Abstract
The phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2)-Ca2+ signaling system is important for cell activation in response to various extracellular stimuli. This signaling system is initiated by receptor-induced hydrolysis of PI(4,5)P2 in the plasma membrane (PM) to generate the soluble second messenger inositol 1,4,5-trisphosphate (IP3). IP3 subsequently triggers the release of Ca2+ from the endoplasmic reticulum (ER) store to the cytosol to activate Ca2+-mediated responses, such as secretion and proliferation. The consumed PM PI(4,5)P2 and ER Ca2+ must be quickly restored to sustain signaling responses, and to maintain the homeostasis of PI(4,5)P2 and Ca2+. Since phosphatidylinositol (PI), the precursor lipid for PM PI(4,5)P2, is synthesized in the ER membrane, and a Ca2+ influx across the PM is required to refill the ER Ca2+ store, efficient communications between the ER and the PM are critical for the homeostatic regulation of the PI(4,5)P2-Ca2+ signaling system. This review describes the major findings that established the framework of the PI(4,5)P2-Ca2+ signaling system, and recent discoveries on feedback control mechanisms at ER-PM junctions that sustain the PI(4,5)P2-Ca2+ signaling system. Particular emphasis is placed on the characterization of ER-PM junctions where efficient communications between the ER and the PM occurs, and the activation mechanisms of proteins that dynamically localize to ER-PM junctions to provide the feedback control during PI(4,5)P2-Ca2+ signaling, including the ER Ca2+ sensor STIM1, the extended synaptotagmin E-Syt1, and the PI transfer protein Nir2. This review is part of a Special Issue entitled The Cellular Lipid Landscape.
1. Introduction
Phosphatidylinositol 4,5-bisphosphate, or PI(4,5)P2, is an inositol-containing phospholipid that is enriched at the inner leaflet of the plasma membrane (PM) [1]. PI(4,5)P2 governs many cellular functions including membrane trafficking, ion channel and transporter activity, and cytoskeleton-PM interactions [2, 3]. Aberrant metabolism of PI(4,5)P2 has been implied in diseases such as cancers, oculocerebrorenal syndrome of Lowe (OCRL), and infectious diseases [1, 4]. A remarkable function of PI(4,5)P2 is its ability to mediate Ca2+ signaling following stimulation of cell surface receptors [5]. Receptor-induced signal is transduced from the PM to the endoplasmic reticulum (ER) by the soluble second messenger inositol 1,4,5-trisphosphate (IP3), generated via PI(4,5)P2 hydrolysis by receptor-activated phospholipase C (PLC). IP3 binds and opens a Ca2+ channel, namely the IP3 receptor (IP3R), in the ER membrane to release Ca2+ from the ER store into the cytosol. The increase in cytosolic Ca2+ levels activates an extensive array of Ca2+ effectors in the cell. This PI(4,5)P2-Ca2+ signaling system is fundamental to many cellular processes in various cell types [6].
PI(4,5)P2 at the PM and Ca2+ in the ER are consumed to generate IP3 and cytosolic Ca2+ signals in the PI(4,5)P2-Ca2+ signaling system. It is important to quickly restore PM PI(4,5)P2 and ER Ca2+ levels in order to maintain the homeostasis and sustain Ca2+ signaling in stimulated cells. Following IP3-induced depletion of the ER Ca2+ store, a mechanism is activated to open PM Ca2+ channels and bring in Ca2+ from the extracellular space to sustain cytosolic Ca2+ levels, and to refill the ER Ca2+ store [7]. This homeostatic mechanism is called store-operated Ca2+ entry (SOCE).Defective SOCE causes human diseases including severe combined immunodeficiency and muscular dysplasia [8]. The molecular basis of SOCE activation was identified in recent years. It involves the interaction of the ER Ca2+ sensor STIM1 and the PM Ca2+ channel Orai1 at ER-PM junctions where the ER forms close appositions with the PM. The observation of STIM1 translocation to ER-PM junctions to activate SOCE following ER Ca2+ depletion revived interests in understanding these membrane junctions and their roles beyond excitation-contraction (E-C) coupling in muscles [9, 10]. The functions and regulation of ER-PM junctions have since become areas of intense investigation [11-13].
The homeostasis of PM PI(4,5)P2 in stimulated cells is maintained by a process called the PI cycle that promotes the resynthesis of PI(4,5)P2 after agonist-induced breakdown [1]. After receptor-induced hydrolysis and the release of IP3 to the cytosol, the remaining diacylglycerol (DAG) part of PI(4,5)P2 in the PM is converted to phosphatidic acid (PA). PA is utilized to generate PI by enzymes resided in the ER for resynthesis of PI(4,5)P2 in the PM. Although the first evidence of the PI cycle, known as the “phospholipid effect”, was reported in 1953 by Lowell and Mabel Hokin [14-16], the inter-organelle lipid exchange pathway of the PI cycle was not revealed until 2015, when the PI transfer protein (PITP) Nir2 was identified as the PI-PA exchanger [17-19]. Similar to STIM1, Nir2 translocates to ER-PM junctions to support PI(4,5)P2-Ca2+ signaling following receptor stimulation [18, 20, 21]. Thus, ER-PM junctions are pivotal in the homeostatic regulation of PM PI(4,5)P2 and ER Ca2+ in stimulated cells.
This review begins with a brief historical overview of selected milestones in the research of the PI(4,5)P2-Ca2+ signaling system to provide context for more recent mechanistic studies. The detailed history of studies in the field can be found in three recent reviews [1, 6, 22]. Thereafter, the focus is on recent developments that contribute to our current understanding of the molecular and cellular mechanisms underlying SOCE and the PI cycle that maintain the homeostasis of Ca2+ and PI(4,5)P2, respectively. A special emphasis on advances in the study of ER-PM junctions, the hubs for SOCE and the PI cycle, is also included.
2. The PI(4,5)P2-Ca2+ Signaling System: an Historical Overview
2.1 PI, PI4P, and PI(4,5)P2
PI is the most abundant inositol-containing lipid in eukaryotic cells [1]. Each PI molecule contains an sn-1,2-DAG backbone linked to the hydroxyl group at position 1 of a myo-inositol (cis-1,2,3,5-trans-4,6-cyclohexanehexol) via a phosphodiester bond. One marked feature of PI in animal cells is the enrichment of stearic acid (18:0) at the sn-1 position and arachidonic acid (20:4) at the sn-2 position of its DAG backbone [1, 22, 23]. It remains unclear how this enrichment takes place and whether it has a functional significance. PI is synthesized from CDP-DAG and myo-inositol by PI synthase in the ER and in ER-derived mobile compartments, and is delivered to the PM and other organelles via vesicular transport and PITPs [24-26]. After delivery, PI is subsequently phosphorylated at the hydroxyl groups at positions 3, 4, and/or 5 by various kinases at different membranes to generate seven structurally distinct phosphoinositides, providing membrane identities for selective recruitment of proteins to support organelle functions [1, 3, 27].
PI4P and PI(4,5)P2 were the first two discovered phosphoinositides and were called diphosphoinositide and triphosphoinositide, respectively, in some early literatures [28-30]. PI4P is found in multiple organelles, including the Golgi apparatus, endosomes, and the PM [31]. The enrichment of PI4P in these organelles has recently been shown to provide a driving force for lipid export from the ER via lipid transfer proteins (LTPs) at organelle junctions [32-35]. PI(4,5)P2 is predominantly found at the inner leaflet of the PM. PI(4,5)P2 at the PM can be generated from PI via sequential phosphorylation at the 4- and 5- positions in the inositol head group by kinases localized at the PM [1, 36]. The roles of PI, PI4P, and PI(4,5)P2 in receptor-induced signaling were revealed many years after the initial characterizations of these phospholipids.
2.2 The PI Cycle in Stimulated Cells
In 1953, Mabel and Lowell Hokin first discovered that enzyme secretion stimulated by cholinergic agonists in pancreas slices was accompanied by an increase in the incorporation of 32P into phospholipids, which they called the “phospholipid effect” [14]. In subsequent studies, the Hokins found that 32P was incorporated primarily into PI and PA [15, 37]. Moreover, they found that the rate of 3H-inositol incorporation into PI was significantly enhanced upon stimulation, but the rate of 14C incorporation into the DAG backbone was only slightly increased. These results indicated that the phosphoryl-inositol group of PI is renewed in response to extracellular stimuli, whereas the DAG backbone is recycled. Based on these results and other findings, the Hokins proposed a cycle of reactions leading to the increases in PI and PA turnover following stimulation [38]. As illustrated later by Robert Michell (Figure 1A), this PI cycle begins with the cleavage of the phosphoryl-inositol group of PI accompanied by the production of DAG. PA is generated from DAG by DAG kinases (DGKs); 32P incorporation occurs at this step. PA is then converted into CDP-DAG, and PI is synthesized from CDP-DAG; it is at this step that 3H-inositol incorporation takes place [38, 39]. The causal relationship between the enhanced PI turnover and stimulated secretion of enzymes remained unclear at the time.
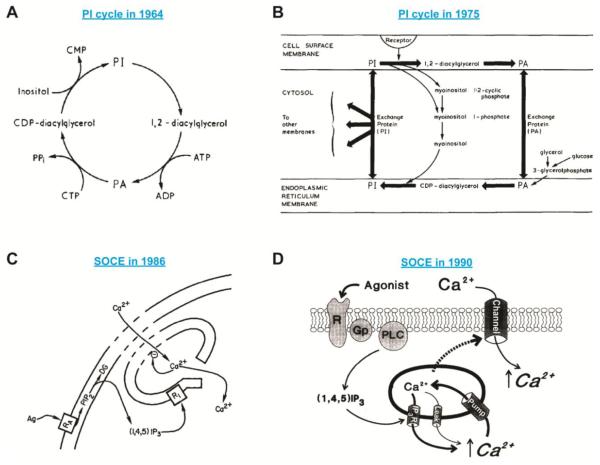
Figure 1.
Models of the PI cycle and SOCE.
(A) An illustration of the first PI cycle model proposed by Mabel and Lowell Hokin in 1964. © Michell and BBA, 1975. Originally published in Biochim Biophys Acta, 415 (1975) 81-147.
(B) An advanced version of the PI cycle model proposed by Robert Michell in 1975. The ideas that the PI cycle is an inter-organelle pathway and that cytosolic exchange proteins mediate PI/PA shuttling between the ER and the PM were incorporated into this model. © Michell and BBA, 1975. Originally published in Biochim Biophys Acta, 415 (1975) 81-147.
(C) The first model of SOCE proposed by James Putney in 1986. Ag, agonist; RA, receptor; PIP2, PI(4,5)P2; (1,4,5)IP3, IP3; RI, IP3R. © Putney and Cell Calcium, 1986. Originally published in Cell Calcium, 7 (1986) 1-12.
(D) A revised model of SOCE proposed by James Putney in 1990. The dashed arrow indicates an unknown mechanism that links ER Ca2+ depletion to the opening of a Ca2+ channel at the PM. Ca2+ first enters the cytosol and is then pumped into the ER lumen to refill the ER Ca2+ store. R, receptor; GP; G-protein. © Putney and Cell Calcium, 1990. Originally published in Cell Calcium, 11 (1990) 611-624.
One intriguing phenomena later noted in the field was that most stimuli that enhanced PI turnover also elicited Ca2+-mediated responses, even though the requirement of extracellular Ca2+ for the enhanced PI turnover was under debate [1, 39]. One common feature of these stimuli was that their effects on target cells were mediated through cell surface receptors. In 1973, Lapetina and Michell identified a PM-associated PLC that mediates PI breakdown and suggested that this enzyme might be controlled by cell surface receptors [40]. Based on these observations, together with other accumulative evidence, Michell proposed an advanced version of the PI cycle in his seminal review in 1975 (Figure 1B) [39]. In this model, Michell suggested that receptor activation-mediated PI breakdown at the PM is the key to trigger PI turnover. Michell also noted that mechanisms must exist to facilitate lipid interchange between the PM and the ER in order to feed PA generated at the PM into the PI synthesis machinery in the ER membrane as well as to provide the PM with PI. Based on the findings of soluble phospholipid exchange proteins [41], Michell hypothesized that cytosolic lipid exchange/transfer proteins mediate the PI cycle by quickly shuttling lipids between the PM and the ER. This model provided substantial conceptual advances and prompted further research to resolve the relationship between PI turnover and Ca2+-mediated responses. The next question was whether PI turnover was simply in the upstream of Ca2+-mediated responses, or these two were independent signaling events following receptor stimulation.
2.3 PI(4,5)P2 Hydrolysis, IP3 Production, and Ca2+ Release from the ER Store
In 1979, Fain and Berridge clarified the relationship between the enhanced PI turnover and Ca2+-mediated responses by directly measuring trans-epithelial 45Ca2+ flux induced by serotonin in the blowfly salivary gland [42]. They demonstrated that PI breakdown and trans-epithelial Ca2+ flux were kinetically coupled, whereas Ca2+-mediated responses, such as fluid secretion, could be un-coupled from PI breakdown under certain conditions. In addition, by carefully removing Ca2+ from intracellular stores and extracellular space, they found that the presence of Ca2+ was not required for PI breakdown. These observations suggested that PI breakdown is involved in gating Ca2+ flux. A few months later, the same authors presented evidence that Ca2+ flux was downstream of PI resynthesis after its breakdown during prolonged receptor activation [43]. The next question became evident: what is the mechanism that links PI breakdown to Ca2+ flux?
