used to develop the genotype predictor score. Statistical analyses were carried out using JMP 10.0 statistical software (SAS Institute Inc, Cary, NC, USA.).
Results
Overall, 564 patients were not only diagnosed with HCM using standard clinical, ECG, and echocardiographic criteria between January 2005 and June 2014, but were also seen by a genetic counselor. Demographic information for the cohort is summarized in Table 1. Genetic testing was offered to all the patients, but was performed in only 198 of 564 patients (35 %), serving as the study cohort for validation of the genotype predictor score. The most common reasons cited by patients who declined clinical genetic testing were the financial cost of the genetic test, lack of insurance reimbursement, and concerns regarding genetic privacy and fear of future misuse of genetic information. However, full dissection of the reasons for genetic test declinations was beyond the scope of this study.
Table 1.
Cohort demographics and comparison between patients who did (study cohort) and did not pursue genetic testing
| N (%) | All patients 564 |
No genetic testing 366 (65) |
Study cohort 198 (35) |
p value |
|---|---|---|---|---|
| Sex M/F, n (% male) | 327/237 (58) | 219/147 (60) | 108/90 (55) | 0.3 |
| Age at diagnosis (years) | 47.9± 18.1 | 46.1 ± 17.8 | 50.0± 19.7 | 0.02 |
| FH HCM, n (%) | 170 (30) | 98 (27) | 72 (36) | 0.02 |
| FH SCD, n (%) | 108 (19) | 44 (12) | 64 (32) | <0.001 |
| FH HCM and/or SCD, n (%) | 214 (38) | 111 (30) | 103 (52) | <0.001 |
| Hx of hypertension, n (%) | 177 (32) | 116 (32) | 61 (31) | 0.9 |
| Symptoms at diagnosis, n (%) | 445 (79) | 291 (80) | 154 (78) | 0.7 |
| MLVWT (mm) | 18.9± 6.0 | 18.4 ±5.8 | 19.8± 6.3 | <0.001 |
| LVOT gradient (mm Hg) | 69.8±7.5 | 71.3 ±37.7 | 67.0 ±36.9 | 0.2 |
| Septal shape, n (%) | ||||
| Sigmoid | 100 (51) | |||
| Reverse curve | 48 (24) | |||
| Neutral | 21 (11) | |||
| Apical | 20 (10) | |||
| Myectomy, n (%) | 281 (50) | 192 (52) | 89 (45) | 0.1 |
| ICD, n (%) | 155 (27) | 100 (27) | 55 (28) | 0.9 |
Clinical demographics of the patients who underwent genetic testing (n=198) are summarized in Table 1. In brief, just over half the patients undergoing testing were male (55 %) with a mean age of HCM diagnosis of 50.0 ± 19.7 years. FHHCM was present in 72 patients (36 %) and a FHSCD in 64 (32 %). A sigmoid septum (51 % of patients) was the most frequent septal shape seen on cardiac imaging followed by reverse septal curvature (24 %). The genetic testing was carried out by six commercial providers, and genetic panels ranged from single gene to 31 genes included summarized in Supplemental Table 1. The majority of patients (91 %) underwent comprehensive genetic testing for at least nine HCM-associated genes.
Additionally, the patients who chose to forgo genetic testing (n = 366; 60 % male) are summarized in Table 1. Compared to the patients in the study cohort, these patients were younger at diagnosis (46.1 ± 17.8 vs. 50 ± 19.7 years; p =0.02). Conversely, the patients in the study cohort had higher likelihood of family history of HCM (36 vs. 27 %; p =0.02), family history of SCD (32 vs. 12 %; p<0.01), or family history of HCM and/or SCD (52 vs. 30 %; p<0.01), respectively.
Overall, 101 patients (51 %) were genotype positive for an HCM-associated mutation in one or more HCM-associated genes (55 % male; mean age at diagnosis, 41.9±17.9 years). Demographic information is summarized in Table 2. Similar to previous observations, mutations were most commonly found in MYBPC3 (53 % of genotype positive patients) and MYH7 (29 %), distantly followed by TNNT2 (5 %). Six patients (6 %) had ≥1 disease-associated mutations (Table 3). Of the 82 mutations, 29 (35 %) were classified on the genetic test report as variants of uncertain significance (VUS). All specific variants and yield of these variants among individual patients are summarized in Supplemental Table 2 complete with data on population frequency (ExAC) as well as results from various in silico mutation prediction tools.
Table 2.
Cohort demographics and comparison between genotype positive and genotype negative patients
| Genotype positive | Genotype negative | p value | |
|---|---|---|---|
| N (%) | 101 (51) | 97 (49) | |
| Sex M/F, n (% male) | 56/45 (55) | 52/45 (54) | 0.9 |
| Age at diagnosis (years) | 41.9± 17.9 | 58.3± 18.0 | <0.001 |
| FH HCM, n (%) | 55 (54) | 17 (18) | <0.001 |
| FH SCD, n (%) | 40 (40) | 24 (25) | 0.03 |
| FH HCM and/or SCD, n (%) | 26 (26) | 7 (7) | <0.001 |
| Hx of hypertension, n (%) | 23 (23) | 38 (39) | 0.01 |
| Symptoms at diagnosis, n (%) | 75 (74) | 79 (81) | 0.2 |
| MLVWT (mm) | 21.3 ±7.5 | 18.3 ±4.4 | <0.001 |
| LVOT gradient (mm Hg) | 62.4±39.5 | 71.2±33.9 | 0.2 |
| Septal shape, n (%) | |||
| Sigmoid | 37 (37) | 63 (65) | <0.001 |
| Reverse curve | 37 (37) | 11 (11) | <0.001 |
| Neutral | 12 (12) | 9 (9) | 0.7 |
| Apical | 10 (10) | 10 (10) | 0.8 |
| Myectomy, n (%) | 46 (46) | 43 (44) | 0.9 |
| ICD, n (%) | 42 (42) | 13 (13) | <0.001 |
Table 3.
