Coat protein complex II (COPII) is responsible for recruiting and packaging proteins into vesicles for export from the endoplasmic reticulum. Recruitment by COPII does not ensure subsequent packaging into vesicles, suggesting that cargo recruitment and cargo packaging are distinct rather than concomitant processes.
Abstract
In addition to its role in forming vesicles from the endoplasmic reticulum (ER), the coat protein complex II (COPII) is also responsible for selecting specific cargo proteins to be packaged into COPII transport vesicles. Comparison of COPII vesicle formation in mammalian systems and in yeast suggested that the former uses more elaborate mechanisms for cargo recognition, presumably to cope with a significantly expanded repertoire of cargo that transits the secretory pathway. Using proTGFα, the transmembrane precursor of transforming growth factor α (TGFα), as a model cargo protein, we demonstrate in cell-free assays that at least one auxiliary cytosolic factor is specifically required for the efficient packaging of proTGFα into COPII vesicles. Using a knockout HeLa cell line generated by CRISPR/Cas9, we provide functional evidence showing that a transmembrane protein, Cornichon-1 (CNIH), acts as a cargo receptor of proTGFα. We show that both CNIH and the auxiliary cytosolic factor(s) are required for efficient recruitment of proTGFα to the COPII coat in vitro. Moreover, we provide evidence that the recruitment of cargo protein by the COPII coat precedes and may be distinct from subsequent cargo packaging into COPII vesicles.
INTRODUCTION
The endoplasmic reticulum (ER) is the entry point for newly synthesized proteins into the secretory pathway, and the coat protein complex II (COPII) is the membrane coat that is responsible for recruiting and transporting cargo from the ER via COPII vesicles. The COPII coat consists of five core components: the small GTPase Sar1, the Sec23/24 heterodimer, and the Sec13/31 heterotetramer. Cytosolic Sar1 is activated when it exchanges bound GDP for GTP, a process mediated by the guanine nucleotide exchange factor (GEF) Sec12, which is an ER- resident protein (Nakano et al., 1988; Barlowe and Schekman, 1993). Activated Sar1 extends an amphipathic N-terminal α-helix and associates with the ER membrane, to which it recruits the Sec23/24 heterodimer, forming the prebudding complex (Kuehn et al., 1998; Huang et al., 2001). After assembly of the prebudding complex, the Sec13/31 heterotetramer is recruited to the ER, where it polymerizes to guide membrane curvature and vesicle formation (Antonny et al., 2001).
The selection and enrichment of COPII cargo proteins is achieved by interaction with the COPII coat, mainly via the Sec24 subunit (Miller et al., 2002, 2003). In principle, transmembrane cargo proteins can interact directly with Sec24 via their cytoplasmic domains. Indeed, this has been observed for a number of well-characterized COPII cargo proteins (Mossessova et al., 2003; Wendeler et al., 2007). In the course of investigating the molecular requirements for COPII packaging of certain mammalian transmembrane proteins, however, we detected the involvement of one or more cytosolic factors in addition to the COPII coat.
Transforming growth factor α (TGFα) was identified as a secreted peptide that had cell-transforming properties. TGFα is synthesized as a small type I transmembrane protein with a short cytoplasmic C-terminus that has a PDZ-binding motif (PBM) at the C-terminus. The N-terminal luminal domain contains glycosylation sites and an epidermal growth factor (EGF)–like domain. At the cell surface, proTGFα undergoes two sequential proteolytic cleavages, which result in the release of the soluble TGFα ligand into the extracellular space (Singh and Coffey, 2014). In HeLa cells, proTGFα was reported to interact in the ER with Cornichon-1 (hereafter referred to as Cornichon and abbreviated as CNIH for simplicity). A luminal loop of CNIH was shown to interact with the luminal EGF-like domain of proTGFα in the ER (Castro et al., 2007). CNIH was proposed to act as a cargo receptor for proTGFα, but overexpression of CNIH–hemagglutinin (HA) paradoxically led to more retention of proTGFα in the ER, making it unclear whether CNIH plays a direct role in the ER export of proTGFα in vivo (Castro et al., 2007). In addition, mutation of the cytoplasmic C-terminal valine to glycine (V160G) was shown in multiple studies to significantly impede the ER export kinetics of proTGFα, suggesting a cytoplasmic component involved in proTGFα trafficking (Bosenberg et al., 1992; Fernández-Larrea et al., 1999; Ureña et al., 1999). Using a cell-free reaction that reproduces the cargo-selective packaging of mammalian membrane cargo proteins into COPII vesicles, we probed the requirement for auxiliary cytosolic factors in the ER export of specific mammalian cargo, as well as the function of CNIH in mammalian cells.
RESULTS
Evidence for existence of auxiliary cytosolic factors
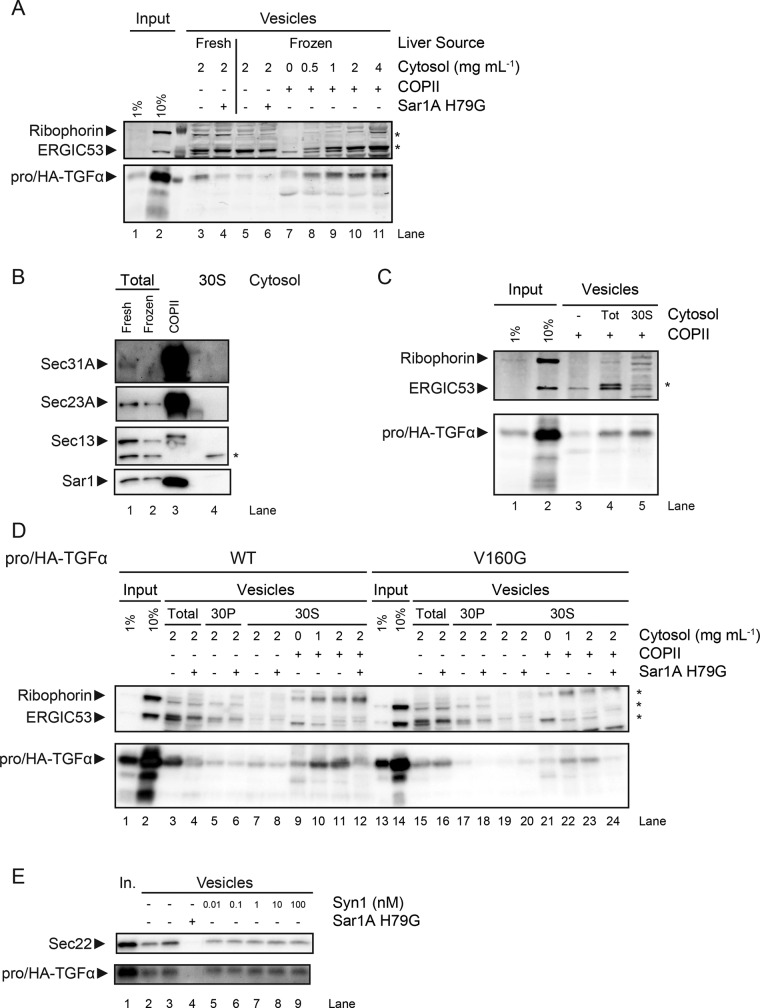
Cytosol prepared from fresh rat livers was sufficient to support packaging of both pro/HA-TGFα and ERGIC53, a lectin that cycles between the ER and ER-Golgi intermediate compartment (ERGIC), into COPII vesicles made in a cell-free vesicle budding reaction. The control protein ribophorin I, an ER-resident protein and component of the eukaryotic oligosaccharide transferase complex, was absent in the vesicle fractions, indicating the preservation of ER membrane integrity (Figure 1A, lanes 3 and 4). Cytosol prepared from frozen rat livers (livers were flash-frozen in liquid nitrogen and then thawed slowly at 4°C), in contrast, was inactive in the budding reaction (Figure 1A, lanes 5 and 6). Purified COPII components supported the efficient packaging of ERGIC53 but not pro/HA-TGFα (Figure 1A, lane 7), suggesting the requirement of a cargo-selective cytosolic factor(s) for the packaging of the latter. Addition of freeze-thaw–inactivated cytosol and purified COPII components restored the packaging of pro/HA-TGFα into COPII vesicles (Figure 1A, lanes 8–11). Thus we speculated that the freeze-thaw–inactivated cytosol might have lost COPII activity but retained an additional factor essential for the packaging of proTGFα.
