Abstract
Objective
Abnormal metabolic activities of chondrocytes may cause articular cartilage (AC) degradation, but key transcription factors regulating metabolic activities in AC of aging individuals remain unknown. This study aimed to investigate the role of transcription factor NFAT1 in regulating the expression of anabolic and catabolic molecules in AC of aged mice.
Methods
The hip, knee, and shoulder joints of BALB/c mice were harvested at 6, 12, 15, 18, and 24 months of age for histopathological and immunohistochemical analyses. Total RNA was isolated from AC for gene expression. Genomic DNA and chromatin were prepared from AC for methylated DNA immunoprecipitation (MeDIP) and chromatin immunoprecipitation (ChIP) assays.
Results
NFAT1 expression in AC of mice was significantly decreased after 12 months of age, which was associated with reduced proteoglycan staining, decreased expression of chondrocyte markers, and increased expression of interleukin-1β. Forced Nfat1 expression in chondrocytes from aged mice significantly reversed the abnormal metabolic activities. ChIP assays confirmed that NFAT1 bound to the promoter of the Acan, Col2a1, Col9a1, Col11a1, Il1b, Mmp13 and Tnfa genes in articular chondrocytes of aged mice. ChIP and MeDIP assays revealed that reduced NFAT1 expression in AC of aged mice was regulated by epigenetic histone methylation at the promoter region and was correlated with increased DNA methylation at introns 1 and 10 of the Nfat1 gene.
Conclusion
NFAT1 is a transcriptional regulator of multiple anabolic and catabolic genes in AC of aged mice. Epigenetically mediated reduction of NFAT1 expression causes imbalanced metabolic activities of articular chondrocytes in aged mice.
Keywords: NFAT1, chondrocyte, aging, osteoarthritis, epigenetics
Introduction
Age is an independent risk factor for osteoarthritis (OA)1, 2. It has been well recognized that younger individuals with osteochondral defects have a lower incidence of posttraumatic OA than the elderly3. However, the biological mechanisms underlying the age-related predisposition to OA are poorly understood. Previous studies suggest that senescent changes associated with telomere erosion in chondrocytes may play a role in the pathogenesis of OA and in limiting cartilage repair4. Increased oxidative stress with aging may reduce chondrocyte survival and responsiveness to growth factors5, 6. Abnormal anabolic and catabolic activities of articular chondrocytes in older individuals may lead to articular cartilage (AC) degradation7, 8, but key transcription factors that regulate the expression of anabolic and catabolic molecules in AC of the elderly remain to be identified.
NFAT1 (NFATc2/NFATp) is a member of the family of nuclear factor of activated T cells NFAT/Nfat) transcription factors. NFAT1 was first identified as a repressor of the immune response in mice, as Nfat1-deficient mice displayed an enhanced immune response and dysregulation of interleukin-expression9, 10. In resting immune cells, NFAT1 protein exists in the cytoplasm as phosphorylated species. Sustained increases in cytoplasmic calcium activate calcineurin, a calcium/calmodulin-dependent protein phosphatase, which dephosphorylates NFAT1, resulting in nuclear translocation of NFAT1 to drive gene transcription. Dephosphorylated NFAT1 is then rephosphorylated and translocated back to the cytoplasm11. Our recent study showed that mice lacking Nfat1 exhibited normal skeletal development but began to show articular chondrocyte dysfunction and early OA-like changes as young adults12, 13. More recently, Greenblatt et al. reported that NFATc1 (NFAT2) expression was down-regulated in human AC with OA lesions; but mice lacking Nfat2 in cartilage displayed no evidence of OA14. However, mice lacking both Nfat2 and Nfat1 (Nfatc2) in cartilage developed early joint instability and subsequent OA14, suggesting that NFAT1 may be more important than NFAT2 for cartilage homeostasis and prevention of OA.
All previously published in vivo studies on Nfat deficiency-mediated OA have focused on the OA phenotype of Nfat knockout mice. Such animal models with OA-like changes in the developing stage14 or young adult stage13 caused by a complete loss of function of an NFAT member(s) in cartilage, however, do not accurately mimic the condition seen in OA cartilage of elderly humans, in which NFAT expression is reduced but not absent14. Moreover, our previous studies indicated that NFAT1 expression in AC was low in the embryonic and newborn stages but high in the young adult stage (2–6 months of age)13.
The objective of this study was to investigate whether NFAT1 expression level in AC changes after the young adult stage and to determine whether NFAT1 plays a role in regulating the expression of anabolic and catabolic molecules in AC of aged wild-type mice.
Materials and methods
Animals
Wild-type BALB/c mice at 6, 12, 15, 18, and 24 months of age were used for histopathological cellular, and molecular biological analyses. These ages were chosen because 6-month-old mice are considered to be young adults with a high level of NFAT1 expression in their AC13, 12-month-old mice are considered middle-aged, and 15 to 24-month-old mice are considered aged15, 16. The breeder pairs of Nfat1−/− mice were a gift from Dr. Laurie Glimcher (Harvard University). All animal procedures were approved by the Institutional Animal Care and Use Committee at the University of Kansas Medical Center in compliance with federal and state laws and regulations.
Histological analyses
After euthanasia of animals, the hip, knee, and shoulder joints were harvested and fixed in 2% paraformaldehyde, decalcified in 25% formic acid, embedded in paraffin and sectioned for histochemical staining to evaluate age-dependent histological changes. Safranin-O and fast green stains were used to identify chondrocytes and cartilage matrix. Tissue sections which had been decalcified in 10% EDTA were used for NFAT1 immunohistochemical staining (Santa Cruz). Ten to twelve mice (5–6 mice per gender) were examined at each time point.
Collection of AC
Because AC of the femoral and humeral heads is much thicker and more readily removed than AC in other joints, AC from these two sites was collected from 6- to 18-month-old mice for isolation of RNA, chromatin, and chondrocytes. AC of 24-month-old mice was not collected because the AC at that age was too thin to be peeled off. The morphology of collected AC and the ability of isolated articular chondrocytes to express proteoglycans were confirmed by safranin-O and Alcian blue staining as described previously13.
Quantitative real-time reverse transcription-polymerase chain reaction (qPCR)
Total RNA was isolated from femoral head AC or cultured articular chondrocytes using TRIzol reagent (Invitrogen). cDNA was synthesized using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) and qPCR reactions were performed in triplicate using a 7500 Real-Time qPCR system and SYBR Green reagents (Applied Biosystems). Specific primers used for qPCR gene expression assays are listed in Table 1. Gapdh expression levels were used as internal controls. Gene expression levels were quantified using 2−ΔΔCt methods as described previously12, 17.
Table 1.
Specific primers used for quantitative real-time RT–PCR
| Gene | Forward | Reverse | GenBank No. |
|---|---|---|---|
| Mmp13 | TCACCTGATTCTTGCGTGCTA | CAGATGGACCCCATGTTTGC | NM_008607 |
| Acan | TGGGATCTACCGCTGTGAAGT | CTCGTCCTTGTCACCATAGCAA | NM_007424 |
| Col2α1 | CGAGATCCCCTTCGGAGAGT | TGAGCCGCGAAGTTCTTTTC | NM_031163 |
| Col9α1 | TCTTAAGCGTCGTGCAAGATTTC | CTTGGGACACAGTTCACTTCCA | NM_007740 |
| Col11α1 | CACAAAACCCCTCGATAGAAGTG | CCTGTGATCAGGAACTGCTGAA | NM_007729 |
| IL1b | GCTTCCTTGTGCAAGTGTCTGA | TCAAAAGGTGGCATTTCACAGT | NM_008361 |
| Nfat1 | GTGCAGCTCCACGGCTACAT | GCGGCTTAAGGATCCTCTCA | NM_010899 |
| Lsd1 | GCAGCCTGTTTCCCAGACA | CTGCAATGTGCGATTCCTGAT | NM_133872 |
| Tnfa | AGGGATGAGAAGTTCCCAAATG | GGCTTGTCACTCGAATTTTGAGA | NM_013693 |
| Gapdh | AGGTTGTCTCCTGCGACTTCA | CCAGGAAATGAGCTTGACAAAG | NM_008084 |
Isolation and cultivation of articular chondrocytes
Articular chondrocytes were isolated from pooled femoral head AC samples using collagenase D (1.5 mg/ml, Roche Diagnostics). Isolated cells were cultivated in DMEM/F12 medium in a humidified incubator with 5% CO2 at 37°C in monolayer. Because primary chondrocytes (P0) displayed uneven growth rate and cell density, passage 1 (P1) of cultured articular chondrocytes was used for forced Nfat1 expression and siRNA-mediated knockdown experiments.
