Abstract
Repositioning of approved drugs has recently gained new momentum for rapid identification and development of new therapeutics for diseases that lack effective drug treatment. Reported repurposing screens have increased dramatically in number in the past five years. However, many newly identified compounds have low potency; this limits their immediate clinical applications because the known, tolerated plasma drug concentrations are lower than the required therapeutic drug concentrations. Drug combinations of two or more compounds with different mechanisms of action are an alternative approach to increase the success rate of drug repositioning.
Keywords: Drug combination, drug repurposing, drug repositioning, high-throughput screening
Introduction
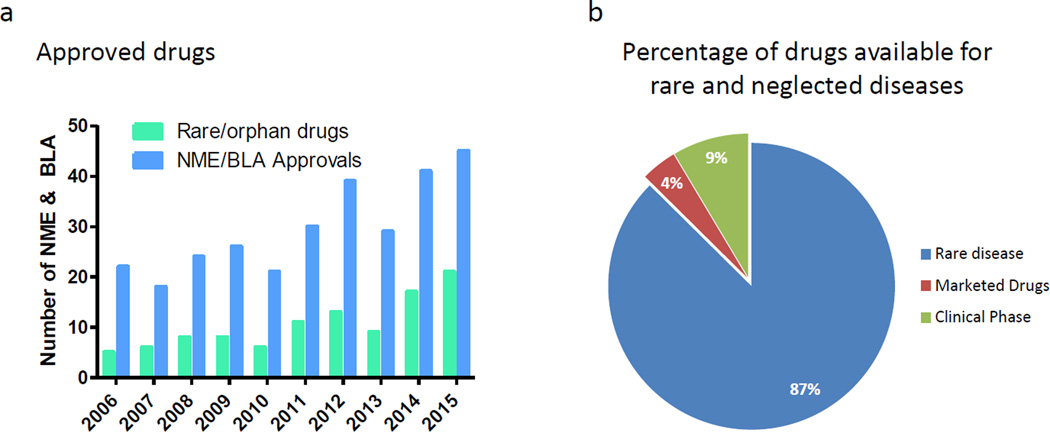
Although the pharmaceutical industry spends billions of dollars on R&D [1], the number of new drugs approved has been around 40 per year over the past five years (http://www.fda.gov/Drugs/DevelopmentApprovalProcess/DrugInnovation) (Figure 1a). The success rate of new drug discovery and development does not satisfactorily address the unmet clinical need for disease treatments. Common diseases such as Alzheimer’s disease (AD), Parkinson’s disease, congestive heart failure and pulmonary hypertension still lack effective therapeutics. In addition, there are over 7800 rare and neglected diseases (https://rarediseases.info.nih.gov), most of which lack approved drug treatments. Although there are 281 approved drugs for these orphan diseases and 600 compounds in clinical trials, there are still approximately 7000 diseases without drug treatment (Figure 1b). Despite an increase in FDA approvals for drugs for use in rare or orphan diseases in 2014 and 2015 [2] (Figure 1a), alternative approaches to speed up the drug development for these 7000 diseases are urgently needed.
Figure 1.
The gap between current drug development, untreated rare diseases and growth of drug repositioning screens. (a) Number of new molecular entities (NMEs) and biologics license applications (BLAs) approved by the Center for Drug Evaluation and Research (CDER) from 2006 to 2015. Data from Drugs@FDA(http://www.fda.gov/Drugs/DevelopmentApprovalProcess/DrugInnovation). (b) The percentage of currently approved drugs and investigational drugs for rare diseases. Data from http://www.prnewswire.com/news-releases/global-orphan-drug-market-to-reach-us-120-billion-by-2018-244195511.html.