FIGURE 1:
ProTGFα requires an auxiliary cytosolic factor for efficient COPII packaging. HeLa cells were transfected with plasmids expressing pro/HA-TGFα, and membranes were harvested the next day for use in in vitro budding reactions. (A) Quantitative infrared fluorescence immunoblot (LiCor) analysis of in vitro budding reaction. Rat liver cytosol was prepared from fresh or frozen rat livers as indicated. Sar1A H79G is a GTPase mutant that is a dominant-negative inhibitor of in vitro budding. (B) Immunoblot of various cytosol fractions and purified COPII samples. (C) LiCor analysis of in vitro budding reaction. (D, E) Immunoblots of in vitro budding reactions. Total, total cytosol extract; 30P, pellet fraction of cytosol after 30% ammonium sulfate precipitation; 30S, supernatant fraction of cytosol after 30% ammonium sulfate precipitation. Syn1, purified recombinant human syntenin-1. Asterisks indicate unrelated bands.
Although cytosol prepared from frozen rat livers displayed minimal budding activity, it still contained significant amounts of the COPII subunits (Figure 1B, lanes 1 and 2). To rule out the possibility that the pro/HA-TGFα–specific packaging activity was due to the residual COPII components, we mixed proteins from the inactive cytosol with 30% ammonium sulfate under conditions in which COPII proteins are precipitated (Kim et al., 2005), which reduced COPII levels below the detection limit (Figure 1B, lane 4). Vesicle budding reactions supplemented with a dialyzed aliquot of the 30% supernatant fraction (30S) showed specific enhancement of pro/HA-TGFα packaging (Figure 1C, compare lanes 3 and 5), indicating that the pro/HA-TGFα–specific activity was not attributable to known COPII components but possibly to at least one auxiliary cytosolic factor specifically required in the packaging of pro/HA-TGFα. Of note, whereas ERGIC53 packaging was enhanced by total cytosol extract, the 30S cytosol fraction did not have an effect in this regard (Figure 1C, lanes 4 and 5).
Further experiments were conducted to confirm that the observed pro/HA-TGFα packaging activity was a COPII-dependent process. The 30S cytosol fraction was unable to support packaging of either ERGIC53 or pro/HA-TGFα in the absence of purified COPII (Figure 1D, lane 7). Supplementing purified COPII components with the 30S fraction greatly enhanced pro/HA-TGFα packaging efficiency (Figure 1D, lanes 9–11), and this activity was inhibited when Sar1A H79G, a dominant-negative inhibitor of COPII budding in vitro, was included in the reaction (Figure 1D, lane 12), thus confirming that the pro/HA-TGFα–specific activity of the 30S fraction was dependent on COPII.
To test the physiological relevance of our in vitro observations, we examined the behavior of the V160G mutant form of proTGFα, which exhibited trafficking defects (Briley et al., 1997). When tested in the in vitro budding assay, pro/HA-TGFα V160G was packaged inefficiently into vesicles (Figure 1D, compare lane 15 and lanes 3 and 16). As expected, supplementing purified COPII with the 30S cytosolic fraction resulted in inefficient packaging of pro/HA-TGFα V160G as well (Figure 1D, compare lanes 22 and 23 and lanes 10 and 11), and thus the mutant behaved in vitro as it did in vivo.
The PDZ domain–containing protein syntenin-1 (Syn1) was previously identified as an interaction partner of proTGFα. Syn1 interacted with the wild-type (WT) C-terminus of proTGFα but not the V160G mutant form. Hence it was proposed to function in the early steps of proTGFα trafficking (Fernández-Larrea et al., 1999). In light of the foregoing observations, we tested whether Syn1 acted as the auxiliary cytosolic factor. Purified recombinant Syn1 was supplemented to in vitro budding reactions, but no significant effect on pro/HA-TGFα budding was observed (Figure 1E). Thus our in vitro evidence does not support a role for Syn1 as the auxiliary factor. However, it is possible that the purified Syn1 is inactive, and our results do not rule out the possibility that Syn1 might act in coordination with other cytosolic factors.
Cornichon functions as a cargo receptor for proTGFα
In Drosophila, Cornichon has been implicated in the ER export of Gurken (Roth et al., 1995; Bökel et al., 2006), a single-pass transmembrane protein that also contains an extracellular EGF-like domain. The Cornichon homologue in yeast, Erv14p, has been characterized to function as a cargo receptor for the ER export of Axl2p (Powers and Barlowe, 1998, 2002), among other client proteins (Nakanishi et al., 2007; Herzig et al., 2012).