Preparation of Nfat1 expression lentivirus
To enforce Nfat1 expression in cultured articular chondrocytes, Nfat1 coding sequences with the stop codon were subcloned into pCDH-EF1-MCS-T2A-copGFP cDNA Cloning and Expression Lentivector (System Biosciences). Lentiviral particles were produced in 293FT cells (Invitrogen) with packaging plasmid psPAX2 (Addgene) and envelope plasmid pMD2.G (Addgene). Lentiviral qPCR Titer Kit (Applied Biological Materials) was used for lentiviral titration.
Lentiviral transduction
P1 articular chondrocytes isolated from mice were seeded in 24-well tissue culture plates at 8 × 104 cells/well and cultured for 24 hours, at which time ~70% of the cells were confluent. Chondrocytes were transduced with Nfat1 cDNA lentivirus at the ratio of 5 viruses per cell in the presence of 5 µg/ml of Polybrene (Santa Cruz). Empty cDNA expression lentiviral vectors were used as controls. After cultivation for 72 hours, total RNA was isolated for gene and protein expression analyses.
DNA methylation assay
Genomic DNA was purified from AC of the femoral and humeral heads and was sonicated. Methylated DNA immunoprecipitation (MeDIP) assays were carried out using a CpG MethylQuest DNA isolation Kit (Millipore). qPCR was performed to quantify the methylation level at the CpG island of the Nfat1 promoter and specific introns, using the primers listed in Table 2. The relative MeDIP enrichment was presented as a ratio to the level of input DNA.
Table 2.
Specific primers used for Methylated DNA immunoprecipitation (MeDIP) and Chromatin immunoprecipitation (ChIP)
| Assay/Gene | Forward | Reverse | GenBank No. |
|---|---|---|---|
| (1) MeDIP | |||
| Nfat1-TSS | AGGTGCCTTGAGCAGGAA | GGATGCGGGTTCGTATAGAG | NM_010899 |
| Nfat1-intron1 | GGAATGTAAGGGGTCAGGCT | TCTGCTGTCCCATCTCAAGG | NM_010899 |
| Nfat1-intron10 | CCTGAGGTCATGAACTGGGT | GGCTTCTCTCCCTTCCCTTT | NM_010899 |
| (2) NFAT1 binding ChIP | |||
| Acan | AAATCACTGTATGCTGCAACTAGC | GCCTGGCCTCTACAACACTC | NM_007424 |
| Col2α1 | ATGGGTTAGGGAGGCTGTG | GGGGACAGTTTTCCAGACCT | NC_000081 |
| Col9α1 | CTCTGGTTAAATCCCCCACA | CTGAAGGAGGGACTGAATGG | NC_000067 |
| Col11α1 | TGCTTCTCCCTTTCATCCAG | TCACTGAGGGGCTTGTCTG | NM_007729 |
| IL1b | TCCCCTAAGAATTCCCATCA | GCTGTGAAATTTTCCCTTGG | NM_008361 |
| Mmp13 | CTGCTGCTTCTCCCCACTAT | GAGGGAAATGGAAAATGAAGG | NM_008607 |
| Tnfa | CGATGGAGAAGAAACCGAGA | CTCATTCAACCCTCGGAAAA | NM_013693 |
| (3) Histone methylation ChIP | |||
| Nfat1-P1 | AGGCCCAGACAACTTCTGCTT | CGCAGGCAACAGCTTCTGA | NM_010899 |
| Nfat1-P2 | GCTATGGCTCATGCCAAAGTG | CAGGCCTGGTCAATTGAGTTTAG | NM_010899 |
| Nfat1-P3 | GGGATGCAAATTCGTATAGA | CAGGTGCCTTGAGCAGGAA | NM_010899 |
Chromatin immunoprecipitation (ChIP) assay
Chromatins were prepared from AC or cultured articular chondrocytes and cross-linked with 1% formaldehyde. ChIP assays were performed according to the manual of Magna ChIP A/G kit (Millipore) using a ChIP grade NFAT1 antibody (Santa Cruz, Sc-13034). To identify potential NFAT1-binding target genes, specific primers were designed from Acan, Col2a1, Col9a1, Col11a1, Ilb1, Mmp13, and Tnfa promoter sequences encompassing NFAT1 binding sites (Table 2). Histone methylation ChIP assays were performed using chromatins prepared from articular chondrocytes with dimethylated histone 3 lysine 4 (H3K4me2, Millipore) or H3K9me2 (Millipore) antibodies. Normal mouse IgG was used as a baseline control to confirm the specificity of the ChIP assays. Chromatin prepared from femoral head AC of Nfat1−/− mice was used as a negative control for the NFAT1 ChIP assay. Input, immunoprecipitated and control samples were analyzed by qPCR using specific primers designed around the transcription start site (TSS) of the Nfat1 gene (Table 2). The relative enrichment levels of ChIPed samples were first normalized to input samples and were then compared to the relative enrichment levels of IgG control samples.
siRNA-mediated knockdown of lysine specific demethylase 1 (lsd1)
To determine whether histone demethylases regulate NFAT1 expression in articular chondrocytes of aged mice, pooled target-specific 19–25 nt siRNA sequences designed to knock down the expression of mouse Lsd1 (Santa Cruz) was used for gene silencing experiments. Control siRNA (Santa Cruz) was used as a negative control. Transfection of siRNA was performed in cultured articular chondrocytes using previously described methods18. P1 articular chondrocytes were seeded in 24-well tissue culture plates with DMEM/F12 medium at 5 ×104 cells/well. After incubation for 24 hours, subconfluent chondrocytes were transfected using Lipofectamine RNAiMAX (Invitrogen). For each well, 30 pmoles of siRNA and 1.5 µl of Lipofectamine RNAiMAX were mixed gently in 100 µl of Opti-MEM I medium without serum (Gibco/Invitrogen). After incubation for 72 hours, total RNA was isolated for gene and protein expression analyses.
Western blot
Total protein was extracted from cultured cells and quantified by the Bradford method (Bio-Rad). 10µg total protein of each sample was separated on 12% SDS-PAGE (30-µl wells, Bio-Rad) and were then transferred to PVDF membrane (Millipore) at 100V for 1 hour. After being blocked in 5% milk at 4°C overnight, the membrane was incubated with antibodies against LSD1 (Cell signaling) or NFAT1 (Santa Cruz) for 2 hours at room temperature. Horseradish peroxidase (HRP)-conjugated anti-rabbit or anti-mouse IgG (Santa Cruz) was used as the secondary antibody. β-actin (Sigma) was used for the loading control. The protein bands were detected in Pierce 1-Step Ultra TMB Blotting Solution (Thermo Scientific).
Statistics
Quantitative data were presented as the mean ± 95% confidence intervals from at least three independent repeats for each experimental condition. Significant differences between experimental groups were determined by Student’s t-tests or ANOVA. Multivariable logistic regression was used to determine the association between age-dependent Nfat1 expression and DNA methylation levels. Statistical analysis was conducted using Microsoft Excel software (Version 2010). A P value of less than 0.05 was considered statistically significant. Graphs with quantitative data were constructed with GraphPad Prism software (version 6.0).