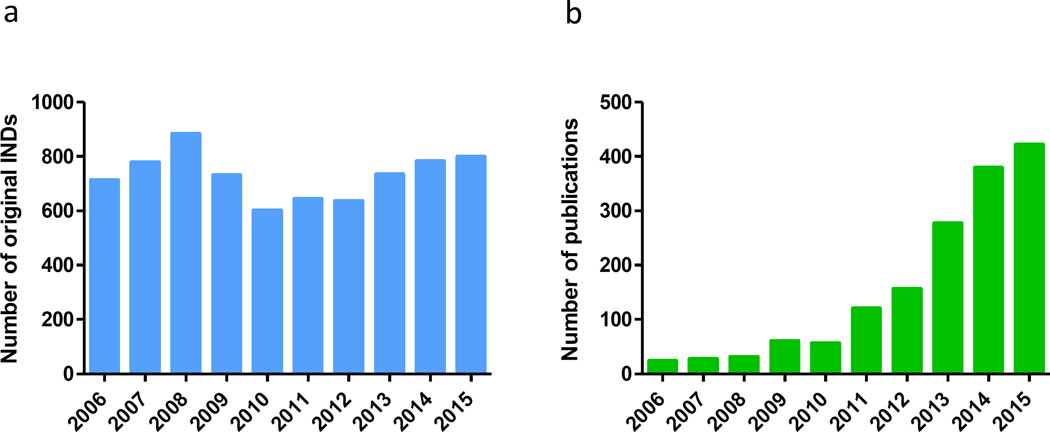
In the past decade, new technologies such as induced pluripotent stem cells (iPSCs), clustered regularly interspaced short palindromic repeats (CRISPR) gene editing, proteomics and next-generation sequencing have emerged and greatly enhanced research for target identification, disease modeling and drug discovery. Phenotypic screening has regained momentum and has been extensively used in drug discovery and development. However, the translation rate from basic research and drug discovery to approved drugs remains rather disappointing. In the 10-year period from 2006 to 2015, the number of original investigational new drug (IND) applications submitted was stable at around 700 per year (http://www.fda.gov/Drugs/DevelopmentApprovalProcess/) (Figure 2a); but over this period only 20 to 40 new drugs were approved each year (Figure 1a), a less than 6% success rate. Development of new drug therapies remains time-consuming and costly. New strategies, new approaches and new technologies are needed to accelerate new drug discovery and to improve the success rate of drug development. Repositioning existing drugs and drug candidates offers an alternative approach to develop new therapeutics quickly for many diseases that currently do not have treatments.
Figure 2.
(a) Number of original investigational new drug (IND) applications received by the FDA from commercial sources Data from http://www.fda.gov/Drugs/DevelopmentApprovalProcess/. (b) Growth of drug repositioning (repurposing) screens indexed in PubMed from 2006 to 2015. In January 2016 we searched PubMed with key words of ‘drug repositioning’ and ‘drug repurposing’ with publication dates from 01 January 2006 to 31 December 2015. Search for each year was started from 01 January and ended with 31 December.
Different types of drug repurposing
Historically, a number of drugs have been repurposed based on clinical results. Sildenafil (Viagra®) was initially studied for the treatment of hypertension and angina pectoris by Pfizer in the 1980s. It failed for angina, but unexpectedly showed erectile effects. This compound was then marketed as the first oral treatment for erectile dysfunction in the USA [3]. This is an example of drugs that were originally meant to treat one malady but were discovered during clinical trials to have other effects.
The second mode of drug repurposing is arrived at via compound screening using approved drug collections. This type of drug repurposing has been boosted in the past five years owing to the availability of drug collections and improved screening technologies as evidenced by publications that increase from under 100 to over 400 per year (Figure 2b). In 2007, Chong et al. identified itraconazole as a potent hit for inhibiting angiogenesis from a screen of the Johns Hopkins Drug Library (JHDL) [4]. In the follow-up preclinical studies, itraconazole showed promising results in several cancer models. It directly entered into several Phase II studies and showed positive results in advanced lung cancer, prostate cancer and basal cell carcinoma trials [5].
The third method of drug repurposing elucidation is a recent program initiated by the National Center for Advancing Translational Sciences (NCATS) at the National Institutes of Health (NIH). Launched in 2012, this initiative connected academic researchers and eight of the largest pharma companies for the opportunity to repurpose 58 unsuccessful investigational drugs for new disease indications. Huge amounts of effort and resources had been spent for advancing these compounds into clinical trials. By making available these drugs to academic researchers, the hope is that novel therapeutic indications might be found for these abandoned compounds. For example, in 2015 AstraZeneca’s AZD0530, a failed new drug for solid tumors, exhibited Fyn kinase activity and is a promising therapeutic candidate for the treatment of AD [6,7]. Currently, a Phase IIa clinical trial of AZD0530 for treating patients with AD is underway [6]. This development demonstrates the utility of these previously failed drug candidates and a great shortening of drug development times by eliminating preclinical drug development and further Phase I clinical trials.