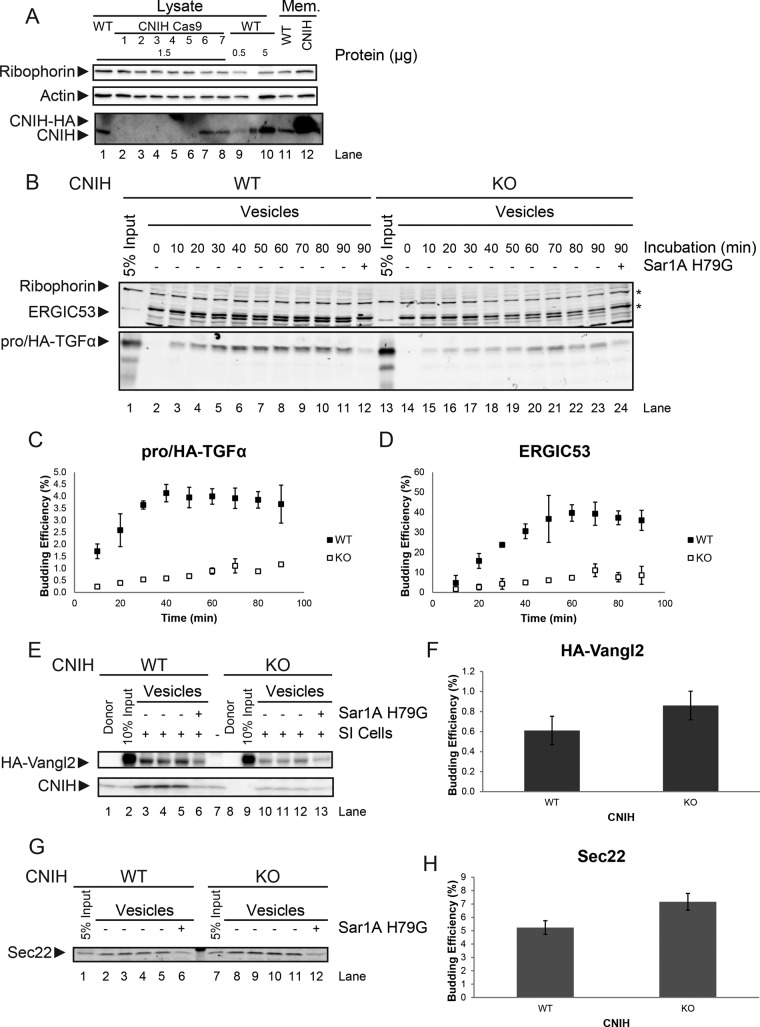
CNIH has been proposed to perform a similar function in mammalian cells, but previous characterization of its role in trafficking in vivo was inconclusive (Castro et al., 2007). To ask whether CNIH might indeed play such a role, we generated a CNIH-knockout (KO) cell line in HeLa cells using genome editing with clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9. Seven clones were expanded and analyzed for expression of CNIH (Figure 2A). Genomic sequencing confirmed that clone #3 was a bona fide genomic knockout of CNIH, and this cell line was used in subsequent experiments.
FIGURE 2:
Loss of CNIH impairs ER export of proTGFα. (A) Immunoblot of HeLa cell samples. Lysate, total cell lysate; WT, wild type; CNIH Cas9, cells treated for modification at the CNIH locus by CRISPR/Cas9. Mem., permeabilized WT cells that were either untreated (WT) or transfected with a plasmid encoding CNIH-HA (CNIH). (B) LiCor analysis of an in vitro budding reaction. (C, D) Quantification of budding efficiency of pro/HA-TGFα (C) and ERGIC53 (D). (E) Immunoblot of in vitro budding reactions. Donor, permeabilized HeLa cells that were not subjected to in vitro translation. (G) LiCor analysis of in vitro budding reactions. (F, H) Quantification of budding efficiency of HA-Vangl2 (F) and Sec22 (H). Quantifications are represented as mean ± SEM (p > 0.05 in both cases). Asterisks indicate unrelated bands.
We first assessed the effect of CNIH KO on proTGFα trafficking. CNIH KO resulted in diminished budding efficiency of pro/HA-TGFα but also in diminished ERGIC53 budding efficiency (Figure 2, B–D). However, CNIH KO did not significantly affect the trafficking of Sec22, a vesicle soluble N-ethylmaleimide–sensitive factor attachment protein receptor (SNARE) that cycles between the ER and ERGIC, and Vangl2, a transmembrane protein involved in cell signaling in the planar cell polarity pathway (Figure 2, E–H), suggesting that CNIH KO did not result in general disruption of COPII vesicle formation. Of importance, CNIH itself was also packaged very efficiently into COPII vesicles (Figure 2E). Together these data point to a role for CNIH in the ER export of proTGFα.
Reintroducing cornichon into CNIH KO HeLa cells partially rescues the proTGFα budding phenotype
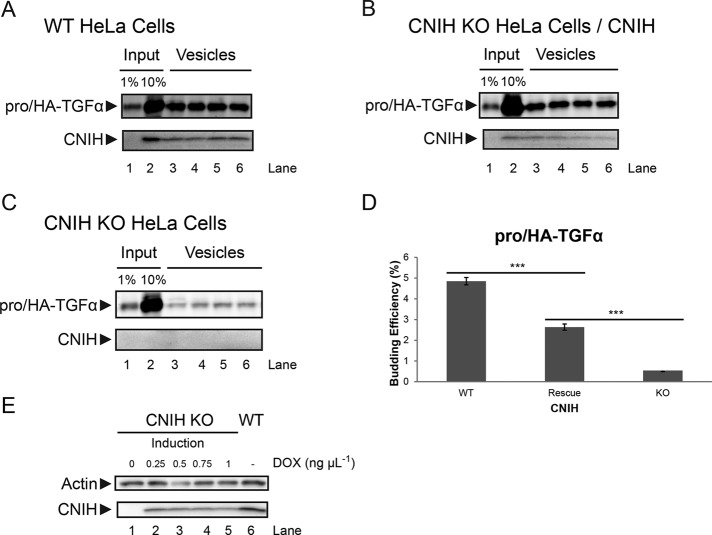
To confirm that the proTGFα budding defects observed in CNIH KO membranes could be attributed to the absence of CNIH, we performed rescue experiments. Full-length human CNIH was put under the control of a tetracycline-inducible promoter and stably integrated into CNIH KO HeLa cells. A moderate level of CNIH expression was induced by addition of doxycycline (Figure 3E). Cells were then processed for use in the in vitro budding reactions.
FIGURE 3:
Expression of CNIH in CNIH KO HeLa cells partially rescues proTGFα budding phenotype. (A–C) In vitro budding reactions. CNIH was detected using immunoblotting, and pro/HA-TGFα was detected using LiCor. Membranes were prepared from wild-type HeLa cells expressing pro/HA-TGFα (A), CNIH KO HeLa cells expressing pro/HA-TGFα and CNIH induced by 1 ng/μl doxycycline (B), or CNIH KO HeLa cells expressing pro/HA-TGFα without doxycycline induction (C). Lanes 3–6 in A–C are quadruplicate budding reactions. (D) Quantification of proTGFα budding efficiency. Results are represented as mean ± SEM (***p < 0.001). (E) Immunoblot showing the induction of CNIH expression. Cells were treated with the indicated amounts of doxycycline (DOX) for 24 h, and then lysates were harvested for analysis.
In membranes prepared from WT HeLa cells, pro/HA-TGFα was efficiently exported from the ER, as was CNIH (Figure 3A). In membranes prepared from CNIH KO cells, packaging of pro/HA-TGFα into COPII vesicles was significantly impaired (Figure 3C). When a moderate level of CNIH expression was reintroduced into CNIH KO cells (rescue), pro/HA-TGFα budding efficiency was significantly increased above KO levels, although it was not restored to WT levels (Figure 3, B and D).
The difference in proTGFα budding efficiency in WT and rescue membranes may be attributed to the lower CNIH expression levels in the rescue cells. Higher expression levels of CNIH led to ER retention (Supplemental Figure S1), making it challenging to approximate WT levels of CNIH expression in CNIH KO cells. Despite this drawback, the efficiency of proTGFα budding showed a clear dependence on CNIH levels in the donor membranes (Figure 3D).