Yield of genetic testing
| Gene | Number of patients N = 198 | Yield among all patients (%) |
Yield among genotype positive patients (%) |
|---|---|---|---|
| Genotype positive | 101 | 51 | |
| MYBPC3 | 54 | 27 | 53 |
| MYH7 | 29 | 15 | 29 |
| TNNT2 | 5 | 3 | 5 |
| Multiple | 6 | 3 | 5 |
| MYL2 | 2 | 1 | 2 |
| ACTC1 | 1 | 1 | 1 |
| MYL3 | 1 | 1 | 1 |
| TNNI3 | 1 | 1 | 1 |
| LAMP2 | 1 | 1 | 1 |
| PRKAG2 | 1 | 1 | 1 |
Overall and akin to previous observations, genotype positive patients were more severely affected than genotype negative patients with respect to age at diagnosis, FHHCM, FHSCD, and MLVWT (Table 2). Genotype positive patients were significantly younger than the genotype negative patients (41.9 ± 17.9 vs. 58.3 ± 18.0 years, respectively; p < 0.001). A FHHCM was present in a majority of genotype positive patients (54 %) compared to genotype negative patients (18 %; p < 0.001) similar to FHSCD (40 versus 25 %, respectively; p = 0.03). Conversely, a history of systemic hypertension was a more prevalent in genotype negative patients (39 %) compared to genotype positive patients (23 %; p =0.01). Sigmoid septal shape was the most common morphology in all genetically tested participants, though reverse curve was significantly more common in the genotype positive patients compared to genotype negative patients (37 and 11 % respectively, p < 0.001). Lastly, genotype positive patients showed significantly more hypertrophy (MLVWT 21.3±7.5 mm) compared to genotype negative patients (18.3 ± 4.4 mm; p < 0.001; Table 2).
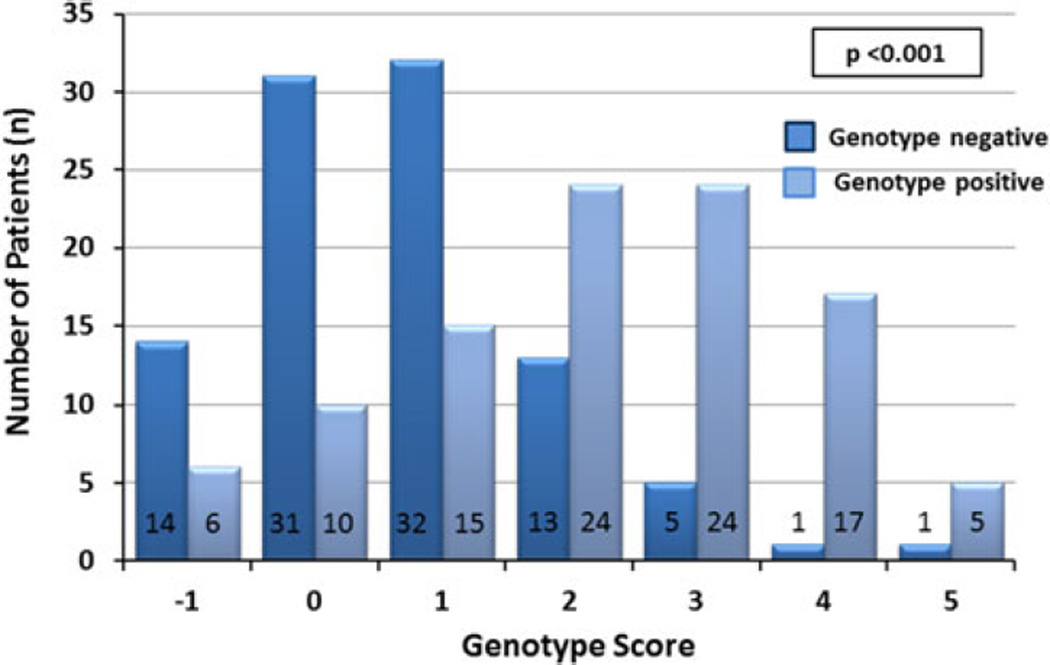
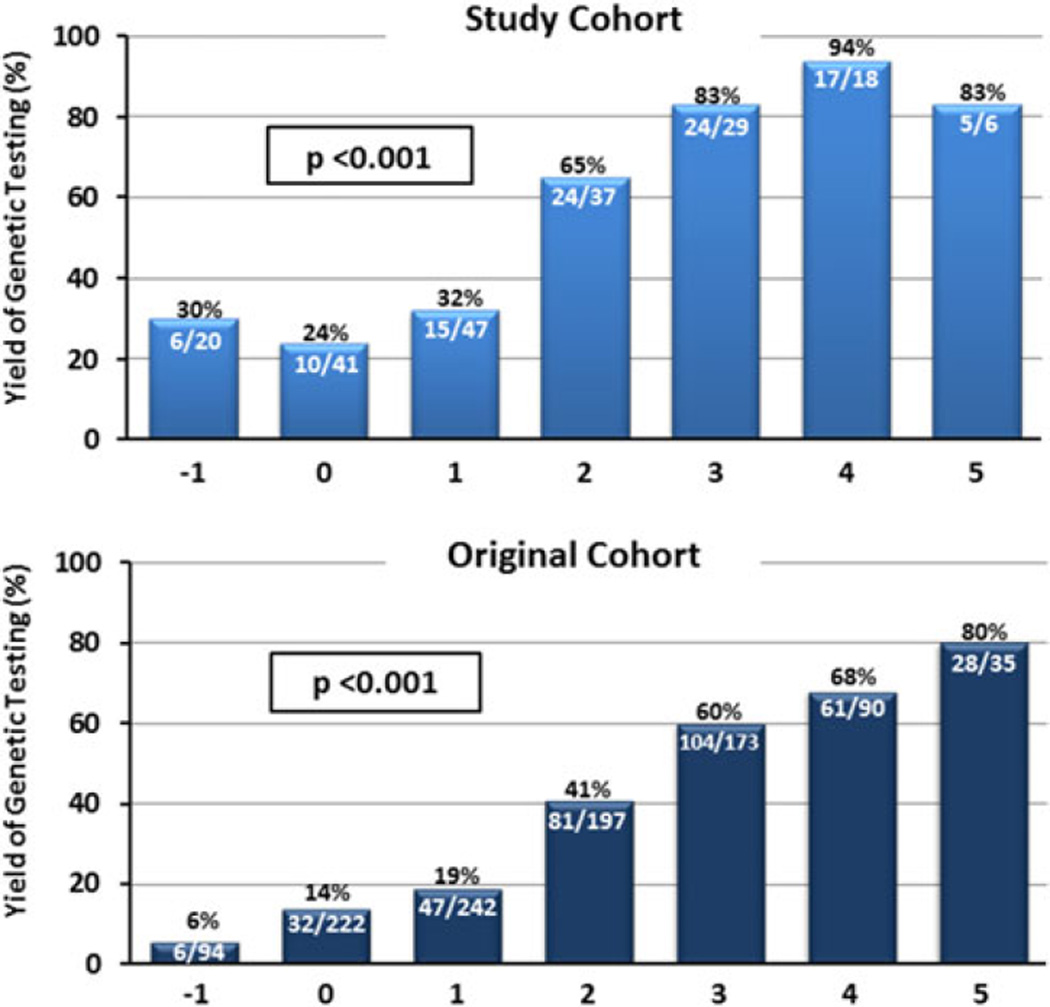
Akin to our sentinel paper, in this cohort evaluated at the same institution, the genotype predictor score strongly correlated with the yield of genetic testing. A significant shift to the right could be seen for genotype positive patients with many of the patients (n=46; 45 %) having a score of 3 or greater. Conversely, a large proportion of genotype negative patients had a score of 0 or less (n=45; 45 %; p<0.001; Fig. 1). For overall yield of genetic testing, the genotype predictor score correlated with the yield of genetic testing as seen previously (Fig. 2a). Patients scoring −1 on the genotype predictor score, meaning there was a history of systemic hypertension in the absence of other at-risk markers for a positive genetic test result, had the lowest genetic mutation yield (6/20 patients; 30 %). From