Abstract
The list of protein aggregation-associated degenerative diseases is long and growing, while the portfolio of disease-modifying strategies is very small. In this review and perspective, we assess what has worked to slow the progression of an aggregation-associated degenerative disease, covering the underlying mechanism of pharmacologic action and what we have learned about the etiology of the transthyretin amyloid diseases and likely amyloidoses in general. Finally, we introduce emerging therapies that should apply more generally to protein misfolding and/or aggregation diseases that rely on adapting the protein homeostasis or proteostasis network for disease amelioration.
Introduction
Amyloidogenesis refers to the concentration-dependent, multi-step process of protein aggregation–resulting either from the alteration of the tertiary structure of a natively folded protein or the linked conformational changes and misassembly of an intrinsically disordered protein [1,2]. Compelling evidence suggests that the process of aggregation from one or several of more than 30 amyloidogenic proteins causes unique degenerative diseases [3]. These include Alzheimer’s and Parkinson’s diseases, the transthyretin amyloidoses, and light chain amyloidosis that result from the abnormal proliferation of cancer cells that secrete amyloidogenic light chains. Amyloidogenesis can occur intra- and/or extracellularly, affording numerous distinct aggregate structures including insoluble, fibrous cross-β-sheet assemblies, known as amyloid fibrils, on one end of the structural continuum [4] and at the other end, soluble oligomers exhibiting a range of secondary structures, including β-sheet-rich structures [2,5]. The process of amyloidogenesis is known to compromise the function of post-mitotic tissues, such as the heart or the peripheral, autonomic or central nervous systems by mechanisms under intense investigation [2,6]. While amyloid fibrils are a distinctive feature of amyloid diseases, there is no consensus on what drives the pathology of these maladies. Some investigators hypothesize that soluble, non-native, β-sheet-rich oligomers may be the major proteotoxic structure, more problematic than the amyloid fibrils themselves [7,8].
The precise factors that trigger amyloid pathology in humans remain incompletely characterized [9,10]. While inherited mutations generally make proteins prone to misfolding and aggregation [11,12], it is not clear why the process of amyloidogenesis only results in pathology later in life. One explanation may be aging-associated deficiencies in stress-responsive signaling pathways that regulate cellular proteostasis network capacity [13,14]. It has been hypothesized that the attenuated ability to activate stress-responsive signaling pathways upon aging results in increased protein aggregation that becomes pathogenic, triggering the onset of many age-onset sporadic and inherited amyloid diseases [14,15].
Current therapeutic approaches for the treatment of human amyloid diseases target various aspects of the amyloidogenesis cascade. A proven approach for ameliorating light chain amyloidosis is to use chemotherapy agents to kill the plasma cells secreting the light chains that aggregate. Antisense and RNAi approaches under development reduce the amyloidogenic protein concentration by mRNA degradation, whereas another approach removes aggregates and amyloid fibrils by antibody-mediated clearance [2,16]. Herein, we cover a related strategy—blocking the protein misfolding that commences the amyloidogenic cascade. The amyloidogenic protein that is the focus of the first part of this review is transthyretin which is associated with the transthyretin amyloidoses [12,17–19].
Transthyretin as an amyloidogenic protein
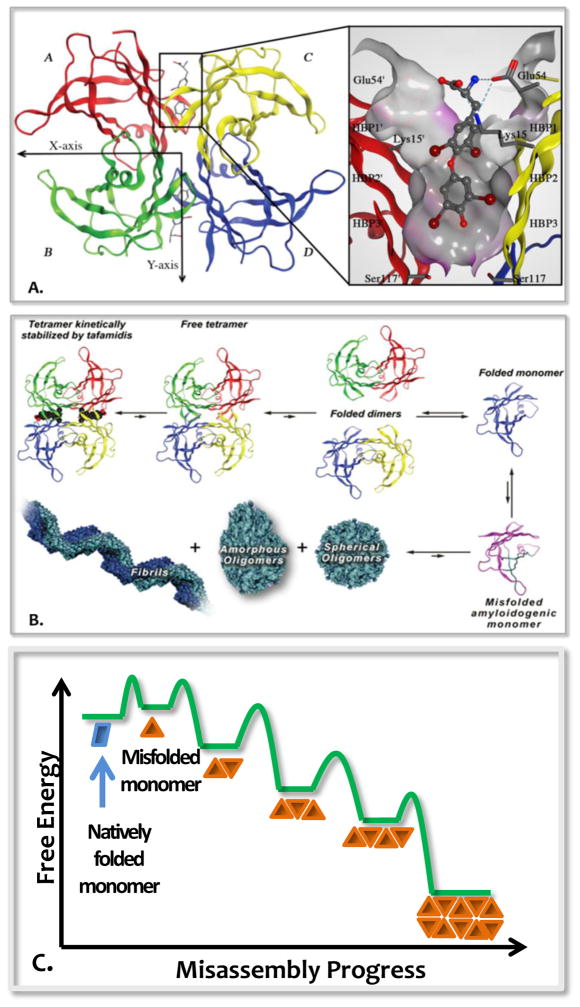
Transthyretin (TTR) is a protein produced in and secreted by the liver, by the choroid plexus and by the retinal pigment epithelial cells [20–22]. TTR is a 55 kDa tetramer made up of identical subunits (Figure 1A), each adopting a β-sheet-rich secondary structure comprising 127 amino acids [23]. The quaternary structure of transthyretin is characterized by two distinct dimer-dimer interfaces, the least stable of which, bisected by the X-axis, forms two C2-symmetric funnel-like binding sites for thyroxine (T4) [24,25]. In blood, TTR transports ≈ 0.5 eq. of holo-retinol binding protein and a very small amount of T4, due to the presence of other T4 carrier proteins.
Figure 1.
Transthyretin structure and amyloidogenesis cascade. (A) Ribbon diagram depiction of the structure of homo-tetrameric TTR with T4 occupying the ligand-binding site. (B) TTR amyloidogenic cascade. (C) Free energy diagram consistent with TTR aggregation.
In order for TTR to form amyloid fibrils, its tetramer must undergo rate-limiting dissociation to natively folded monomers, but this is not sufficient (Figure 1B) [18,26]. Partial monomer denaturation enables misassembly of TTR into structurally diverse aggregates, including amyloid fibrils—provided the concentration is high enough [27,28]. This process is best described as a downhill polymerization, wherein the misfolded monomer misassembles spontaneously, affording progressively more stable aggregate structures (Figure 1C) [29]. Three sequence-dependent factors determine the rate of TTR aggregation: 1) the rate of TTR homo-or heterotetramer (mutation carrier) dissociation, which is almost always rate-limiting [12,18,26,30]; 2) the rate of monomer misfolding, the extent of which is influenced by the thermodynamic stability of the natively folded monomers [26,29,31]; and 3) the total extracellular TTR concentration, which is controlled by the cells’ proteostasis network capacity that determines the partitioning of TTR between degradation or folding and secretion by sensing the stability of TTR [12].
In the autosomal dominantly inherited or mutation-associated TTR amyloidoses—including familial amyloid polyneuropathy (FAP) [32], and familial amyloid cardiomyopathy (FAC) [33,34]—aggregation of both mutant and wild type (WT) TTR leads to the degenerative phenotype. FAP compromises the function of the peripheral and autonomic nervous systems and initially manifests as a loss of temperature and pain sensation in the feet [35]. The progression of FAP leads to cachexia and often death within 10 years, if untreated. The onset of the familial forms of TTR amyloidosis can be as early as age 20 [36]. Aggregation of exclusively WT-TTR is the TTR amyloid disease affecting the most patients, typically men over age 60 [37,38]. This disease, called senile systemic amyloidosis (SSA), is a cardiomyopathy that is fatal approximately 5 years after initial diagnosis, if untreated. Hereditary amyloid FAP and/or FAC can also be caused by the aggregation of other proteins, such as apolipoprotein A1 [39,40].