Compound collections for drug repurposing screens
As of 31 December 2015, 1539 drugs had been approved by the FDA since its establishment in 1938. Every year another 20 to 40 new drugs will accumulate in this pool with current trends. In 2015, WHO announced 409 essential medicines [8]. In addition, there is a pool of drug candidates that are either in active clinical trials or have failed in different stages because of insufficient efficacy. Clinical studies registered in the USA as of 14 January 2016 numbered 78 140 (Clinicaltrials.gov.), and 15 130 of them are currently at the patient recruitment stage. Approximately half of these clinical studies are registered as drug or biologic interventions. All these approved drugs and drug candidates have passed the preclinical drug development stage with appropriate profiles of animal efficacy, pharmacokinetics (PK) and toxicology. Most of them include rich information on clinical pharmacology and toxicology.
Repositioning of approved drugs has emerged as an alternative approach to identify new treatments for diseases that lack effective treatments. In January 2016 we searched PubMed for literature regarding drug reposition with the keywords ‘drug repositioning’ and ‘drug repurposing’ with publication dates from 01 January 2006 to 31 December 2015. In the past ten years, there has been a significant increase in published papers for drug repositioning and/or repurposing (Figure 2b). The increase in the number of accessible approved drug collections combined with the drug repurposing screening efforts by academia, government and industry has contributed greatly to the increase in drug-reposition-related publications.
Three groups of compounds are usually included in screening collections for drug repurposing. The first one comprises drugs approved for marketing by the FDA or other regulatory agencies; these are available in pharmacies. The second one consists of drugs that were previously approved but that are no longer used, and that need to be accessed by customized synthesis or purchased from commercial vendors. The third group comprises clinical investigational compounds that could be obtained from pharmaceutical companies, commercial vendors or by customized synthesis. Table 1 shows a list of drug libraries available from academic and government organizations; many commercial libraries are also available.
Table 1.
List of various FDA-approved and other-approved drug collections and the number of compounds in each
| Drug collection | Number of drugs in collection |
|---|---|
| Johns Hopkins Drug Library (JHDL) [64] | 1600 |
| National Chemical Genomics Centera [65] | ~2750 |
| National Institutes of Health (NIH) clinical collectionb [66] | ~450 |
| National Institute of Neurological Disorders and Stroke (NINDS 1040) [67] | 1040 |
This pharmaceutical collection is at the National Center for Advancing Translational Sciences (NCATS), which is also known as NCGC Pharmaceutical Collection (NPC).
This clinical collection can be accessed commercially through the company Evotec.
Phenotypic screening assays
Phenotypic screens have a new momentum in drug discovery [9,10]. Different from molecular-target-based ones, phenotypic screens do not require detailed understanding of the disease targets and networks. Phenotypic screens offer the advantage of identifying potential treatments for complicated diseases, where there might be difficulty in identifying the primary therapeutic targets. Executing this approach requires a characteristic phenotype associated with the disease that is known. Cell-based phenotypic assays usually use primary cells [11], isolated pathogens [12], engineered cell lines [13] or the recently emerged iPSC-derived cells including neuronal cells, cardiomyocytes, hepatocytes and epithelial cells [14–16]. As an example of this, Eggan, Woolf and co-workers discovered hyperexcitability as a result of a reduced delayed-rectifier potassium channel as a disease phenotype in iPSC-derived motor neurons from amyotrophic lateral sclerosis (ALS) patients [16]. An approved anticonvulsant, retigabine was then shown to correct the phenotype and improve the in vitro survival of motor neurons derived from ALS patients. Because retigabine is an approved drug, a Phase II clinical trial of retigabine in ALS subjects was immediately started in 2015. This report indicates that iPSC-derived disease models can provide an alternative to animal models for drug screening and drug efficacy tests before human clinical trials.