Cytosol prepared from CNIH KO HeLa cells showed enhanced COPII activity
In the in vitro budding reactions, pro/HA-TGFα budding efficiency was significantly reduced in membranes prepared from CNIH KO HeLa cells (Figures 2 and 3). However, immunofluorescence microscopy showed no significant difference in steady-state proTGFα localization in WT and CNIH KO cells. In addition, no significant difference in proTGFα trafficking kinetics was observed between WT and CNIH KO cells (Supplemental Figure S2).
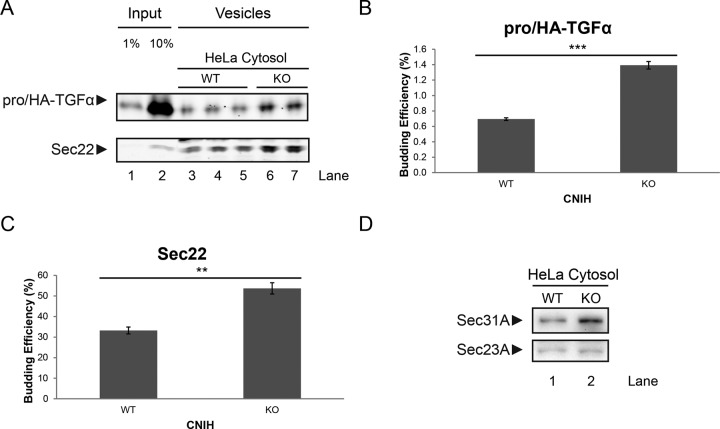
Of note, the experiments in Figures 2 and 3 were done using HeLa cells as membrane source and rat liver cytosol as COPII source. The observations made in vitro and in vivo suggested the possibility of some compensatory effect attributable to the cytosolic protein content of CNIH KO HeLa cells as an explanation of this discrepancy.
To test this idea, we prepared cytosol from WT and CNIH KO HeLa cells and used it in the in vitro budding reaction. As shown in Figure 4, cytosol prepared from CNIH KO cells displayed significantly higher COPII activity than that prepared from WT cells (Figure 4A). When incubated with CNIH KO membranes, cytosol prepared from CNIH KO cells significantly increased the budding efficiency of pro/HA-TGFα compared with WT cytosol (Figure 4B). Sec22 budding was also enhanced but to a lesser degree (Figure 4C).
FIGURE 4:
Cytosol from CNIH KO HeLa cells displays higher COPII activity in vitro. (A) LiCor analysis of an in vitro budding reaction. WT HeLa cells were transiently transfected with plasmids encoding pro/HA-TGFα, and membranes were harvested for use in the in vitro budding reaction. Cytosol was harvested either from WT HeLa cells or CNIH KO HeLa cells and used in the in vitro budding reaction. Lanes 3–5 are triplicates of budding reactions done using cytosol prepared from WT HeLa cells. Lanes 6 and 7 are duplicates of budding reactions done using cytosol prepared from CNIH KO HeLa cells. (B, C) Quantification of budding efficiency of pro/HA-TGFα (B) and Sec22 (C). Quantifications are represented as mean ± SEM (***p < 0.001; **p < 0.01). (D) Immunoblot of cytosol prepared from WT or CNIH KO HeLa cells.
Analysis of cytosolic protein content showed that the expression of Sec31A was increased in CNIH KO cells compared with WT, whereas Sec23A levels remained similar (Figure 4D). This observation also supports the idea that CNIH KO cells might have undergone adaptation in response to the loss of CNIH.
proTGFα recruitment to prebudding complex requires cooperation of cytosolic factor(s) and CNIH
The apparent involvement of both a transmembrane cargo receptor and a cytosolic factor(s) in the ER export of proTGFα prompted us to investigate the mechanism of proTGFα recruitment to the prebudding complex using the COPII recruitment assay, which allowed us to isolate the prebudding complex and analyze its contents.
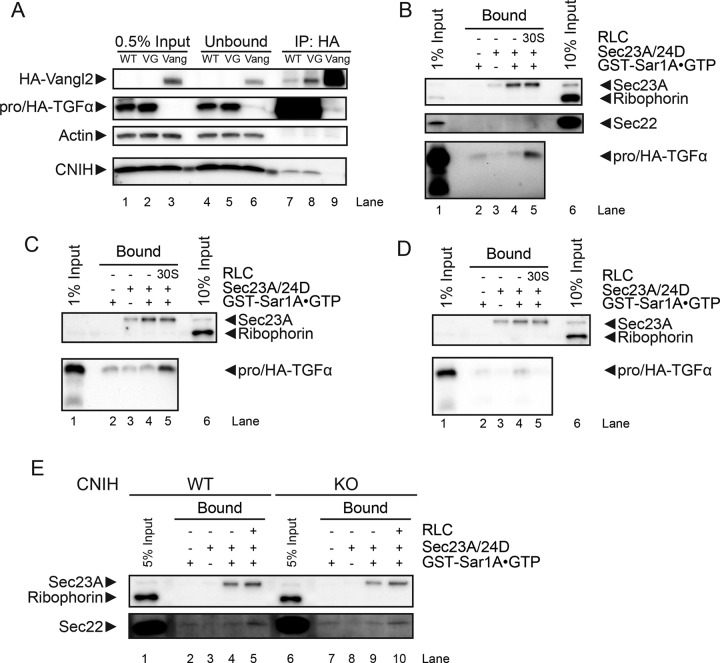
To confirm that the trafficking defect of pro/HA-TGFα V160G was not a result of defective binding to CNIH, we performed coimmunoprecipitation experiments and demonstrated that pro/HA-TGFα V160G bound to CNIH as efficiently as wild type. The interaction between CNIH and pro/HA-TGFα was specific, as no interaction was detected between CNIH and HA-Vangl2 (Figure 5A, lanes 7–9; in lanes 7 and 8, an unrelated band appears at an apparent mobility similar to HA-Vangl2).
FIGURE 5:
CNIH and cytosolic factor(s) required for proTGFα recruitment to the prebudding complex. (A) HeLa cells were transiently transfected with plasmids expressing pro/HA-TGFα wild type (WT), V160G mutant (VG), or HA-Vangl2 (Vang) as indicated, and immunoprecipitation was performed using anti-HA antibodies. (B–E) Immunoblots of COPII recruitment assays. Recruitment of pro/HA-TGFα was examined using membranes prepared from WT HeLa cells expressing proTGFα WT (B) or V160G (C) or using membranes prepared from CNIH KO HeLa cells expressing proTGFα WT (D). (E) Recruitment of Sec22 compared in HeLa WT or CNIH KO membranes.