Milestone discoveries
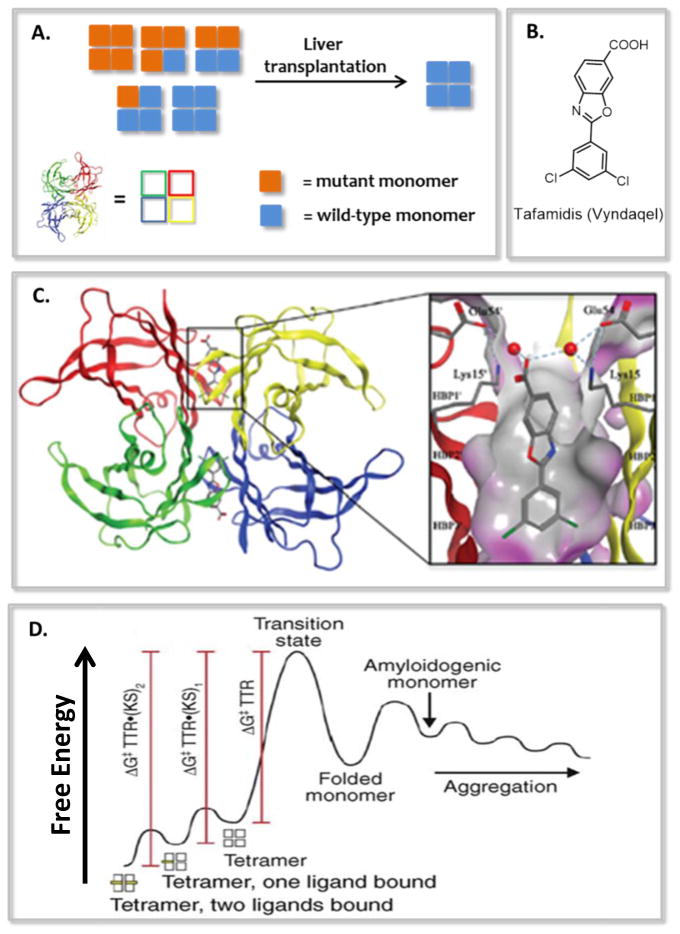
Liver transplantation–mediated TTR gene therapy was the first therapeutic approach for the treatment of V30M FAP [41,42]. In this strategy, a liver secreting heterotetrameric TTR, comprising mutated and/or wild-type monomers, was replaced with a donor liver secreting the more stable homotetrameric WT-TTR tetramers (Figure 2A, and 3). Although profoundly innovative, liver transplantation as a treatment strategy involves risks, including 10% surgery-related mortality and the requirement of life-long immunosuppression (increasing the risk of infection). Moreover, it was not anticipated that WT-TTR would continue to deposit after liver transplantation, leading to cardiomyopathy [43].
Figure 2.
Summary of key findings that led to a tafamidis-based kinetic stabilization strategy for the treatment and/or prevention of transthyretin-related amyloidoses. (A) Original liver transplantation-based strategy for replacing kinetically less stable heterotetrameric TTR with kinetically more stable wild type homotetrameric TTR for slowing the progression of familial amyloid polyneuropathy. (B) Line drawing of the tafamidis structure. (C) Structure of the (tafamidis)2•TTR complex that dissociates slowly. (D) Mechanism of TTR stabilization by tafamidis.
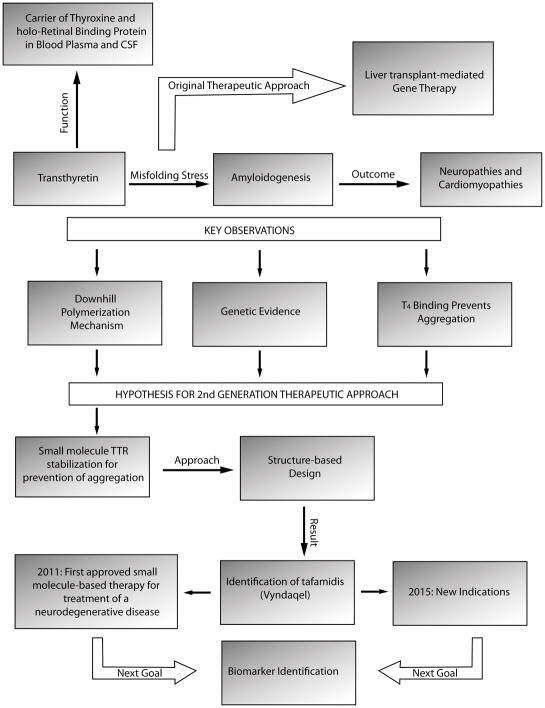
Figure 3.
Discovery path to tafamidis. Numerous observations summarized on this flow chart led to the discovery of tafamidis, its regulatory agency approval, and continuing biomarker discovery efforts.
In 1992, Coelho and coworkers described a Portuguese family carrying the V30M mutation that developed a rather benign type of FAP, if they developed the disease at all [44,45]. These individuals produced the V30M-TTR mutation from allele 1 and the T119M-TTR from allele 2, affording tetramers comprising a statistical distribution of V30M and T119M subunits (in contrast to the normal heterozygous FAP patient whose tetramers are a statistical mix of V30M and WT subunits) [44,45]. T119M-TTR subunit incorporation into tetramers composed of disease-associated TTR sequences kinetically stabilized the resulting tetramers by increasing the activation energy required for tetramer dissociation. The activation barrier increases proportional to the number of T119M subunits making up the tetramer [19,46]. These findings suggested that strategies which increase the tetramer dissociation barrier have the potential to slow disease progression.
Building on the observation that TTR aggregation can be accelerated under acidic conditions (pH=4–5) [47,48], leading to aggregation on a convenient laboratory time scale (72 h), our laboratory evaluated the influence of T4 binding to TTR with regard to selectively stabilizing the native tetramer over the dissociation transition state to slow tetramer dissociation—the rate limiting step of amyloidogenesis. We found that formation of a TTR•T4 complex prevented TTR aggregation by kinetically stabilizing TTR, i.e., by dramatically slowing tetramer dissociation [17]. Furthermore, we demonstrated that numerous ligands can bind to the tetramer and kinetically stabilize TTR, proportional to their binding constants [49–52].
Structure-based design of TTR kinetic stabilizers
This logic motivated us to design and synthesize > 1000 candidate small molecule kinetic stabilizers of TTR [53,54]. The exact process of structure-based design has been explained previously [51–53]. Briefly, small molecules that inhibited acid-mediated TTR aggregation were co-crystalized with TTR, leading to crystal structures of TTR bound to kinetic stabilizers. These structures guided small molecule modifications that have the potential to enhance binding affinity and selectivity. Promising candidates were defined as those which reduced TTR aggregation to less than 10% of vehicle control in the acid-mediated TTR aggregation assay [55–58] and did not exhibit nonsteroidal anti-inflammatory (NSAID) activity (a contraindicated activity for cardiomyopathy as NSAIDs further restrict renal blood flow) [55–57]. A candidate kinetic stabilizer’s ability to bind to TTR over the 4000+ additional proteins in plasma, including albumin, was assessed and over the years a few different methods have been utilized, each exhibiting some limitations [59–61]. The TTR subunit exchange assay, carried out in human plasma, is currently the best method to assess binding selectivity [61]. This method also quantifies kinetic stabilization under physiological conditions.
In 2003, our laboratory described the synthesis and evaluation of a library of benzoxazoles as candidate TTR kinetic stabilizers [58]. Of the 28 compounds envisioned by structure-based design, 11 prevented TTR aggregation in the acid-accelerated fibril formation assay, and a further six were found to selectively bind to TTR in blood plasma. These TTR kinetic stabilizers were among those considered for development by FoldRx Pharmaceuticals into an orally available therapeutic for the treatment of TTR-related amyloidoses. 2-(3,5-dichloro-phenyl)-benzoxazole-6-carboxylic acid, later named tafamidis, was one of those compounds (Figure 2B) [62]. Tafamidis proved to be orally bioavailable and it prevented TTR tetramer dissociation of wild-type and pathogenic TTR variants under fibril-promoting and physiological conditions [62]. Tafamidis was found to bind to TTR with high affinity (Kd1= 3 nM, Kd2= 278 nM), which undoubtedly contributed to its high binding selectivity towards TTR in complicated biological environments like blood [62]. Importantly, tafamidis lacked NSAID activity.