High IC50 values of identified compounds: a bottleneck in repurposing screens
An emerging challenge for drug repurposing screens is the inability to identify clinically useful compounds for new indications. This could be because of either weak potency of the identified hits, with effective concentration for 50% of the maximum response (IC50) values higher than the safely achievable plasma concentrations in humans, or a simple lack of active compounds. In a malaria repurposing screen [17], 27 of 32 hits identified had high IC50 values (>10 nM) compared with dihydroartemisinin (IC50 <10nM) [18], a standard drug. According to the non-profit foundation Medicines for Malaria Venture (MMV), for a candidate to be considered as a late lead the IC50 or IC90 of a compound needs to be less than 10 nM for potency in erythrocyte assays. The weak potency of these hits prevented their use as a single drug therapy for malaria infection. Weak potency of hits from repurposing screens was also reported for other diseases, such as chronic lymphocytic leukemia [19] and metabolic disorders [20].
In another repositioning screen for identification of compounds that block Ebola virus entry [21], 53 hits had been found but only four of them have efficacies (IC90) at or below their maximum serum concentration (Cmax) values in human blood. If a drug’s achievable blood concentration is below its efficacy value for the new indication, this newly identified compound obviously cannot be used in patients. Although these hits can provide new chemical scaffolds and potential new targets for drug development, the low efficacy and/or limited PK profiles of hit compounds hamper rapid drug repositioning efforts. This problem has become a bottleneck in the use of drug repurposing screens for the identification of clinically useful compounds.
Drug combination reduces drug concentrations of individual drugs
From the literature search and our own recent practice, we have found that drug combination therapy using two-to-three compounds with different mechanisms of action can overcome the above described drug repurposing screen challenge. The use of drugs in combination can produce a synergistic effect if each of the drugs impinges on a different target or signaling pathway that results in reduction of required drug concentrations for each individual drug. Therefore, use of drug combinations could increase the success rate of drug repurposing screens. This can be achieved in two steps: the drug repurposing screen with approved drug collections to identify hit compounds and then a drug combination test with the identified hit compounds to find effective drug combinations for clinical use with a new indication.
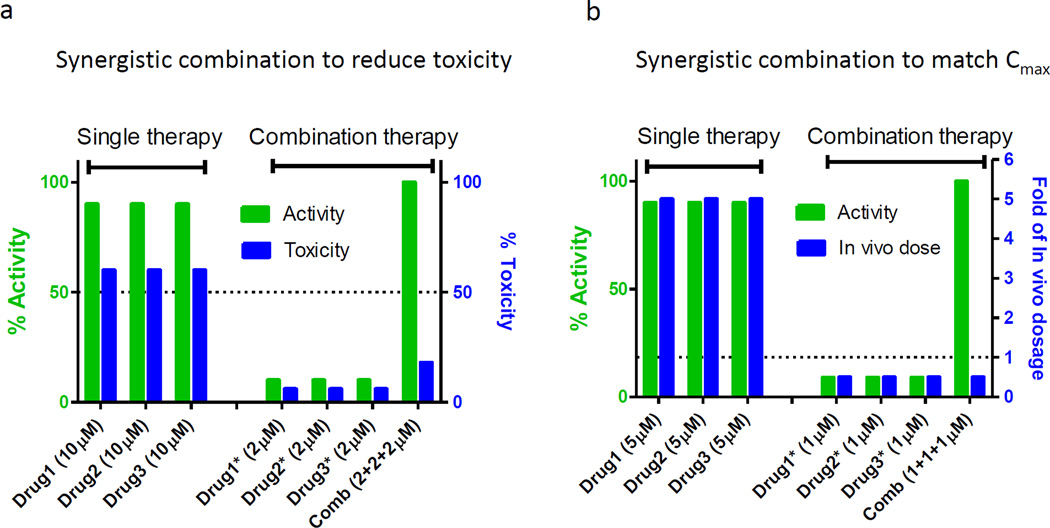
Based on our experience we propose two approaches to finding synergistic combinations using hits from drug repositioning screens. The first type of synergistic combination will address the issue related to weak compound potency relative to toxicity. As shown in Figure 3a, the concentrations of drug 1, drug 2 or drug 3 exhibit 90% efficacy at a concentration that would trigger severe cytotoxicity if used by themselves. However, the synergistic effect of a three-drug combination enables the reduction of each of the individual drug concentrations to 1/10 of the concentration used in the single drug treatment.
Figure 3.