HeLa cells were transiently transfected with plasmids encoding pro/HA-TGFα WT, and membranes were harvested the next day for use in the COPII recruitment assay. The prebudding complex was captured using GST-Sar1A-GTP–restricted mutant protein and isolated by GST pull down (for details, see Materials and Methods). When the prebudding complex was assembled using purified Sec23A/24D heterodimer, pro/HA-TGFα was not recruited to the prebudding complex (Figure 5B, lane 4). Supplementing the 30% supernatant fraction (30S) to the reaction resulted in recruitment of pro/HA-TGFα WT to the prebudding complex, demonstrating the requirement of cytosolic factor(s) in this process (Figure 5B, lane 5). Of note, whereas pro/HA-TGFα was recruited to the prebudding complex, Sec22 was not, indicating cargo specificity. HeLa cells were then transiently transfected with plasmids encoding pro/HA-TGFα V160G and processed for use in the COPII recruitment assay. Of interest, although the V160G mutant was defective for packaging into COPII vesicles, it was still recruited to the prebudding complex in the presence of purified Sec23A/24D and the 30S fraction (Figure 5C, lane 5). Finally, HeLa CNIH KO cells were transiently transfected with plasmids encoding pro/HA-TGFα WT and processed for use in the COPII recruitment assay. Removal of CNIH from HeLa membranes prevented pro/HA-TGFα WT from being recruited to the prebudding complex (Figure 5D, lane 5). As a control, the effect of CNIH KO on Sec22 recruitment was analyzed. Whereas supplementing the 30S fraction to purified Sec23A/24D did not support Sec22 recruitment to the prebudding complex, supplementing total rat liver cytosol did (Figure 5E, lane 5). This can be explained by the fact that Sec22 specifically interacts with isoforms A and B of Sec24 (Mancias and Goldberg, 2008). Total rat liver cytosol contained all four Sec24 isoforms and thus was able to support Sec22 recruitment to the prebudding complex. In contrast, the 30S fraction did not contain Sec24, and thus Sec22 was not recruited to the prebudding complex. Of greater importance, CNIH KO did not affect Sec22 recruitment to the prebudding complex, supporting a specific role for CNIH in proTGFα recruitment (Figure 5E, lane 10).
DISCUSSION
In vertebrates, there are four homologues of Cornichon: CNIH-1–4. CNIH-2 and -3 were shown to play a role in regulating the subunit composition of AMPA receptors, presumably by regulating trafficking of the receptor components from the ER (Kato et al., 2010; Herring et al., 2013). CNIH-4 was shown to play a role in the ER export of G protein–coupled receptors (GPCRs) in HeLa cells (Sauvageau et al., 2014). These observations suggest that Cornichon homologues play an evolutionarily conserved role as cargo receptors in ER export. However, overexpression of CNIH in HeLa cells increases the retention of proTGFα in the ER (Castro et al., 2007). This may be explained by our observation that overexpression of CNIH results in its ER retention, which in turn traps proTGFα in the ER (Supplemental Figure S1). By analogy, this might also explain the observation that CNIH-4 overexpression in HeLa cells similarly results in the ER retention of GPCRs (Sauvageau et al., 2014).
We showed that the efficient recruitment and packaging of proTGFα requires the cooperation of CNIH with one or more cytosolic factors. The yeast homologue, Erv14p, is required for the ER export of the transmembrane protein Yor1p, even though Yor1p interacts directly with Sec24p. The requirement of dual ER export signals is known for other Erv14p client proteins as well, suggesting that this phenomenon was not restricted to Yor1p (Pagant et al., 2015). Our observations suggest that this might also be the case for CNIH client proteins in mammals. This might be due to the weak and transient nature of interactions between cargo and coats, which requires multiple sites of interaction to stabilize the prebudding complex for efficient incorporation into COPII vesicles.
Although a defect in pro/HA-TGFα recruitment and packaging was observed in vitro, a significant defect in pro/HA-TGFα steady-state localization or ER export kinetics was not detected in CNIH KO cells (Supplemental Figure S2). We did find evidence of a change in the cytosolic protein content of CNIH KO cells, which resulted in elevated COPII activity in vitro (Figure 4A), which supports the idea that the cells had undergone adaptation in response to a loss of CNIH. However, the cause of this change is unclear. Of note, most known client proteins of Erv14p, the yeast homologue of CNIH, are single-pass transmembrane proteins that ultimately localize to the plasma membrane (Powers and Barlowe, 1998; Herzig et al., 2012). These plasma membrane–localized proteins possess transmembrane domains that are longer than those of ER- or Golgi-resident proteins (Sharpe et al., 2010). Thus it was proposed that Erv14p might also act as a chaperone in the ER to shield the hydrophobic transmembrane domains of its client proteins (D’Arcangelo et al., 2013). Indeed, deletion of the ERV14 gene was found to enhance the unfolded protein response (UPR) in yeast, which leads to up-regulation of many genes involved in the ER-to-Golgi trafficking pathway, including SEC12, SEC13, and SEC24 (Travers et al., 2000; Jonikas et al., 2009). It is possible that CNIH plays a similar role in mammalian cells and loss of CNIH might lead to a moderate UPR in mammalian cells that enhances COPII activity.
The moderate UPR triggered by loss of CNIH might also potentially explain the unexpected impairment of ERGIC53 export (Figure 2D). ERGIC53 oligomerization is required for its efficient export from the ER (Nufer et al., 2003). Whereas the COPII interaction motif resides in the cytoplasmic C-terminus of ERGIC53 (Kappeler et al., 1997; Wendeler et al., 2007), the luminal domain plays a crucial role in mediating oligomerization (Neve et al., 2005). Thus it is possible that CNIH KO resulted in changes in the ER lumen that affected ERGIC53 oligomerization, resulting in the diminished budding efficiency observed.
Of note, the V160G mutant form of proTGFα, although packaged inefficiently into COPII vesicles, is recruited normally to the prebudding complex when CNIH and the cytosolic factor(s) are present. This shows that efficient recruitment of cargo to the prebudding complex does not guarantee efficient cargo packaging into COPII vesicles, suggesting that cargo recruitment and cargo packaging are distinct processes.
Also noteworthy is the lack of correlation between cargo recruitment and budding efficiency. In the in vitro budding reactions, ERGIC53 was very efficiently packaged into COPII vesicles. However, it was undetectable in the COPII-binding assays (unpublished data). In contrast, Sec22 was also very efficiently packaged into COPII vesicles and was detected at reasonable levels in the COPII-binding assays (Figure 5E). This might be explained by the stable association of Sec22 with Sec23/24 upon formation of a fusogenic SNARE complex with Bos1 and Sed5, as shown with yeast proteins (Sato and Nakano, 2005). The apparently different binding affinities of two efficiently packaged cargo proteins to the prebudding complex again supports the notion that cargo recruitment and cargo packaging are two distinct events in the process of COPII vesicle formation.
Prebudding complexes are maintained in a highly dynamic manner. Interaction between Sec24 and cargo persists even after Sar1 hydrolyzes its bound GTP, whereas Sec12 activity dynamically maintains the interaction between Sec24 and cargo, presumably by reloading Sar1 with GTP (Sato and Nakano, 2005). This suggests that multiple rounds of GTP hydrolysis by Sar1 might be required for productive packaging of cargo into COPII vesicles. The established methods used to isolate prebudding complexes use either nonhydrolyzable GTP analogues or GTPase-defective Sar1 mutants and thus are unable to capture the dynamic nature of this assembly intermediate and might explain the extremely low efficiency of its recovery (Aridor et al., 1998; Kuehn et al., 1998).
