Abstract
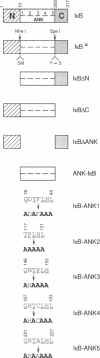
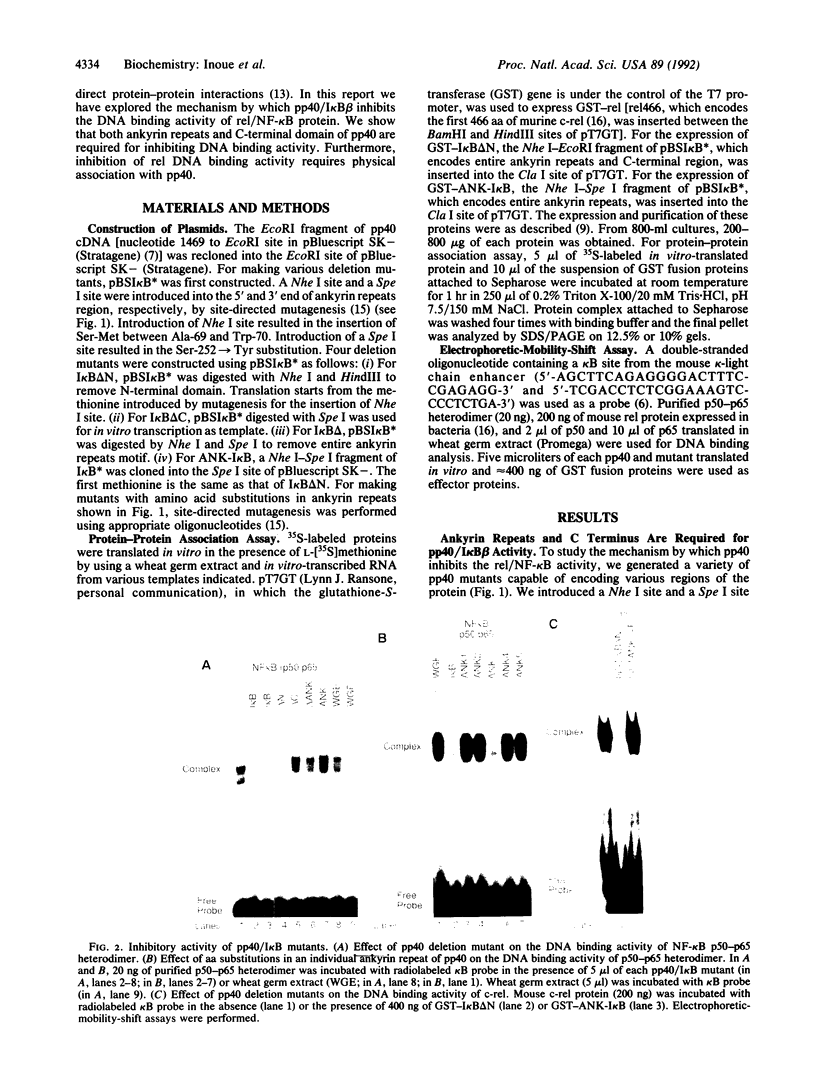
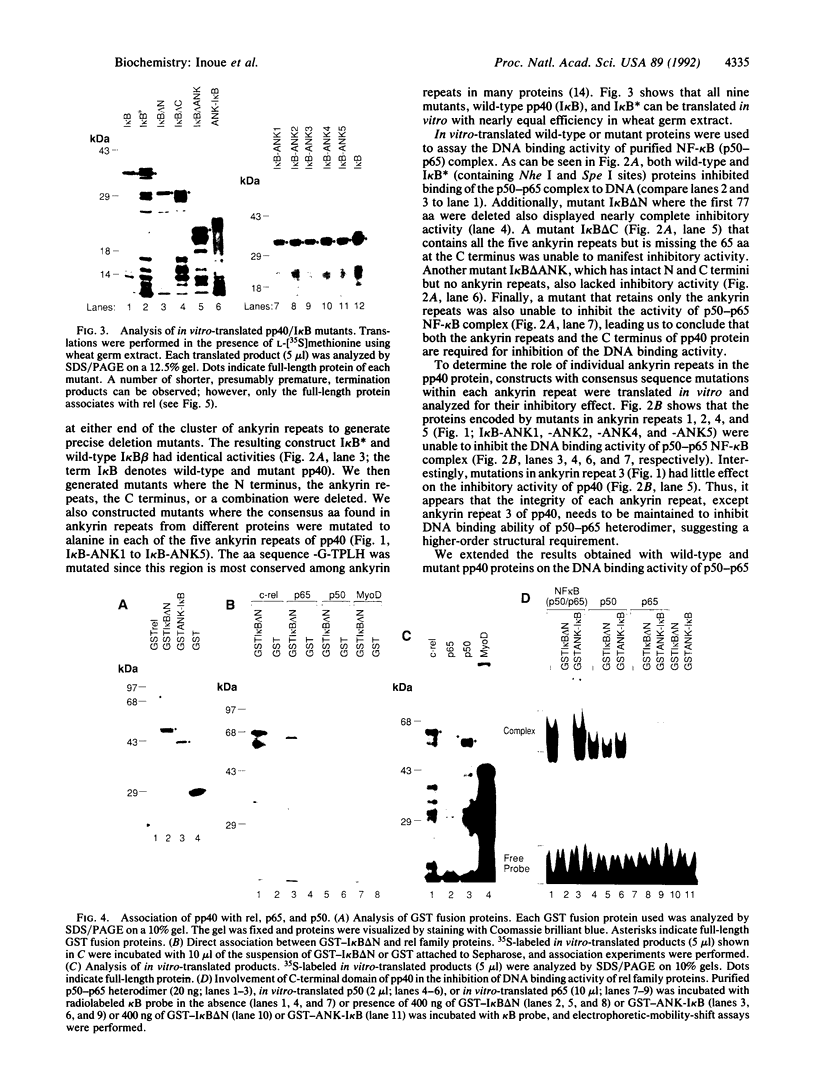
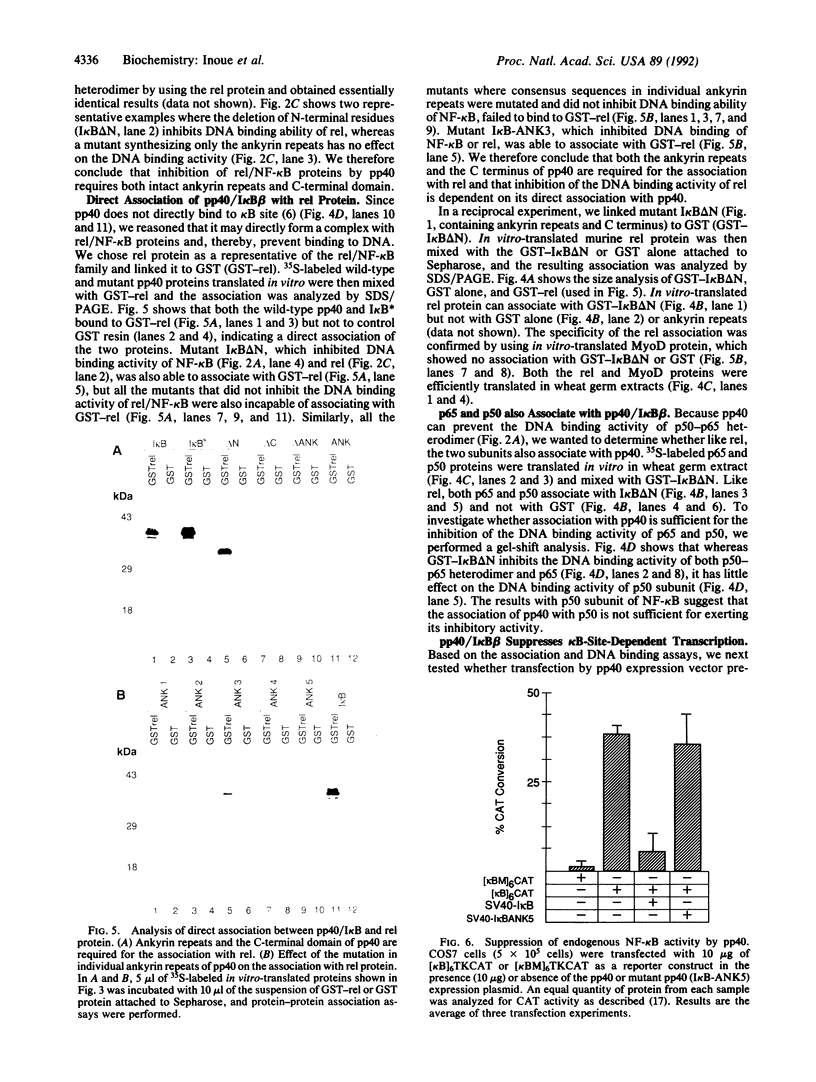
To understand the mechanism by which pp40/I kappa B beta inhibits DNA binding activity of the rel/NF-kappa B family of transcription factors, we have investigated the role of ankyrin repeats on the biological function of pp40 by deleting or mutating conserved residues. We show that (i) ankyrin repeats alone are not sufficient to manifest biological activity but require the C-terminal region of the pp40 protein; (ii) four out of the five ankyrin repeats are essential for inhibiting the DNA binding activity; (iii) pp40 mutants that do not inhibit DNA binding of rel protein also do not associate with rel; (iv) although pp40 can associate with the p65 and p50 subunits of NF-kappa B, pp40 inhibits the DNA binding activity of only the p50-p65 heterodimer and the p65 homodimer; and (v) pp40 inhibits the transcription of genes linked to kappa B site; however, mutants that do not affect DNA binding have no effect. We propose that the ankyrin repeats and the C-terminal region of pp40 form a structure that associates with the rel homology domain to inhibit DNA binding activity.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andrews B. J., Herskowitz I. The yeast SWI4 protein contains a motif present in developmental regulators and is part of a complex involved in cell-cycle-dependent transcription. Nature. 1989 Dec 14;342(6251):830–833. doi: 10.1038/342830a0. [DOI] [PubMed] [Google Scholar]
- Aves S. J., Durkacz B. W., Carr A., Nurse P. Cloning, sequencing and transcriptional control of the Schizosaccharomyces pombe cdc10 'start' gene. EMBO J. 1985 Feb;4(2):457–463. doi: 10.1002/j.1460-2075.1985.tb03651.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeuerle P. A., Baltimore D. Activation of DNA-binding activity in an apparently cytoplasmic precursor of the NF-kappa B transcription factor. Cell. 1988 Apr 22;53(2):211–217. doi: 10.1016/0092-8674(88)90382-0. [DOI] [PubMed] [Google Scholar]