there, an increasing yield of genetic testing could be observed with each increase of the genotype score to a yield of genetic testing of 83, 94, and 83 % in patients with a predictor score of 3, 4, and 5, respectively. Although compared to the original study, percentages at both ends of the score are slightly higher for patients with a score of −1 and slightly less between a score of 4 and 5, this is likely caused by the lower number of genotype positive patients in the validation cohort (N = 101) compared to the original study cohort (n = 359). In fact, Cochran-Armitage trend analyses demonstrated for both the studies that there was a significant trend associating a higher phenotype-based genotype prediction score with likelihood of a positive genetic test result validating the developed model scores (p < 0.001; Fig. 2a, b).
Fig. 1.
Distribution of genotype score between genotype positive and genotype negative patients
Fig. 2.
Yield of genetic testing in each scored subgroup in validation cohort (top) and original data cohort [20] (bottom) with Cochran-Armitage analysis showing a significant correlation for trend in genotype score for both the studies (p < 0.001)
Designation of genotype status and correlation with genotype predictor score for this study was based on the result as reported on the original (commercial) genetic test report (Supplemental Table 2). Additionally, all variants were retrospectively cross-referenced with literature, genomic population databases, and in silico prediction tools to further stratify variants. Results for each variant are summarized in Supplemental Table 2. In doing so, 11 variants previously reported as VUS, disease-associated or (likely) pathogenic, would by current knowledge be demoted to VUS-(likely) benign status, especially because of a minor allele frequency >0.01 % in population controls (ExAC) [20, 50]. Interestingly, close inspection of the patients hosting these demoted variants revealed that all of these patients had a low genotype score based on clinical phenotype suggesting an a priori low outcome of positive genetic test. In fact, all patients had a score ranging from −1 to 2 with no score of 3 or higher observed (data not shown). As a result, compared to the results in the study cohort, the yield of genetic testing for patients with a genotype score −1, 0, 1, or 2 decreased to 16, 20, 26, and 61 %, respectively (p < 0.001; Supplemental Figure 1).