To seek the link, the field started to re-evaluate previous observations of PI(4,5)P2 breakdown after receptor activation. It has been shown in early studies that PI(4,5)P2 has a high basal rate of 32P incorporation, suggesting that it may have important functions [44]. In 1964, PI(4,5)P2-specific PLC generating DAG and IP3 was isolated and characterized [45]. In the same year, PI(4,5)P2 breakdown after receptor activation was reported by the Hokins’ group [46]. They found that 32P incorporation into PI(4,5)P2 decreased under the same experimental conditions that resulted in enhanced PI and PA labeling. A later study demonstrated that inositol-bisphosphate, which can only be generated by PI4P or PI(4,5)P2 breakdown, accumulated following cell stimulation [47]. The breakdown of PI(4,5)P2 following receptor stimulation was examined in details in the late 1970s [48]. Nevertheless, most studies of receptor-stimulated effects on the turnover of inositol-containing lipids at that time were focused on PI, and PI(4,5)P2 was generally ignored because it is of low abundance in most tissues, and was difficult to extract due to its unusual chemical properties [49]. These factors also led to contradictory results of PI(4,5)P2 metabolism from 32P labeling experiments following receptor activation.
In 1983, several reports demonstrated a rapid accumulation of IP3 after receptor-induced breakdown of PI(4,5)P2 [50-52]. In the same year, the collaboration of Schulz, Irvine, and Berridge elegantly demonstrated that the addition of exogenous IP3 to permeabilized rat pancreatic acinar cells was sufficient to induce Ca2+ release from a non-mitochondrial Ca2+ store [53]. A few years later in 1989, several groups identified the IP3R in the ER membrane as the Ca2+ release channel activated following receptor stimulation [54-56]. Together, these findings uncovered the initial steps of the PI(4,5)P2-Ca2+ signaling system, and linked receptor-induced PI(4,5)P2 hydrolysis at the PM to the release of Ca2+ from the ER store by IP3 production.
2.4 Store-Operated Ca2+ Entry (SOCE)
Cytosolic Ca2+ levels are tightly controlled at a low concentration (~100 nM, which is 5,000-20,000 times lower than Ca2+ concentrations in the ER store or in the extracellular space) in resting cells by the activities of sarco/endoplasmic reticulum Ca2+-ATPase (SERCA) and plasma membrane Ca2+-ATPase (PMCA) in the ER and the PM, respectively [57]. It was well recognized in the 1980s that receptor stimulation triggered a biphasic cytosolic Ca2+ response comprising of an initial transient increase caused by the release of an intracellular Ca2+ store, and a subsequent sustained response resulted from activation of Ca2+ entry across the PM [58]. Since the intracellular store provides finite sources of Ca2+, it is important to activate Ca2+ influx to sustain Ca2+ signaling and refill the Ca2+ store for cell homeostasis. Early studies indicated a mechanism for coordinated regulation of the initial intracellular Ca2+ release and the subsequent Ca2+ influx [59]. Based on the observation that the Ca2+ level of the intracellular store negatively operated Ca2+ influx across the PM in agoist-stimulated cells, the first model of this mechanism, later recognized as SOCE, was proposed by James Putney in 1986 (Figure 1C) [7]. Subsequent studies using thapsigargin (TG), a SERCA inhibitor that empties the ER Ca2+ store without activating the IP3R [60], demonstrated that ER Ca2+ depletion is necessary and sufficient to activate SOCE [61]. These observations revealed the requirement of an ER-to-PM signaling pathway that links the reduction of ER Ca2+ levels to Ca2+ influx across the PM as depicted by James Putney in the revised model of SOCE (Figure 1D) [62]. Additional evidence in support of SOCE came from the detection of a Ca2+ current corresponding to Ca2+ influx activated in agonist-stimulated mast cells and T cells [63, 64]. This current, which is highly selective for Ca2+ over monovalent cations and has extremely low conductance, was named as Ca2+ release-activated Ca2+ current, abbreviated as ICRAC [65]. Many models were proposed to explain this important ER-to-PM signaling pathway that activates SOCE or ICRAC [66]. Nonetheless, the molecular mechanisms of SOCE had remained mysterious for nearly two decades since the first model was proposed.
Overall, the studies described in this chapter established the framework of the PI(4,5)P2-Ca2+ signaling system: receptor stimulation induces the hydrolysis of PM PI(4,5)P2 by PLC to generate IP3 that triggers Ca2+ release from the ER for cell activation. In this system, PM PI(4,5)P2 can be resynthesized after hydrolysis via the PI cycle, and ER Ca2+ is refilled after depletion via SOCE to sustain receptor-induced signaling. Both the PI cycle and SOCE provide homeostatic regulation of the PI(4,5)P2-Ca2+ signaling system and require fast communications between the ER and the PM in receptor-stimulated cells. In the next chapters, we will focus on recent discoveries of molecular mechanisms underlying SOCE and the PI cycle. These discoveries revealed the important role of ER-PM junctions, where the ER and the PM are within 10-20 nm from each other (Figure 2A), in the PI(4,5)P2-Ca2+ signaling system.
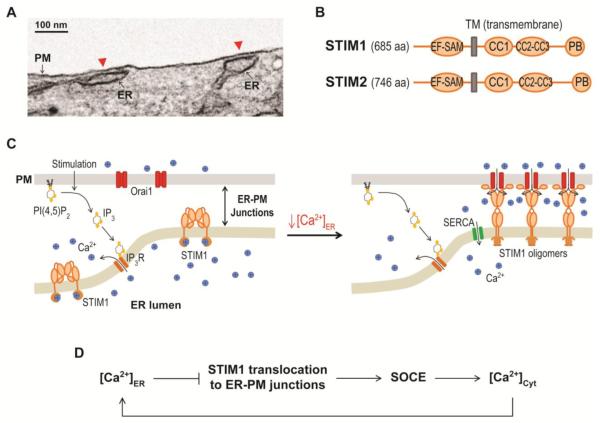
Figure 2.
Activation of SOCE by STIM1 and Orai1 at ER-PM junctions.
(A) An electron microscopy image of ER-PM junctions (red arrow heads) in a MAPPER-expressing HeLa cell.
(B) Diagrams of the domain structure of human STIM1 and STIM2. EF-SAM, EF hand and sterile alpha motif; TM, transmembrane; CC1, CC2, and CC3, coiled coil domain 1,2, and 3; PB, polybasic.
(C) STIM1 translocation to ER-PM junctions induced by a decrease in ER Ca2+ levels during PI(4,5)P2-Ca2+ signaling resulting in SOCE by Orai1 activation.
(D) Schematic representation of feedback regulation of ER Ca2+ levels mediated by dynamic translocation of STIM1 to ER-PM junctions.
3. Regulation of Ca2+ Homeostasis via SOCE at ER-PM Junctions
3.1 Activation of SOCE by STIM1 and Orai1
A major breakthrough in understanding the molecular and cellular mechanisms of SOCE was the identification of STIM1 and Orai1 [9, 67-70]. STIM1 and its homolog STIM2 were identified as the ER Ca2+ sensor of the ER-to-PM SOCE pathway using RNA interference screens in 2005 [9, 67]. In the subsequent year, Orai1 was identified as the PM Ca2+ channel that synergistically activates SOCE with STIM1 [68-75]. Both STIM1 and STIM2 are single transmembrane ER proteins with an N-terminal Ca2+-sensing EF hand-SAM (EF-SAM) domain in the ER lumen, and a C-terminal cytosolic region with three coiled-coil domains and a polybasic tail (Figure 2B) [76]. The minimal Orai1-activating domain of STIM1, roughly corresponding to the CC2-CC3 domain, was identified by several group [77-79]. In the resting state, inactive STIM1 binds Ca2+ via its luminal EF hand motif, and localizes throughout the ER membrane (Figure 2C) [9]. Following stimulation-induced ER Ca2+ depletion, STIM1 undergoes conformational changes and rapidly forms oligomers due to loss of Ca2+ binding [80, 81]. The exposed polybasic tail in STIM1 oligomers binds PM phosphoinositides, resulting in STIM1 redistribution to ER-PM junctions [80, 82-84]. The translocation of STIM1 to ER-PM junctions was sometimes referred as puncta formation based on the initial finding that cells overexpressing YFP-tagged STIM1 showed bright fluorescence signals in numerous puncta near the cell periphery following ER Ca2+ depletion [9]. The close appositions between the ER and the PM at ER-PM junctions allow STIM1 in the ER to trap and activate the PM-localized Orai1 to elicit SOCE [85]. Overall, STIM1 mediates a feedback mechanism that maintains ER Ca2+ homeostasis during PI(4,5)P2-Ca2+ signaling by dynamically translocating to ER-PM junctions upon IP3-induced drop of ER Ca2+ levels to activate SOCE, and bringing in Ca2+ from the extracellular space to the cytosol to refill the ER store (Figure 2D). Similar to STIM1, STIM2 activates Ca2+ influx via activation of Orai1 [86]. Interestingly, STIM2 translocates to ER-PM junctions and activates Ca2+ influx upon smaller decreases in ER Ca2+ comparing with STIM1, suggesting a role of STIM2 in maintaining basal Ca2+ homeostasis. Recent studies identified several proteins, including junctate, CRACR2A, α-SNAP, SARAF, Surf4, POST, septins, and STIMATE, that modulate SOCE mediated by STIM1 and Orai1 at ER-PM junctions [87-94]. Many of these proteins directly bind to STIM1 and/or Orai1, suggesting that the interaction of STIM1 and Orai1 may be regulated by complex mechanisms. Detailed description of studies on STIM1, Orai1, and SOCE can be found in a recent review [8].
3.2 ER-PM Junctions: the Hubs for SOCE and More
ER-PM junctions were first described by Porter and Palade in 1957 in muscle cells studied using electron microscopy (EM) [95]. Similar structures were later reported in neurons [96, 97]. In general, the distance between the ER and the PM at ER-PM junctions is in the range of 10 to 20 nm [10, 98-101]. The narrow distance allows proteins in one compartment to directly interact with proteins or lipids in the other; however, membrane fusion between these two compartments has not been reported at these junctions. The ER and the PM are often aligned in parallel over lengths of approximately 200 nm at ER-PM junctions in cells without overexpressing ER-PM tethering proteins [10, 99]. In addition, ER-PM junctions in many non-excitable cell types occupy less than 1% of the PM [99, 101]. The detection of these small and sparse membrane junctions using conventional microscopy had been a challenge that was recently overcome by developments of genetically-encoded fluorescence markers of ER-PM junctions [20, 92, 102, 103].
In the literature throughout the last half century, a variety of nomenclatures have been used to describe ER-PM junctions in different cell types and organisms. These terms include dyad and triad junctions (or dyads and triads), E-C units, and peripheral couplings in muscle cells [104]; subsurface cisterns in neurons (SSC’s) [96]; PM-associated ER and cortical ER in yeast [105, 106]; subrhabdomeric cisternae (SRC) in Drosophila photoreceptor cells [107]; and ER-PM contacts or ER-PM contact sites. We prefer the term junctions as this nomenclature emphasizes that these are the loci at which the ER and the PM are joined in both physical proximity and functional coupling [11].
The roles of dyads and triads in muscle contractile functions were widely accepted in the 1980s, but the functional relevance of ER-PM junctions in non-excitable cells had remained elusive despite their structural similarities to dyads and triads in muscles [98]. The discovery of the universal SOCE signaling pathway activated by the direct interaction of STIM1 in the ER and Orai1 in the PM at ER-PM junctions indicated that these minute subcellular structures are fundamental to inter-organelle communications in most, if not all, cells. Indeed, multiple lines of evidence recently demonstrated the importance of ER-PM junctions in the metabolism of several lipids [17-20, 33, 34, 108, 109]. This review will focus on the findings of ER-PM junctions relevant to the PI(4,5)P2-Ca2+ signaling system.
STIM1 overexpression caused a significant expansion of ER-PM junctions which greatly facilitated their detection in EM studies [10, 99, 100]. This observation was expected as STIM1 contains both the ER and the PM targeting motifs, and thus can mediate ER-PM tethering. Nevertheless, a requirement for STIM1 in the formation of ER-PM junctions has not been demonstrated in cells lacking STIM1. Intriguingly, STIM1 puncta formed repeatedly in almost identical positions in live cells upon multiple rounds of stimulation [110], suggesting that ER-PM junctions pre-exist, and that proteins other than STIM1 maintain ER-PM junctions in resting cells.