Results
Decreased NFAT1 expression in AC is associated with chondrocyte dysfunction in aged mice
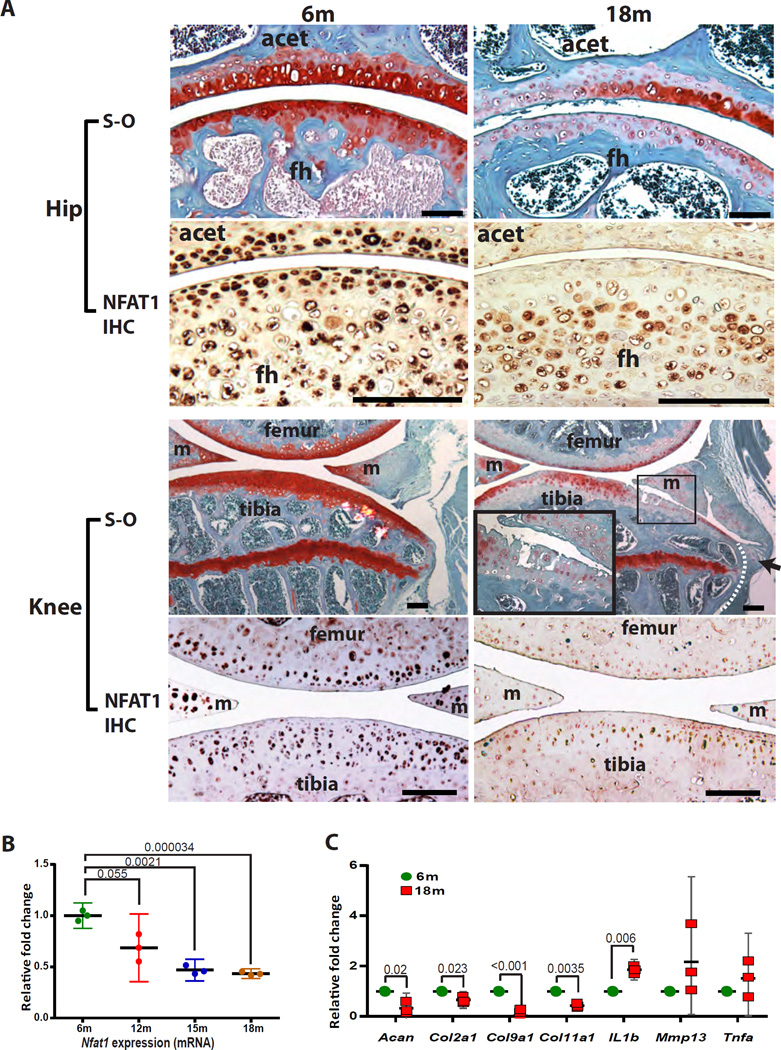
In 6 to 12-month-old wild-type mice, all hip, knee, and shoulder joints showed normal articular structures with safranin-O stained proteoglycans (red in Figure 1A) throughout the AC. At 15, 18 and 24 months (aged mice)15, 16, however, focal or generalized loss of safranin-O staining was observed in the vast majority of joints examined, suggestive of reduced synthesis and/or increased catabolism of proteoglycans in aged AC. However, typical OA changes (e.g., loss and fibrillation of AC, osteophytes, subchondral bone changes)19 were observed in only a minority of the knee joints (Figure 1A) and were not apparent until 18 months (2/10 mice) and 24 months (2/10 mice). Histological changes in the hip and knee joints of 6-month-old and 18-month-old mice are shown in Figure 1A. Typical OA changes were not seen in any of the hip or shoulder joints, suggesting that a majority of aged joints with loss of proteoglycans in the AC do not progress to classic OA even by 24 months of age. No gender differences were observed.
Figure 1.
Chondrocyte dysfunction is associated with decreased NFAT1 expression in AC of aged wild-type mice. (A) Hip joints: A hip joint from a 6-month-old (6m) mouse shows normal articular structure, with safranin-O (S-O) staining (red) of proteoglycans throughout the AC. A hip joint from an 18-month-old (18m) mouse shows extensive depletion of safranin-O staining in the AC of femoral head (fh) and acetabulum (acet). Immunohistochemistry (IHC) shows a reduction of NFAT1 protein expression (particularly in the superficial zone) in the hip AC of an 18m mouse, in comparison with the hip AC of a 6-month-old mouse. In order to clearly demonstrate the zonal difference in NFAT1 expression, the hip joints were sectioned at about 30° to the longitudinal axis of the femoral neck so that the AC appears to be thicker than those in the upper panels. Knee joints: A knee joint from a 6m mouse shows normal joint structure and safranin-O staining in the AC and growth plate, whereas a knee joint from an 18m mouse shows OA changes with fibrillation of articular surface (in magnified rectangle) and osteophyte formation at the joint margin (arrow). IHC shows reduced number and intensity of NFAT1-expressing chondrocytes in the AC and menisci of an 18m mouse. m = meniscus, the dotted line indicates the original bone surface. Safranin-O and fast green staining, counterstained with haematoxylin. Scale bar = 100 µm. (B) qPCR analyses showing age-dependent reduction of Nfat1 mRNA expression in articular chondrocytes. (C) qPCR analyses showing a significant decrease in expression of Acan, Col2a1, Col9a1, and Col11a1 and a significant increase in expression of Il1b in the AC of 18m mice. The expression levels in the AC of 6m mice were normalized to 1.0. N = 3, each pooled sample prepared from the AC of 6 femoral heads (3 mice per sample).
We next evaluated the expression level of NFAT1 and some metabolic genes in AC of aged mice. Immunohistochemical (IHC) analysis demonstrated a substantial reduction of NFAT1 protein expression in AC of aged hip, knee, and shoulder joints, especially in the superficial zone of AC of aged hip and knee joints (Figure 1A). qPCR analyses revealed that Nfat1 mRNA expression in the AC of wild-type mice was gradually decreased from 12 to 18 months of age (Figure 1B). Expression levels of the anabolic genes such as Acan, Col2a1, Col9a1, and Col11a1 were significantly decreased whereas mRNA for interleukin-1 beta (Il1b, catabolic gene) was significantly increased in AC of 18-month-old mice compared to that of 6-momth-old mice, suggesting diminished anabolic and increased catabolic activities in the aged AC. These findings indicate that decreased NFAT1 expression is associated with dysfunction of articular chondrocytes in aged wild-type mice.