Two synergistic combination models to optimize hits from drug repositioning screens. (a) Synergistic combination to reduce toxicity when drug 1, drug 2 or drug 3 showed 90% efficacy at single use, the drug concentration will trigger severe toxicity (e.g., 60%). Through synergistic combination, the combined efficacy is not reduced but each drug concentration was reduced to 1/10, leading to a lower toxicity (e.g., 6%). (b) Synergistic combination to match in vivo dosage. When drug 1, drug 2 or drug 3 showed 90% efficacy at single use, the drug concentration is fivefold of their allowed in vivo dosages (Cmean or Cmax). Through synergistic combination, the combined efficacy is not reduced but each drug concentration was reduced to 1/10, reaching the allowed in vivo concentration (1/2 of Cmean or Cmax).
The second type of synergistic combination is to correlate the effective drug concentrations with the reported achievable drug concentration in human blood. As shown in Figure 3b, the effective concentration of drug 1, drug 2 or drug 3 as a single use medication is fivefold higher than their achievable human blood concentration (Cmean or Cmax). It is obvious that these drugs individually are not practically useful for clinical application. The effective drug combinations can be identified by screening of different sets of three-drug combinations with individual drug concentrations equal to or below their achievable human blood concentrations (Figure 3b).
Polypharmacology: another factor for drug combination therapy
Recently, the importance of multifactor and polygenic pathologies is being recognized for many diseases including neurodegenerative diseases, cancer, diabetes and hypertension. The high attrition rates of drug candidates in clinical trials could partly result from underestimation of the complexity of the pathophysiology in these diseases [22,23]. These diseases might not be caused by a single factor or genetic variant but rather are associated with multiple factors or genetic determinants. In addition, disease manifestations can be affected by many other factors, such as age, tissue type and environmental stimuli. Therefore, simultaneous targeting of different proteins or signaling pathways of a disease network might be necessary to combat these diseases when previous efforts using single drugs have failed to produce effective therapeutics.
One of the polypharmacology approaches is a single drug that binds to and modulates the functions of multiple targets related to disease pathophysiology [24]. Another polypharmacology approach is the use of multiple drugs that interact with different targets related to the disease pathogenesis. A combination of two-to-three drugs to treat a disease, also known as drug combination therapy, has been more broadly used in clinical practice. Although it is still up for debate whether a single multitarget drug or a combination with selective drugs is a better treatment strategy, drug combination therapy is clearly a practical useful approach to extending the clinical applications of drug repositioning.
Computational approach for polypharmacology
Computer-aided drug design is also useful for development of multitargeted drugs or combination therapies. Structure-based methods, ligand-based approaches, QSAR or docking simulation and deep learning are well documented virtual screening technologies [25,26]. The Connectivity map (CMAP) established the first collection for genome-wide transcriptional expression data from small-molecule-treated human cells and simple pattern-matching algorithms [27]. Butte and colleagues performed a large-scale correlation analysis for chemical structures and gene expression from PubChem and Library of Integrated Network-based Cellular Signatures (LINCS) [28]. By using the expression data from CMAP and LINCS, the connection among small molecules, genes and diseases could guide future drug repurposing efforts. Recently, Reutlinger and co-workers developed a quantitative polypharmacology model for 640 human drug targets [29]. Rodrigues et al. designed quantitative bioactivity models to achieve structurally novel, selective, potent and ligand-efficient 5-hydroxytryptamine 2B receptor (5-HT2B) modulators with sustained cell-based effects [30]. Gujral et al. combined elastic net regulation with mRNA expression profiling of 32 kinase inhibitors to reveal kinases that are important for epithelial and mesenchymal cell migration [31]. The RCSB Protein Data Bank (RCSB PDB; http://www.rcsb.org) provides 3D structures of macromolecules. The use of structure-based methods to search second- and off-targets will improve the understanding of diseases and repurposed drugs [32]. A retrospective cohort study showed that population-based electronic healthcare records (EHRs) could provide early drug safety profiles [33].