In yeast, ER-PM junctions are maintained by multiple families of proteins, including tricalbins, Scs2/Scs22, and Ist2 [111]. The orthologs of tricalbins, namely extended-synaptotagmins (E-Syts), were shown to mediate ER-PM tethering in mammalian cells [112]. E-Syts were named after synaptotagmin because of the similarity in domain architecture [113]. E-Syts contain an N-terminal hydrophobic stretch that is anchored in the ER membrane (Figure 3A). These proteins also contain a cytosolic SMP (synaptotagmin-like mitochondrial lipid binding protein) domain followed by three to five C2 domains. The SMP domain of E-Syts belongs to a conserved superfamily of lipid-binding domains [114]. The SMP domain of E-Syt2 has been shown to bind phospholipids, implying a role of E-Syts in shuttling lipids at ER-PM junctions [115]. The very C-terminal C2 domain of E-Syt2 and E-Syt3 mediate PI(4,5)P2-dependent PM targeting and enable ER-PM tethering (Figure 3B) [112, 113]. When fluorescent protein-tagged E-Syt2 and E-Syt3 were overexpressed in cells, they were enriched at ER-PM junctions and caused an expansion of ER-PM junctions [112]. Consistently, knockdown of all three E-Syts resulted in a significantly reduction in the number of ER-PM junctions. Nonetheless, mice lacking both E-Syt2 and ESyt3 were viable and developed normally [116], suggesting that components in addition to E-Syts provide ER-PM tethering in resting cells to support functions occurring at ER-PM junctions. Intriguingly, E-Syt2 also facilitated activated FGF receptor endocytosis to support FGF signaling [117, 118]. In combination with the finding that ER-PM junctions may serve as membrane protein trafficking hubs [119], these observations suggest a functional role of ER-PM junctions in receptor recycling.
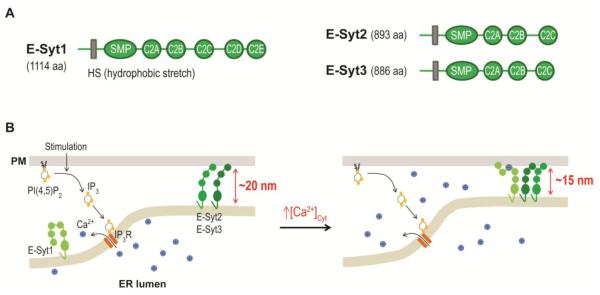
Figure 3.
Regulation of ER-PM junctions by E-Syts.
(A) Diagrams of human E-Syt1, E-Syt2, and E-Syt3 domain structures. HS, hydrophobic stretch; SMP, synaptotagmin-like-mitochondrial-lipid binding protein domain.
(B) E-Syt2 and E-Syt3 provide ER-PM tethering in resting cells. The increase in cytosolic Ca2+ levels during PI(4,5)P2-Ca2+ signaling triggers E-Syt1 translocation to ER-PM junctions, resulting in a reduction in the gap distance.
In contrast to the enrichment of E-Syt2 and E-Syt3 at ER-PM junctions, VAP-A (vesicle-associated membrane protein-associated protein A) and VAP-B, the mammalian orthologs of Scs2/Scs22, are distributed throughout the ER. VAP proteins can bind the FFAT (an acronym for two phenylalanines (FF) in an Acidic Tract) motif present in several regulators of phospholipids [120], and thus can support the targeting of proteins, such as Nir2 and OSBP, to ER-PM junctions and ER-Golgi junctions, respectively [20, 35].
Junctate, another ER membrane protein, was found to contribute to the formation of ER-PM junctions [121]. Overexpression of junctate resulted in a significant increase in the number and size of ER-PM junctions, whereas knockdown of junctate had the opposite effect. Overexpressed junctate displayed a diffuse localization throughout the ER, suggesting that junctate interacts with other components, such as the IP3R or STIM1-Orai1 complexes, to provide ER-PM tethering [91, 121, 122]. Additional molecular components contributing to the formation of ER-PM junctions likely exist, and are remained to be identified.
3.3 Dynamic Regulation of ER-PM Junctions during Ca2+ Signaling
Since ER-PM junctions are indispensable platforms for the communications between the ER and the PM, the regulation of the number, size, and/or shape of ER-PM junctions is expected to have significant functional impacts. Several studies demonstrated that the number of ER-PM junctions is dynamically regulated during Ca2+ signaling. It was first reported in the EM study by Gardiner and Grey in 1983, which showed that the abundance of ER-PM junctions correlates with Ca2+ signaling events during Xenopus oocyte maturation [98]. In addition, an increase in the number of ER-PM junctions in Jurkat T cells and in HeLa cells following ER Ca2+ depletion by TG was detected by EM [10, 99]. Using a genetically-encoded marker, MAPPER, that selectively labels ER-PM, the increase in the number of ER-PM junctions can be monitored during Ca2+ signaling in live cells [20]. Notably, the size and shape of ER-PM junctions labeled by MAPPER remained similar during Ca2+ signaling as measured using stimulated emission depletion (STED) super-resolution microscopy (supplemental info in [20]). Interestingly, the number of ER-PM junctions in pancreatic acinar cells remained unchanged following TG treatment [100]. These results suggested that the abundance of ER-PM junctions may be differentially regulated depending on physiological needs of the cell.
The regulation of the gap distance between the ER and the PM at ER-PM junctions is also expected to have a functional significance [11]. Extending the gap distance at ER-PM junctions by ~6 nm using an engineered Sec22b protein led to impaired neurite growth [123]. By contrast, artificial constraint of the gap at ER-PM junctions restricted the accessibility of STIM1-Orai1 protein complexes to ER-PM junctions [102]. These observations imply that changes in the physical space at ER-PM junctions may lead to differential accommodation of proteins and ions enabling distinct cellular functions.
Recent studies demonstrated that E-Syt1 translocation to ER-PM junctions triggered a decrease in the gap distance at ER-PM junctions (Figure 3B) [20, 124]. Unlike E-Syt2 and E-Syt3, fluorescence protein-tagged E-Syt1 displayed a diffuse ER localization in resting cells. E-Syt1 translocates to ER-PM junctions following an increase in cytosolic Ca2+ to the lower micromolar range [20, 112, 125]. The translocation of E-Syt1 requires its cytosolic C2C domain for Ca2+ binding and its C2E domain for PM PI(4,5)P2 binding [20, 112]. The E-Syt1-triggered decrease in the gap distance at ER-PM junctions was first suggested in live HeLa cells using TIRF (total internal reflection fluorescence) microscopy by applying MAPPER and MAPPER-s, a short form of MAPPER that restricts the gap distance of ER-PM junctions to 10 nm or less [20]. A recent cryo-electron tomography study further confirmed that the gap distance was reduced from 21.8 nm to 14.8 nm following TG treatment in COS-7 cells overexpressing E-Syt1 [124]. This study also demonstrated that overexpression of different ER-PM tethering proteins resulted in differences in the gap distance and in architecture at ER-PM junctions, suggesting the existence of structurally, and likely functionally, distinct ER-PM junctions. E-Syt1 may mediate the decrease in the gap distance through the interaction with other E-Syts at ER-PM junctions since they form hetero-oligomers [112, 118]. In support of this hypothesis, a model of E-Syt2 mediating a decrease in gap at ER-PM junctions was proposed in a recent structural study [126]. Further investigation is required to understand the mechanism underlying the decrease in the gap distance following E-Syt1 translocation to ER-PM junctions.
A decrease in the gap distance at ER-PM junctions mediated by E-Syt1 was shown to be important for PI(4,5)P2-Ca2+ signaling [20]. Knockdown of E-Syt1 resulted in defective PI(4,5)P2 replenishment following receptor-induced hydrolysis. This defect was rescued by overexpression of wild-type E-Syt1 but not that of E-Syt1-D724A, a mutant incapable of translocating to ER-PM junctions to regulate the gap distance. Consistently, expression of MAPPER-s enhanced the replenishment of PI(4,5)P2. Moreover, receptor-induced Ca2+ signaling was profoundly reduced when E-Syt1 knockdown cells were challenged with periodic stimulation. Given that knockdown of all three E-Syts failed to disrupt the onset of SOCE [112], the reduced Ca2+ signaling in E-Syt1 knockdown cells was likely due to a decrease in IP3-mediated Ca2+ release resulting from a defective replenishment of PM PI(4,5)P2 and the subsequent IP3 generation. Overall, these observations suggest that the decrease in the gap distance at ER-PM junctions by E-Syt1 translocation facilitates PI(4,5)P2 replenishment during PI(4,5)P2-Ca2+ signaling.
Both STIM1 and E-Syt1 are ER membrane proteins that translocate to ER-PM junctions by binding to PI(4,5)P2 in the PM in stimulated cells. STIM1 translocation is induced by ER Ca2+ depletion, whereas E-Syt1 translocation can be independently induced by an increase in cytosolic Ca2+. It seems paradoxical that STIM1 and E-Syt1 translocate to ER-PM junctions in stimulated cells via PM PI(4,5)P2 binding while PM PI(4,5)P2 is being hydrolyzed. It is possible that the pool of PM PI(4,5)P2 utilized for PLC-mediated hydrolysis is different from that for the recruitment of STIM1 and E-Syt1 to ER-PM junctions. A recent study suggested that ER-PM tethering by E-Syt1 may form PI(4,5)P2-enriched micro-domains for STIM1 targeting [127]. Nevertheless, direct evidence for the enrichment of PI(4,5)P2 at ER-PM junctions has not been demonstrated. Another explanation is that there is enough PI(4,5)P2 at the PM for IP3 generation and for the targeting of STIM1 and E-Syt1 to ER-PM junctions in stimulated cells.
4. Regulation of PI(4,5)P2 Homeostasis via the PI Cycle
4.1 A Non-vesicular Mechanism for PI(4,5)P2 Replenishment
It was observed more than 30 years ago that the level of PI(4,5)P2 quickly recovered after the initial breakdown even in the presence of continuous receptor stimulation [50]. This observation indicated that a feedback mechanism is promptly activated to replenish PM PI(4,5)P2 levels. The resulting near steady PM PI(4,5)P2 levels may support the recruitment of STIM1 and E-Syt1 to ER-PM junctions as well as other PI(4,5)P2-dependent functions in stimulated cells. The PM only contains a limited pool of PI for PI(4,5)P2 re-synthesis [128]. Thus, cells must rely on PI synthesized in the ER to sustain PI(4,5)P2 levels in the PM during PI(4,5)P2-Ca2+ signaling. This hypothesis is supported by the findings that constitutive or inducible removal of PI from the ER by a bacterial PI-PLC resulted in defective replenishment of PI(4,5)P2 in the PM [17, 24]. To maintain PM PI(4,5)P2 levels, PI in the ER must be transported to the PM within a few minutes or less after receptor stimulation. Since diacylglycerol lipids are highly hydrophobic, the rate of spontaneous lipid transfer across an aqueous medium is extremely slow [129]. Blocking vesicular trafficking or depletion of PI(4,5)P2 precursor lipids in the Golgi apparatus had minimal effects on PI(4,5)P2 replenishment during PI(4,5)P2-Ca2+ signaling [17, 130, 131]. These findings suggest that a non-vesicular mechanism is involved in the rapid delivery of PI in the ER to the PM for PI(4,5)P2 replenishment.
Early on, Michell had hypothesized that cytosolic LTPs participate in the lipid shuttling process between the ER and the PM in the PI cycle [39]. The ability of LTPs to mediate lipid transport between membranes was demonstrated in numerous in vitro studies using radiolabeled or fluorescent lipids [132]. This inter-membrane lipid transport is likely caused by LTPs binding and facilitating lipid desorption from membranes [133]. It was proposed that lipid transfer between membranes via LTPs in vivo would be most efficient when LTPs function at membrane contact sites where LTPs can simultaneously bind two membrane compartments (Figure 4A) [26, 132, 134, 135]. It is likely that efficient shuttling of PI and PA between the ER and the PM in the PI cycle is mediated by LTPs docking at ER-PM junctions. Consistent with this hypothesis, the decrease in the gap distance via E-Syt1 translocation or MAPPER-s expression facilitated the docking of Nir2, a PITP, at ER-PM junctions during PI(4,5)P2-Ca2+ signaling [20].
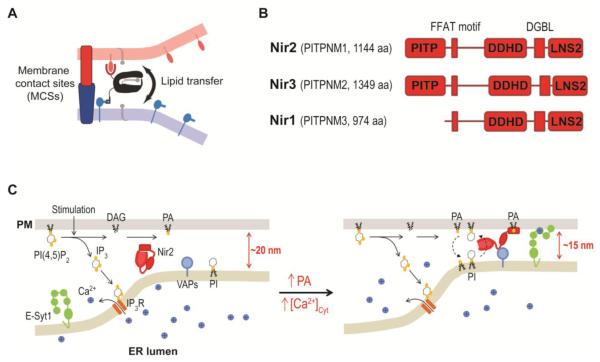
Figure 4.
PI(4,5)P2 homeostasis mediated by Nir2 at ER-PM junctions during PI(4,5)P2-Ca2+ signaling.
(A) Efficient lipid transfer at membrane contact sites illustrated by Tim Levine in 2004. © Levine and TICB, 2004. Originally published in Trends in Cell Biology, 14 (2004) 483-490.
(B) Domain structures of human Nir2, Nir3, and Nir1.
(C) Nir2 is a feedback regulator for PI(4,5)P2 homeostasis. Nir2 senses PA produced by PI(4,5)P2 hydrolysis and mediates PI/PA transfer at ER-PM junctions to support PI(4,5)P2 replenishment during PI(4,5)P2-Ca2+ signaling. The decrease in the gap distance via E-Syt1 translocation facilitates the accumulation of Nir2 at ER-PM junctions.