Forced Nfat1 expression rescues dysfunction of articular chondrocytes from aged mice
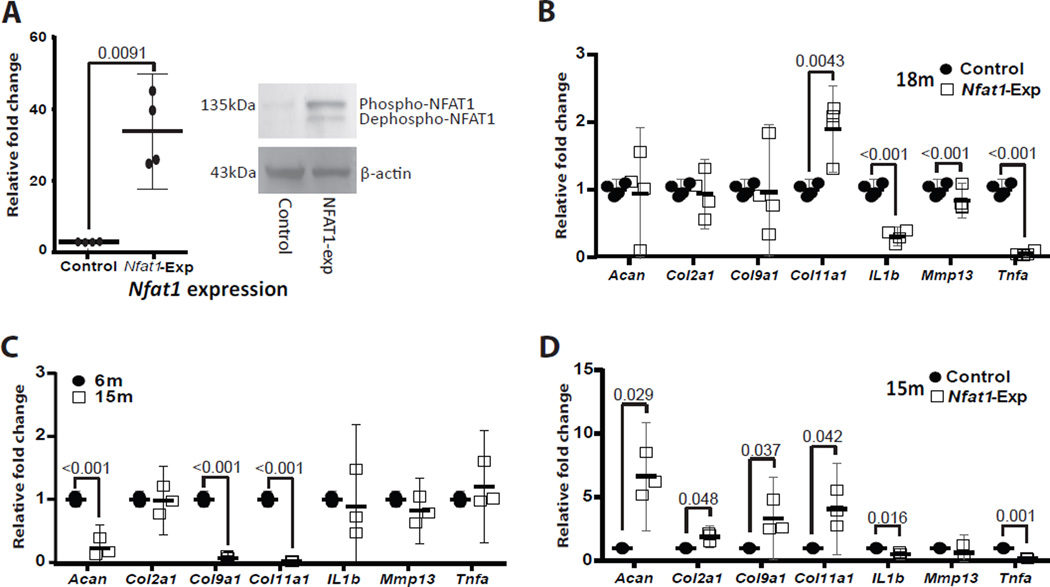
To determine whether the imbalanced metabolic activities of articular chondrocytes of aged mice are induced by decreased NFAT1 expression, cultured chondrocytes isolated from AC of 18-month-old wild-type mice were transduced with Nfat1 expression lentivectors. The expression efficiency of Nfat1 mRNA and protein was confirmed by qPCR and Western blot, respectively (Figure 2A). Forced Nfat1 expression significantly increased Col11a1 expression and decreased the expression of Il1b, Mmp13 and Tnfα (Figure 2B), but no significant effect was seen on the expression of Acan, Col2a1, and Col9a1. To investigate whether the rescue effect of NFAT1 overexpression can be improved in chondrocytes from younger mice, forced Nfat1 expression was performed in cultured chondrocytes isolated from AC of 15-month-old wild-type mice in which the expression level of Acan, Col9a1 and Col11a1 was decreased, in comparison with that in 6-month-old mice (Figure 2C). NFAT1 overexpression significantly up-regulated the expression of Acan, Col2a1, Col9a1 and Col11a1 and down-regulated the expression of Il1b and Tnfa (Figure 2D), even though the expression of IL1b and Tnfa was not increased in AC of 15-month-old mice, suggesting that decreased NFAT1 expression is an important cause of the imbalanced metabolic activities of articular chondrocytes in aged mice.
Figure 2.
Rescue effect of forced Nfat1 expression on the dysfunction of cultured articular chondrocytes. (A) Confirmation of the efficiency of forced Nfat1 expression on Nfat1 mRNA (left panel) and NFAT1 protein (right panel) in lentiviral transduced articular chondrocytes derived from 18-month-old (18m) mice. Phosphorylated- (Phosph-) and dephosphorylated-NFAT1 (Dephospho-NFAT1) were detected by Western blot (WB). The photographic conditions for NFAT1 WB and β-actin WB were different, resulting in different background for each WB. (B) Rescue effect of forced Nfat1 expression in cultured articular chondrocytes from 18-month-old mice transduced with empty lentiviral vectors (Control) or Nfat1 cDNA lentiviral vectors (Nfat1-exp). (C) Aberrant gene expression in articular chondrocytes of 15-month-old (15m) mice. (D) Rescue effect of forced Nfat1 expression in cultured articular chondrocytes from 15-month-old mice transduced with Control or Nfat1 overexpression lentiviral vectors. N = 3 cultures.
NFAT1 directly binds to the promoter of specific anabolic and catabolic genes in articular chondrocytes
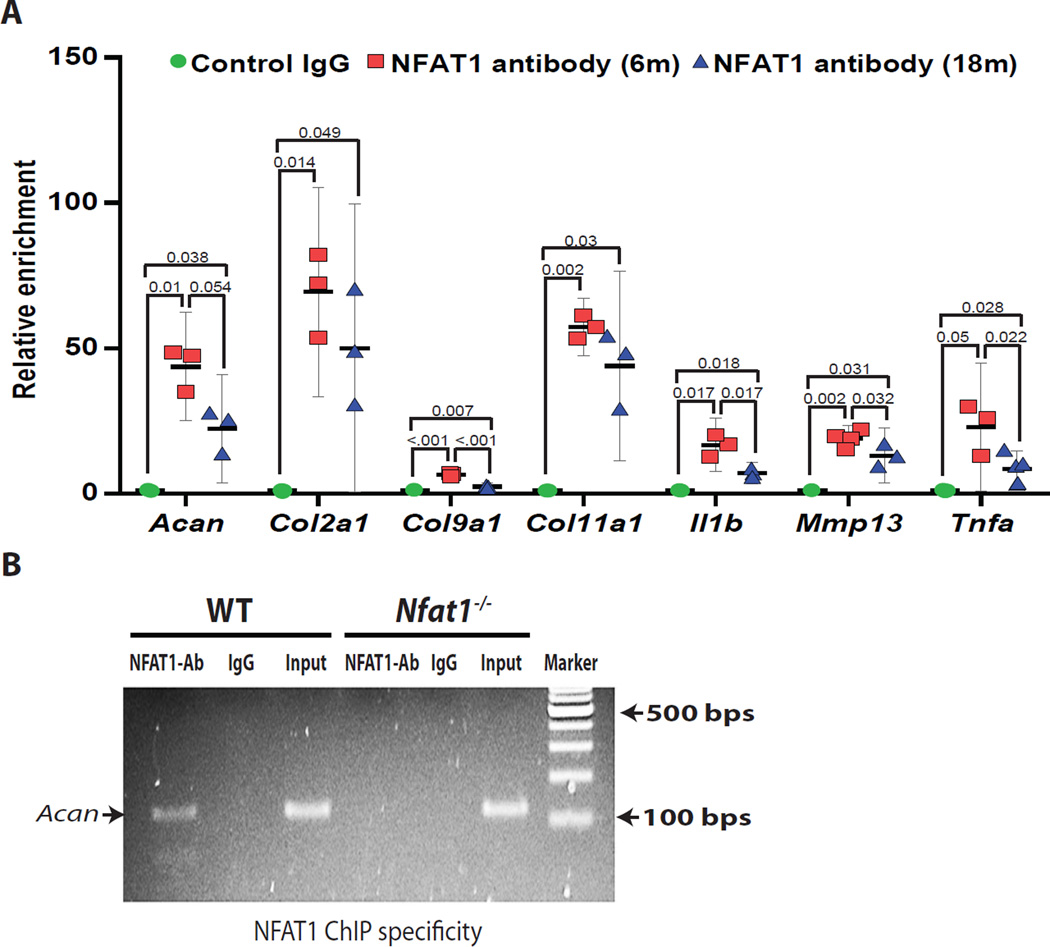
To validate whether NFAT1 binds to the promoter region of the aberrantly expressed genes observed in AC of aged mice, we performed ChIP analysis using the chromatins isolated from AC of young adult and aged mice. NFAT1 bound to the promoter region of the Acan, Col2a1, Col9a1, Col11a1, Il1b, Mmp13 and Tnfa genes. For most of the genes tested, NFAT1 binding levels were higher in 6-month-old mice than in 18-month-old mice (Figure 3A). The specificity of the NFAT1 ChIP assay was confirmed by using chromatins prepared from Nfat1−/− AC as negative controls (Figure 3B). These results suggest that NFAT1 is likely to be an upstream regulator of these anabolic and catabolic genes. Therefore, decreased NFAT1 expression may cause aberrant expression of these specific genes in AC of aged mice.
Figure 3.
Binding of NFAT1 to the promoter region of its candidate target genes. (A) NFAT1 ChIP assay followed by qPCR quantification using chromatin prepared from the AC of 6- and 18-month-old (6m and 18m) mice. Normal mouse IgG was used as a baseline control. (B) Representative agarose gel image of the PCR products of Acan primers verifies the specificity of NFAT1 ChIP assay. The chromatin isolated from the AC of Nfat1−/− mice was used as a negative control. N = 3.