The X-ray crystal structure revealed that tafamidis occupied the T4-binding sitesas designed, and its affinity for TTR was driven by a combination of hydrophobic and electrostatic interactions (Figure 2C) [62]. Tafamidis binding strengthened the weaker dimer-dimer interface via multi-subunit hydrophobic and electrostatic interactions, resulting in the kinetic stabilization of TTR tetramer (Figure 2D). Binding of only one tafamidis molecule per TTR tetramer is sufficient to achieve enough TTR kinetic stabilization to arrest aggregation [63].
In 2011, the results of a phase II/III 18-month randomized double-blind placebo-controlled clinical trial of tafamidis for the treatment of FAP were reported, revealing that the predetermined endpoints were met in the efficacy evaluable population [64]. Tafamidis delayed neuropathic progression in these heterozygous patients harboring the V30M mutation. In comparison to placebo controls, these individuals exhibited 52% less neurologic deterioration demonstrated by 53% and 80% preservation of large- and small-nerve fiber function, respectively. Additionally, tafamdis treatment significantly prevented weight loss, a factor used in the assessment of a change in the quality of life of patients undergoing therapy. A subsequent 12-month, open-label extension study further demonstrated the long-term safety of tafamidis, its tolerability and efficacy in slowing disease progression [65]. The European Medicines Agency, later joined by the Japanese Pharmaceuticals and Medical Devices Agency as well as other regulatory agencies, approved tafamidis for the treatment of FAP. A multicenter, placebo-controlled, double-blinded clinical trial is well underway to determine the efficacy of tafamidis for the treatment of patients diagnosed with transthyretin-related cardiomyopathy (clinicaltrials.gov) [66].
Therapeutics of the Future
Despite immense effort from the biotechnology and pharmaceutical industries, as well as academia, the list of regulatory agency approved therapies that modify the course of protein aggregation-associated degenerative diseases remains very short [2]. It seems likely that the design of clinical trials focusing on patients already exhibiting substantial neuronal loss is contributing to the high failure rate of drug candidates [2]. Moreover, it is now becoming clear that the pathology of some degenerative diseases may be due to aggregation of more than one protein. For example, multiple proteins are aggregating in Alzheimer’s disease, not just Aβ and Tau. Thus repairing the ability to maintain the proteome [67–69] and performing more sophisticated trials might together contribute to increasing the list of efficacious drugs.
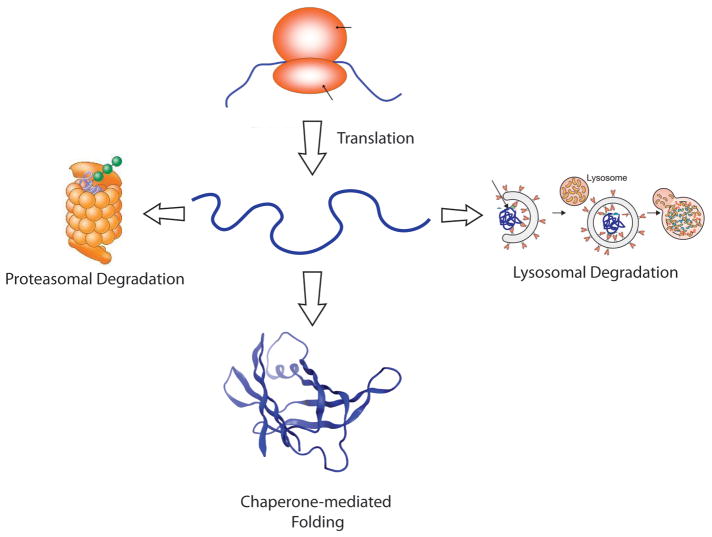
Towards this end, one promising future therapeutic strategy involves manipulation of the cellular protein homeostasis (proteostasis) network to minimize the accumulation of harmful aggregated proteins [67–69]. The proteostasis network comprises a variety of competing and integrated biological pathways, including chaperone folding pathways, as well as the ubiquitin proteasome and autophagy degradation pathways. The basic function of the cellular proteostasis network is to make a decision to either facilitate the folding of a given protein or to degrade it (Figure 4). Various stressors—a fever, a viral infection, the presence of ingested oxidants or oxidants produced by dysfunctional mitochondria, etc.—can cause protein misfolding and/or aggregation. We hypothesize that when we are young, protein aggregation and misfolding caused by such stresses are countered by stress-responsive signaling pathways, which increase proteostasis network capacity to meet demand and remedy misfolding and/or aggregation, required for normal organismal function [70–73]. In the cytosol and nucleus of cells, the heat shock response (HSR) stress-responsive signaling pathway, mediated by the transcription factor HSF1, induces ~500 and represses ~1000 genes to ensure the maintenance of proteostasis in those cellular compartments [74]. The unfolded protein response (UPR) stress-responsive signaling pathway regulates proteostasis within the secretory pathway of eukaryotic cells and also contributes to extracellular proteostasis maintenance [75,76]. A growing body of evidence suggests that the inability to activate stress-responsive signaling pathways as humans age leads to exacerbated improper folding and aggregation [13–15]. Thus, a therapeutic strategy that restores normal regulation of the cellular proteostasis network(s) could potentially have a broad, positive impact on cellular and organismal function, and hopefully could halt the progression of aggregation and protein misfolding diseases [77–79].
Figure 4.
The main decision to be made by the protein homeostasis, or proteostasis, network is to fold or refold a protein or degrade it by the ubiquitin proteasome system or by one of several lysosomal degradation pathways (autophagy is shown). While it is clear that intrinsically disordered proteins are degraded both by the proteasome and by the lysosome, it is less clear which proteostasis network components engage this class of proteins to keep them soluble and functional.
Recent publications demonstrate the potential benefits of proteostasis network manipulation as a therapeutic strategy to ameliorate diseases of protein conformation. Small-molecule mediated manipulation of Hsp90–co-chaperone machinery or the Hsp70–co-chaperone pathway in the cytosol has been shown to impede the effects of misfolding and aggregation of various proteins associated with the onset of a number of neurodegenerative diseases including Parkinson’s, Huntington’s and Alzheimer’s diseases [80–85].
While many uncertainties about the role of stress-responsive signaling remain to be addressed [86–88], multiple preclinical studies indicate that selective modulation of stress-responsive signaling pathways reduces the damaging effects of protein misfolding and aggregation [14]. Selective enhancement of either the HSR or the UPR has been shown to extend life span in various misfolding disease animal models [79,89,90]. In a recent report, Das and coworkers demonstrated that inhibition of stress-induced phosphatase PPP1R15A, a component of the PERK arm of the UPR, ameliorated degeneration in both cell and animal models of Charcot-Marie-Tooth 1B disease and amyotrophic lateral sclerosis [91]. Interestingly, the small molecule Sephin1 identified in this study did not bind to related and constitutive phosphatase PPP1R15B, sparing undesired pro-apoptotic stress response activation [91]. Cellular studies demonstrate that activation of a specific arm(s) of the UPR stress-responsive signaling pathway increases secretion of functional misfolding-prone proteins associated with loss-of-function diseases [92,93] while preventing secretion of dysfunctional proteins [94]. Moreover, arm-selective UPR activation has been demonstrated to achieve intracellular degradation of the mutant aggregation-prone variants while simultaneously allowing for the proper folding and secretion of functional wild type amyloidogenic proteins in heterozygotes [78,95,96]. In the extracellular space, UPR activation reduces the concentration of misfolding-prone proteins, which should reduce concentration-dependent amyloidogenesis [95,97]. A reduction in aggregation is also likely achieved owing to the secretion of the Hsp40 co-chaperone ERdj3, as a consequence of activation of the ATF6 arm of the UPR [98]. Small molecule ligands that modulate the activity of specific arms of stress-responsive signaling pathways are envisioned to be valuable [75,79,88,99].