Discussion
Genetic testing of unselected patients with HCM diagnosed using standard clinical, electrocardiographic, and echocardiographic criteria has a reported yield for sarcomeric gene mutations in the range of 30 to 50 % [9–15, 20]. This leads to practical problems as current guidelines recommend all clinically diagnosed HCM patients to pursue genetic testing in addition to clinical screening of first degree relatives [9–15, 20]. If this testing reveals specific, disease-causing mutations, first degree relatives can undergo mutation-specific confirmatory genetic testing. Herein, phenotype negative relatives who do not host the family’s HCM-causative mutation need not be clinically screened further whereas family members with the variant (phenotype positive or negative) must be followed more closely.
To improve management and genetic counseling, several investigators have attempted to identify clinical markers that can predict the likelihood of a positive genetic test [5, 6, 14, 19–21]. As a positive genetic test provides a useful diagnostic test for further screening of relatives of the HCM index cases, a valid tool to determine the outcome of the genetic test would have significance not only for the individual patient but also for their relatives.
We recently developed a genotype predictor score [20], that based on six easy-to-assess clinical parameters provides the a priori yield of the HCM genetic test and can thereby help providers, patients, and their family members decide whether to pursue genetic testing. However, this study was performed in a single cohort of patients referred for research genetic testing in the era of research-based genetic testing. Herein, we set out to validate the genotype predictor score in a subsequent cohort of unique patients with HCM evaluated by Mayo Clinic, who underwent commercially available, clinical genetic testing. In fact, in the new cohort, we were able to successfully validate the previously established and published Mayo HCM Genotype Predictor Score, demonstrating a similar increased yield of genetic testing for patients with a higher genotype predictor score as exemplified by significant Cochran-Armitage values.
Aside from validation of the Mayo Clinic Genotype Predictor, we made a number of additional observations along the way. First of all, utilization of the genetic testing for patients at Mayo Clinic was relatively low. Although over 500 patients were evaluated over a 9-year period (years 2005 to 2014) and had a genetic counseling session with a licensed genetic counselor as part of their comprehensive evaluation, nearly two-thirds declined HCM genetic testing, a percentage that has stayed consistent over the years. Indeed, there were observed phenotypic differences between the patients who chose to pursue genetic testing and those who did not. Among these, patients who did have a genetic test done had a significantly higher frequency of family history of HCM and/or SCD, which might explain some of the motivation to pursue HCM genetic testing. However, it must be stressed that the philosophy of Mayo Clinic genetic counselors on genetic testing is the patient’s choice and theirs alone. This is emphasized by the fact that still 30 % of patients that declined genetic testing had a family history of HCM and/or SCD. In addition, despite using the same cardiac genetic counselors in our genetic heart diseases program, the uptake for HCM genetic testing among patients with HCM has been lower than the uptake for long QTsyndrome (LQTS) genetic testing suggesting differences in value—whether perceived or real—of the genetic test for patients affected with one of these two diseases (data not shown). Therefore, the clinical phenotype and culminating genotype predictor score can be used as an additional educational tool to aid patients, their family members as well as their providers to decide whether to pursue genetic testing and the anticipated likelihood that the HCM genetic test will return a positive result.
When compared to our initial study [20], there were some notable differences between the original patient cohort in whom the Mayo Genotype Predictor Score was developed and the current validation cohort, especially in regard to the yield of genetic testing between the two cohorts (51 % in the validation cohort vs. 34 % in the original cohort). Most importantly, the patients in the validation cohort showed a stronger family history of disease evidenced by a higher frequency of reported FHSCD and/or FHHCM (52 % of the validation cohort as compared to 37 % of those in the earlier study; p < 0.001; data not shown). This may explain the larger yield of genetic testing in the current study. In addition, after reclassification of the variants by applying the same stringent classification guidelines, and taking into account that the PRKAG2 and LAMP2 genes were not analyzed in the original cohort, the yield would in fact be closer to 44 % and therefore more comparable to the original study.
The reasons for declining or accepting HCM genetic testing are complex and are dependent on individual patient psychological, social, and family values as well as extrinsic insurance, financial, and privacy concerns. The specific reasons were not prospectively assessed during patients’ visit, and comprehensive analysis of low uptake was therefore beyond the scope of this paper. Several investigators have reported previously on genetic screening issues and have highlighted the importance of patient autonomy as a central tenet of genetic counseling, and although it may be medically and scientifically beneficial in the provider’s opinion to know an individual’s genetic mutation status, each patient has the absolute right to decide for themselves on this issue of paramount personal importance [19, 51]. Reasons commonly cited for refusal of genetic testing, especially among asymptomatic individuals, include genetic privacy, concerns over misuse of genetic information by employers or healthcare organizations, and financial concerns. Further, specifically in regards to the HCM genetic test, the absence of a genotype-guided therapies might be a reason underlying the low utilization of HCM genetic testing in contrast to the much higher utilization seen in our long QT syndrome clinic that utilizes the same cardiogenetic counselor (data not shown).