The selection of appropriate cargo proteins to include into nascent vesicles is a critical function of the COPII coat, in addition to its role in vesicle formation. This is the fundamental mechanism to ensure proper functioning of the eukaryotic endomembrane system and, by extension, to ensure cell viability. During the course of evolution, novel mechanisms might have emerged to cope with the expanding repertoire of cargo proteins that pass through the endomembrane system. New methods are needed to capture the transient and dynamic nature of cargo recruitment and packaging. The identification and characterization of the auxiliary cytosolic factor(s) required for efficient COPII packaging of proTGFα will also provide important information to help elucidate the molecular mechanisms for cargo recognition and packaging in mammalian cells.
MATERIALS AND METHODS
Antibodies
Anti-Sec13 and anti-Sec22 sera were prepared as previously described (Merte et al., 2010). Anti-Sec31A was from BD Transduction Laboratories (612350; San Jose, CA). Anti-Sec23A, anti-Sar1A/B, and anti-ERGIC53 sera were prepared as previously described (Fromme et al., 2007). Anti–ribophorin I serum was provided by Peter Walter (University of California, San Francisco, San Francisco, CA). Rabbit anti-HA monoclonal antibody was from Cell Signaling (C29F4; Danvers, MA). Anti-CNIH was from Sigma-Aldrich (SAB1304796; St. Louis, MO). Anti-mouse and anti-rabbit horseradish peroxidase conjugates were from GE Healthcare (NXA931 and NA934V, respectively; Little Chalfont, United Kingdom).
Preparation of rat liver cytosol
Where indicated, rat livers were preserved by flash freezing with liquid N2 and stored at −80°C. Frozen livers were then thawed in 400 ml of phosphate-buffered saline (PBS) at 4°C. Fresh livers were used unless otherwise indicated. Rat liver cytosol was prepared in buffer E as described in Kim et al. (2005), typically at a protein concentration of 30–40 mg/ml.
Preparation of HeLa cytosol
Wild-type or CNIH-KO HeLa cells were grown in 10 150-mm plates to 90% confluency, washed with PBS, and collected using a cell scraper into 10 ml B88 buffer containing 1× protease inhibitors. Cells were rendered permeable by exposure to 80 μg/ml digitonin in B88 with protease inhibitors for 30 min at 4°C with gentle rocking. Bio-Beads SM-2 (2 g; Bio-Rad, Hercules, CA) resin was hydrated and washed with 35 ml of B88 buffer. Permeabilized cells were centrifuged at 300 × g for 5 min at 4°C, and the supernatant (10 ml) was collected and incubated with Bio-Beads overnight at 4°C with gentle rocking to remove cell debris and digitonin. Bio-Beads were subsequently removed by centrifugation at 300 × g for 5 min at 4°C. The resulting supernatant was further centrifuged at 135,000 × g for 30 min at 4°C to remove remaining insoluble material or protein aggregates. The supernatant was then transferred to Amicon tubes and concentrated to a final volume of ∼500 μl. Protein concentration was then measured, and the final cytosol was distributed into aliquots, flash frozen, and stored at −80°C.
Preparation of permeabilized cells
The procedure for preparing permeabilized cells and subsequent in vitro translation was performed as previously described (Merte et al., 2010), with slight modifications. In detail, HeLa cells were grown in a 100-mm dish to 90% confluency, washed with 5 ml of PBS, trypsinized at room temperature for 5 min, and washed again in 6 ml of KHM buffer containing 10 μg/ml soybean trypsin inhibitor. Cells were then permeabilized in 40 μg/ml digitonin for 5 min in 6 ml of ice-cold KHM buffer. Permeabilized cells were then washed once with B88-LiCl buffer and once in B88 buffer and finally resuspended in B88 buffer such that the optical density at 600 nm of membranes in B88 buffer was 0.750. The resuspension was aliquoted and stored at −80°C.
For use in in vitro translation, semi-intact cells were washed once in KHM buffer after permeabilization with digitonin and resuspended in 100 μl of KHM buffer.
In vitro translation
In vitro translation was performed as previously described (Merte et al., 2010), with slight modifications. In detail, 1 mM CaCl2 and 10 μg/ml micrococcal nuclease were added to 100 μl of permeabilized cells prepared as described to remove endogenous RNA. After incubation at room temperature for 12 min, 4 mM ethylene glycol tetraacetic acid (EGTA) was then added to terminate the reaction. Membranes were then centrifuged at 10,000 × g at 4°C for 15 s and resuspended in KHM buffer such that the optical density at 600 nm of 5-μl membranes in 500 μl of KHM buffer was 0.060. These membranes were added to in vitro translation reactions containing rabbit reticulocyte lysate (Flexi; Promega, Madison, WI), amino acids, KCl, and mRNA encoding HA-Vangl2, which was incubated at 30°C for 60 min (Merte et al., 2010). Donor membranes were centrifuged at 2700 × g at 4°C for 3 min and resuspended with 1 ml of B88-LiCl buffer. Donor membranes were then washed twice with B88 buffer and resuspended in 20 μl of B88 buffer per reaction.
In vitro COPII vesicle budding assay
Vesicle formation and purification was performed as described previously (Kim et al., 2005), with slight modifications. In detail, donor membranes were incubated where indicated with rat liver cytosol (2 mg/ml), recombinant COPII proteins (10 ng/μl Sar1A, 10 ng/μl Sec23A/24D, 10 ng/μl Sec13/31A), ATP regeneration system, 0.3 mM GTP, and Sar1A H79G (10 ng/μl) in a final volume of 100 μl. The reaction was carried out at 30°C for 1 h and terminated by incubation on ice for 5 min. Donor membranes were removed by centrifugation at 14,000 × g for 12 min at 4°C. The resulting supernatant was centrifuged at 115,000 × g for 25 min at 4°C to collect COPII vesicles. Vesicle pellets were then resuspended in 15 μl of buffer C (10 mM Tris-HCl, pH 7.6, 100 mM NaCl, 1% Triton X-100) and Laemmli sample buffer and heated at 55°C for 15 min. Vesicle samples and original donor membrane samples were resolved by SDS–PAGE, transferred onto polyvinylidene fluoride (PVDF) membranes, and subjected to immunoblotting to detect various ER-resident proteins and COPII cargo proteins.