Removal of dysfunctional and aggregated proteins through direct activation of cellular degradation pathways is being explored intensively as a therapeutic strategy for the amelioration of protein misfolding/aggregation diseases [100–104]. Animals appear to lack the dedicated disaggregase that yeast have. Instead, proteins can be degraded by the ubiquitin proteasome system or aggregates can be cleared by acid denaturation and proteolysis in the lysosome, which can be accessed through multiple pathways, including autophagy routes [102,105,106]. In some organelles, especially the mitochondria, specialized proteases exist to degrade the proteome [107].
Activating the proteasome is an attractive approach for enhancing the degradation capacity of the cytosolic proteostasis network, an approach that is currently being investigated preclinically in aggregation-associated degenerative diseases [108,109]. The proteasome-associated deubiquitination enzymes, or the DUBs, negatively regulate the proteasome by removing ubiquitin from a subset of proteasome clients, reducing the efficiency of degradation. Thus, DUB inhibitors are being developed by several biotechnology and pharmaceutical companies to enhance the degradation of client proteins associated with neurodegenerative disorders, including α-synuclein and tau, to treat Parkinson’s disease and the tauopathies including Alzheimer’s disease [108,109].
Degradation of soluble and aggregated proteins in the lysosome is a very attractive strategy to remedy aging-linked degenerative diseases that appear to be caused by the aggregation of one or more proteins. Numerous research groups are focused on discovering autophagy activating small molecules—this field is vast and cannot be adequately reviewed here [105,110–124]. However, there is enough preliminary data to motivate extensive studies to determine whether enhancing proteostasis by activating aggregate clearance mechanisms will be useful for the treatment of human degenerative diseases.
Conclusion and Perspectives
The list of disease-modifying strategies for human amyloid diseases is very short. Besides directly stabilizing an amyloidogenic protein, as we have done in the case of the transthyretin amyloidoses, we envision that therapies that rely on adapting the proteostasis network will also be very useful for ameliorating amyloid diseases. We envision that mechanistically distinct drugs will be used in combination with a plethora of additional potential strategies, including activation of the immune system to clear distinct aggregate types [2].
Highlights.
Protein misfolding and misassembly leads to amyloid diseases and tissue degeneration.
Genetic and aggregation insights led to a drug for the transthyretin amyloidoses.
Tafamidis prevents the progression of the otherwise fatal transthyretin amyloidoses.
An understanding of the cellular proteostasis networks is emerging.
Proteostasis regulators should be useful for treating protein misfolding diseases.
Acknowledgments
JWK thanks the Skaggs Institute for Chemical Biology and the National Institutes of Health (DK046335 and DK046495) for financial support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kelly JW. The alternative conformations of amyloidogenic proteins and their multi-step assembly pathways. Curr Opin Struct Biol. 1998;8:101–106. doi: 10.1016/s0959-440x(98)80016-x. [DOI] [PubMed] [Google Scholar]
- 2•.Eisele YS, Monteiro C, Fearns C, Encalada SE, Wiseman RL, Powers ET, Kelly JW. Targeting protein aggregation for the treatment of degenerative diseases. Nat Rev Drug Discovery. 2015;14:759–780. doi: 10.1038/nrd4593. Much more comprehensive review on the strategies for ameliorating degenerative amyloid diseases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3•.Knowles TP, Vendruscolo M, Dobson CM. The amyloid state and its association with protein misfolding diseases. Nat Rev Mol Cell Biol. 2014;15:384–396. doi: 10.1038/nrm3810. Nice general review of amyloidosis. [DOI] [PubMed] [Google Scholar]
- 4.Nelson R, Sawaya MR, Balbirnie M, Madsen AO, Riekel C, Grothe R, Eisenberg D. Structure of the cross-beta spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laganowsky A, Liu C, Sawaya MR, Whitelegge JP, Park J, Zhao ML, Pensalfini A, Soriaga AB, Landau M, Teng PK, et al. Atomic View of a Toxic Amyloid Small Oligomer. Science. 2012;335:1228–1231. doi: 10.1126/science.1213151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6•.Eisenberg D, Jucker M. The amyloid state of proteins in human diseases. Cell. 2012;148:1188–1203. doi: 10.1016/j.cell.2012.02.022. Nice general review of amyloid diseases and seeding as a mechanism for the spread of pathology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caughey B, Lansbury PT. Protofibrils, pores, fibrils, and neurodegeneration: separating the responsible protein aggregates from the innocent bystanders. Annu Rev Neurosci. 2003;26:267–298. doi: 10.1146/annurev.neuro.26.010302.081142. [DOI] [PubMed] [Google Scholar]
- 8.Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 9.Bourgault S, Solomon JP, Reixach N, Kelly JW. Sulfated glycosaminoglycans accelerate transthyretin amyloidogenesis by quaternary structural conversion. Biochemistry. 2011;50:1001–1015. doi: 10.1021/bi101822y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pepys MB, Herbert J, Hutchinson WL, Tennent GA, Lachmann HJ, Gallimore JR, Lovat LB, Bartfai T, Alanine A, Hertel C, et al. Targeted pharmacological depletion of serum amyloid P component for treatment of human amyloidosis. Nature. 2002;417:254–259. doi: 10.1038/417254a. [DOI] [PubMed] [Google Scholar]
- 11.Chiti F, Stefani M, Taddei N, Ramponi G, Dobson CM. Rationalization of the effects of mutations on peptide and protein aggregation rates. Nature. 2003;424:805–808. doi: 10.1038/nature01891. [DOI] [PubMed] [Google Scholar]
- 12.Sekijima Y, Wiseman RL, Matteson J, Hammarstrom P, Miller SR, Sawkar AR, Balch WE, Kelly JW. The biological and chemical basis for tissue-selective amyloid disease. Cell. 2005;121:73–85. doi: 10.1016/j.cell.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 13.Ben-Zvi A, Miller EA, Morimoto RI. Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc Natl Acad Sci U S A. 2009;106:14914–14919. doi: 10.1073/pnas.0902882106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haigis MC, Yankner BA. The aging stress response. Mol Cell. 2010;40:333–344. doi: 10.1016/j.molcel.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cuanalo-Contreras K, Mukherjee A, Soto C. Role of protein misfolding and proteostasis deficiency in protein misfolding diseases and aging. Int J Cell Biol. 2013;2013:638083. doi: 10.1155/2013/638083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohen FE, Kelly JW. Therapeutic approaches to protein-misfolding diseases. Nature. 2003;426:905–909. doi: 10.1038/nature02265. [DOI] [PubMed] [Google Scholar]
- 17.Miroy GJ, Lai Z, Lashuel HA, Peterson SA, Strang C, Kelly JW. Inhibiting transthyretin amyloid fibril formation via protein stabilization. Proc Natl Acad Sci U S A. 1996;93:15051–15056. doi: 10.1073/pnas.93.26.15051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hammarstrom P, Jiang X, Hurshman AR, Powers ET, Kelly JW. Sequence-dependent denaturation energetics: A major determinant in amyloid disease diversity. Proc Natl Acad Sci U S A. 2002;99(Suppl 4):16427–16432. doi: 10.1073/pnas.202495199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19•.Hammarstrom P, Wiseman RL, Powers ET, Kelly JW. Prevention of transthyretin amyloid disease by changing protein misfolding energetics. Science. 2003;299:713–716. doi: 10.1126/science.1079589. This paper explains and exemplifies the concept of kinetic stabilization. [DOI] [PubMed] [Google Scholar]