The final study cohort of 198 patients comprised those HCM patients who agreed to clinical genetic testing and was used to validate the Mayo Genotype Predictor Score developed from an earlier and separate population of Mayo Clinic HCM patients [20]. When compared to our initial study [20], there were some notable differences between the earlier model patient cohort in whom the Mayo Genotype Predictor Score was developed and the current validation patient cohort. Our validation cohort has a stronger family history of disease evidenced by a higher frequency of reported FHSCD and/or FHHCM (52 % of the current cohort as compared to 37 % of those in the earlier study; p < 0.001). Additionally, it would seem that the yield of genetic testing is higher in the validation cohort compared to that in the original study (51 % in the validation cohort and 34 % in the original). That being said, if the validation cohort genetic yield is calculated with the exclusion of patients with variants reclassified as likely benign and those positive for LAMP2 and PRKAG2 variants (not tested for in the original study), the yield is 45 %, more in line with the yield of the original study.
However, despite phenotypic differences and ultimately different yield in genetic testing, our study successfully validated the Mayo Clinic HCM Genotype Predictor showing a significant Cochran-Armitage score for trend testing in both the models. The difference in disease characteristics of the model and validation HCM populations reinforces the general validity of the Mayo Genotype Predictor model in HCM populations which have been reported previously to differ significantly based on differences between HCM seen in the community and in a referral HCM practice. To further confirm this predictive tool’s generalizable utility, future studies involving patients seen at other HCM centers are necessary. For patients with HCM seen at our center, implementation of the genotype predictor score has already aided both providers in genetic testing counseling as well as patients in decision making.
Though many patients found cost of testing to be a limiting factor, appropriately selected patient genetic testing could stand to avoid stressful and costly screening of family members with negative proband testing.
Limitations of the current study include the significant number of HCM patients who declined genetic testing. It is possible that patients who opted for genetic studies had self-selected for a higher genetic positivity than unselected patients. However, within the patient group who consented to genetic testing, there was a graded level of genetic positivity that correlated with the individual patient phenotype score. We speculate that the ability to select HCM patient more likely to be genetically positive for HCM genetic mutations will further the adoption of genetic testing in HCM patients and offer a greater economic rationale for genetic screening of both the patients and their first degree relatives. At minimum, this simple phenotype-based genotype predictor now provides essential information for the genetic counseling session to enable the providers and the patients to truly engage in shared decision making.
Conclusions
The Mayo HCM predictor score provides an easy-to-use, clinical tool to aid in determining the a priori yield of the HCM genetic test. In our clinic, approximately 65 % of patients referred for genetic counseling opted not to or were unable to pursue genetic testing. Implementation of the genotype predictor score should aid providers in genetic testing counseling as well as patients in decision making.
Supplementary Material
Acknowledgments
Sources of Funding This work was supported by the Mayo Clinic Windland Smith Rice Comprehensive Sudden Cardiac Death Program (MJA). This project was supported by Grant Number UL1 TR000135 from the National Center for Advancing Translational Sciences (NCATS). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institute of Health (NIH).
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s12265-016-9681-5) contains supplementary material, which is available to authorized users.
Compliance with Ethical Standards
Conflict of Interest The authors declare that they have no competing interests.
Human Subjects/Informed Consent Statement All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2000 (5). Informed consent was obtained from all the patients for being included in the study. No animal studies were carried out by the authors for this article.
References
- 1.Maron BJ. Hypertrophic cardiomyopathy: a systematic review. JAMA. 2002;287:1308–1320. doi: 10.1001/jama.287.10.1308. [DOI] [PubMed] [Google Scholar]