COPII recruitment assay
The membrane recruitment of COPII components using recombinant Sar1A-GTP–restricted mutant was performed as previously described (Aridor et al., 1995), with modifications. In detail, donor membranes were mixed where indicated with rat liver cytosol (2 mg/ml), recombinant COPII components (20 ng/μl Sec23A/24D), ATP regeneration system, 1 mM GTP, and GST-Sar1A-GTP–restricted mutant protein (Sar1A H79G with an N-terminal GST tag, 30 ng/μl) in a total volume of 100 μl. The mixture was incubated at 30°C for 30 min, after which the reaction was terminated by transfer onto ice. Membranes were collected by centrifugation at 20,000 × g for 10 min at 4°C. The supernatant was discarded, and membranes were dissolved by incubation with 1 ml of solubilization buffer (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES], pH 7.2, 1 mM magnesium acetate, 1% digitonin) on ice for 30 min, with occasional mixing. Insoluble material was removed by centrifugation at 163,000 × g for 30 min at 4°C. The resulting supernatant was then incubated with glutathione agarose beads (Pierce, Waltham, MA) at 4°C for 30 min with rotational rocking. Subsequently, the beads were collected by centrifugation (200 × g) and washed three times with 200 μl of solubilization buffer. Beads were eluted by heating in Laemmli sample buffer at 55°C for 15 min. Samples were resolved by SDS–PAGE, transferred onto PVDF membranes, and subjected to immunoblotting to detect various ER-resident proteins, COPII cargo proteins, and Sec23A.
Purification of recombinant proteins
Purification of human Sar1A WT and H79G was performed as described (Kim et al., 2005). Purification of human GST-Sar1A-GTP–restricted form was performed as described (Kim et al., 2005), with modification. Briefly, instead of thrombin cleavage, the GST-tagged recombinant protein was directly eluted with 5 mM glutathione in PBS buffer, dialyzed overnight in HKM-G buffer, distributed into aliquots, flash frozen in liquid N2, and stored at −80°C. Purification of human FLAG-Sec23A/His-Sec24D was performed according to previously described procedures (Kim et al., 2005).
Immunoprecipitation
HeLa cells grown in 10-cm plates were transiently transfected with 2 μg of plasmids encoding pro/HA-TGFα wild type or V160G mutant, or HA-Vangl2, using Lipofectamine 2000 according to the manufacturer’s recommended protocol. Twenty-four hours later, cells were washed twice in PBS. Cells were then solubilized in 1 ml of immunoprecipitation (IP) lysis buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM ethylenediaminetetraacetic acid [EDTA], 0.5% Triton X-100, 1× protease inhibitors) on ice for 15 min. Cell lysates were cleared by centrifugation at 20,817 × g for 15 min at 4°C. An aliquot (10 μl) of the supernatant fraction was reserved in a separate tube and mixed with Laemmli sample buffer. An aliquot (30 μl) of 50% monoclonal anti–HA-agarose (Sigma-Aldrich) slurry was washed three times with IP lysis buffer, added to cleared cell lysates, and incubated at 4°C for 1 h. The monoclonal anti–HA-agarose beads were centrifuged at 200 × g and washed three times with IP lysis buffer. The beads were then resuspended in 15 μl of 1× Laemmli sample buffer and heated to 55°C for 15 min, and samples were resolved by SDS–PAGE, transferred onto PVDF membranes, and subjected to immunoblotting.
Supplemental methods
The methods used to generate the images shown in Supplemental Figure S2 can be found online in the Supplemental Materials.
Supplementary Material
Acknowledgments
We thank Lixon Wang, Lazar Dimitrov, and Kartoosh Heydari for assistance in preparing CNIH-KO HeLa cells and Ann Fisher at the Berkeley Tissue Culture Facility for assistance with cell culture. The plasmid encoding CNIH-HA was a generous gift from Carolina Castro and Rik Derynck. R.S. is an investigator of the Howard Hughes Medical Institute.
Abbreviations used:
- CNIH
Cornichon
- COPII
coat protein complex II
- EGF
epidermal growth factor
- ER
endoplasmic reticulum
- GEF
guanine nucleotide exchange factor
- PBM
PDZ-binding motif
- TGFα
transforming growth factor α.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-02-0090) on April 27, 2016.
REFERENCES
- Antonny B, Madden D, Hamamoto S, Orci L, Schekman R. Dynamics of the COPII coat with GTP and stable analogues. Nat Cell Biol. 2001;3:531–537. doi: 10.1038/35078500. [DOI] [PubMed] [Google Scholar]
- Aridor M, Bannykh SI, Rowe T, Balch WE. Sequential coupling between COPII and COPI vesicle coats in endoplasmic reticulum to Golgi transport. J Cell Biol. 1995;131:875–893. doi: 10.1083/jcb.131.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aridor M, Weissman J, Bannykh S, Nuoffer C, Balch WE. Cargo selection by the COPII budding machinery during export from the ER. J Cell Biol. 1998;141:61–70. doi: 10.1083/jcb.141.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe C, Schekman R. SEC12 encodes a guanine-nucleotide-exchange factor essential for transport vesicle budding from the ER. Nature. 1993;365:347–349. doi: 10.1038/365347a0. [DOI] [PubMed] [Google Scholar]
- Bökel C, Dass S, Wilsch-Bräuninger M, Roth S. Drosophila Cornichon acts as cargo receptor for ER export of the TGFalpha-like growth factor Gurken. Development. 2006;133:459–470. doi: 10.1242/dev.02219. [DOI] [PubMed] [Google Scholar]
- Bosenberg MW, Pandiella A, Massagué J. The cytoplasmic carboxy-terminal amino acid specifies cleavage of membrane TGF alpha into soluble growth factor. Cell. 1992;71:1157–1165. doi: 10.1016/s0092-8674(05)80064-9. [DOI] [PubMed] [Google Scholar]
- Briley GP, Hissong MA, Chiu ML, Lee DC. The carboxyl-terminal valine residues of proTGF alpha are required for its efficient maturation and intracellular routing. Mol Biol Cell. 1997;8:1619–1631. doi: 10.1091/mbc.8.8.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro CP, Piscopo D, Nakagawa T, Derynck R. Cornichon regulates transport and secretion of TGFalpha-related proteins in metazoan cells. J Cell Sci. 2007;120:2454–2466. doi: 10.1242/jcs.004200. [DOI] [PubMed] [Google Scholar]
- D’Arcangelo JG, Stahmer KR, Miller EA. Vesicle-mediated export from the ER: COPII coat function and regulation. Biochim Biophys Acta. 2013;1833:2464–2472. doi: 10.1016/j.bbamcr.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Larrea J, Merlos-Suárez A, Ureña JM, Baselga J, Arribas J. A role for a PDZ protein in the early secretory pathway for the targeting of proTGF-alpha to the cell surface. Mol Cell. 1999;3:423–433. doi: 10.1016/s1097-2765(00)80470-0. [DOI] [PubMed] [Google Scholar]