DNA methylation is associated with decreased NFAT1 expression in AC of aged mice
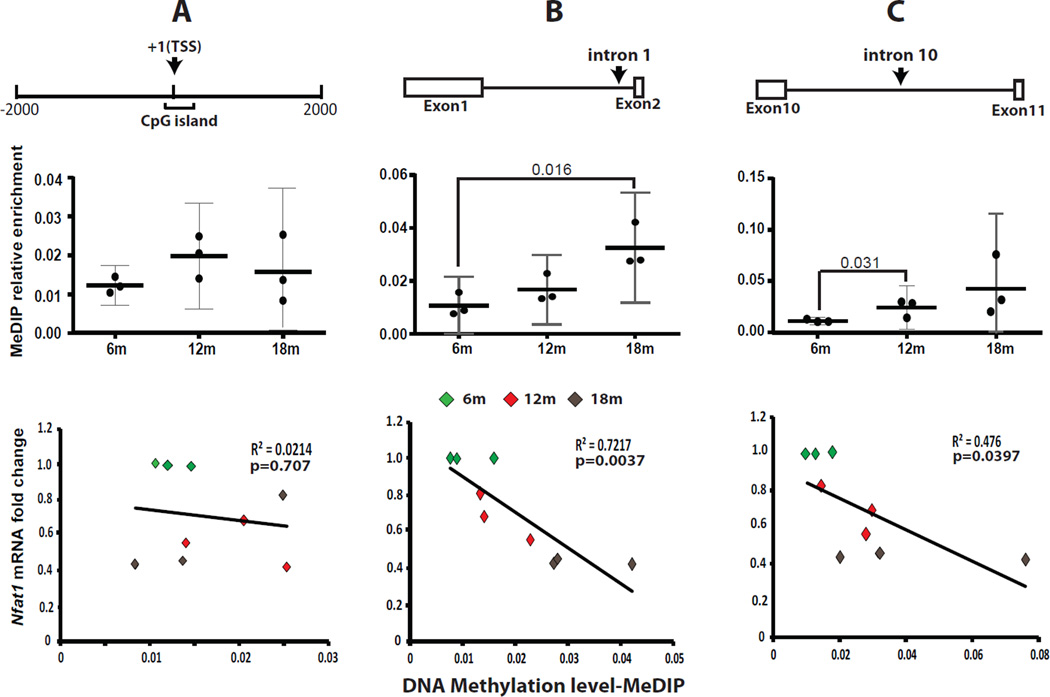
To explore the potential epigenetic regulatory mechanisms for age-dependent NFAT1 expression, we first performed DNA methylation immunoprecipitation (MeDIP) assays. One CpG island (398 bp) covering the transcription start site (TSS) of the mouse Nfat1 gene was identified (Figure 4A, top panel) using the MethyPrimer program (http://www.urogene.org/methprimer/index1.html). The overall DNA methylation status of the Nfat1 promoter region was low (<2%) in AC from 6-, 12-, and 18-month-old mice, with no significant difference in the DNA methylation level between the 3 age groups (Figure 4A, middle panel); no correlation between Nfat1 expression levels and the DNA methylation status at Nfat1 promoter region was observed (Figure 4A, bottom panel). This suggests that DNA methylation in the Nfat1 promoter region is unlikely to be involved in the regulation of age-dependent NFAT1 expression.
Figure 4.
Correlation of age-dependent Nfat1 expression with DNA methylation in AC. (A) Top panel: A diagram showing a CpG island around the transcription start site (TSS) of the Nfat1 gene. Middle panel: No significant difference in DNA methylation level between the 6-, 12- and 18-month-old (6m, 12m and 18m) age groups. Bottom panel: No correlation between age-dependent Nfat1 expression and DNA methylation around TSS. (B) Top panel: Nfat1 intron 1 region for evaluation of DNA methylation. Middle panel: There is a significant difference in the DNA methylation level between 6m and 18m. Bottom panel: Negative correlation between age-dependent Nfat1 expression and DNA methylation levels. (C) Top panel: Nfat1 intron 10 region for evaluation of DNA methylation. Middle panel: There is a significant difference in DNA methylation between 6m and 12m. Bottom panel: Negative correlation between age-dependent Nfat1 expression and DNA methylation levels. For all bottom panels, correlations of Nfat1 expression with DNA methylation were determined by multivariable logistic regression analyses. The Nfat1 mRNA expression level of each of the 6m samples has been normalized to 1. The relative enrichment of MeDIP is normalized to input. N = 3.
We next examined DNA methylation levels at two regions located in “intron 1” and “intron 10” of the Nfat1 gene (Figure 4B–C, top panels). We chose to evaluate these regions because enhancer activity has been identified in these regions for the Nfat2 gene and the gene structure of Nfat1 and Nfat2 is highly conserved20, 21. As shown in Figure 4B (middle panel), DNA methylation levels of intron 1 were increased with age. Multivariable logistic regression analyses showed a negative correlation between the age-dependent Nfat1 expression levels and the DNA methylation status at introns 1 and 10 (Figure 4B–C, bottom panels).
Histone methylation is associated with decreased NFAT1 expression in AC of aged mice
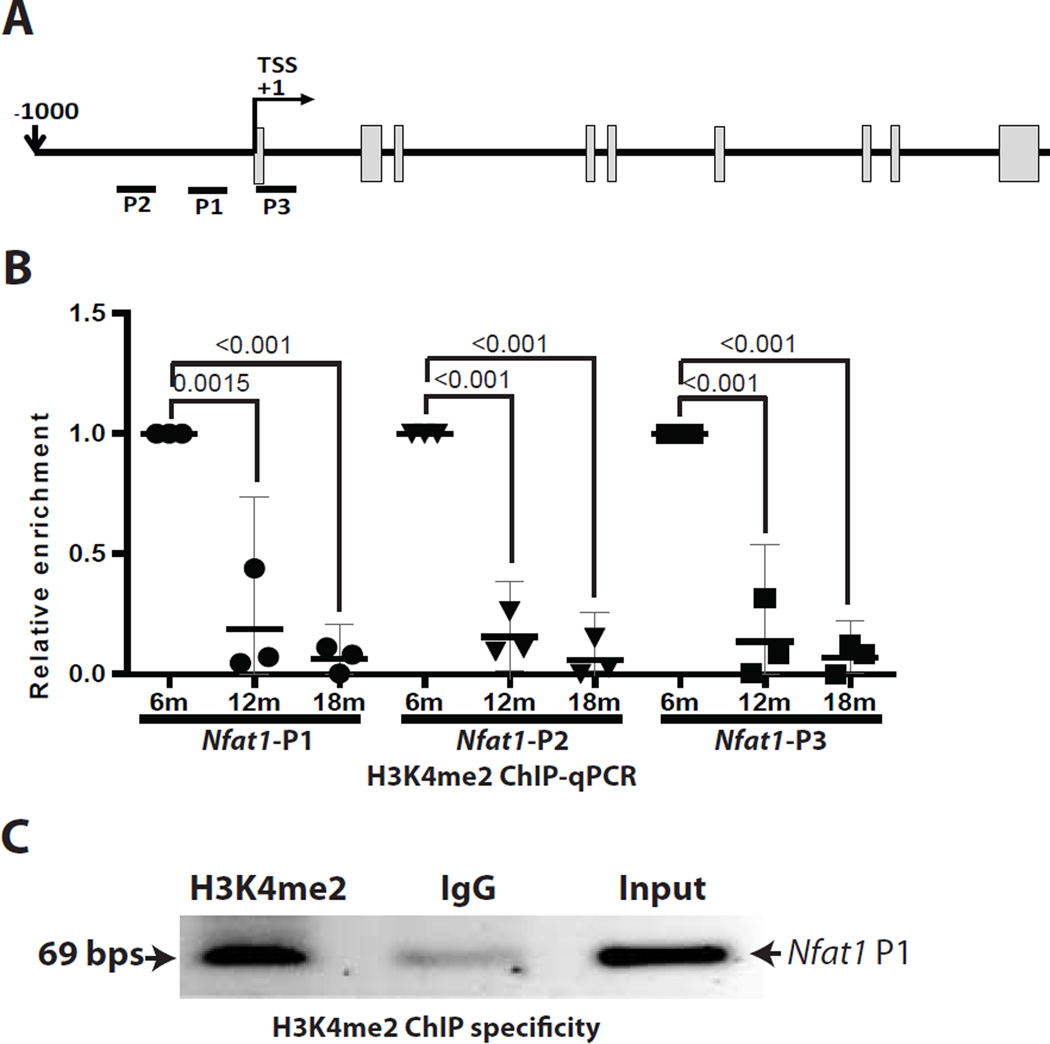
Next, we examined the histone methylation status in the Nfat1 promoter region by ChIP assays. qPCR analysis of the ChIP samples, using 3 different primers designed around TSS of the Nfat1 gene (Figure 5A), showed that the enrichment levels of H3K4me2 in the Nfat1 promoter of 12- and 18- month-old mice were significantly decreased in comparison with the level in 6-momth-old mice (Figure 5B). Age-dependent changes in H3K9me2 (a histone code linked to transcriptional repression) were not significant (data not shown). These findings suggest that decreased NFAT1 expression in articular chondrocytes of aged mice is associated with a decrease in H3K4me2, a histone code linked to transcriptional activation. IgG from normal mice was used as a negative control to validate the specificity of the ChIP assays (Figure 5C).