4.2 PI Transfer Proteins
There are five PITPs in the human genome which are grouped into two classes based on sequence homology [26]. Class I PITPs consisting of PITPα and PITPβ were first purified in the 1970s [136]. These two Class I PITPs only contain the conserved PITP domain that catalyzes the transfer of PI and phosphatidylcholine (PC) between membranes. Thus, Class I PITPs are PI-PC exchangers. The crystal structures of PITPα and PITPβ revealed a hydrophobic cavity in the PITP domain that can be occupied by either PI or PC [137, 138]. This cavity is closed by a lid-like α-helix, and the phospholipid is completely buried in this cavity with the polar head group located deep in the cavity. Four residues, threonine 59, lysine 61, glutamate 86, and asparagine 90, in this cavity make hydrogen bonds with the inositol group of PI. These residues are conserved in many, if not all, PITPs in animal cells [137]. Mutation of any one of these four residues resulted in defective PI binding and transfer [139, 140]. Another conserved residue, cysteine 95, was shown to be important for PC binding and transfer [140, 141]. PITPα was identified as a soluble component essential for reconstitution of PLC signaling in permeabilized cells [142-144]. Subsequent studies demonstrated that both PITPα and PITPβ support PI(4,5)P2-related functions [26]. Mice without PITPα died shortly after birth and exhibited spinocerebellar degeneration, intestinal and hepatic fat accumulation, and hypoglycemia; while ablation of PITPβ expression in mice resulted in embryonic lethality [145-147]. These results indicated the functional significance of PITPα and PITPβ in physiology and development. Nonetheless, the molecular basis of how PITPα and PITPβ support PLC signaling and PI(4,5)P2-related functions remain unclear.
Class II PITPs are consisted of three proteins: PITPNC1 (also known as RdgBβ) that only contains the PITP domain; Nir2 (also known as PITPNM1 or RdgBαI) that contains several C-terminal domains following the N-terminal PITP domain (Figure 4B); and Nir3 (also known as PITPNM2 or RdgBαII) that shares a similar domain architecture with Nir2. Early genetic studies revealed the importance of RdgBα, the Drosophila ortholog of Nir2 and Nir3, in photo-transduction [26]. In Drosophila photoreceptor cells, PI(4,5)P2 is hydrolyzed by light-activated PLC during photo-transduction. Flies lacking RdgBα exhibited defective photo-transduction and retinal degeneration [148]. Moreover, electrophysiological studies using Kir channels as a PI(4,5)P2 indicator demonstrated defective PI(4,5)P2 replenishment following light exposure in the RdgBα mutant photoreceptor cells [149]. This result was recently confirmed by pseudopupil imaging using PLCδ-PH domain to detect PI(4,5)P2 levels in Drosophila photoreceptors [19]. The PITP domain of RdgBα, but not PITPα, was sufficient to rescue the defects in light response and retinal degeneration in RdgBα mutant flies [19, 150]. These findings suggested that the PITP domain of RdgBα contains activities essential for maintaining PM PI(4,5)P2 levels during photo-transduction, and that the activities cannot be provided by a PI-PC exchanger.
4.3 PI-PA Exchange Mediated by Nir2 at ER-PM Junctions
Although early genetic data indicated the role of RdgBα in PI(4,5)P2 replenishment following light-induced hydrolysis, the mechanistic basis of how the PI cycle is mediated by PITPs remained unclear until recent studies of a human ortholog of RdgBα, namely Nir2. Nir2 is expressed in most tissues, and is named as the PYK2 N-terminal domain-interacting receptor (Nir) because it was identified as a PYK2-binding protein [151]. A role of Nir2 has been implicated in many cellular functions, including cytokinesis and secretion [152, 153]. Recent studies showed that Nir2 regulates PI(4,5)P2-Ca2+ signaling [17-21]. Knockdown of Nir2 significantly reduced PI(4,5)P2 replenishment following receptor-induced hydrolysis, whereas Nir2 overexpression had an enhancing effect. Moreover, Ca2+ signaling and other PI(4,5)P2-dependent functions were abolished in cells depleted of Nir2 [17, 18, 21]. The PITP activity of Nir2 is required for PI(4,5)P2 replenishment as mutations in the conserved inositol-interacting residues or deletion of the PITP domain led to defective PI(4,5)P2 replenishment [20]. The PI transfer ability of the PITP domains of Nir2 and RdgBα has been shown in vitro [19, 150, 154]. Nonetheless, it is very difficult to demonstrate PI transfer among different membrane compartments in vivo, because PI specific biosensor is not currently available, and it is believed that PI is quickly converted to PI4P or PI(4,5)P2 by kinases once it reaches the PM. Despite the technical difficulties, a recent study demonstrated that Nir2-enhanced PI(4,5)P2 replenishment is dependent on PI transfer from the ER to the PM by employing an inducible approach to acutely remove PI in the ER membrane while monitoring PM PI(4,5)P2 replenishment following receptor stimulation in live cells [17]. This finding strongly suggests that Nir2 mediates PI transfer from the ER to the PM to support PI(4,5)P2 replenishment in vivo.
Besides the PITP domain, Nir2 possesses targeting motifs for the ER and the PM (Figure 4B). The FFAT motif of Nir2 interacts with the ER membrane proteins VAPs [155]. A C-terminal region containing the DAG-binding DGBL domain and the PA-binding LNS2 domain is important for PM targeting [17, 18, 21]. Overexpressed Nir2 was mainly cytosolic with moderate ER association in resting cells, indicating that Nir2 adopts a partially closed conformation obscuring the ER- and the PM-targeting motifs [17, 18]. The detection of Nir2 in membrane fractions of HeLa cells and Drosophila head extract likely reflects the exposure of the ER- and/or the PM-targeting motifs during biochemical preparations in vitro [19, 156]. During PI(4,5)P2-Ca2+ signaling, Nir2 rapidly translocated to ER-PM junctions [17, 18, 20]. This observation is consistent with EM studies showing that RdgBα localized to SRC, a specialized ER domain juxtaposed to the PM, in Drosophila photoreceptor cells [107, 157]. Nir2 translocation to ER-PM junctions was abolished in the Nir2-FM-D1128A mutant containing point mutations disrupting its VAP- and PA-binding motifs [17]. In addition, inhibition of PA production following PI(4,5)P2 hydrolysis by pharmacological inhibitors of PLC or DGK significantly reduced Nir2 translocation to ER-PM junctions. Consistently, addition of exogenous PA triggered Nir2 translocation in the absence of receptor stimulation. These observations indicate that Nir2 detects receptor-induced PI(4,5)P2 hydrolysis by binding to PA in the PM via its C-terminal LNS2 domain. A recent report showed that addition of DiC8-DAG also induced binding of Nir2 to the PM, suggesting that Nir2 interacts with the PM via combined DAG and PA binding [18]. The accumulation of Nir2 at ER-PM junctions was further facilitated by the decrease in the gap distance at ER-PM junctions by E-Syt1 translocation [20]. These findings suggest that Nir2 translocation to ER-PM junctions is tightly gated during PI(4,5)P2-Ca2+ signaling by DAG and PA production via PI(4,5)P2 hydrolysis and E-Syt1 translocation. Intriguingly, Nir2 has been shown to localize in the Golgi apparatus and to maintain the Golgi DAG pool [152, 156]. It would be of importance to understand how Nir2 functions in the Golgi apparatus.
The enhanced PI(4,5)P2 replenishment mediated by Nir2 appears to be dependent on its localization at ER-PM junctions. Unlike wild-type Nir2, overexpression of the Nir2-FM-D1128A mutant failed to enhance PI(4,5)P2 replenishment [17]. Interestingly, overexpression of the Nir2 PITP domain alone, which distributed in the cytosol, was able to promote PI(4,5)P2 replenishment in receptor-stimulated cells albeit with much slower kinetics comparing with those overexpressing the full-length Nir2 [17]. These observations are consistent with genetic studies in Drosophila showing that the PITP domain of RdgBα is sufficient to rescue retinal defects [19, 150], and support the notion that docking at ER-PM junctions enhances the efficiency of lipid transfer by Nir2. Given that the Nir2-FM-D1128A mutant contains the PITP domain but is unable to enhance PI(4,5)P2 replenishment, the functional PITP domain appeared to be structurally hindered in resting cells. The binding of Nir2 to PA in the PM and/or VAPs in the ER following receptor stimulation likely opens up the structure and exposes the functional PITP domain to mediate lipid transport at ER-PM junctions. Based on these results, it was proposed that Nir2 is a feedback regulator for PI(4,5)P2 homeostasis during PI(4,5)P2-Ca2+ signaling by sensing PA production following PI(4,5)P2 hydrolysis and translocating to ER-PM junctions to support PI(4,5)P2 replenishment by mediating ER-to-PM PI transfer (Figure 4C) [17]. Ca2+-induced E-Syt1 translocation to ER-PM junctions further couples Ca2+ signaling to PI(4,5)P2 homeostasis by ensuring efficient docking of Nir2 at ER-PM junctions [20].
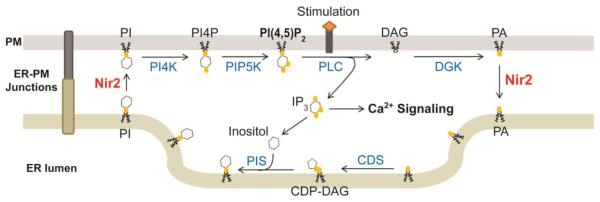
Moreover, recent studies demonstrated that Nir2 facilitates the clearance of PA from the PM during PI(4,5)P2-Ca2+ signaling, and the PITP domain of Nir2 mediates PA transfer in vitro [18, 19]. Thus, Nir2 is a PI-PA exchanger that shuttles lipids across the membranes at ER-PM junctions to complete the PI cycle. PITPNC1 can also transfer PA, indicating that PA transfer is a conserved function in Class II PITPs [154]. PM-to-ER PA transfer seems to be important for the PI cycle since inhibition of PA production at PM resulted in a reduced rate of PI resynthesis [26]. The fact that reconstitution of PITPα, a PI-PC exchanger, failed to rescue the defects caused by RdgBα mutation in Drosophila also suggests the importance of the PM-to-ER PA transfer in PI(4,5)P2 replenishment [19, 150]. Interestingly, PA generated by phospholipase D2 (PLD2) from PC failed to support CDP-DAG production in the PI cycle despite that it triggered Nir2 translocation to ER-PM junctions (unpublished data mentioned in [18]). This observation is consistent with a previous report showing that only PA derived from the action of PLC, but not that of PLD, was converted into PI in permeabilized neutrophils [158]. These findings suggest that Nir2 preferentially transfers a subgroup of PA molecules generated by PLC-induced PI(4,5)P2 hydrolysis. It is of importance to investigate whether Nir2 contributes to the enrichments of stearic acid (18:0) at the sn-1 position and arachidonic acid (20:4) at the sn-2 position in the DAG backbone of PI. A revised PI cycle showing that Nir2 supports PI(4,5)P2-Ca2+ signaling by facilitating PI-PA exchange at ER-PM junctions is depicted in summary of recent discoveries made by multiple groups [17-21] (Figure 5).
Figure 5.
updated model of the PI cycle with Nir2 mediating PI-PA exchange at ER-PM junctions. PI4K, PI-4 kinase; PIP5K, PI4P-5 kinase; CDS, CDP-DAG synthase; PIS, PI synthase.
Nir2 translocation to ER-PM junctions may be regulated by a self-inactivation mechanism involving binding to PA in the PM by the C-terminal LNS2 domain, and transferring PA in the PM to the ER by the N-terminal PITP domain. Thus, when PA levels at the PM drop below the critical concentration for LNS2 binding to the PM, Nir2 reverts to the cytosol and PI(4,5)P2 replenishment is subsequently inactivated. This elaborated mechanism is exemplified by recent work on OSBP, a multi-domain LTP that binds PI4P at the Golgi membrane via the N-terminal PH domain and transfers PI4P from the Golgi apparatus to the ER via its C-terminal lipid transfer domain to mediate sterol-PI4P exchange at ER-Golgi contact sites [35].
4.4 Functions of other Nir Proteins
Nir3, a homolog of Nir2, is expressed in most human tissues [151], although it may be mainly expressed in the retina and the dentate gyrus [159]. Similar to Nir2, Nir3 translocates to ER-PM junctions and mediates PI(4,5)P2 replenishment following receptor stimulation [17]. Intriguingly, analysis of domain-swapped chimeras of Nir2 and Nir3 demonstrated that Nir3 harbors weaker PITP activity toward PI(4,5)P2 replenishment, but is more sensitive to PA production than Nir2. These properties allow Nir3 to sense small amounts of PA production, and translocate to ER-PM junctions to maintain basal PM PI(4,5)P2 levels. By contrast, Nir2 mediates robust PI(4,5)P2 replenishment, but only following receptor stimulation. Thus, Nir2 and Nir3 work in tandem to support PI(4,5)P2 homeostasis in response to different physiological conditions. These observations may explain why reconstitution of Nir2, but not Nir3, fully rescued the defects in RdgBα mutant flies [159, 160], since photoreceptors are subjected to intense light stimulation and require ample amounts of PI that would be provided by the stronger PITP Nir2 to support the signaling machinery. In addition, the domain-swapping experiment also indicated that the PITP domain and the ER-PM junction targeting motifs work primarily as modules. It will be important to identify the residues responsible for the substantial differences in activities of Nir2 and Nir3.