- Fromme JC, Ravazzola M, Hamamoto S, Al-Balwi M, Eyaid W, Boyadjiev SA, Cosson P, Schekman R, Orci L. The genetic basis of a craniofacial disease provides insight into COPII coat assembly. Dev Cell. 2007;13:623–634. doi: 10.1016/j.devcel.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herring BE, Shi Y, Suh YH, Zheng C-YY, Blankenship SM, Roche KW, Nicoll RA. Cornichon proteins determine the subunit composition of synaptic AMPA receptors. Neuron. 2013;77:1083–1096. doi: 10.1016/j.neuron.2013.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzig Y, Sharpe HJ, Elbaz Y, Munro S, Schuldiner M. A systematic approach to pair secretory cargo receptors with their cargo suggests a mechanism for cargo selection by Erv14. PLoS Biol. 2012;10:e1001329. doi: 10.1371/journal.pbio.1001329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Weissman JT, Beraud-Dufour S, Luan P, Wang C, Chen W, Aridor M, Wilson IA, Balch WE. Crystal structure of Sar1-GDP at 1.7 A resolution and the role of the NH2 terminus in ER export. J Cell Biol. 2001;155:937–948. doi: 10.1083/jcb.200106039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonikas MC, Collins SR, Denic V, Oh E, Quan EM, Schmid V, Weibezahn J, Schwappach B, Walter P, Weissman JS, Schuldiner M. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 2009;323:1693–1697. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappeler F, Klopfenstein DR, Foguet M, Paccaud J-P, Hauri H-P. The recycling of ERGIC-53 in the early secretory pathway: ERGIC-53 carries a cytosolic endoplasmic reticulum-exit determinant interacting with COPII. J Biol Chem. 1997;272:31801–31808. doi: 10.1074/jbc.272.50.31801. [DOI] [PubMed] [Google Scholar]
- Kato AS, Gill MB, Ho MT, Yu H, Tu Y, Siuda ER, Wang H, Qian YW, Nisenbaum ES, Tomita S, Bredt DS. Hippocampal AMPA receptor gating controlled by both TARP and cornichon proteins. Neuron. 2010;68:1082–1096. doi: 10.1016/j.neuron.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Hamamoto S, Ravazzola M, Orci L, Schekman R. Uncoupled packaging of amyloid precursor protein and presenilin 1 into coat protein complex II vesicles. J Biol Chem. 2005;280:7758–7768. doi: 10.1074/jbc.M411091200. [DOI] [PubMed] [Google Scholar]
- Kuehn MJ, Herrmann JM, Schekman R. COPII-cargo interactions direct protein sorting into ER-derived transport vesicles. Nature. 1998;391:187–190. doi: 10.1038/34438. [DOI] [PubMed] [Google Scholar]
- Mancias JD, Goldberg J. Structural basis of cargo membrane protein discrimination by the human COPII coat machinery. EMBO J. 2008;27:2918–2928. doi: 10.1038/emboj.2008.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merte J, Jensen D, Wright K, Sarsfield S, Wang Y, Schekman R, Ginty DD. Sec24b selectively sorts Vangl2 to regulate planar cell polarity during neural tube closure. Nat Cell Biol. 2010;12:41–46. doi: 10.1038/ncb2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller E, Antonny B, Hamamoto S, Schekman R. Cargo selection into COPII vesicles is driven by the Sec24p subunit. EMBO J. 2002;21:6105–6113. doi: 10.1093/emboj/cdf605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller EA, Beilharz TH, Malkus PN, Lee MC, Hamamoto S, Orci L, Schekman R. Multiple cargo binding sites on the COPII subunit Sec24p ensure capture of diverse membrane proteins into transport vesicles. Cell. 2003;114:497–509. doi: 10.1016/s0092-8674(03)00609-3. [DOI] [PubMed] [Google Scholar]
- Mossessova E, Bickford LC, Goldberg J. SNARE selectivity of the COPII coat. Cell. 2003;114:483–495. doi: 10.1016/s0092-8674(03)00608-1. [DOI] [PubMed] [Google Scholar]
- Nakanishi H, Suda Y, Neiman AM. Erv14 family cargo receptors are necessary for ER exit during sporulation in Saccharomyces cerevisiae. J Cell Sci. 2007;120:908–916. doi: 10.1242/jcs.03405. [DOI] [PubMed] [Google Scholar]
- Nakano A, Brada D, Schekman R. A membrane glycoprotein, Sec12p, required for protein transport from the endoplasmic reticulum to the Golgi apparatus in yeast. J Cell Biol. 1988;107:851–863. doi: 10.1083/jcb.107.3.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neve EP, Lahtinen U, Pettersson RF. Oligomerization and interacellular localization of the glycoprotein receptor ERGIC-53 is independent of disulfide bonds. J Mol Biol. 2005;354:556–568. doi: 10.1016/j.jmb.2005.09.077. [DOI] [PubMed] [Google Scholar]
- Nufer O, Kappeler F, Guldbrandsen S, Hauri H-PP. ER export of ERGIC-53 is controlled by cooperation of targeting determinants in all three of its domains. J Cell Sci. 2003;116:4429–4440. doi: 10.1242/jcs.00759. [DOI] [PubMed] [Google Scholar]
- Pagant S, Wu A, Edwards S, Diehl F, Miller EA. Sec24 is a coincidence detector that simultaneously binds two signals to drive ER export. Curr Biol. 2015;25:403–412. doi: 10.1016/j.cub.2014.11.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers J, Barlowe C. Transport of axl2p depends on erv14p, an ER-vesicle protein related to the Drosophila cornichon gene product. J Cell Biol. 1998;142:1209–1222. doi: 10.1083/jcb.142.5.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers J, Barlowe C. Erv14p directs a transmembrane secretory protein into COPII-coated transport vesicles. Mol Biol Cell. 2002;13:880–891. doi: 10.1091/mbc.01-10-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S, Neuman-Silberberg FS, Barcelo G, Schüpbach T. cornichon and the EGF receptor signaling process are necessary for both anterior-posterior and dorsal-ventral pattern formation in Drosophila. Cell. 1995;81:967–978. doi: 10.1016/0092-8674(95)90016-0. [DOI] [PubMed] [Google Scholar]
- Sato K, Nakano A. Dissection of COPII subunit-cargo assembly and disassembly kinetics during Sar1p-GTP hydrolysis. Nat Struct Mol Biol. 2005;12:167–174. doi: 10.1038/nsmb893. [DOI] [PubMed] [Google Scholar]
- Sauvageau E, Rochdi MD, Oueslati M, Hamdan FF, Percherancier Y, Simpson JC, Pepperkok R, Bouvier M. CNIH4 interacts with newly synthesized GPCR and controls their export from the endoplasmic reticulum. Traffic. 2014;15:383–400. doi: 10.1111/tra.12148. [DOI] [PubMed] [Google Scholar]
- Sharpe HJ, Stevens TJ, Munro S. A comprehensive comparison of transmembrane domains reveals organelle-specific properties. Cell. 2010;142:158–169. doi: 10.1016/j.cell.2010.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B, Coffey RJ. From wavy hair to naked proteins: the role of transforming growth factor alpha in health and disease. Semin Cell Dev Biol. 2014;28:12–21. doi: 10.1016/j.semcdb.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- Ureña JM, Merlos-Suárez A, Baselga J, Arribas J. The cytoplasmic carboxy-terminal amino acid determines the subcellular localization of proTGF-(alpha) and membrane type matrix metalloprotease (MT1-MMP) J Cell Sci. 1999;112:773–784. doi: 10.1242/jcs.112.6.773. [DOI] [PubMed] [Google Scholar]
- Wendeler MW, Paccaud J-PP, Hauri H-PP. Role of Sec24 isoforms in selective export of membrane proteins from the endoplasmic reticulum. EMBO Rep. 2007;8:258–264. doi: 10.1038/sj.embor.7400893. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.