Figure 5.
ChIP assay of histone methylation at the Nfat1 promoter region. (A) Genomic structure of the Nfat1 gene with locations of three different qPCR primers (Nfat1 P1-3); (B) H3K4me2 ChIP followed by qPCR quantification showing significant reduction of H3K4me2 in the Nfat1 promoter of 12- and 18- month-old (12m and 18m) mice, in comparison with that of 6-month-old (6m) mice. (C) Representative image of conventional PCR agarose gel using Nfat1 P1 primers confirmed the specificity of the ChIP assays. N = 3.
LSD1 is a key regulator of NFAT1 expression in articular chondrocytes of aged mice
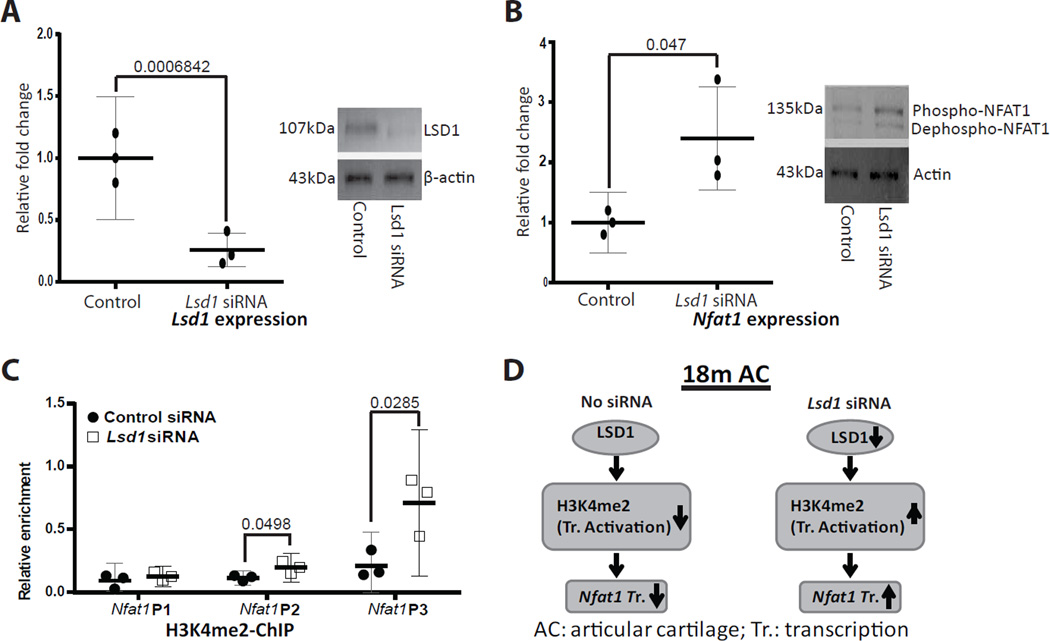
KDM1a (LSD1) has been previously identified as a key H3K4me2 demethylase22, 23 responsible for the low NFAT1 expression in cartilage of mouse embryos13. To examine whether LSD1 regulates NFAT1 expression in articular chondrocytes of aged mice, we performed siRNA-mediated knockdown of Lsd1 in cultured articular chondrocytes isolated from the AC of 18-month-old wild-type mice. The efficiency of Lsd1 siRNA knockdown was verified by qPCR (Figure 6A, left panel) and Western blot (Figure 6A, right panel). Upon Lsd1 knockdown, Nfat1 mRNA expression was increased by 2.4 fold, with increased NFAT1 protein expression (Figure 6B). However, this level of increase in NFAT1 expression did not rescue the abnormal expression of Acan, Col2a1, Col9a1, Col11a1, Il1b, Mmp13 and Tnfa (data not shown). Finally, ChIP assays using the Lsd1 siRNA-treated articular chondrocytes revealed that knockdown of Lsd1 increased H3K4me2 enrichment at Nfat1 promoter regions (Figure 6C). These results suggest that transcription of the Nfat1 gene in articular chondrocytes of aged mice is negatively regulated by LSD1 through demethylation of H3K4me2 at the Nfat1 promoter regions (Figure 6D).
Figure 6.
siRNA knockdown of Lsd1 in articular chondrocytes from18-month-old (18m) mice. (A) Confirmation of the Lsd1-siRNA knockdown efficiency by qPCR (left panel) and Western blot (right panel. (B) Up-regulation of Nfat1 mRNA expression by qPCR (left panel) and NFAT1 protein by Western blotting (right panel) after Lsd1 knockdown. Phospho = Phosphorylated, Dephospho = dephosphorylated. The photographic conditions for LSD1, NFAT1 and β-actin WB were different, resulting in different background for each WB. (C) ChIP-qPCR assays using chromatins from cultured articular chondrocytes treated with Lsd1 siRNA show increased H3K4me2 at specific Nfat1 promoter regions. N = 3 independent cultures per group. (D) A schematic summary of LSD1-mediated NFAT1 expression in AC of 18m mice.
Discussion
The present study has revealed for the first time that spontaneous reduction of NFAT1 expression may cause an imbalance of anabolic and catabolic activities in AC of aged wild-type mice. This is consistent with our previous study showing that knockdown of Nfat1 resulted in dysfunction of cultivated articular chondrocytes from young adult wild-type mice13. Forced expression of Nfat1 in cultured chondrocytes isolated from aged mice is able to reverse aberrant expression of specific anabolic and catabolic genes. It is noteworthy that forced expression of Nfat1 in cultured chondrocytes derived from 18-month-old mice rescues aberrant expression of collagen type-XI, IL-1β, MMP13, and TNFα, but not aggrecan and collagen types-II and -IX. However, the rescuable effect on the expression of aggrecan and collagen types-II and -IX is significantly improved in the cultured chondrocytes from 15- month-old mice, suggesting that decreased expression of some anabolic genes may be more reversible in relatively young mice. Thus, therapeutic efficacy of NFAT1 on aging-related dysfunction of articular chondrocytes appears to be more effective in younger mice than in older ones.
A recent editorial review24 highlighted a role of NFAT family members in maintaining cartilage and joint health and emphasized that more work is required to elucidate both upstream and downstream components of the NFAT pathway. In the current study, our ChIP assays confirmed that NFAT1 binds the promoter regions of Acan, Col2a1, Col9a1, Col11a1, Il1b, Mmp13 and Tnfa genes, indicating that NFAT1 is able to directly regulate the expression of both anabolic and catabolic genes in articular chondrocytes. In addition, IL-1β and TNFα are proinflammatory cytokines which may stimulate the expression of MMPs and suppress the expression of chondrocyte markers such as type-II collagen and aggrecan25. These results suggest that NFAT1 plays an important role in regulating the transcription of specific anabolic and catabolic genes in AC of aged mice.