- Baeuerle P. A., Baltimore D. I kappa B: a specific inhibitor of the NF-kappa B transcription factor. Science. 1988 Oct 28;242(4878):540–546. doi: 10.1126/science.3140380. [DOI] [PubMed] [Google Scholar]
- Baeuerle P. A. The inducible transcription activator NF-kappa B: regulation by distinct protein subunits. Biochim Biophys Acta. 1991 Apr 16;1072(1):63–80. doi: 10.1016/0304-419x(91)90007-8. [DOI] [PubMed] [Google Scholar]
- Breeden L., Nasmyth K. Similarity between cell-cycle genes of budding yeast and fission yeast and the Notch gene of Drosophila. Nature. 1987 Oct 15;329(6140):651–654. doi: 10.1038/329651a0. [DOI] [PubMed] [Google Scholar]
- Davis N., Ghosh S., Simmons D. L., Tempst P., Liou H. C., Baltimore D., Bose H. R., Jr Rel-associated pp40: an inhibitor of the rel family of transcription factors. Science. 1991 Sep 13;253(5025):1268–1271. doi: 10.1126/science.1891714. [DOI] [PubMed] [Google Scholar]
- Ghosh S., Baltimore D. Activation in vitro of NF-kappa B by phosphorylation of its inhibitor I kappa B. Nature. 1990 Apr 12;344(6267):678–682. doi: 10.1038/344678a0. [DOI] [PubMed] [Google Scholar]
- Ghosh S., Gifford A. M., Riviere L. R., Tempst P., Nolan G. P., Baltimore D. Cloning of the p50 DNA binding subunit of NF-kappa B: homology to rel and dorsal. Cell. 1990 Sep 7;62(5):1019–1029. doi: 10.1016/0092-8674(90)90276-k. [DOI] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haskill S., Beg A. A., Tompkins S. M., Morris J. S., Yurochko A. D., Sampson-Johannes A., Mondal K., Ralph P., Baldwin A. S., Jr Characterization of an immediate-early gene induced in adherent monocytes that encodes I kappa B-like activity. Cell. 1991 Jun 28;65(7):1281–1289. doi: 10.1016/0092-8674(91)90022-q. [DOI] [PubMed] [Google Scholar]