- 2.Golbus JR, Puckelwartz MJ, Fahrenbach JP, Dellefave-Castillo LM, Wolfgeher D, McNally EM. Population-based variation in cardiomyopathy genes. Circulation. Cardiovascular Genetics. 2012;5:391–399. doi: 10.1161/CIRCGENETICS.112.962928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tester DJ, Ackerman MJ. Genetic testing for potentially lethal, highly treatable inherited cardiomyopathies/ channelopathies in clinical practice. Circulation. 2011;123:1021–1037. doi: 10.1161/CIRCULATIONAHA.109.914838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Landstrom AP, Ackerman MJ. Mutation type is not clinically useful in predicting prognosis in hypertrophic cardiomy-opathy. Circulation. 2010;122:2441–2449. doi: 10.1161/CIRCULATIONAHA.110.954446. discussion 2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gruner C, Ivanov J, Care M, Williams L, Moravsky G, Yang H, et al. Toronto hypertrophic cardiomyopathy genotype score for prediction of a positive genotype in hypertrophic cardio-myopathy. Circulation. Cardiovascular Genetics. 2013;6:19–26. doi: 10.1161/CIRCGENETICS.112.963363. [DOI] [PubMed] [Google Scholar]
- 6.Ingles J, Sarina T, Yeates L, Hunt L, Macciocca I, McCormack L, et al. Clinical predictors of genetic testing outcomes in hypertrophic cardiomyopathy. Genetics in Medicine. 2013;15:972–977. doi: 10.1038/gim.2013.44. [DOI] [PubMed] [Google Scholar]
- 7.Bos JM, Theis JL, Tajik AJ, Gersh BJ, Ommen SR, Ackerman MJ. Relationship between sex, shape, and substrate in hypertrophic cardiomyopathy. American Heart Journal. 2008;155:1128–1134. doi: 10.1016/j.ahj.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ackerman MJ, VanDriest SL, Ommen SR, Will ML, Nishimura RA, Tajik AJ, et al. Prevalence and age-dependence of malignant mutations in the beta-myosin heavy chain and troponin T genes in hypertrophic cardiomyopathy: a comprehensive outpatient perspective. Journal of the American College of Cardiology. 2002;39:2042–2048. doi: 10.1016/s0735-1097(02)01900-9. [DOI] [PubMed] [Google Scholar]
- 9.Andersen PS, Havndrup O, Hougs L, Sorensen KM, Jensen M, Larsen LA, et al. Diagnostic yield, interpretation, and clinical utility of mutation screening of sarcomere encoding genes in Danish hypertrophic cardiomyopathy patients and relatives. Human Mutation. 2009;30:363–370. doi: 10.1002/humu.20862. [DOI] [PubMed] [Google Scholar]
- 10.Millat G, Bouvagnet P, Chevalier P, Dauphin C, Jouk PS, Da Costa A, et al. Prevalence and spectrum of mutations in a cohort of 192 unrelated patients with hypertrophic cardiomyopathy. European Journal of Medical Genetics. 2010;53:261–267. doi: 10.1016/j.ejmg.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 11.Olivotto I, Girolami F, Ackerman MJ, Nistri S, Bos JM, Zachara E, et al. Myofilament protein gene mutation screening and outcome of patients with hypertrophic cardiomyop-athy. Mayo Clinic Proceedings. 2008;83:630–638. doi: 10.4065/83.6.630. [DOI] [PubMed] [Google Scholar]
- 12.Van Driest SL, Ommen SR, Tajik AJ, Gersh BJ, Ackerman MJ. Yield of genetic testing in hypertrophic cardiomyopathy. Mayo Clinic Proceedings. 2005;80:739–744. doi: 10.1016/S0025-6196(11)61527-9. [DOI] [PubMed] [Google Scholar]
- 13.Erdmann J, Daehmlow S, Wischke S, Senyuva M, Werner U, Raible J, et al. Mutation spectrum in a large cohort of unrelated consecutive patients with hypertrophic cardiomyopathy. Clinical Genetics. 2003;64:339–349. doi: 10.1034/j.1399-0004.2003.00151.x. [DOI] [PubMed] [Google Scholar]
- 14.Binder J, Ommen SR, Gersh BJ, Van Driest SL, Tajik AJ, Nishimura RA, et al. Echocardiography-guided genetic testing in hypertrophic cardiomyopathy: septal morphological features predict the presence of myofilament mutations. Mayo Clinic Proceedings. 2006;81:459–467. doi: 10.4065/81.4.459. [DOI] [PubMed] [Google Scholar]
- 15.Van Driest SL, Ommen SR, Tajik AJ, Gersh BJ, Ackerman MJ. Sarcomeric genotyping in hypertrophic cardiomyopathy. Mayo Clinic Proceedings. 2005;80:463–469. doi: 10.1016/S0025-6196(11)63196-0. [DOI] [PubMed] [Google Scholar]
- 16.Gersh BJ, Maron BJ, Bonow RO, Dearani JA, Fifer MA, Link MS, et al. 2011 ACCF/AHA guideline for the diagnosis and treatment of hypertrophic cardiomyopathy: executive summary: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. Circulation. 2011;124:2761–2796. doi: 10.1161/CIR.0b013e318223e230. [DOI] [PubMed] [Google Scholar]
- 17.Ackerman MJ, Priori SG, Willems S, Berul C, Brugada R, Calkins H, et al. HRS/EHRA Expert Consensus Statement on the State of Genetic Testing for the Channelopathies and Cardiomyopathies: this document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA) Heart Rhythm. 2011;8:1308–1339. doi: 10.1016/j.hrthm.2011.05.020. [DOI] [PubMed] [Google Scholar]
- 18.Gersh BJ, Maron BJ, Bonow RO, Dearani JA, Fifer MA, Link MS, et al. 2011 ACCF/AHA guideline for the diagnosis and treatment of hypertrophic cardiomyopathy: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. Journal of Thoracic and Cardiovascular Surgery. 2011;142:e153–e203. doi: 10.1016/j.jtcvs.2011.10.020. [DOI] [PubMed] [Google Scholar]