We previously showed that Nfat1−/− mice exhibit normal skeletal development but begin to show dysfunction of articular chondrocytes in young adults13. The present study revealed that NFAT1 continues to regulate articular chondrocyte function after middle-age. These findings suggest that NFAT1 is not required for skeletal development, but is essential for the homeostasis of AC in adult and aged mice. This is in contrast to other transcription factors (e.g., Sox9, Runx2/Cbfa1, and β-catenin) whose deletion causes defective skeletal cell differentiation and bone/cartilage formation26–28.
This study revealed that the age-dependent decrease in Nfat1 expression in AC after 6 months of age is regulated primarily by epigenetic histone methylation. Reduced enrichment of H3K4me2 (a histone code linked to transcriptional activation) in the Nfat1 promoter region is associated with decreased Nfat1 expression in articular chondrocytes of aged mice. Histone methylation was dynamically regulated by histone methytransferases and histone lysine demethylases (KDMs). KDM1 and KDM5 both target histone 3 lysine 4 di- or tri-methylation22, 23, 29. In the present study, siRNA-mediated knockdown of Lsd1 (encoding LSD1/KDM1a) in articular chondrocytes from 18-month-old mice up-regulated Nfat1 expression concomitant with increased H3K4me2 at the Nfat1 promoter, indicating that LSD1 is a key demethylase responsible for reduced NFAT1 expression in AC of aged mice.
DNA methylation is another well-studied epigenetic regulatory mechanism of gene transcription. Loss of DNA methylation at CpG sites in the promoters of IL-1β and specific MMPs as well as in the NF-kappaB enhancer elements of inducible nitric oxide synthase was found responsible for expression of these genes in human articular chondrocytes30–32. The current study demonstrated that age-dependent reduction of NFAT1 expression is not associated with the level of DNA methylation in the Nfat1 promoter region. However, a negative correlation between the age-dependent Nfat1 expression levels and DNA methylation status at introns 1 and 10 of the Nfat1 gene was detected. Since gene body DNA methylation can regulate gene expression33, 34, DNA methylation at these introns may play a role in the age-dependent Nfat1 expression in AC of mice. Further studies are needed to fully elucidate the mechanisms of DNA methylation in regulation of Nfat1 expression in AC of mice.
Age-related changes, including cellular senescence and reduced responsiveness to growth factors by chondrocytes, impair the ability of chondrocytes to maintain cartilage homeostasis and regenerate damaged cartilage4, 5, 35, 36. In addition, aging has a deleterious effect on the extracellular matrix of AC due to the changes in size, structure, and function of proteoglycan and collagen, compromising the biomechanical properties of AC in aged individuals37, 38. Furthermore, the age-related decline in muscle strength and reduced flexibility of ligaments and tendons may compromise the ability of elderly subjects to handle the mechanical stress (force/unit area) on their AC39. In the present study, however, only 20% of aged wild-type mice (with reduced NFAT1 expression) displayed OA changes in their knee joints. In contrast, the majority of Nfat1−/− mice (with complete loss of NFAT1 function) showed severe OA in multiple joints (knee, hip, shoulder) around 12 months of age12. These observations suggest that the level of NFAT1 expression in AC affects the onset and severity of OA in mice.
It has been suggested that loss of proteoglycans (loss of safranin-O staining) in AC is an early sign of OA40. In this study, focal or generalized loss of proteoglycans in AC was seen in almost all aged mice. Eighty percent of these mice, however, did not show OA-like structural changes19 even by 24 months of age. In a dog model of joint disuse, loss of safranin-O staining of AC that resulted from immobilization of the knee was fully reversible after removal of the cast and return to load-bearing for 3 weeks41. Therefore, although loss of safranin-O staining in AC is a sign of cartilage degradation, it is not a specific indicator of OA. Nonetheless, AC of aged mice with significant loss of proteoglycans due to dysfunction of articular chondrocytes should be considered an abnormal joint tissue that predisposes the joint to OA under mechanical stress that the metabolically abnormal cartilage cannot adequately withstand. This could explain, at least in part, the high incidence of OA in aging individuals.
In summary, the present study has revealed that NFAT1 plays an important role in regulating the transcription of specific anabolic and catabolic genes. Histone methylation-regulated and DNA methylation associated spontaneous reduction of NFAT1 expression in AC of aged mice results in imbalanced metabolic activities of articular chondrocytes. AC of aged mice, therefore, is an abnormal joint tissue that may predispose the joint to OA under mechanical stress. These findings may provide new insights into the mechanisms underlying the aging of AC. The results of this pre-clinical study justify further investigation of the relevance of age-dependent expression of NFAT1 in human joint tissues to development of OA in older humans.
Acknowledgments
Some 15-month-old mice used in this study were gifted from the National Institute on Aging (NIA, Bethesda, MD). The authors thank Dr. Jennifer McEllin and Mr. Andrew Miller for graphic design and editorial assistance.
Role of the funding source
Research reported in this article was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health (NIH) under Award Number R01 AR059088 (to J. Wang), the Mary A. & Paul R. Harrington Distinguished Professorship Endowment, and the Asher Orthopedic Research Endowment.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authors’ contributions
Study design: JW and MZ. Study conduct: MZ, QL, BE, and JW. Data analysis and interpretation: MZ, JW, XZ, and KB. Drafting manuscript: MZ, JW, XZ, and KB. MZ and JW take responsibility for the integrity of the data analysis.
Competing interests
The authors declare that they have no competing interests.
Contributor Information
Mingcai Zhang, Email: mzhang@kumc.edu.
Qinghua Lu, Email: qlu@kumc.edu.
Brian Egan, Email: began@kumc.edu.
Xiao-bo Zhong, Email: xiaobo.zhong@uconn.edu.
Kenneth Brandt, Email: kenbrandt@yahoo.com.