Besides Nir2 and Nir3, another Nir protein called Nir1 (PITPNM3) has been identified. Nir1 contains the ER- and the PM-targeting motifs but lacks the PITP domain (Figure 4B) [151, 161]. Thus, Nir1 may serve as an ER-PM tethering protein at ER-PM junctions. The functional significance of Nir1 has been demonstrated in a human disease called autosomal dominant cone dystrophy [162]. Patients with a mutation in the gene encoding Nir1 have defective color vision, low visual acuity, and abnormal cone responses. In addition, Nir1 has been shown to bind to CCL18 to promote the phosphorylation of Akt, and its expression is associated with lymph node and distant metastasis in patients with invasive ductal carcinoma [163]. In RdgBα mutant flies, reconstitution of Nir1 slowed the retina degeneration phenotype, but failed to restore photo-transduction [161]. It is of interest to further examine the subcellular localization and the functions of Nir1.
Previous studies have shown that Nir proteins bind to Ca2+ but not Mg2+ via their acidic regions [151]. Notably, the VAP-binding FFAT motif resides in this acidic region. This raises interesting questions of whether activation of Nir2 and Nir3 requires Ca2+ binding to the acidic regions, and whether Ca2+ binding affects the ER targeting via the VAP-FFAT interaction. Answers to these questions may further expand our understanding of the crosstalk between Ca2+ and PI(4,5)P2 at ER-PM junctions. In addition, Nir proteins were identified as PYK2-binding partners, suggesting a correlation of ER-PM junctions and receptor signaling. Moreover, the importance of Nir2 at the organismal level remains unclear because of contradictory results derived from Nir2-deficient mice [164, 165]. Further studies are needed to resolve this controversy. Finally, mice lacking Nir3 are fertile and appear healthy [164]. Generating mice with deficiencies in both Nir2 and Nir3 is likely to reveal the importance of these functional redundant PITPs during development.
5. Concluding Remarks
The PI(4,5)P2-Ca2+ signaling system is fundamental for cells to respond to extracellular stimuli. Inspired by the initial observation of the “phospholipid effect” in 1953, tremendous research efforts eventually established the framework of the PI(4,5)P2-Ca2+ signaling system in 1983. Since then, signaling events and cellular functions activated by the PI(4,5)P2-Ca2+ signaling system have been studied extensively. Nonetheless, the molecular mechanisms underlying the feedback control that resets this signaling system for further activation and maintain PI(4,5)P2/Ca2+ homeostasis have remained unresolved until recently. In the past decade, the discovery of STIM1 that mediates SOCE by its dynamic translocation to ER-PM junctions has revolutionized our understanding of the feedback regulation of Ca2+ during PI(4,5)P2-Ca2+ signaling. Moreover, the identification of E-Syt1 and Nir2 that dynamically translocate to ER-PM junctions has revealed the feedback mechanisms maintaining PM PI(4,5)P2 homeostasis during PI(4,5)P2-Ca2+ signaling. The feedback control of the PI(4,5)P2-Ca2+ signaling system mediated by STIM1, E-Syt1, and Nir2 at ER-PM junctions starts quickly following receptor stimulation and persists during PI(4,5)P2-Ca2+ signaling. Remarkably, STIM1, E-Syt1, and Nir2 translocate to ER-PM junctions during PI(4,5)P2-Ca2+ signaling to mediate their functions in response to signals generated in different subcellular compartments: ER Ca2+ depletion for STIM1, cytosolic Ca2+ increase for E-Syt1, and PA production at the PM for Nir2. These three proteins all have homologs that target to ER-PM junctions by mechanisms conserved within their protein families. Notably, STIM2 and Nir3 have been shown to work in tandem with their homologs STIM1 and Nir2, respectively, at ER-PM junctions to achieve distinct levels of feedback under different physiological states. Further understanding of these three families of proteins as well as ER-PM junctions, the hubs for the homeostatic regulation of PI(4,5)P2-Ca2+ signaling, will shed light on our knowledge of other membrane junctions/contact sites and the spatiotemporal regulation of cell signaling.
Highlights.
Key discoveries established the PI(4,5)P2-Ca2+ signaling system are summarized
The PI cycle and SOCE mediate homeostatic regulation of the PI(4,5)P2-Ca2+ signaling system.
SOCE is mediated by STIM1-Orai1interaction at ER-PM junctions.
The PI cycle requires Nir2-mediated inter-organelle PI-PA exchange at ER-PM junctions
ER-PM junctions are hubs for homeostatic regulation of the PI(4,5)P2-Ca2+ signaling system
Acknowledgement
The authors’ research is supported by NIH grant 1 R01 GM113079-01A1 and Welch foundation grant I-1789. Jen Liou is a Sowell Family Scholar in Medical Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Balla T. Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiological reviews. 2013;93:1019–1137. doi: 10.1152/physrev.00028.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Hilgemann DW, Feng S, Nasuhoglu C. The complex and intriguing lives of PIP2 with ion channels and transporters. Science's STKE : signal transduction knowledge environment. 2001;2001:re19. doi: 10.1126/stke.2001.111.re19. [DOI] [PubMed] [Google Scholar]
- [3].Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- [4].Staiano L, De Leo MG, Persico M, De Matteis MA. Mendelian disorders of PI metabolizing enzymes. Biochim Biophys Acta. 2015;1851:867–881. doi: 10.1016/j.bbalip.2014.12.001. [DOI] [PubMed] [Google Scholar]
- [5].Berridge MJ, Bootman MD, Roderick HL. Calcium signalling: dynamics, homeostasis and remodelling. Nat Rev Mol Cell Biol. 2003;4:517–529. doi: 10.1038/nrm1155. [DOI] [PubMed] [Google Scholar]
- [6].Berridge MJ. Inositol trisphosphate and calcium signalling mechanisms. Biochim Biophys Acta. 2009;1793:933–940. doi: 10.1016/j.bbamcr.2008.10.005. [DOI] [PubMed] [Google Scholar]
- [7].Putney JW., Jr. A model for receptor-regulated calcium entry. Cell Calcium. 1986;7:1–12. doi: 10.1016/0143-4160(86)90026-6. [DOI] [PubMed] [Google Scholar]
- [8].Prakriya M, Lewis RS. Store-Operated Calcium Channels. Physiological reviews. 2015;95:1383–1436. doi: 10.1152/physrev.00020.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Liou J, Kim ML, Heo WD, Jones JT, Myers JW, Ferrell JE, Jr., Meyer T. STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr Biol. 2005;15:1235–1241. doi: 10.1016/j.cub.2005.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wu MM, Buchanan J, Luik RM, Lewis RS. Ca2+ store depletion causes STIM1 to accumulate in ER regions closely associated with the plasma membrane. J Cell Biol. 2006;174:803–813. doi: 10.1083/jcb.200604014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Carrasco S, Meyer T. STIM proteins and the endoplasmic reticulum-plasma membrane junctions. Annu Rev Biochem. 2011;80:973–1000. doi: 10.1146/annurev-biochem-061609-165311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Stefan CJ, Manford AG, Emr SD. ER-PM connections: sites of information transfer and inter-organelle communication. Curr Opin Cell Biol. 2013;25:434–442. doi: 10.1016/j.ceb.2013.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Henne WM, Liou J, Emr SD. Molecular mechanisms of inter-organelle ER-PM contact sites. Curr Opin Cell Biol. 2015;35:123–130. doi: 10.1016/j.ceb.2015.05.001. [DOI] [PubMed] [Google Scholar]
- [14].Hokin MR, Hokin LE. Enzyme secretion and the incorporation of P32 into phospholipides of pancreas slices. J Biol Chem. 1953;203:967–977. [PubMed] [Google Scholar]
- [15].Hokin LE, Hokin MR. Effects of acetylcholine on the turnover of phosphoryl units in individual phospholipids of pancreas slices and brain cortex slices. Biochim Biophys Acta. 1955;18:102–110. doi: 10.1016/0006-3002(55)90013-5. [DOI] [PubMed] [Google Scholar]
- [16].Hokin LE. Receptors and phosphoinositide-generated second messengers. Annu Rev Biochem. 1985;54:205–235. doi: 10.1146/annurev.bi.54.070185.001225. [DOI] [PubMed] [Google Scholar]
- [17].Chang CL, Liou J. Phosphatidylinositol 4,5-Bisphosphate Homeostasis Regulated by Nir2 and Nir3 Proteins at Endoplasmic Reticulum-Plasma Membrane Junctions. J Biol Chem. 2015;290:14289–14301. doi: 10.1074/jbc.M114.621375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kim YJ, Guzman-Hernandez ML, Wisniewski E, Balla T. Phosphatidylinositol-Phosphatidic Acid Exchange by Nir2 at ER-PM Contact Sites Maintains Phosphoinositide Signaling Competence. Developmental cell. 2015;33:549–561. doi: 10.1016/j.devcel.2015.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yadav S, Garner K, Georgiev P, Li M, Gomez-Espinosa E, Panda A, Mathre S, Okkenhaug H, Cockcroft S, Raghu P. RDGBalpha, a PtdIns-PtdOH transfer protein, regulates G-protein-coupled PtdIns(4,5)P2 signalling during Drosophila phototransduction. Journal of cell science. 2015;128:3330–3344. doi: 10.1242/jcs.173476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Chang CL, Hsieh TS, Yang TT, Rothberg KG, Azizoglu DB, Volk E, Liao JC, Liou J. Feedback regulation of receptor-induced Ca2+ signaling mediated by E-Syt1 and Nir2 at endoplasmic reticulum-plasma membrane junctions. Cell reports. 2013;5:813–825. doi: 10.1016/j.celrep.2013.09.038. [DOI] [PubMed] [Google Scholar]
- [21].Kim S, Kedan A, Marom M, Gavert N, Keinan O, Selitrennik M, Laufman O, Lev S. The phosphatidylinositol-transfer protein Nir2 binds phosphatidic acid and positively regulates phosphoinositide signalling. EMBO reports. 2013;14:891–899. doi: 10.1038/embor.2013.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Michell RH. First came the link between phosphoinositides and Ca2+ signalling, and then a deluge of other phosphoinositide functions. Cell Calcium. 2009;45:521–526. doi: 10.1016/j.ceca.2009.03.007. [DOI] [PubMed] [Google Scholar]
- [23].Epand RM. Recognition of polyunsaturated acyl chains by enzymes acting on membrane lipids. Biochim Biophys Acta. 2012;1818:957–962. doi: 10.1016/j.bbamem.2011.07.018. [DOI] [PubMed] [Google Scholar]
- [24].Kim YJ, Guzman-Hernandez ML, Balla T. A highly dynamic ER-derived phosphatidylinositol-synthesizing organelle supplies phosphoinositides to cellular membranes. Dev Cell. 2011;21:813–824. doi: 10.1016/j.devcel.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Agranoff BW, Bradley RM, Brady RO. The enzymatic synthesis of inositol phosphatide. J Biol Chem. 1958;233:1077–1083. [PubMed] [Google Scholar]
- [26].Cockcroft S, Garner K. Potential role for phosphatidylinositol transfer protein (PITP) family in lipid transfer during phospholipase C signalling. Advances in biological regulation. 2013;53:280–291. doi: 10.1016/j.jbior.2013.07.007. [DOI] [PubMed] [Google Scholar]
- [27].Hammond GR, Balla T. Polyphosphoinositide binding domains: Key to inositol lipid biology. Biochim Biophys Acta. 2015;1851:746–758. doi: 10.1016/j.bbalip.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Folch J. Complete fractionation of brain cephalin; isolation from it of phosphatidyl serine, phosphatidyl ethanolamine, and diphosphoinositide. J Biol Chem. 1949;177:497–504. [PubMed] [Google Scholar]
- [29].Dittmer JC, Dawson RM. The isolation of a new lipid, triphosphoinositide, and monophosphoinositide from ox brain. Biochem J. 1961;81:535–540. doi: 10.1042/bj0810535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Tomlinson RV, Ballou CE. Complete characterization of the myo-inositol polyphosphates from beef brain phosphoinositide. J Biol Chem. 1961;236:1902–1906. [PubMed] [Google Scholar]
- [31].Hammond GR, Machner MP, Balla T. A novel probe for phosphatidylinositol 4-phosphate reveals multiple pools beyond the Golgi. J Cell Biol. 2014;205:113–126. doi: 10.1083/jcb.201312072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].von Filseck J. Moser, Vanni S, Mesmin B, Antonny B, Drin G. A phosphatidylinositol-4-phosphate powered exchange mechanism to create a lipid gradient between membranes. Nat Commun. 2015;6:6671. doi: 10.1038/ncomms7671. [DOI] [PubMed] [Google Scholar]