- 19.Ingles J, Zodgekar PR, Yeates L, Macciocca I, Semsarian C, Fatkin D. Guidelines for genetic testing of inherited cardiac disorders. Heart, Lung & Circulation. 2011;20:681–687. doi: 10.1016/j.hlc.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 20.Bos JM, Will ML, Gersh BJ, Kruisselbrink TM, Ommen SR, Ackerman MJ. Characterization of a phenotype-based genetic test prediction score for unrelated patients with hypertrophic cardiomyopathy. Mayo Clinic Proceedings. 2014;89:727–737. doi: 10.1016/j.mayocp.2014.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hengstenberg C, Erdmann J, Charron P. Outcome of clinical versus genetic family screening in hypertrophic cardiomy-opathy with focus on cardiac beta-myosin gene mutations: prediction of clinical status—is molecular genetics a new tool for the management of hypertrophic cardiomyopathy in clinical practice? Cardiovascular Research. 2003;57:298–301. doi: 10.1016/s0008-6363(02)00781-2. [DOI] [PubMed] [Google Scholar]
- 22.Exome Aggregation Consortium (ExAC)C,MA. 2015 Dec; (URL: http://exac.broadinstitute.org)
- 23.Van Driest SL, Vasile VC, Ommen SR, Will ML, Tajik AJ, Gersh BJ, et al. Myosin binding protein C mutations and compound heterozygosity in hypertrophic car-diomyopathy. Journal of the American College of Cardiology. 2004;44:1903–1910. doi: 10.1016/j.jacc.2004.07.045. [DOI] [PubMed] [Google Scholar]
- 24.Niimura H, Bachinski LL, Sangwatanaroj S, Watkins H, Chudley AE, McKenna W, et al. Mutations in the gene for cardiac myosin-binding protein C and late-onset familial hypertrophic cardiomyopathy. New England Journal of Medicine. 1998;338:1248–1257. doi: 10.1056/NEJM199804303381802. [DOI] [PubMed] [Google Scholar]
- 25.Kaski JP, Syrris P, Shaw A, Alapi KZ, Cordeddu V, Esteban MT, et al. Prevalence of sequence variants in the RAS-mitogen activated protein kinase signaling pathway in pre-adolescent children with hypertrophic cardiomyopathy. Circulation. Cardiovascular Genetics. 2012;5:317–326. doi: 10.1161/CIRCGENETICS.111.960468. [DOI] [PubMed] [Google Scholar]
- 26.Wilson MG, Chandra N, Papadakis M, O’Hanlon R, Prasad SK, Sharma S. Hypertrophic cardiomyopathy and ultra-endurance running—two incompatible entities? Journal of Cardiovascular Magnetic Resonance. 2011;13:77. doi: 10.1186/1532-429X-13-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gruner C, Care M, Siminovitch K, Moravsky G, Wigle ED, Woo A, et al. Sarcomere protein gene mutations in patients with apical hypertrophic cardiomyopathy. Circulation. Cardiovascular Genetics. 2011;4:288–295. doi: 10.1161/CIRCGENETICS.110.958835. [DOI] [PubMed] [Google Scholar]
- 28.Kapa S, Tester DJ, Salisbury BA, Harris-Kerr C, Pungliya MS, Alders M, et al. Genetic testing for long-QT syndrome: distinguishing pathogenic mutations from benign variants. Circulation. 2009;120:1752–1760. doi: 10.1161/CIRCULATIONAHA.109.863076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andersen PS, Havndrup O, Bundgaard H, Larsen LA, Vuust J, Pedersen AK, et al. Genetic and phenotypic characterization of mutations in myosin-binding protein C (MYBPC3) in 81 families with familial hypertrophic cardiomyop-athy: total or partial haploinsufficiency. European Journal of Human Genetics. 2004;12:673–677. doi: 10.1038/sj.ejhg.5201190. [DOI] [PubMed] [Google Scholar]
- 30.Girolami F, Ho CY, Semsarian C, Baldi M, Will ML, Baldini K, et al. Clinical features and outcome of hypertrophic cardiomyopathy associated with triple sarcomere protein gene mutations. Journal of the American College of Cardiology. 2010;55:1444–1453. doi: 10.1016/j.jacc.2009.11.062. [DOI] [PubMed] [Google Scholar]
- 31.Xin B, Puffenberger E, Tumbush J, Bockoven JR, Wang H. Homozygosity for a novel splice site mutation in the cardiac myosin-binding protein C gene causes severe neonatal hypertrophic cardiomyopathy. American Journal of Medical Genetics Part A. 2007;143A:2662–2667. doi: 10.1002/ajmg.a.31981. [DOI] [PubMed] [Google Scholar]
- 32.Waldmuller S, Muller M, Rackebrandt K, Binner P, Poths S, Bonin M, et al. Array-based resequencing assay for mutations causing hypertrophic cardiomyopathy. Clinical Chemistry. 2008;54:682–687. doi: 10.1373/clinchem.2007.099119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ababou A, Rostkova E, Mistry S, Le Masurier C, Gautel M, Pfuhl M. Myosin binding protein C positioned to play a key role in regulation of muscle contraction: structure and interactions of domain C1. Journal of Molecular Biology. 2008;384:615–630. doi: 10.1016/j.jmb.2008.09.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nanni L, Pieroni M, Chimenti C, Simionati B, Zimbello R, Maseri A, et al. Hypertrophic cardiomyopathy: two homo-zygous cases with “typical” hypertrophic cardiomyopathy and three new mutations in cases with progression to dilated cardiomy-opathy. Biochemical and Biophysical Research Communications. 2003;309:391–398. doi: 10.1016/j.bbrc.2003.08.014. [DOI] [PubMed] [Google Scholar]