- Inoue J., Kerr L. D., Kakizuka A., Verma I. M. I kappa B gamma, a 70 kd protein identical to the C-terminal half of p110 NF-kappa B: a new member of the I kappa B family. Cell. 1992 Mar 20;68(6):1109–1120. doi: 10.1016/0092-8674(92)90082-n. [DOI] [PubMed] [Google Scholar]
- Inoue J., Kerr L. D., Ransone L. J., Bengal E., Hunter T., Verma I. M. c-rel activates but v-rel suppresses transcription from kappa B sites. Proc Natl Acad Sci U S A. 1991 May 1;88(9):3715–3719. doi: 10.1073/pnas.88.9.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr L. D., Inoue J., Davis N., Link E., Baeuerle P. A., Bose H. R., Jr, Verma I. M. The rel-associated pp40 protein prevents DNA binding of Rel and NF-kappa B: relationship with I kappa B beta and regulation by phosphorylation. Genes Dev. 1991 Aug;5(8):1464–1476. doi: 10.1101/gad.5.8.1464. [DOI] [PubMed] [Google Scholar]
- Kieran M., Blank V., Logeat F., Vandekerckhove J., Lottspeich F., Le Bail O., Urban M. B., Kourilsky P., Baeuerle P. A., Israël A. The DNA binding subunit of NF-kappa B is identical to factor KBF1 and homologous to the rel oncogene product. Cell. 1990 Sep 7;62(5):1007–1018. doi: 10.1016/0092-8674(90)90275-j. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- LaMarco K., Thompson C. C., Byers B. P., Walton E. M., McKnight S. L. Identification of Ets- and notch-related subunits in GA binding protein. Science. 1991 Aug 16;253(5021):789–792. doi: 10.1126/science.1876836. [DOI] [PubMed] [Google Scholar]
- Lux S. E., John K. M., Bennett V. Analysis of cDNA for human erythrocyte ankyrin indicates a repeated structure with homology to tissue-differentiation and cell-cycle control proteins. Nature. 1990 Mar 1;344(6261):36–42. doi: 10.1038/344036a0. [DOI] [PubMed] [Google Scholar]