- [33].Chung J, Torta F, Masai K, Lucast L, Czapla H, Tanner LB, Narayanaswamy P, Wenk MR, Nakatsu F, De Camilli P, INTRACELLULAR TRANSPORT PI4P/phosphatidylserine countertransport at ORP5- and ORP8-mediated ER-plasma membrane contacts. Science. 2015;349:428–432. doi: 10.1126/science.aab1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].von Filseck J. Moser, Copic A, Delfosse V, Vanni S, Jackson CL, Bourguet W, Drin G, INTRACELLULAR TRANSPORT Phosphatidylserine transport by ORP/Osh proteins is driven by phosphatidylinositol 4-phosphate. Science. 2015;349:432–436. doi: 10.1126/science.aab1346. [DOI] [PubMed] [Google Scholar]
- [35].Mesmin B, Bigay J, von Filseck J. Moser, Lacas-Gervais S, Drin G, Antonny B. A four-step cycle driven by PI(4)P hydrolysis directs sterol/PI(4)P exchange by the ER-Golgi tether OSBP. Cell. 2013;155:830–843. doi: 10.1016/j.cell.2013.09.056. [DOI] [PubMed] [Google Scholar]
- [36].Nakatsu F, Baskin JM, Chung J, Tanner LB, Shui G, Lee SY, Pirruccello M, Hao M, Ingolia NT, Wenk MR, De Camilli P. PtdIns4P synthesis by PI4KIIIalpha at the plasma membrane and its impact on plasma membrane identity. The Journal of cell biology. 2012;199:1003–1016. doi: 10.1083/jcb.201206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Hokin LE, Hokin MR. Phosphoinositides and protein secretion in pancreas slices. J Biol Chem. 1958;233:805–810. [PubMed] [Google Scholar]
- [38].Hokin MR, Hokin LE. Interconversions of phosphatidylinositol and phosphatidic acid involved in the responses to acetylcholine in the salt gland. Wiley; New York: 1964. [Google Scholar]
- [39].Michell RH. Inositol phospholipids and cell surface receptor function. Biochim Biophys Acta. 1975;415:81–47. doi: 10.1016/0304-4157(75)90017-9. [DOI] [PubMed] [Google Scholar]
- [40].Lapetina EG, Michell RH. A membrane-bound activity catalysing phosphatidylinositol breakdown to 1,2-diacylglycerol, D-myoinositol 1:2-cyclic phosphate an D-myoinositol 1-phosphate. Properties and subcellular distribution in rat cerebral cortex. The Biochemical journal. 1973;131:433–442. doi: 10.1042/bj1310433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Wirtz KW, Zilversmit DB. Exchange of phospholipids between liver mitochondria and microsomes in vitro. J Biol Chem. 1968;243:3596–3602. [PubMed] [Google Scholar]
- [42].Fain JN, Berridge MJ. Relationship between hormonal activation of phosphatidylinositol hydrolysis, fluid secretion and calcium flux in the blowfly salivary gland. Biochem J. 1979;178:45–58. doi: 10.1042/bj1780045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Fain JN, Berridge MJ. Relationship between phosphatidylinositol synthesis and recovery of 5-hydroxytryptamine-responsive Ca2+ flux in blowfly salivary glands. Biochem J. 1979;180:655–661. doi: 10.1042/bj1800655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Brockerhoff H, Ballou CE. Phosphate incorporation in brain phosphionositides. J Biol Chem. 1962;237:49–52. [PubMed] [Google Scholar]
- [45].Thompson W, Dawson RM. The triphosphoinositide phosphodiesterase of brain tissue. Biochem J. 1964;91:237–243. doi: 10.1042/bj0910237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Santiago-Calvo E, Mule S, Redman CM, Hokin MR, Hokin LE. The Chromatographic Separation of Polyphosphoinositides and Studies on Their Turnover in Various Tissues. Biochim Biophys Acta. 1964;84:550–562. doi: 10.1016/0926-6542(64)90125-8. [DOI] [PubMed] [Google Scholar]
- [47].Durell J, Garland JT. Acetylcholine-stimulated phosphodiesteratic cleavage of phosphoinositides: hypothetical role in membrane depolarization. Ann N Y Acad Sci. 1969;165:743–754. [PubMed] [Google Scholar]
- [48].Abdel-Latif AA, Akhtar RA, Hawthorne JN. Acetylcholine increases the breakdown of triphosphoinositide of rabbit iris muscle prelabelled with [32P] phosphate. Biochem J. 1977;162:61–73. doi: 10.1042/bj1620061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Downes P, Michell RH. Phosphatidylinositol 4-phosphate and phosphatidylinositol 4,5-bisphosphate: lipids in search of a function. Cell Calcium. 1982;3:467–502. doi: 10.1016/0143-4160(82)90031-8. [DOI] [PubMed] [Google Scholar]
- [50].Creba JA, Downes CP, Hawkins PT, Brewster G, Michell RH, Kirk CJ. Rapid breakdown of phosphatidylinositol 4-phosphate and phosphatidylinositol 4,5-bisphosphate in rat hepatocytes stimulated by vasopressin and other Ca2+-mobilizing hormones. Biochem J. 1983;212:733–747. doi: 10.1042/bj2120733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Berridge MJ. Rapid accumulation of inositol trisphosphate reveals that agonists hydrolyse polyphosphoinositides instead of phosphatidylinositol. Biochem J. 1983;212:849–858. doi: 10.1042/bj2120849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Berridge MJ, Dawson RM, Downes CP, Heslop JP, Irvine RF. Changes in the levels of inositol phosphates after agonist-dependent hydrolysis of membrane phosphoinositides. Biochem J. 1983;212:473–482. doi: 10.1042/bj2120473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Streb H, Irvine RF, Berridge MJ, Schulz I. Release of Ca2+ from a nonmitochondrial intracellular store in pancreatic acinar cells by inositol-1,4,5-trisphosphate. Nature. 1983;306:67–69. doi: 10.1038/306067a0. [DOI] [PubMed] [Google Scholar]
- [54].Ross CA, Meldolesi J, Milner TA, Satoh T, Supattapone S, Snyder SH. Inositol 1,4,5-trisphosphate receptor localized to endoplasmic reticulum in cerebellar Purkinje neurons. Nature. 1989;339:468–470. doi: 10.1038/339468a0. [DOI] [PubMed] [Google Scholar]
- [55].Furuichi T, Yoshikawa S, Miyawaki A, Wada K, Maeda N, Mikoshiba K. Primary structure and functional expression of the inositol 1,4,5-trisphosphate-binding protein P400. Nature. 1989;342:32–38. doi: 10.1038/342032a0. [DOI] [PubMed] [Google Scholar]
- [56].Mignery GA, Sudhof TC, Takei K, Camilli P. De. Putative receptor for inositol 1,4,5-trisphosphate similar to ryanodine receptor. Nature. 1989;342:192–195. doi: 10.1038/342192a0. [DOI] [PubMed] [Google Scholar]
- [57].Berridge MJ, Lipp P, Bootman MD. The versatility and universality of calcium signalling. Nat Rev Mol Cell Biol. 2000;1:11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- [58].Putney JW., Jr. Stimulus-permeability coupling: role of calcium in the receptor regulation of membrane permeability. Pharmacol Rev. 1978;30:209–245. [PubMed] [Google Scholar]
- [59].Putney JW, Jr., Poggioli J, Weiss SJ. Receptor regulation of calcium release and calcium permeability in parotid gland cells. Philos Trans R Soc Lond B Biol Sci. 1981;296:37–45. doi: 10.1098/rstb.1981.0169. [DOI] [PubMed] [Google Scholar]
- [60].Thastrup O, Dawson AP, Scharff O, Foder B, Cullen PJ, Drobak BK, Bjerrum PJ, Christensen SB, Hanley MR. Thapsigargin, a novel molecular probe for studying intracellular calcium release and storage. Agents and actions. 1989;27:17–23. doi: 10.1007/BF02222186. [DOI] [PubMed] [Google Scholar]
- [61].Takemura H, Hughes AR, Thastrup O, Putney JW., Jr. Activation of calcium entry by the tumor promoter thapsigargin in parotid acinar cells. Evidence that an intracellular calcium pool and not an inositol phosphate regulates calcium fluxes at the plasma membrane. J Biol Chem. 1989;264:12266–12271. [PubMed] [Google Scholar]
- [62].Putney JW., Jr. Capacitative calcium entry revisited. Cell Calcium. 1990;11:611–624. doi: 10.1016/0143-4160(90)90016-n. [DOI] [PubMed] [Google Scholar]
- [63].Lewis RS, Cahalan MD. Mitogen-induced oscillations of cytosolic Ca2+ and transmembrane Ca2+ current in human leukemic T cells. Cell regulation. 1989;1:99–112. doi: 10.1091/mbc.1.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Penner R, Matthews G, Neher E. Regulation of calcium influx by second messengers in rat mast cells. Nature. 1988;334:499–504. doi: 10.1038/334499a0. [DOI] [PubMed] [Google Scholar]
- [65].Hoth M, Penner R. Depletion of intracellular calcium stores activates a calcium current in mast cells. Nature. 1992;355:353–356. doi: 10.1038/355353a0. [DOI] [PubMed] [Google Scholar]
- [66].Lewis RS. Store-operated calcium channels: new perspectives on mechanism and function. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a003970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Roos J, DiGregorio PJ, Yeromin AV, Ohlsen K, Lioudyno M, Zhang S, Safrina O, Kozak JA, Wagner SL, Cahalan MD, Velicelebi G, Stauderman KA. STIM1, an essential and conserved component of store-operated Ca2+ channel function. J Cell Biol. 2005;169:435–445. doi: 10.1083/jcb.200502019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Feske S, Gwack Y, Prakriya M, Srikanth S, Puppel SH, Tanasa B, Hogan PG, Lewis RS, Daly M, Rao A. A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function. Nature. 2006;441:179–185. doi: 10.1038/nature04702. [DOI] [PubMed] [Google Scholar]
- [69].Zhang SL, Yeromin AV, Zhang XH, Yu Y, Safrina O, Penna A, Roos J, Stauderman KA, Cahalan MD. Genome-wide RNAi screen of Ca(2+) influx identifies genes that regulate Ca(2+) release-activated Ca(2+) channel activity. Proc Natl Acad Sci U S A. 2006;103:9357–9362. doi: 10.1073/pnas.0603161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Vig M, Peinelt C, Beck A, Koomoa DL, Rabah D, Koblan-Huberson M, Kraft S, Turner H, Fleig A, Penner R, Kinet JP. CRACM1 is a plasma membrane protein essential for store-operated Ca2+ entry. Science. 2006;312:1220–1223. doi: 10.1126/science.1127883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Yeromin AV, Zhang SL, Jiang W, Yu Y, Safrina O, Cahalan MD. Molecular identification of the CRAC channel by altered ion selectivity in a mutant of Orai. Nature. 2006;443:226–229. doi: 10.1038/nature05108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Prakriya M, Feske S, Gwack Y, Srikanth S, Rao A, Hogan PG. Orai1 is an essential pore subunit of the CRAC channel. Nature. 2006;443:230–233. doi: 10.1038/nature05122. [DOI] [PubMed] [Google Scholar]
- [73].Peinelt C, Vig M, Koomoa DL, Beck A, Nadler MJ, Koblan-Huberson M, Lis A, Fleig A, Penner R, Kinet JP. Amplification of CRAC current by STIM1 and CRACM1 (Orai1) Nat Cell Biol. 2006;8:771–773. doi: 10.1038/ncb1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Mercer JC, Dehaven WI, Smyth JT, Wedel B, Boyles RR, Bird GS, Putney JW., Jr. Large store-operated calcium selective currents due to co-expression of Orai1 or Orai2 with the intracellular calcium sensor, Stim1. J Biol Chem. 2006;281:24979–24990. doi: 10.1074/jbc.M604589200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Soboloff J, Spassova MA, Tang XD, Hewavitharana T, Xu W, Gill DL. Orai1 and STIM reconstitute store-operated calcium channel function. J Biol Chem. 2006;281:20661–20665. doi: 10.1074/jbc.C600126200. [DOI] [PubMed] [Google Scholar]
- [76].Feske S, Prakriya M. Conformational dynamics of STIM1 activation. Nat Struct Mol Biol. 2013;20:918–919. doi: 10.1038/nsmb.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Park CY, Hoover PJ, Mullins FM, Bachhawat P, Covington ED, Raunser S, Walz T, Garcia KC, Dolmetsch RE, Lewis RS. STIM1 clusters and activates CRAC channels via direct binding of a cytosolic domain to Orai1. Cell. 2009;136:876–890. doi: 10.1016/j.cell.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Yuan JP, Zeng W, Dorwart MR, Choi YJ, Worley PF, Muallem S. SOAR and the polybasic STIM1 domains gate and regulate Orai channels. Nat Cell Biol. 2009;11:337–343. doi: 10.1038/ncb1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Kawasaki T, Lange I, Feske S. A minimal regulatory domain in the C terminus of STIM1 binds to and activates ORAI1 CRAC channels. Biochem Biophys Res Commun. 2009;385:49–54. doi: 10.1016/j.bbrc.2009.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Liou J, Fivaz M, Inoue T, Meyer T. Live-cell imaging reveals sequential oligomerization and local plasma membrane targeting of stromal interaction molecule 1 after Ca2+ store depletion. Proc Natl Acad Sci U S A. 2007;104:9301–9306. doi: 10.1073/pnas.0702866104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Zhou Y, Srinivasan P, Razavi S, Seymour S, Meraner P, Gudlur A, Stathopulos PB, Ikura M, Rao A, Hogan PG. Initial activation of STIM1, the regulator of store-operated calcium entry. Nat Struct Mol Biol. 2013;20:973–981. doi: 10.1038/nsmb.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Ercan E, Momburg F, Engel U, Temmerman K, Nickel W, Seedorf M. A conserved, lipid-mediated sorting mechanism of yeast Ist2 and mammalian STIM proteins to the peripheral ER. Traffic. 2009;10:1802–1818. doi: 10.1111/j.1600-0854.2009.00995.x. [DOI] [PubMed] [Google Scholar]