- 20.Schreiber G, Richardson SJ. The evolution of gene expression, structure and function of transthyretin. Comp Biochem Physiol B Biochem Mol Biol. 1997;116:137–160. doi: 10.1016/s0305-0491(96)00212-x. [DOI] [PubMed] [Google Scholar]
- 21.Herbert J, Wilcox JN, Pham KTC, Fremeau RT, Zeviani M, Dwork A, Soprano DR, Makover A, Goodman DS, Zimmerman EA, et al. Transthyretin - A choroid plexus-specific transport protein in human brain. Neurology. 1986;36:900–911. doi: 10.1212/wnl.36.7.900. [DOI] [PubMed] [Google Scholar]
- 22.Cavallaro T, Martone RL, Dwork AJ, Schon EA, Herbert J. The retinal pigment epithelium is the unique site of transthyretin synthesis in the rat eye. Invest Ophthalmol Vis Sci. 1990;31:497–501. [PubMed] [Google Scholar]
- 23.Blake CC, Geisow MJ, Oatley SJ, Rerat B, Rerat C. Structure of prealbumin: secondary, tertiary and quaternary interactions determined by Fourier refinement at 1. 8 A. J Mol Biol. 1978;121:339–356. doi: 10.1016/0022-2836(78)90368-6. [DOI] [PubMed] [Google Scholar]
- 24.Foss TR, Kelker MS, Wiseman RL, Wilson IA, Kelly JW. Kinetic stabilization of the native state by protein engineering: implications for inhibition of transthyretin amyloidogenesis. J Mol Biol. 2005;347:841–854. doi: 10.1016/j.jmb.2005.01.050. [DOI] [PubMed] [Google Scholar]
- 25•.Foss TR, Wiseman RL, Kelly JW. The pathway by which the tetrameric protein transthyretin dissociates. Biochemistry. 2005;44:15525–15533. doi: 10.1021/bi051608t. This paper reveals that transthyretin dissociates about the interface comprising the thyroxine or tafamidis binding site. [DOI] [PubMed] [Google Scholar]
- 26.Hurshman Babbes AR, Powers ET, Kelly JW. Quantification of the thermodynamically linked quaternary and tertiary structural stabilities of transthyretin and its disease-associated variants: the relationship between stability and amyloidosis. Biochemistry. 2008;47:6969–6984. doi: 10.1021/bi800636q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lashuel HA, Lai Z, Kelly JW. Characterization of the transthyretin acid denaturation pathways by analytical ultracentrifugation: implications for wild-type, V30M, and L55P amyloid fibril formation. Biochemistry. 1998;37:17851–17864. doi: 10.1021/bi981876+. [DOI] [PubMed] [Google Scholar]
- 28.Lashuel HA, Wurth C, Woo L, Kelly JW. The most pathogenic transthyretin variant, L55P, forms amyloid fibrils under acidic conditions and protofilaments under physiological conditions. Biochemistry. 1999;38:13560–13573. doi: 10.1021/bi991021c. [DOI] [PubMed] [Google Scholar]
- 29.Hurshman AR, White JT, Powers ET, Kelly JW. Transthyretin aggregation under partially denaturing conditions is a downhill polymerization. Biochemistry. 2004;43:7365–7381. doi: 10.1021/bi049621l. [DOI] [PubMed] [Google Scholar]
- 30.Hammarstrom P, Sekijima Y, White JT, Wiseman RL, Lim A, Costello CE, Altland K, Garzuly F, Budka H, Kelly JW. D18G transthyretin is monomeric, aggregation prone, and not detectable in plasma and cerebrospinal fluid: a prescription for central nervous system amyloidosis? Biochemistry. 2003;42:6656–6663. doi: 10.1021/bi027319b. [DOI] [PubMed] [Google Scholar]
- 31.Jiang X, Buxbaum JN, Kelly JW. The V122I cardiomyopathy variant of transthyretin increases the velocity of rate-limiting tetramer dissociation, resulting in accelerated amyloidosis. Proc Natl Acad Sci U S A. 2001;98:14943–14948. doi: 10.1073/pnas.261419998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andrade C. A peculiar form of peripheral neuropathy; familiar atypical generalized amyloidosis with special involvement of the peripheral nerves. Brain. 1952;75:408–427. doi: 10.1093/brain/75.3.408. [DOI] [PubMed] [Google Scholar]
- 33.Cornwell GG, 3rd, Sletten K, Johansson B, Westermark P. Evidence that the amyloid fibril protein in senile systemic amyloidosis is derived from normal prealbumin. Biochem Biophys Res Commun. 1988;154:648–653. doi: 10.1016/0006-291x(88)90188-x. [DOI] [PubMed] [Google Scholar]
- 34.Westermark P, Sletten K, Johansson B, Cornwell GG., 3rd Fibril in senile systemic amyloidosis is derived from normal transthyretin. Proc Natl Acad Sci U S A. 1990;87:2843–2845. doi: 10.1073/pnas.87.7.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coelho T. Familial amyloid polyneuropathy: new developments in genetics and treatment. Curr Opin Neurol. 1996;9:355–359. [PubMed] [Google Scholar]
- 36.Jacobson DR, McFarlin DE, Kane I, Buxbaum JN. Transthyretin Pro55, a variant associated with early-onset, aggressive, diffuse amyloidosis with cardiac and neurologic involvement. Hum Genet. 1992;89:353–356. doi: 10.1007/BF00220559. [DOI] [PubMed] [Google Scholar]
- 37.Dharmarajan K, Maurer MS. Transthyretin Cardiac Amyloidoses in Older North Americans. J Am Geriat Soc. 2012;60:765–774. doi: 10.1111/j.1532-5415.2011.03868.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Falk RH, Elkayam U. Cardiomyopathy: the importance of recognizing the uncommon diagnosis. Prog Cardiovasc Dis. 2010;52:262–263. doi: 10.1016/j.pcad.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 39.Gursky O, Mei X, Atkinson D. The crystal structure of the C-terminal truncated apolipoprotein A-I sheds new light on amyloid formation by the N-terminal fragment. Biochemistry. 2012;51:10–18. doi: 10.1021/bi2017014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hamidi Asl L, Liepnieks JJ, Hamidi Asl K, Uemichi T, Moulin G, Desjoyaux E, Loire R, Delpech M, Grateau G, Benson MD. Hereditary amyloid cardiomyopathy caused by a variant apolipoprotein A1. Am J Pathol. 1999;154:221–227. doi: 10.1016/S0002-9440(10)65268-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holmgren G, Ericzon BG, Groth CG, Steen L, Suhr O, Andersen O, Wallin BG, Seymour A, Richardson S, Hawkins PN. Clinical improvement and amyloid regression after liver transplantation in hereditary transthyretin amyloidosis. Lancet. 1993;341:1113–1116. doi: 10.1016/0140-6736(93)93127-m. [DOI] [PubMed] [Google Scholar]
- 42.Herlenius G, Wilczek HE, Larsson M, Ericzon BG. Ten years of international experience with liver transplantation for familial amyloidotic polyneuropathy: results from the Familial Amyloidotic Polyneuropathy World Transplant Registry. Transplantation. 2004;77:64–71. doi: 10.1097/01.TP.0000092307.98347.CB. [DOI] [PubMed] [Google Scholar]
- 43.Olofsson BO, Backman C, Karp K, Suhr OB. Progression of cardiomyopathy after liver transplantation in patients with familial amyloidotic polyneuropathy, Portuguese type. Transplantation. 2002;73:745–751. doi: 10.1097/00007890-200203150-00015. [DOI] [PubMed] [Google Scholar]
- 44.Coelho T, Carvalho M, Saraiva MJ, Alves I, Almeida MR, Costa PP. A strikingly benign evolution of FAP in an individual found to be a compound heterozygote for two TTR mutations: TTR MET 30 and TTR MET 119. J Rheumatol. 1993;20:179. [Google Scholar]
- 45.Coelho T, Chorao R, Sausa A, Alves I, Torres MF, Saraiva MJ. Compound heterozygotes of transthyretin Met30 and transthyretin Met119 are protected from the devastating effects of familial amyloid polyneuropathy. Neuromusc Disord. 1996;6:27. [Google Scholar]
- 46•.Hammarstrom P, Schneider F, Kelly JW. Trans-suppression of misfolding in an amyloid disease. Science. 2001;293:2459–2462. doi: 10.1126/science.1062245. The genetic evidence that kinetic stabilization of transthyretin should ameliorate transthyretin amyloidosis is presented. [DOI] [PubMed] [Google Scholar]