- 35.Richard P, Charron P, Carrier L, Ledeuil C, Cheav T, Pichereau C, et al. Hypertrophic cardiomyopathy: distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation. 2003;107:2227–2232. doi: 10.1161/01.CIR.0000066323.15244.54. [DOI] [PubMed] [Google Scholar]
- 36.Fokstuen S, Munoz A, Melacini P, Iliceto S, Perrot A, Ozcelik C, et al. Rapid detection of genetic variants in hypertrophic cardiomyopathy by custom DNA resequencing array in clinical practice. Journal of Medical Genetics. 2011;48:572–576. doi: 10.1136/jmg.2010.083345. [DOI] [PubMed] [Google Scholar]
- 37.Arai S, Matsuoka R, Hirayama K, Sakurai H, Tamura M, Ozawa T, et al. Missense mutation of the beta-cardiac myosin heavy-chain gene in hypertrophic cardiomyopathy. American Journal of Medical Genetics. 1995;58:267–276. doi: 10.1002/ajmg.1320580314. [DOI] [PubMed] [Google Scholar]
- 38.Perrot A, Schmidt-Traub H, Hoffmann B, Prager M, Bit-Avragim N, Rudenko RI, et al. Prevalence of cardiac beta-myosin heavy chain gene mutations in patients with hypertro-phic cardiomyopathy. Journal of Molecular Medicine (Berl) 2005;83:468–477. doi: 10.1007/s00109-005-0635-7. [DOI] [PubMed] [Google Scholar]
- 39.Alpert NR, Mohiddin SA, Tripodi D, Jacobson-Hatzell J, Vaughn-Whitley K, Brosseau C, et al. Molecular and phenotypic effects of heterozygous, homozygous, and compound heterozygote myosin heavy-chain mutations. American Journal of Physiology Heart C. 2005;288:H1097–H1102. doi: 10.1152/ajpheart.00650.2004. [DOI] [PubMed] [Google Scholar]
- 40.Cuda G, Fananapazir L, Zhu WS, Sellers JR, Epstein ND. Skeletal muscle expression and abnormal function of beta-myosin in hypertrophic cardiomyopathy. Journal of Clinical Investigation. 1993;91:2861–2865. doi: 10.1172/JCI116530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Epstein ND, Cohn GM, Cyran F, Fananapazir L. Differences in clinical expression of hypertrophic cardiomyopathy associated with two distinct mutations in the beta-myosin heavy chain gene. A 908Leu—Va l m u ta tio n a n d a 4 0 3 A r g —Gln mutation. Circulation. 1992;86:345–352. doi: 10.1161/01.cir.86.2.345. [DOI] [PubMed] [Google Scholar]
- 42.Morita H, Larson MG, Barr SC, Vasan RS, O’Donnell CJ, Hirschhorn JN, et al. Single-gene mutations and increased left ventricular wall thickness in the community: the Framingham Heart Study. Circulation. 2006;113:2697–2705. doi: 10.1161/CIRCULATIONAHA.105.593558. [DOI] [PubMed] [Google Scholar]
- 43.Frisso G, Limongelli G, Pacileo G, Del Giudice A, Forgione L, Calabro P, et al. A child cohort study from southern Italy enlarges the genetic spectrum of hypertrophic cardiomyopa-thy. Clinical Genetics. 2009;76:91–101. doi: 10.1111/j.1399-0004.2009.01190.x. [DOI] [PubMed] [Google Scholar]
- 44.Kimura A. Molecular basis of hereditary cardiomyop-athy: abnormalities in calcium sensitivity, stretch response, stress response and beyond. Journal of Human Genetics. 2010;55:81–90. doi: 10.1038/jhg.2009.138. [DOI] [PubMed] [Google Scholar]
- 45.Rosenzweig A, Watkins H, Hwang DS, Miri M, McKenna W, Traill TA, et al. Preclinical diagnosis of familial hypertrophic cardiomyopathy by genetic analysis of blood lymphocytes. New England Journal of Medicine. 1991;325:1753–1760. doi: 10.1056/NEJM199112193252501. [DOI] [PubMed] [Google Scholar]
- 46.Watkins H, Rosenzweig A, Hwang DS, Levi T, McKenna W, Seidman CE, et al. Characteristics and prognostic implications of myosin missense mutations in familial hypertrophic cardiomyopathy. New England Journal of Medicine. 1992;326:1108–1114. doi: 10.1056/NEJM199204233261703. [DOI] [PubMed] [Google Scholar]
- 47.Thierfelder L, Watkins H, MacRae C, Lamas R, McKenna W, Vosberg HP, et al. Alpha-tropomyosin and cardiac tropo-nin T mutations cause familial hypertrophic cardiomyopathy: a disease of the sarcomere. Cell. 1994;77:701–712. doi: 10.1016/0092-8674(94)90054-x. [DOI] [PubMed] [Google Scholar]
- 48.Torricelli F, Girolami F, Olivotto I, Passerini I, Frusconi S, Vargiu D, et al. Prevalence and clinical profile of troponin T mutations among patients with hypertrophic cardio-myopathy in tuscany. American Journal of Cardiology. 2003;92:1358–1362. doi: 10.1016/j.amjcard.2003.08.031. [DOI] [PubMed] [Google Scholar]
- 49.Theopistou A, Anastasakis A, Miliou A, Rigopoulos A, Toutouzas P, Stefanadis C. Clinical features of hypertrophic cardiomyopathy caused by an Arg278Cys missense mutation in the cardiac troponin T gene. American Journal of Cardiology. 2004;94:246–249. doi: 10.1016/j.amjcard.2004.03.077. [DOI] [PubMed] [Google Scholar]
- 50.Kapplinger JD, Landstrom AP, Bos JM, Salisbury BA, Callis TE, Ackerman MJ. Distinguishing hypertro-phic cardiomyopathy-associated mutations from background genetic noise. Journal of Cardiovascular Translational Research. 2014;7:347–361. doi: 10.1007/s12265-014-9542-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ingles J, Yeates L, Semsarian C. The emerging role of the cardiac genetic counselor. Heart Rhythm. 2011;8:1958–1962. doi: 10.1016/j.hrthm.2011.07.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.