- [83].Korzeniowski MK, Popovic MA, Szentpetery Z, Varnai P, Stojilkovic SS, Balla T. Dependence of STIM1/Orai1-mediated calcium entry on plasma membrane phosphoinositides. J Biol Chem. 2009;284:21027–21035. doi: 10.1074/jbc.M109.012252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Walsh CM, Chvanov M, Haynes LP, Petersen OH, Tepikin AV, Burgoyne RD. Role of phosphoinositides in STIM1 dynamics and store-operated calcium entry. Biochem J. 2010;425:159–168. doi: 10.1042/BJ20090884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Wu MM, Covington ED, Lewis RS. Single-molecule analysis of diffusion and trapping of STIM1 and Orai1 at endoplasmic reticulum-plasma membrane junctions. Molecular biology of the cell. 2014;25:3672–3685. doi: 10.1091/mbc.E14-06-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Brandman O, Liou J, Park WS, Meyer T. STIM2 is a feedback regulator that stabilizes basal cytosolic and endoplasmic reticulum Ca2+ levels. Cell. 2007;131:1327–1339. doi: 10.1016/j.cell.2007.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Krapivinsky G, Krapivinsky L, Stotz SC, Manasian Y, Clapham DE. POST, partner of stromal interaction molecule 1 (STIM1), targets STIM1 to multiple transporters. Proc Natl Acad Sci U S A. 2011;108:19234–19239. doi: 10.1073/pnas.1117231108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Fujii Y, Shiota M, Ohkawa Y, Baba A, Wanibuchi H, Kinashi T, Kurosaki T, Baba Y. Surf4 modulates STIM1-dependent calcium entry. Biochem Biophys Res Commun. 2012;422:615–620. doi: 10.1016/j.bbrc.2012.05.037. [DOI] [PubMed] [Google Scholar]
- [89].Palty R, Raveh A, Kaminsky I, Meller R, Reuveny E. SARAF inactivates the store operated calcium entry machinery to prevent excess calcium refilling. Cell. 2012;149:425–438. doi: 10.1016/j.cell.2012.01.055. [DOI] [PubMed] [Google Scholar]
- [90].Miao Y, Miner C, Zhang L, Hanson PI, Dani A, Vig M. An essential and NSF independent role for alpha-SNAP in store-operated calcium entry. eLife. 2013;2:e00802. doi: 10.7554/eLife.00802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Srikanth S, Jew M, Kim KD, Yee MK, Abramson J, Gwack Y. Junctate is a Ca2+-sensing structural component of Orai1 and stromal interaction molecule 1 (STIM1) Proc Natl Acad Sci U S A. 2012;109:8682–8687. doi: 10.1073/pnas.1200667109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Jing J, He L, Sun A, Quintana A, Ding Y, Ma G, Tan P, Liang X, Zheng X, Chen L, Shi X, Zhang SL, Zhong L, Huang Y, Dong MQ, Walker CL, Hogan PG, Wang Y, Zhou Y. Proteomic mapping of ER-PM junctions identifies STIMATE as a regulator of Ca influx. Nat Cell Biol. 2015 doi: 10.1038/ncb3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Sharma S, Quintana A, Findlay GM, Mettlen M, Baust B, Jain M, Nilsson R, Rao A, Hogan PG. An siRNA screen for NFAT activation identifies septins as coordinators of store-operated Ca2+ entry. Nature. 2013;499:238–242. doi: 10.1038/nature12229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Srikanth S, Jung HJ, Kim KD, Souda P, Whitelegge J, Gwack Y. A novel EF-hand protein, CRACR2A, is a cytosolic Ca2+ sensor that stabilizes CRAC channels in T cells. Nat Cell Biol. 2010;12:436–446. doi: 10.1038/ncb2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Porter KR, Palade GE. Studies on the endoplasmic reticulum. III. Its form and distribution in striated muscle cells. The Journal of biophysical and biochemical cytology. 1957;3:269–300. doi: 10.1083/jcb.3.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Rosenbluth J. Subsurface cisterns and their relationship to the neuronal plasma membrane. J Cell Biol. 1962;13:405–421. doi: 10.1083/jcb.13.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Henkart M, Landis DM, Reese TS. Similarity of junctions between plasma membranes and endoplasmic reticulum in muscle and neurons. J Cell Biol. 1976;70:338–347. doi: 10.1083/jcb.70.2.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Gardiner DM, Grey RD. Membrane junctions in Xenopus eggs: their distribution suggests a role in calcium regulation. J Cell Biol. 1983;96:1159–1163. doi: 10.1083/jcb.96.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Orci L, Ravazzola M, Coadic M. Le, Shen WW, Demaurex N, Cosson P. From the Cover: STIM1-induced precortical and cortical subdomains of the endoplasmic reticulum. Proc Natl Acad Sci U S A. 2009;106:19358–19362. doi: 10.1073/pnas.0911280106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Lur G, Haynes LP, Prior IA, Gerasimenko OV, Feske S, Petersen OH, Burgoyne RD, Tepikin AV. Ribosome-free terminals of rough ER allow formation of STIM1 puncta and segregation of STIM1 from IP(3) receptors. Curr Biol. 2009;19:1648–1653. doi: 10.1016/j.cub.2009.07.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Perni S, Dynes JL, Yeromin AV, Cahalan MD, Franzini-Armstrong C. Nanoscale patterning of STIM1 and Orai1 during store-operated Ca2+ entry. Proc Natl Acad Sci U S A. 2015;112:E5533–5542. doi: 10.1073/pnas.1515606112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Varnai P, Toth B, Toth DJ, Hunyady L, Balla T. Visualization and manipulation of plasma membrane-endoplasmic reticulum contact sites indicates the presence of additional molecular components within the STIM1-Orai1 Complex. J Biol Chem. 2007;282:29678–29690. doi: 10.1074/jbc.M704339200. [DOI] [PubMed] [Google Scholar]
- [103].Tsai FC, Seki A, Yang HW, Hayer A, Carrasco S, Malmersjo S, Meyer T. A polarized Ca2+, diacylglycerol and STIM1 signalling system regulates directed cell migration. Nature cell biology. 2014;16:133–144. doi: 10.1038/ncb2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Franzini-Armstrong C, Jorgensen AO. Structure and development of E-C coupling units in skeletal muscle. Annual review of physiology. 1994;56:509–534. doi: 10.1146/annurev.ph.56.030194.002453. [DOI] [PubMed] [Google Scholar]
- [105].West M, Zurek N, Hoenger A, Voeltz GK. A 3D analysis of yeast ER structure reveals how ER domains are organized by membrane curvature. J Cell Biol. 2011;193:333–346. doi: 10.1083/jcb.201011039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Loewen CJ, Young BP, Tavassoli S, Levine TP. Inheritance of cortical ER in yeast is required for normal septin organization. J Cell Biol. 2007;179:467–483. doi: 10.1083/jcb.200708205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Vihtelic TS, Goebl M, Milligan S, O'Tousa JE, Hyde DR. Localization of Drosophila retinal degeneration B, a membrane-associated phosphatidylinositol transfer protein. J Cell Biol. 1993;122:1013–1022. doi: 10.1083/jcb.122.5.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Gatta AT, Wong LH, Sere YY, Calderon-Norena DM, Cockcroft S, Menon AK, Levine TP. A new family of StART domain proteins at membrane contact sites has a role in ER-PM sterol transport. eLife. 2015;4 doi: 10.7554/eLife.07253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Stefan CJ, Manford AG, Baird D, Yamada-Hanff J, Mao Y, Emr SD. Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell. 2011;144:389–401. doi: 10.1016/j.cell.2010.12.034. [DOI] [PubMed] [Google Scholar]
- [110].Smyth JT, Dehaven WI, Bird GS, Putney JW., Jr. Ca2+-store-dependent and -independent reversal of Stim1 localization and function. J Cell Sci. 2008;121:762–772. doi: 10.1242/jcs.023903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Manford AG, Stefan CJ, Yuan HL, Macgurn JA, Emr SD. ER-to-plasma membrane tethering proteins regulate cell signaling and ER morphology. Dev Cell. 2012;23:1129–1140. doi: 10.1016/j.devcel.2012.11.004. [DOI] [PubMed] [Google Scholar]
- [112].Giordano F, Saheki Y, Idevall-Hagren O, Colombo SF, Pirruccello M, Milosevic I, Gracheva EO, Bagriantsev SN, Borgese N, De Camilli P. PI(4,5)P(2)-dependent and Ca(2+)-regulated ER-PM interactions mediated by the extended synaptotagmins. Cell. 2013;153:1494–1509. doi: 10.1016/j.cell.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Min SW, Chang WP, Sudhof TC. E-Syts, a family of membranous Ca2+-sensor proteins with multiple C2 domains. Proc Natl Acad Sci U S A. 2007;104:3823–3828. doi: 10.1073/pnas.0611725104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Kopec KO, Alva V, Lupas AN. Homology of SMP domains to the TULIP superfamily of lipid-binding proteins provides a structural basis for lipid exchange between ER and mitochondria. Bioinformatics. 2010;26:1927–1931. doi: 10.1093/bioinformatics/btq326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Schauder CM, Wu X, Saheki Y, Narayanaswamy P, Torta F, Wenk MR, De Camilli P, Reinisch KM. Structure of a lipid-bound extended synaptotagmin indicates a role in lipid transfer. Nature. 2014;510:552–555. doi: 10.1038/nature13269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Herdman C, Tremblay MG, Mishra PK, Moss T. Loss of Extended Synaptotagmins ESyt2 and ESyt3 does not affect mouse development or viability, but in vitro cell migration and survival under stress are affected. Cell Cycle. 2014;13:2616–2625. doi: 10.4161/15384101.2014.943573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Jean S, Tremblay MG, Herdman C, Guillou F, Moss T. The endocytic adapter E-Syt2 recruits the p21 GTPase activated kinase PAK1 to mediate actin dynamics and FGF signalling. Biol Open. 2012;1:731–738. doi: 10.1242/bio.2012968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Tremblay MG, Herdman C, Guillou F, Mishra PK, Baril J, Bellenfant S, Moss T. Extended Synaptotagmin Interaction with the Fibroblast Growth Factor Receptor Depends on Receptor Conformation, Not Catalytic Activity. The Journal of biological chemistry. 2015;290:16142–16156. doi: 10.1074/jbc.M115.656918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Fox PD, Haberkorn CJ, Weigel AV, Higgins JL, Akin EJ, Kennedy MJ, Krapf D, Tamkun MM. Plasma membrane domains enriched in cortical endoplasmic reticulum function as membrane protein trafficking hubs. Molecular biology of the cell. 2013;24:2703–2713. doi: 10.1091/mbc.E12-12-0895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Loewen CJ, Roy A, Levine TP. A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. The EMBO journal. 2003;22:2025–2035. doi: 10.1093/emboj/cdg201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Treves S, Franzini-Armstrong C, Moccagatta L, Arnoult C, Grasso C, Schrum A, Ducreux S, Zhu MX, Mikoshiba K, Girard T, Smida-Rezgui S, Ronjat M, Zorzato F. Junctate is a key element in calcium entry induced by activation of InsP3 receptors and/or calcium store depletion. J Cell Biol. 2004;166:537–548. doi: 10.1083/jcb.200404079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Treves S, Vukcevic M, Griesser J, Armstrong CF, Zhu MX, Zorzato F. Agonist-activated Ca2+ influx occurs at stable plasma membrane and endoplasmic reticulum junctions. J Cell Sci. 2010;123:4170–4181. doi: 10.1242/jcs.068387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Petkovic M, Jemaiel A, Daste F, Specht CG, Izeddin I, Vorkel D, Verbavatz JM, Darzacq X, Triller A, Pfenninger KH, Tareste D, Jackson CL, Galli T. The SNARE Sec22b has a non-fusogenic function in plasma membrane expansion. Nat Cell Biol. 2014;16:434–444. doi: 10.1038/ncb2937. [DOI] [PubMed] [Google Scholar]