References
- 1.Loeser RF. Aging and osteoarthritis. Curr Opin Rheumatol. 2011;23:492–496. doi: 10.1097/BOR.0b013e3283494005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Y, Jordan JM. Epidemiology of osteoarthritis. Clin Geriatr Med. 2010;26:355–369. doi: 10.1016/j.cger.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nehrer S, Dorotka R, Domayer S, Stelzeneder D, Kotz R. Treatment of full-thickness chondral defects with hyalograft C in the knee: a prospective clinical case series with 2 to 7 years' follow-up. Am J Sports Med. 2009;37(Suppl 1):81s–87s. doi: 10.1177/0363546509350704. [DOI] [PubMed] [Google Scholar]
- 4.Martin JA, Buckwalter JA. The role of chondrocyte senescence in the pathogenesis of osteoarthritis and in limiting cartilage repair. J Bone Joint Surg Am. 2003;85-A(Suppl 2):106–110. doi: 10.2106/00004623-200300002-00014. [DOI] [PubMed] [Google Scholar]
- 5.Guerne PA, Blanco F, Kaelin A, Desgeorges A, Lotz M. Growth factor responsiveness of human articular chondrocytes in aging and development. Arthritis Rheum. 1995;38:960–968. doi: 10.1002/art.1780380712. [DOI] [PubMed] [Google Scholar]
- 6.Carlo MD, Jr, Loeser RF. Increased oxidative stress with aging reduces chondrocyte survival: correlation with intracellular glutathione levels. Arthritis Rheum. 2003;48:3419–3430. doi: 10.1002/art.11338. [DOI] [PubMed] [Google Scholar]
- 7.Mueller MB, Tuan RS. Anabolic/Catabolic balance in pathogenesis of osteoarthritis: identifying molecular targets. PM R. 2011;3:S3–S11. doi: 10.1016/j.pmrj.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 8.Sandell LJ, Aigner T. Articular cartilage and changes in arthritis. An introduction: cell biology of osteoarthritis. Arthritis Res. 2001;3:107–113. doi: 10.1186/ar148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xanthoudakis S, Viola JP, Shaw KT, Luo C, Wallace JD, Bozza PT, et al. An enhanced immune response in mice lacking the transcription factor NFAT1. Science. 1996;272:892–895. doi: 10.1126/science.272.5263.892. [DOI] [PubMed] [Google Scholar]
- 10.Hodge MR, Ranger AM, Charles de la Brousse F, Hoey T, Grusby MJ, Glimcher LH. Hyperproliferation and dysregulation of IL-4 expression in NF-ATp-deficient mice. Immunity. 1996;4:397–405. doi: 10.1016/s1074-7613(00)80253-8. [DOI] [PubMed] [Google Scholar]
- 11.Rao A, Luo C, Hogan PG. Transcription factors of the NFAT family: regulation and function. Annu Rev Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- 12.Wang J, Gardner BM, Lu Q, Rodova M, Woodbury BG, Yost JG, et al. Transcription factor Nfat1 deficiency causes osteoarthritis through dysfunction of adult articular chondrocytes. J Pathol. 2009;219:163–172. doi: 10.1002/path.2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodova M, Lu Q, Li Y, Woodbury BG, Crist JD, Gardner BM, et al. Nfat1 regulates adult articular chondrocyte function through its age-dependent expression mediated by epigenetic histone methylation. J Bone Miner Res. 2011;26:1974–1986. doi: 10.1002/jbmr.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greenblatt MB, Ritter SY, Wright J, Tsang K, Hu D, Glimcher LH, et al. NFATc1 and NFATc2 repress spontaneous osteoarthritis. Proc Natl Acad Sci U S A. 2013;110:19914–19919. doi: 10.1073/pnas.1320036110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kozlowska E, Biernacka M, Ciechomska M, Drela N. Age-related changes in the occurrence and characteristics of thymic CD4(+) CD25(+) T cells in mice. Immunology. 2007;122:445–453. doi: 10.1111/j.1365-2567.2007.02667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishikawa S, Yasoshima A, Doi K, Nakayama H, Uetsuka K. Involvement of sex, strain and age factors in high fat diet-induced obesity in C57BL/6J and BALB/cA mice. Exp Anim. 2007;56:263–272. doi: 10.1538/expanim.56.263. [DOI] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Urita A, Matsuhashi T, Onodera T, Nakagawa H, Hato M, Amano M, et al. Alterations of high-mannose type N-glycosylation in human and mouse osteoarthritis cartilage. Arthritis Rheum. 2011;63:3428–3438. doi: 10.1002/art.30584. [DOI] [PubMed] [Google Scholar]
- 19.Pritzker KPH. Pathology of osteoarthritis. In: Brandt KD, Doherty M, Lohmander LS, editors. Osteoarthritis. New York: Oxford University Press; 2003. pp. 49–58. [Google Scholar]
- 20.Zhou B, Wu B, Tompkins KL, Boyer KL, Grindley JC, Baldwin HS. Characterization of Nfatc1 regulation identifies an enhancer required for gene expression that is specific to pro-valve endocardial cells in the developing heart. Development. 2005;132:1137–1146. doi: 10.1242/dev.01640. [DOI] [PubMed] [Google Scholar]
- 21.Rudolf R, Busch R, Patra AK, Muhammad K, Avots A, Andrau JC, et al. Architecture and expression of the nfatc1 gene in lymphocytes. Front Immunol. 2014;5:21. doi: 10.3389/fimmu.2014.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 23.Kooistra SM, Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol. 2012;13:297–311. doi: 10.1038/nrm3327. [DOI] [PubMed] [Google Scholar]
- 24.Beier F. NFATs are good for your cartilage! Osteoarthritis Cartilage. 2014;22:893–895. doi: 10.1016/j.joca.2014.04.011. [DOI] [PubMed] [Google Scholar]
- 25.Goldring SR, Goldring MB. The role of cytokines in cartilage matrix degeneration in osteoarthritis. Clin. Orthop. Relat. Res. 2004;427(Suppl):S27–S36. doi: 10.1097/01.blo.0000144854.66565.8f. [DOI] [PubMed] [Google Scholar]
- 26.Bi W, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B. Sox9 is required for cartilage formation. Nat Genet. 1999;22:85–89. doi: 10.1038/8792. [DOI] [PubMed] [Google Scholar]
- 27.Day TF, Guo X, Garrett-Beal L, Yang Y. Wnt/beta-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev Cell. 2005;8:739–750. doi: 10.1016/j.devcel.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 28.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 29.Rotili D, Mai A. Targeting Histone Demethylases: A New Avenue for the Fight against Cancer. Genes Cancer. 2011;2:663–679. doi: 10.1177/1947601911417976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Andres MC, Imagawa K, Hashimoto K, Gonzalez A, Roach HI, Goldring MB, et al. Loss of methylation in CpG sites in the NF-kappaB enhancer elements of inducible nitric oxide synthase is responsible for gene induction in human articular chondrocytes. Arthritis Rheum. 2013;65:732–742. doi: 10.1002/art.37806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hashimoto K, Oreffo RO, Gibson MB, Goldring MB, Roach HI. DNA demethylation at specific CpG sites in the IL1B promoter in response to inflammatory cytokines in human articular chondrocytes. Arthritis Rheum. 2009;60:3303–3313. doi: 10.1002/art.24882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roach HI, Yamada N, Cheung KS, Tilley S, Clarke NM, Oreffo RO, et al. Association between the abnormal expression of matrix-degrading enzymes by human osteoarthritic chondrocytes and demethylation of specific CpG sites in the promoter regions. Arthritis Rheum. 2005;52:3110–3124. doi: 10.1002/art.21300. [DOI] [PubMed] [Google Scholar]
- 33.Hellman A, Chess A. Gene body-specific methylation on the active X chromosome. Science. 2007;315:1141–1143. doi: 10.1126/science.1136352. [DOI] [PubMed] [Google Scholar]
- 34.Yang X, Han H, De Carvalho DD, Lay FD, Jones PA, Liang G. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. 2014;26:577–590. doi: 10.1016/j.ccr.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blaney Davidson EN, Scharstuhl A, Vitters EL, van der Kraan PM, van den Berg WB. Reduced transforming growth factor-beta signaling in cartilage of old mice: role in impaired repair capacity. Arthritis Res Ther. 2005;7:R1338–R1347. doi: 10.1186/ar1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eltawil NM, De Bari C, Achan P, Pitzalis C, Dell'accio F. A novel in vivo murine model of cartilage regeneration. Age and strain-dependent outcome after joint surface injury. Osteoarthritis Cartilage. 2009;17:695–704. doi: 10.1016/j.joca.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wells T, Davidson C, Morgelin M, Bird JL, Bayliss MT, Dudhia J. Age-related changes in the composition, the molecular stoichiometry and the stability of proteoglycan aggregates extracted from human articular cartilage. Biochem J. 2003;370:69–79. doi: 10.1042/BJ20020968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vaughan-Thomas A, Dudhia J, Bayliss MT, Kadler KE, Duance VC. Modification of the composition of articular cartilage collagen fibrils with increasing age. Connect Tissue Res. 2008;49:374–382. doi: 10.1080/03008200802325417. [DOI] [PubMed] [Google Scholar]
- 39.Loeser RF, Shakoor N. Aging or osteoarthritis: which is the problem? Rheum Dis Clin North Am. 2003;29:653–673. doi: 10.1016/s0889-857x(03)00062-0. [DOI] [PubMed] [Google Scholar]
- 40.Nerlich AG. Histochemical and Immunohistochemical staining of cartilage sections. In: An YH, Martin KL, editors. Handbook of histology methods for bone and cartilage. Totowa, New Jersey: Humana press; 2003. pp. 295–313. [Google Scholar]
- 41.Palmoski MJ, Brandt KD. Running inhibits the reversal of atrophic changes in canine knee cartilage after removal of a leg cast. Arthritis Rheum. 1981;24:1329–1337. doi: 10.1002/art.1780241101. [DOI] [PubMed] [Google Scholar]