- 47.Colon W, Kelly JW. Partial denaturation of transthyretin is sufficient for amyloid fibril formation in vitro. Biochemistry. 1992;31:8654–8660. doi: 10.1021/bi00151a036. [DOI] [PubMed] [Google Scholar]
- 48.Lai Z, Colon W, Kelly JW. The acid-mediated denaturation pathway of transthyretin yields a conformational intermediate that can self-assemble into amyloid. Biochemistry. 1996;35:6470–6482. doi: 10.1021/bi952501g. [DOI] [PubMed] [Google Scholar]
- 49.Baures PW, Oza VB, Peterson SA, Kelly JW. Synthesis and evaluation of inhibitors of transthyretin amyloid formation based on the non-steroidal anti-inflammatory drug, flufenamic acid. Bioorg Med Chem. 1999;7:1339–1347. doi: 10.1016/s0968-0896(99)00066-8. [DOI] [PubMed] [Google Scholar]
- 50.Baures PW, Peterson SA, Kelly JW. Discovering transthyretin amyloid fibril inhibitors by limited screening. Bioorg Med Chem. 1998;6:1389–1401. doi: 10.1016/s0968-0896(98)00130-8. [DOI] [PubMed] [Google Scholar]
- 51.Klabunde T, Petrassi HM, Oza VB, Raman P, Kelly JW, Sacchettini JC. Rational design of potent human transthyretin amyloid disease inhibitors. Nat Struct Biol. 2000;7:312–321. doi: 10.1038/74082. [DOI] [PubMed] [Google Scholar]
- 52.Connelly S, Choi S, Johnson SM, Kelly JW, Wilson IA. Structure-based design of kinetic stabilizers that ameliorate the transthyretin amyloidoses. Curr Opin Struct Biol. 2010;20:54–62. doi: 10.1016/j.sbi.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53••.Johnson SM, Connelly S, Fearns C, Powers ET, Kelly JW. The Transthyretin Amyloidoses: From Delineating the Molecular Mechanism of Aggregation Linked to Pathology to a Regulatory-Agency-Approved Drug. J Mol Biol. 2012;421:185–203. doi: 10.1016/j.jmb.2011.12.060. An in-depth review of the logic and development of tafamidis to ameliorate the transthyretin amyloidoses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johnson SM, Wiseman RL, Sekijima Y, Green NS, Adamski-Werner SL, Kelly JW. Native state kinetic stabilization as a strategy to ameliorate protein misfolding diseases: a focus on the transthyretin amyloidoses. Acc Chem Res. 2005;38:911–921. doi: 10.1021/ar020073i. [DOI] [PubMed] [Google Scholar]
- 55.Johnson SM, Connelly S, Wilson IA, Kelly JW. Toward optimization of the linker substructure common to transthyretin amyloidogenesis inhibitors using biochemical and structural studies. J Med Chem. 2008;51:6348–6358. doi: 10.1021/jm800435s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johnson SM, Connelly S, Wilson IA, Kelly JW. Biochemical and structural evaluation of highly selective 2-arylbenzoxazole-based transthyretin amyloidogenesis inhibitors. J Med Chem. 2008;51:260–270. doi: 10.1021/jm0708735. [DOI] [PubMed] [Google Scholar]
- 57.Johnson SM, Connelly S, Wilson IA, Kelly JW. Toward optimization of the second aryl substructure common to transthyretin amyloidogenesis inhibitors using biochemical and structural studies. J Med Chem. 2009;52:1115–1125. doi: 10.1021/jm801347s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Razavi H, Palaninathan SK, Powers ET, Wiseman RL, Purkey HE, Mohamedmohaideen NN, Deechongkit S, Chiang KP, Dendle MT, Sacchettini JC, et al. Benzoxazoles as transthyretin amyloid fibril inhibitors: synthesis, evaluation, and mechanism of action. Angewandte Chemie. 2003;42:2758–2761. doi: 10.1002/anie.200351179. [DOI] [PubMed] [Google Scholar]
- 59.Sekijima Y, Dendle MA, Kelly JW. Orally administered diflunisal stabilizes transthyretin against dissociation required for amyloidogenesis. Amyloid. 2006;13:236–249. doi: 10.1080/13506120600960882. [DOI] [PubMed] [Google Scholar]
- 60.Choi S, Kelly JW. A competition assay to identify amyloidogenesis inhibitors by monitoring the fluorescence emitted by the covalent attachment of a stilbene derivative to transthyretin. Bioorg Med Chem. 2011;19:1505–1514. doi: 10.1016/j.bmc.2010.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61••.Rappley I, Monteiro C, Novais M, Baranczak A, Solis G, Wiseman RL, Helmke S, Maurer MS, Coelho T, Powers ET, et al. Quantification of Transthyretin Kinetic Stability in Human Plasma Using Subunit Exchange. Biochemistry. 2014;53:1993–2006. doi: 10.1021/bi500171j. A manuscript covering the best way to quantify transthyretin stability in plasma under physiological conditions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62••.Bulawa CE, Connelly S, DeVit M, Wang L, Weigel C, Fleming JA, Packman J, Powers ET, Wiseman RL, Foss TR, et al. Tafamidis, a potent and selective transthyretin kinetic stabilizer that inhibits the amyloid cascade. Proc Natl Acad Sci U S A. 2012;109:9629–9634. doi: 10.1073/pnas.1121005109. S9629/9621-S9629/9629. Focuses on the pre-clinical studies done on tafamidis to qualify it as a drug candidate. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wiseman RL, Johnson SM, Kelker MS, Foss T, Wilson IA, Kelly JW. Kinetic stabilization of an oligomeric protein by a single ligand binding event. J Am Chem Soc. 2005;127:5540–5551. doi: 10.1021/ja042929f. [DOI] [PubMed] [Google Scholar]
- 64••.Coelho T, Maia L, Martins da Silva A, Waddington Cruz M, Planté-Bordeneuve V, Lozeron P, Suhr OB, Campistol JM, Conceição I, Schmidt HH-J, et al. Tafamidis for transthyretin familial amyloid polyneuropathy: a randomized, controlled trial. Neurology. 2012;79:785–792. doi: 10.1212/WNL.0b013e3182661eb1. Reports the outcome of a double-blind placebo-controlled trial for tafamidis in transthyretin polyneuropathy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65••.Coelho T, Maia LF, da Silva AM, Cruz MW, Plante-Bordeneuve V, Suhr OB, Conceicao I, Schmidt HHJ, Trigo P, Kelly JW, et al. Long-term effects of tafamidis for the treatment of transthyretin familial amyloid polyneuropathy. J Neurol. 2013;260:2802–2814. doi: 10.1007/s00415-013-7051-7. Reports 30 month data of FAP patients on tafamidis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66••.Maurer MS, Grogan DR, Judge DP, Mundayat R, Packman J, Lombardo I, Quyyumi AA, Aarts J, Falk RH. Tafamidis in transthyretin amyloid cardiomyopathy: effects on transthyretin stabilization and clinical outcomes. Circ Heart Fail. 2015;8:519–526. doi: 10.1161/CIRCHEARTFAILURE.113.000890. Open-label tafamidis data suggests potential benefit for cardiomyopathy. [DOI] [PubMed] [Google Scholar]
- 67••.Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. This review outlines the potential of protein homeostasis network adaptation. [DOI] [PubMed] [Google Scholar]
- 68••.Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE. Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem. 2009;78:959–991. doi: 10.1146/annurev.biochem.052308.114844. This review outlines the potential of proteostasis network management in more detail. [DOI] [PubMed] [Google Scholar]
- 69••.Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. Outlines the critical role of chaperones in proteostasis management. [DOI] [PubMed] [Google Scholar]
- 70.Morimoto RI. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes & Development. 1998;12:3788–3796. doi: 10.1101/gad.12.24.3788. [DOI] [PubMed] [Google Scholar]
- 71.Lindquist S. The heat shock response. Ann Rev Biochem. 1986;55:1151–1191. doi: 10.1146/annurev.bi.55.070186.005443. [DOI] [PubMed] [Google Scholar]