- [124].Fernandez-Busnadiego R, Saheki Y, De Camilli P. Three-dimensional architecture of extended synaptotagmin-mediated endoplasmic reticulum-plasma membrane contact sites. Proc Natl Acad Sci U S A. 2015;112:E2004–2013. doi: 10.1073/pnas.1503191112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Idevall-Hagren O, Lu A, Xie B, De Camilli P. Triggered Ca2+ influx is required for extended synaptotagmin 1-induced ER-plasma membrane tethering. The EMBO journal. 2015;34:2291–2305. doi: 10.15252/embj.201591565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Xu J, Bacaj T, Zhou A, Tomchick DR, Sudhof TC, Rizo J. Structure and Ca(2)(+)-binding properties of the tandem C(2) domains of E-Syt2. Structure. 2014;22:269–280. doi: 10.1016/j.str.2013.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Maleth J, Choi S, Muallem S, Ahuja M. Translocation between PI(4,5)P2-poor and PI(4,5)P2-rich microdomains during store depletion determines STIM1 conformation and Orai1 gating. Nat Commun. 2014;5:5843. doi: 10.1038/ncomms6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Nasuhoglu C, Feng S, Mao Y, Shammat I, Yamamato M, Earnest S, Lemmon M, Hilgemann DW. Modulation of cardiac PIP2 by cardioactive hormones and other physiologically relevant interventions. American journal of physiology. Cell physiology. 2002;283:C223–234. doi: 10.1152/ajpcell.00486.2001. [DOI] [PubMed] [Google Scholar]
- [129].Holthuis JC, Levine TP. Lipid traffic: floppy drives and a superhighway. Nat Rev Mol Cell Biol. 2005;6:209–220. doi: 10.1038/nrm1591. [DOI] [PubMed] [Google Scholar]
- [130].Dickson EJ, Jensen JB, Hille B. Golgi and plasma membrane pools of PI(4)P contribute to plasma membrane PI(4,5)P2 and maintenance of KCNQ2/3 ion channel current. Proc Natl Acad Sci U S A. 2014;111:E2281–2290. doi: 10.1073/pnas.1407133111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Szentpetery Z, Varnai P, Balla T. Acute manipulation of Golgi phosphoinositides to assess their importance in cellular trafficking and signaling. Proc Natl Acad Sci U S A. 2010;107:8225–8230. doi: 10.1073/pnas.1000157107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Lev S. Non-vesicular lipid transport by lipid-transfer proteins and beyond. Nat Rev Mol Cell Biol. 2010;11:739–750. doi: 10.1038/nrm2971. [DOI] [PubMed] [Google Scholar]
- [133].Lalanne F, Ponsin G. Mechanism of the phospholipid transfer protein-mediated transfer of phospholipids from model lipid vesicles to high density lipoproteins. Biochim Biophys Acta. 2000;1487:82–91. doi: 10.1016/s1388-1981(00)00087-1. [DOI] [PubMed] [Google Scholar]
- [134].Levine T. Short-range intracellular trafficking of small molecules across endoplasmic reticulum junctions. Trends Cell Biol. 2004;14:483–490. doi: 10.1016/j.tcb.2004.07.017. [DOI] [PubMed] [Google Scholar]
- [135].Levine T, Loewen C. Inter-organelle membrane contact sites: through a glass, darkly. Curr Opin Cell Biol. 2006;18:371–378. doi: 10.1016/j.ceb.2006.06.011. [DOI] [PubMed] [Google Scholar]
- [136].Helmkamp GM, Jr., Harvey MS, Wirtz KW, Van Deenen LL. Phospholipid exchange between membranes. Purification of bovine brain proteins that preferentially catalyze the transfer of phosphatidylinositol. J Biol Chem. 1974;249:6382–6389. [PubMed] [Google Scholar]
- [137].Tilley SJ, Skippen A, Murray-Rust J, Swigart PM, Stewart A, Morgan CP, Cockcroft S, McDonald NQ. Structure-function analysis of human [corrected] phosphatidylinositol transfer protein alpha bound to phosphatidylinositol. Structure. 2004;12:317–326. doi: 10.1016/j.str.2004.01.013. [DOI] [PubMed] [Google Scholar]
- [138].Vordtriede PB, Doan CN, Tremblay JM, Helmkamp GM, Jr., Yoder MD. Structure of PITPbeta in complex with phosphatidylcholine: comparison of structure and lipid transfer to other PITP isoforms. Biochemistry. 2005;44:14760–14771. doi: 10.1021/bi051191r. [DOI] [PubMed] [Google Scholar]
- [139].de Brouwer AP, Versluis C, Westerman J, Roelofsen B, Heck AJ, Wirtz KW. Determination of the stability of the noncovalent phospholipid transfer protein-lipid complex by electrospray time-of-flight mass spectrometry. Biochemistry. 2002;41:8013–8018. doi: 10.1021/bi016055a. [DOI] [PubMed] [Google Scholar]
- [140].Carvou N, Holic R, Li M, Futter C, Skippen A, Cockcroft S. Phosphatidylinositol- and phosphatidylcholine-transfer activity of PITPbeta is essential for COPI-mediated retrograde transport from the Golgi to the endoplasmic reticulum. J Cell Sci. 2010;123:1262–1273. doi: 10.1242/jcs.061986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Shadan S, Holic R, Carvou N, Ee P, Li M, Murray-Rust J, Cockcroft S. Dynamics of lipid transfer by phosphatidylinositol transfer proteins in cells. Traffic. 2008;9:1743–1756. doi: 10.1111/j.1600-0854.2008.00794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Cunningham E, Thomas GM, Ball A, Hiles I, Cockcroft S. Phosphatidylinositol transfer protein dictates the rate of inositol trisphosphate production by promoting the synthesis of PIP2. Curr Biol. 1995;5:775–783. doi: 10.1016/s0960-9822(95)00154-0. [DOI] [PubMed] [Google Scholar]
- [143].Thomas GM, Cunningham E, Fensome A, Ball A, Totty NF, Truong O, Hsuan JJ, Cockcroft S. An essential role for phosphatidylinositol transfer protein in phospholipase C-mediated inositol lipid signaling. Cell. 1993;74:919–928. doi: 10.1016/0092-8674(93)90471-2. [DOI] [PubMed] [Google Scholar]
- [144].Kauffmann-Zeh A, Thomas GM, Ball A, Prosser S, Cunningham E, Cockcroft S, Hsuan JJ. Requirement for phosphatidylinositol transfer protein in epidermal growth factor signaling. Science. 1995;268:1188–1190. doi: 10.1126/science.7761838. [DOI] [PubMed] [Google Scholar]
- [145].Alb JG, Jr., Cortese JD, Phillips SE, Albin RL, Nagy TR, Hamilton BA, Bankaitis VA. Mice lacking phosphatidylinositol transfer protein-alpha exhibit spinocerebellar degeneration, intestinal and hepatic steatosis, and hypoglycemia. J Biol Chem. 2003;278:33501–33518. doi: 10.1074/jbc.M303591200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Alb JG, Jr., Phillips SE, Rostand K, Cui X, Pinxteren J, Cotlin L, Manning T, Guo S, York JD, Sontheimer H, Collawn JF, Bankaitis VA. Genetic ablation of phosphatidylinositol transfer protein function in murine embryonic stem cells. Molecular biology of the cell. 2002;13:739–754. doi: 10.1091/mbc.01-09-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Hamilton BA, Smith DJ, Mueller KL, Kerrebrock AW, Bronson RT, van Berkel V, Daly MJ, Kruglyak L, Reeve MP, Nemhauser JL, Hawkins TL, Rubin EM, Lander ES. The vibrator mutation causes neurodegeneration via reduced expression of PITP alpha: positional complementation cloning and extragenic suppression. Neuron. 1997;18:711–722. doi: 10.1016/s0896-6273(00)80312-8. [DOI] [PubMed] [Google Scholar]
- [148].Hotta Y, Benzer S. Genetic dissection of the Drosophila nervous system by means of mosaics. Proc Natl Acad Sci U S A. 1970;67:1156–1163. doi: 10.1073/pnas.67.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Hardie RC, Raghu P, Moore S, Juusola M, Baines RA, Sweeney ST. Calcium influx via TRP channels is required to maintain PIP2 levels in Drosophila photoreceptors. Neuron. 2001;30:149–159. doi: 10.1016/s0896-6273(01)00269-0. [DOI] [PubMed] [Google Scholar]
- [150].Milligan SC, Alb JG, Jr., Elagina RB, Bankaitis VA, Hyde DR. The phosphatidylinositol transfer protein domain of Drosophila retinal degeneration B protein is essential for photoreceptor cell survival and recovery from light stimulation. J Cell Biol. 1997;139:351–363. doi: 10.1083/jcb.139.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Lev S, Hernandez J, Martinez R, Chen A, Plowman G, Schlessinger J. Identification of a novel family of targets of PYK2 related to Drosophila retinal degeneration B (rdgB) protein. Mol Cell Biol. 1999;19:2278–2288. doi: 10.1128/mcb.19.3.2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Litvak V, Dahan N, Ramachandran S, Sabanay H, Lev S. Maintenance of the diacylglycerol level in the Golgi apparatus by the Nir2 protein is critical for Golgi secretory function. Nat Cell Biol. 2005;7:225–234. doi: 10.1038/ncb1221. [DOI] [PubMed] [Google Scholar]
- [153].Litvak V, Tian D, Carmon S, Lev S. Nir2, a human homolog of Drosophila melanogaster retinal degeneration B protein, is essential for cytokinesis. Mol Cell Biol. 2002;22:5064–5075. doi: 10.1128/MCB.22.14.5064-5075.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Garner K, Hunt AN, Koster G, Somerharju P, Groves E, Li M, Raghu P, Holic R, Cockcroft S. Phosphatidylinositol transfer protein, cytoplasmic 1 (PITPNC1) binds and transfers phosphatidic acid. J Biol Chem. 2012;287:32263–32276. doi: 10.1074/jbc.M112.375840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Amarilio R, Ramachandran S, Sabanay H, Lev S. Differential regulation of endoplasmic reticulum structure through VAP-Nir protein interaction. J Biol Chem. 2005;280:5934–5944. doi: 10.1074/jbc.M409566200. [DOI] [PubMed] [Google Scholar]
- [156].Litvak V, Shaul YD, Shulewitz M, Amarilio R, Carmon S, Lev S. Targeting of Nir2 to lipid droplets is regulated by a specific threonine residue within its PI-transfer domain. Curr Biol. 2002;12:1513–1518. doi: 10.1016/s0960-9822(02)01107-7. [DOI] [PubMed] [Google Scholar]
- [157].Trivedi D, Padinjat R. RdgB proteins: functions in lipid homeostasis and signal transduction. Biochim Biophys Acta. 2007;1771:692–699. doi: 10.1016/j.bbalip.2007.04.014. [DOI] [PubMed] [Google Scholar]
- [158].Whatmore J, Wiedemann C, Somerharju P, Swigart P, Cockcroft S. Resynthesis of phosphatidylinositol in permeabilized neutrophils following phospholipase Cbeta activation: transport of the intermediate, phosphatidic acid, from the plasma membrane to the endoplasmic reticulum for phosphatidylinositol resynthesis is not dependent on soluble lipid carriers or vesicular transport. Biochem J. 1999;341(Pt 2):435–444. [PMC free article] [PubMed] [Google Scholar]
- [159].Lu C, Vihtelic TS, Hyde DR, Li T. A neuronal-specific mammalian homolog of the Drosophila retinal degeneration B gene with expression restricted to the retina and dentate gyrus. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1999;19:7317–7325. doi: 10.1523/JNEUROSCI.19-17-07317.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Chang JT, Milligan S, Li Y, Chew CE, Wiggs J, Copeland NG, Jenkins NA, Campochiaro PA, Hyde DR, Zack DJ. Mammalian homolog of Drosophila retinal degeneration B rescues the mutant fly phenotype. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1997;17:5881–5890. doi: 10.1523/JNEUROSCI.17-15-05881.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Elagin VA, Elagina RB, Doro CJ, Vihtelic TS, Hyde DR. Cloning and tissue localization of a novel zebrafish RdgB homolog that lacks a phospholipid transfer domain. Vis Neurosci. 2000;17:303–311. doi: 10.1017/s095252380017213x. [DOI] [PubMed] [Google Scholar]
- [162].Kohn L, Kadzhaev K, Burstedt MS, Haraldsson S, Sandgren O, Golovleva I. Mutation in the PYK2-binding domain of PITPNM3 causes autosomal dominant cone dystrophy (CORD5) in two Swedish families. Advances in experimental medicine and biology. 2008;613:229–234. doi: 10.1007/978-0-387-74904-4_26. [DOI] [PubMed] [Google Scholar]
- [163].Zhang B, Yin C, Li H, Shi L, Liu N, Sun Y, Lu S, Liu Y, Sun L, Li X, Chen W, Qi Y. Nir1 promotes invasion of breast cancer cells by binding to chemokine (C-C motif) ligand 18 through the PI3K/Akt/GSK3beta/Snail signalling pathway. Eur J Cancer. 2013;49:3900–3913. doi: 10.1016/j.ejca.2013.07.146. [DOI] [PubMed] [Google Scholar]
- [164].Lu C, Peng YW, Shang J, Pawlyk BS, Yu F, Li T. The mammalian retinal degeneration B2 gene is not required for photoreceptor function and survival. Neuroscience. 2001;107:35–41. doi: 10.1016/s0306-4522(01)00337-2. [DOI] [PubMed] [Google Scholar]
- [165].Carlisle FA, Pearson S, Steel KP, Lewis MA. Pitpnm1 is expressed in hair cells during development but is not required for hearing. Neuroscience. 2013;248C:620–625. doi: 10.1016/j.neuroscience.2013.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]