- Nolan G. P., Ghosh S., Liou H. C., Tempst P., Baltimore D. DNA binding and I kappa B inhibition of the cloned p65 subunit of NF-kappa B, a rel-related polypeptide. Cell. 1991 Mar 8;64(5):961–969. doi: 10.1016/0092-8674(91)90320-x. [DOI] [PubMed] [Google Scholar]
- Ohno H., Takimoto G., McKeithan T. W. The candidate proto-oncogene bcl-3 is related to genes implicated in cell lineage determination and cell cycle control. Cell. 1990 Mar 23;60(6):991–997. doi: 10.1016/0092-8674(90)90347-h. [DOI] [PubMed] [Google Scholar]
- Schmid R. M., Perkins N. D., Duckett C. S., Andrews P. C., Nabel G. J. Cloning of an NF-kappa B subunit which stimulates HIV transcription in synergy with p65. Nature. 1991 Aug 22;352(6337):733–736. doi: 10.1038/352733a0. [DOI] [PubMed] [Google Scholar]
- Sen R., Baltimore D. Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell. 1986 Aug 29;46(5):705–716. doi: 10.1016/0092-8674(86)90346-6. [DOI] [PubMed] [Google Scholar]
- Thompson C. C., Brown T. A., McKnight S. L. Convergence of Ets- and notch-related structural motifs in a heteromeric DNA binding complex. Science. 1991 Aug 16;253(5021):762–768. doi: 10.1126/science.1876833. [DOI] [PubMed] [Google Scholar]
- Wharton K. A., Johansen K. M., Xu T., Artavanis-Tsakonas S. Nucleotide sequence from the neurogenic locus notch implies a gene product that shares homology with proteins containing EGF-like repeats. Cell. 1985 Dec;43(3 Pt 2):567–581. doi: 10.1016/0092-8674(85)90229-6. [DOI] [PubMed] [Google Scholar]
- Yochem J., Greenwald I. glp-1 and lin-12, genes implicated in distinct cell-cell interactions in C. elegans, encode similar transmembrane proteins. Cell. 1989 Aug 11;58(3):553–563. doi: 10.1016/0092-8674(89)90436-4. [DOI] [PubMed] [Google Scholar]
- Yochem J., Weston K., Greenwald I. The Caenorhabditis elegans lin-12 gene encodes a transmembrane protein with overall similarity to Drosophila Notch. Nature. 1988 Oct 6;335(6190):547–550. doi: 10.1038/335547a0. [DOI] [PubMed] [Google Scholar]