- 72.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Molec Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 73.Schroder M, Kaufman RJ. The mammalian unfolded protein response. Ann Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 74.Guisbert E, Morimoto RI. The regulation and function of the heat shock response. In: Morimoto RIYC, editor. Protein Qualtiy Control in Neurodegenerative Diseases. Springer-Verlag; Berlin Heidelberg: 2013. pp. 1–18. Research and Perspectives in Alzheimer’s Disease. [Google Scholar]
- 75.Ryno LM, Wiseman RL, Kelly JW. Targeting unfolded protein response signaling pathways to ameliorate protein misfolding diseases. Curr Opin Chem Biol. 2013;17:346–352. doi: 10.1016/j.cbpa.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13:89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 77.Shoulders MD, Ryno LM, Cooley CB, Kelly JW, Wiseman RL. Broadly Applicable Methodology for the Rapid and Dosable Small Molecule-Mediated Regulation of Transcription Factors in Human Cells. J Am Chem Soc. 2013;135:8129–8132. doi: 10.1021/ja402756p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78•.Shoulders MD, Ryno LM, Genereux JC, Moresco JJ, Tu PG, Wu CL, Yates JR, Su AI, Kelly JW, Wiseman RL. Stress-Independent Activation of XBP1s and/or ATF6 Reveals Three Functionally Diverse ER Proteostasis Environments. Cell Rep. 2013;3:1279–1292. doi: 10.1016/j.celrep.2013.03.024. This manuscript outlines the utility of destabilized domain technology for post-translational regulation of transcription factor function. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79••.Calamini B, Silva MC, Madoux F, Hutt DM, Khanna S, Chalfant MA, Saldanha SA, Hodder P, Tait BD, Garza D, et al. Small-molecule proteostasis regulators for protein conformational diseases. Nat Chem Biol. 2012;8:185–196. doi: 10.1038/nchembio.763. In this manuscript, the Morimoto lab show that it is possible to use a reporter assay to discover heat shock response transcriptional program activators. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80••.Wang AM, Miyata Y, Klinedinst S, Peng HM, Chua JP, Komiyama T, Li XK, Morishima Y, Merry DE, Pratt WB, et al. Activation of Hsp70 reduces neurotoxicity by promoting polyglutamine protein degradation. Nat Chem Biol. 2013;9:112–118. doi: 10.1038/nchembio.1140. The Gestwicki lab reports the identification of a compound that acts similarly to Hip by allosterically promoting Hsp70 binding, leading to client ubiquitination and degradation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pratt WB, Gestwicki JE, Osawa Y, Lieberman AP. Targeting Hsp90/Hsp70-based protein quality control for treatment of adult onset neurodegenerative diseases. Annu Rev Pharmacol Toxicol. 2015;55:353–371. doi: 10.1146/annurev-pharmtox-010814-124332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82••.Chafekar SM, Wisen S, Thompson AD, Echeverria A, Walter GM, Evans CG, Makley LN, Gestwicki JE, Duennwald ML. Pharmacological Tuning of Heat Shock Protein 70 Modulates Polyglutamine Toxicity and Aggregation. ACS Chem Biol. 2012;7:1556–1564. doi: 10.1021/cb300166p. The Gestwicki lab discovered a small molecule that inhibits Hsp70 ATPase activity and protects against polyQ proteotoxicity by promoting aggregation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83••.Jinwal UK, Akoury E, Abisambra JF, O’Leary JC, Thompson AD, Blair LJ, Jin Y, Bacon J, Nordhues BA, Cockman M, et al. Imbalance of Hsp70 family variants fosters tau accumulation. FASEB J. 2013;27:1450–1459. doi: 10.1096/fj.12-220889. This report demonstrates that Hsp72, but not Hsc70, was able to recruit CHIP, faciltiating the ubiquitination of tau and its proteasomal degradation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84••.Miyata Y, Li XK, Lee HF, Jinwal UK, Srinivasan SR, Seguin SP, Young ZT, Brodsky JL, Dickey CA, Sun DX, et al. Synthesis and Initial Evaluation of YM-08, a Blood-Brain Barrier Permeable Derivative of the Heat Shock Protein 70 (Hsp70) Inhibitor MKT-077, Which Reduces Tau Levels. ACS Chem Neurosci. 2013;4:930–939. doi: 10.1021/cn300210g. These investigators show that inhibiting Hsp70 in the brain using the small molecule YM-08 reduced phosphorylated tau levels in brain slices. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Smith MC, Scaglione KM, Assimon VA, Patury S, Thompson AD, Dickey CA, Southworth DR, Paulson HL, Gestwicki JE, Zuiderweg ERP. The E3 Ubiquitin Ligase CHIP and the Molecular Chaperone Hsc70 Form a Dynamic, Tethered Complex. Biochemistry. 2013;52:5354–5364. doi: 10.1021/bi4009209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hetz C, Mollereau B. Disturbance of endoplasmic reticulum proteostasis in neurodegenerative diseases. Nat Rev Neurosci. 2014;15:233–249. doi: 10.1038/nrn3689. [DOI] [PubMed] [Google Scholar]
- 87.Lamech LT, Haynes CM. The unpredictability of prolonged activation of stress response pathways. J Cell Biol. 2015;209:781–787. doi: 10.1083/jcb.201503107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88••.Narayan P, Ehsani S, Lindquist S. Combating neurodegenerative disease with chemical probes and model systems. Nat Chem Biol. 2014;10:911–920. doi: 10.1038/nchembio.1663. The Lindquist group review the seminal role that chemical biology has played in restoring proteostasis in neurodegenerative disease. [DOI] [PubMed] [Google Scholar]
- 89.Taylor RC, Dillin A. XBP-1 is a cell-nonautonomous regulator of stress resistance and longevity. Cell. 2013;153:1435–1447. doi: 10.1016/j.cell.2013.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cohen E, Bieschke J, Perciavalle RM, Kelly JW, Dillin A. Opposing Activities Protect Against Age-Onset Proteotoxicity. Science. 2006;313:1604–1610. doi: 10.1126/science.1124646. [DOI] [PubMed] [Google Scholar]
- 91••.Das I, Krzyzosiak A, Schneider K, Wrabetz L, D’Antonio M, Barry N, Sigurdardottir A, Bertolotti A. Preventing proteostasis diseases by selective inhibition of a phosphatase regulatory subunit. Science. 2015;348:239–242. doi: 10.1126/science.aaa4484. The authors report the small molecule Sephin1 that selectively binds the PPP1R15A regulatory subunit of protein phosphatase 1, inhibiting it. The prolonged stress responsive signaling benefits the degenerative Charcot-Marie-Tooth phenotype and ALS phenotypes in mouse models. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mu TW, Ong DST, Wang YJ, Balch WE, Yates JR, Segatori L, Kelly JW. Chemical and biological approaches synergize to ameliorate protein-folding diseases. Cell. 2008;134:769–781. doi: 10.1016/j.cell.2008.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hulleman JD, Balch WE, Kelly JW. Translational attenuation differentially alters the fate of disease-associated fibulin proteins. FASEB J. 2012;26:4548–4560. doi: 10.1096/fj.11-202861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94••.Chiang WC, Messah C, Lin JH. IRE1 directs proteasomal and lysosomal degradation of misfolded rhodopsin. Mol Biol Cell. 2012;23:758–770. doi: 10.1091/mbc.E11-08-0663. Chemical-genetic activation of the IRE1 arm of the UPR enhances quality control by directing P23H rhodopsin to degradation while not affecting wild type rhodopsin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95••.Cooley CB, Ryno LM, Plate L, Morgan GJ, Hulleman JD, Kelly JW, Wiseman LR. Unfolded Protein Response Activation Reduces Secretion and Extracellular Aggregation of Amyloidogenic Light Chain. Proc Natl Acad Sci USA. 2014;111:13046–13051. doi: 10.1073/pnas.1406050111. Arm-selective UPR activationreduces the secretion of amyloidogenic light chains and, depending on which arm is activated, determines degradation or not. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Smith SE, Granell S, Salcedo-Sicilia L, Baldini G, Egea G, Teckman JH, Baldini G. Activating transcription factor 6 limits intracellular accumulation of mutant alpha(1)-antitrypsin Z and mitochondrial damage in hepatoma cells. J Biol Chem. 2011;286:41563–41577. doi: 10.1074/jbc.M111.280073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang S, Kaufman RJ. The impact of the unfolded protein response on human disease. J Cell Biol. 2012;197:857–867. doi: 10.1083/jcb.201110131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Genereux JC, Qu S, Zhou M, Ryno LM, Wang S, Shoulders MD, Kaufman RJ, Lasmezas CI, Kelly JW, Wiseman RL. Unfolded protein response-induced ERdj3 secretion links ER stress to extracellular proteostasis. EMBO J. 2015;34:4–19. doi: 10.15252/embj.201488896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jimenez-Sanchez M, Lam W, Hannus M, Sonnichsen B, Imarisio S, Fleming A, Tarditi A, Menzies F, Ed Dami T, Xu C, et al. siRNA screen identifies QPCT as a druggable target for Huntington’s disease. Nat Chem Biol. 2015;11:347–354. doi: 10.1038/nchembio.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ciechanover A, Kwon YT. Degradation of misfolded proteins in neurodegenerative diseases: therapeutic targets and strategies. Exp Mol Med. 2015;47:e147. doi: 10.1038/emm.2014.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101••.King RW, Finley D. Sculpting the proteome with small molecules. Nat Chem Biol. 2014;10:870–874. doi: 10.1038/nchembio.1671. This review elegantly outlines that pharmacology of inhibiting and activating the ubiquitin proteasome system. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 103.Mizushima N, Komatsu M. Autophagy: Renovation of Cells and Tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 104••.Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science. 2004;305:1292–1295. doi: 10.1126/science.1101738. The authors showed that A53T and A30P a-synuclein engage, but are not degraded by, the chaperone-mediated autophagy pathway. [DOI] [PubMed] [Google Scholar]
- 105.Manchon JFM, Uzor NE, Dabaghian Y, Furr-Stimming EE, Finkbeiner S, Tsvetkov AS. Cytoplasmic sphingosine-1-phosphate pathway modulates neuronal autophagy. Scientific Reports. 2015;5 doi: 10.1038/srep15213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schneider JL, Cuervo AM. Autophagy and human disease: emerging themes. Current Opinion in Genetics & Development. 2014;26:16–23. doi: 10.1016/j.gde.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Quiros PM, Langer T, Lopez-Otin C. New roles for mitochondrial proteases in health, ageing and disease. Nature Reviews Molecular Cell Biology. 2015;16:345–359. doi: 10.1038/nrm3984. [DOI] [PubMed] [Google Scholar]
- 108.Villella AT, Malhotra J, Bhalla A, Nokes E, Hurtado-Lorenzo A, Calamani B, Soper J, Gao BB, Giuliano K, Hafiz M, et al. Inhibition of Usp14 Stimulates the Proteolytic Degradation and Clearance of Misfolded Proteins Associated with Neurodegenerative Diseases. FASEB J. 2013;27 [Google Scholar]
- 109••.Lee BH, Lee MJ, Park S, Oh DC, Elsasser S, Chen PC, Gartner C, Dimova N, Hanna J, Gygi SP, et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467:179–U163. doi: 10.1038/nature09299. The USP-14 inhibitors discovered by King and Finley activate the proteasome for the degradation of certain clients. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Karuppagounder SS, Brahmachari S, Lee Y, Dawson VL, Dawson TM, Ko HS. The c-Abl inhibitor, Nilotinib, protects dopaminergic neurons in a preclinical animal model of Parkinson’s disease. Scientific Reports. 2014;4 doi: 10.1038/srep04874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hebron ML, Lonskaya I, Moussa CEH. Tyrosine kinase inhibition facilitates autophagic SNCA/alpha-synuclein clearance. Autophagy. 2013;9:1249–1250. doi: 10.4161/auto.25368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mahul-Mellier AL, Fauvet B, Gysbers A, Dikiy I, Oueslati A, Georgeon S, Lamontanara AJ, Bisquertt A, Eliezer D, Masliah E, et al. c-Abl phosphorylates alpha-synuclein and regulates its degradation: implication for alpha-synuclein clearance and contribution to the pathogenesis of Parkinson’s disease. Human Molecular Genetics. 2014;23:2858–2879. doi: 10.1093/hmg/ddt674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tanabe A, Yamamura Y, Kasahara J, Morigaki R, Kaji R, Goto S. A novel tyrosine kinase inhibitor AMN107 (nilotinib) normalizes striatal motor behaviors in a mouse model of Parkinson’s disease. Frontiers in Cellular Neuroscience. 2014;8 doi: 10.3389/fncel.2014.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gan-Or Z, Dion PA, Rouleau GA. Genetic perspective on the role of the autophagy-lysosome pathway in Parkinson disease. Autophagy. 2015;11:1443–1457. doi: 10.1080/15548627.2015.1067364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lu YH, Yuan XD, Sun QY, Ou Y. Autophagy activator promotes neuronal differentiation of adult adipose-derived stromal cells. Neural Regeneration Research. 2013;8:882–889. doi: 10.3969/j.issn.1673-5374.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang ZH, Ren WY, Zhu L, Hu LJ. Plasminogen Activator Inhibitor-1 Regulates LPS Induced Inflammation in Rat Macrophages through Autophagy Activation. Scientific World Journal. 2014 doi: 10.1155/2014/189168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wong VKW, Ko CB, Law YK. TRI001 is a novel activator of autophagy via MTOR-dependent mechanisms. Eur J Cancer. 2013;49:S4–S4. [Google Scholar]
- 118.Ge D, Han L, Huang SY, Peng N, Wang PC, Jiang Z, Zhao J, Su L, Zhang SL, Zhang Y, et al. Identification of a novel MTOR activator and discovery of a competing endogenous RNA regulating autophagy in vascular endothelial cells. Autophagy. 2014;10:957–971. doi: 10.4161/auto.28363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Liu YQ, Cheng X, Guo LX, Mao C, Chen YJ, Liu HX, Xiao QC, Jiang S, Yao ZJ, Zhou GB. Identification of an Annonaceous Acetogenin Mimetic, AA005, as an AMPK Activator and Autophagy Inducer in Colon Cancer Cells. Plos One. 2012;7 doi: 10.1371/journal.pone.0047049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Vingtdeux V, Chandakkar P, Zhao HT, d’Abramo C, Davies P, Marambaud P. Novel synthetic small-molecule activators of AMPK as enhancers of autophagy and amyloid-beta peptide degradation. FASEB J. 2011;25:219–231. doi: 10.1096/fj.10-167361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang IF, Guo BS, Liu YC, Wu CC, Yang CH, Tsai KJ, Shen CKJ. Autophagy activators rescue and alleviate pathogenesis of a mouse model with proteinopathies of the TAR DNA-binding protein 43. Proc Natl Acad Sci U S A. 2012;109:15024–15029. doi: 10.1073/pnas.1206362109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yang YP, Hu LF, Zheng HF, Mao CJ, Hu WD, Xiong KP, Wang F, Liu CF. Application and interpretation of current autophagy inhibitors and activators. Acta Pharmacologica Sinica. 2013;34:625–635. doi: 10.1038/aps.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Efeyan A, Comb WC, Sabatini DM. Nutrient-sensing mechanisms and pathways. Nature. 2015;517:302–310. doi: 10.1038/nature14190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kang SA, Pacold ME, Cervantes CL, Lim D, Lou HJ, Ottina K, Gray NS, Turk BE, Yaffe MB, Sabatini DM. mTORC1 Phosphorylation Sites Encode Their Sensitivity to Starvation and Rapamycin. Science. 2013;341:1236566. doi: 10.1126/science.1236566. [DOI] [PMC free article] [PubMed] [Google Scholar]