Existing drug combination therapies
Dysregulation of multiple signaling pathways is a hallmark of cancer [34–36]. Targeting multiple proteins such as kinases in the key pathways might be more effective than a drug targeting a single protein. For example, bosutinib, an approved drug for the treatment of chronic myelogenous leukemia (CML), is an ATP-competitive inhibitor of multiple kinases including the breakpoint cluster region-Abelson fusion protein (Bcr-Abl) tyrosine kinase and Src family kinases (Src, Lyn and Hck) [37]. Metformin in combination with temozolomide showed promising synergistic efficacy for treatment of glioblastoma [38]. In addition, in 2014, using a pharmacological drug combination screen of 76 drugs, a combination of repurposed drug AZD with crizotinib showed marked efficacy in vitro and in vivo for cancer treatment and for overcoming cancer drug resistance [39].
The drug combination therapy for HIV infection was developed in the 1990s [40], and is now routinely applied with three- or four-drug cocktails. So far the FDA has approved 13 fixed-dose two-to-four drug combination therapies for the treatment of HIV. In 2014, the FDA approved a four-drug combination therapy (ombitasvir, partaprevir, ritonavir, dasabuvir) for HCV genotype 1 infections [41].
For infection with malaria, in the 1990s WHO recommended artemisinin-based combination therapy as a first-line treatment to overcome drug resistance to the previous gold-standard drug chloroquine, with increasing reports of resistance to artemisinin in Southeast Asia [42]. In 2015, a two-way screening of 13 910 drug pairs identified repurposed drug NVP-BGT226, artemether (ATM) and lumefantrine (LUM) with antimalarial activities in vitro and in vivo [43]. Existing drugs (benznidazole and nifurtimox) for Chagas disease infected with Trypanosoma cruzi are characterized with weak potency and significant toxicities. A repurposing screen of 300 two-way combinations of 24 drugs discovered two pairs of drugs with demonstrated efficacy in vivo [44].
In the world of rare diseases, Acanthamoeba keratitis, a rare disease caused by parasitic infection of the eye, can result in permanent visual impairment or blindness. A combination of miltefosine and olyphexamethylene biguanide is effective for treatment of Acanthamoeba keratitis infection [45]. Langerhans cell histiocytosis (LCH) is a rare disorder predominantly occurring in infants and children. A combination of prednisone and vinblastine and 6-mercatopurine was able to control the disease [46]. Charcot-Marie-Tooth disease type 1A (CMT1A) is a hereditary motor and sensory neuropathy with no treatment available. A combination of three repurposed drugs (baclofen, naltrexone and sorbitol) showed promising results in a Phase II study [47] and its Phase III trial is ongoing.
Drug combination therapy with different antibiotics and/or antifungal reagents is frequently used in the intensive care unit (ICU) for patients with severe infections [48]. In the emergency situation where the exact pathogens are not identified, two-drug (or more) combinations are the practical approach to control the infections. In 2015, the FDA approved the drug combination of avibactam and ceftazidime for treatment of infections caused by multidrug-resistant bacteria. Multidrug-resistant bacteria produce beta-lactamases which break down beta-lactams, causing resistance to ceftazidime and other β-lactam antibiotics. Although avibactam itself has no antibacterial activity, avibactam covalently bonds to and inhibits the activity of β-lactamase [49].
Potential for drug–drug interaction with drug combination therapy
The potential for adverse drug–drug interactions (DDIs) is a concern when selecting and prioritizing drug combinations with synergistic efficacy for clinical applications. Several types of DDI have been identified and characterized. An adverse DDI could be pharmacodynamic (PD) (target) in origin [45]; however the most common and best characterized adverse DDIs are PK (ADME) in origin. An example of a victim–perpetrator pair is the combination of selective estrogen receptor modulator tamoxifen and selective serotonin reuptake inhibitor (SSRI) paroxetine. In the 1990s, doctors prescribed SSRIs to patients with breast cancer to treat depression and reduce the side effects of tamoxifen. Later, it was reported that the paroxetine (the perpetrator) reduced the plasma concentrations of endoxifen, the active metabolite of tamoxifen, through inhibition of the metabolism of tamoxifen (the victim) by cytochrome P450 (CYP)2D6 [50]. In 2010, a study showed that this DDI led to a significant increase in the risk of death from breast cancer in patients who took paroxetine and tamoxifen compared with patients taking only tamoxifen [51].
The most common metabolic enzyme, CYP3A4, is responsible for many PK DDIs [52]. NS3 viral protease inhibitors [boceprevir, telaprevir, simeprevir and faldaprevir for treatment of hepatitis C virus (HCV)] are metabolized by CYP enzymes. Their combination use with CYP3A and CYP3A inhibitors (e.g., ketoconazole) leads to higher toxic levels of the drugs, or inducers like rifampin which lead to lower, less efficacious levels of the drugs [53].
Advantages and shortfalls of drug combination therapy for drug repurposing
As we discussed above, there are three major advantages of drug combination therapy for drug repositioning. The first one is the potential for synergistic effects of a drug combination that significantly reduces required drug concentrations for each of the individual drugs used in the combination. This greatly increases the chances for useful clinical applications of such drugs identified from drug repurposing screens, which are otherwise insufficiently active as single agents. The second benefit is the reduction or delay of the development of drug resistance as a result of the multiple targeting mechanisms of the drug combination. Additionally, it has been reported that partial inhibition of a small number of targets could be more efficient than complete inhibition of a single target [54].
The potential for adverse DDIs should be considered with drug combination therapy. DDIs can increase drug adverse effects or toxicity, or might reduce drug efficacy. Another issue is that formulation of multiple drugs is more complex than individual drugs. In 2009, Merck delayed its three-drug combination for the treatment of high cholesterol because of a formulation issue [55]. For example, one drug might physically or chemically interact with the other drug(s). Differences in drug solubility and stability in the combination also need to be considered.
Useful tools for drug combination therapy
Computation models are useful tools for predicting drug combinations for potential clinical uses. A comprehensive summary of various bioinformatics approaches and databases for drug repositioning studies has been reviewed [56]. A recent study described an efficient combination drug screening method using feedback system control (FSC) [57]. This method used a phenotypic cell viability assay to generate dose–response curves for each drug first. Then, a differential evolution (DE) algorithm was used to predict new combinations from applied drug combinations.
DrugBank provides a drug–target network that reveals the potential target (1466 proteins, metabolizing enzymes, carriers and transporters), off-target, DDIs and side effects of 1486 FDA-approved drugs and 828 investigational drugs that include new molecular entities (NMEs) and repurposed drugs [58]. PharmGKB provides comprehensive information for pharmacogenetics interactions. Other databases are the side effect resource, SIDER, FDA Adverse Event Reporting System (FAERS) and DailyMed. But the data from animals for repurposed drugs are also included in some databases, which might not accurately predict the real situation in humans. For approved or investigational drugs, DDIs can be found in established databases such as DrugBank or SIDER. Several methods have been developed to search existing DDIs rapidly from published results [59,60]. For relatively novel compounds, a recent study reports a similarity-based computational approach to predict PK and PD DDIs [61]. In 2012, hundreds of new DDIs were reported based on the tests of 656 approved drugs and 73 targets [62]. Clinicians are encouraged to report case studies for drug combination therapies, even when they show limited or no efficacy in trials [63].
Concluding remarks
Drug combination therapy with a synergistic effect can increase the success rates of drug repositioning. A phenotypic repurposing screen allows identification of new therapies from approved drug collections without an understanding of the disease pathophysiology. The identification of effective, synergistic drug combinations could lead to an increased understanding of complicated disease pathophysiology and to the design of better treatments for the disease. Drug repositioning offers hope to the many people afflicted with rare diseases with no present therapies.
Highlights.
Publications for drug repositioning have drastically increased in past five years
Different types of drug repositioning and available resources have been summarized
Drug combination increases the success rate of drug repurposing screens
Acknowledgments
This work was supported by the Intramural Research Program of the National Center for Advancing Translational Sciences, National Institutes of Health. The authors would like to thank DeeAnn Visk for reading and critiquing the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mullard A. 2014 FDA drug approvals. Nat. Rev. Drug Discov. 2015;14:77–81. doi: 10.1038/nrd4545. [DOI] [PubMed] [Google Scholar]
- 2.Lindsley CW. 2014 prescription medications in the United States: tremendous growth, specialty/orphan drug expansion, and dispensed prescriptions continue to increase. ACS Chem. Neurosci. 2015;6:811–812. doi: 10.1021/acschemneuro.5b00148. [DOI] [PubMed] [Google Scholar]
- 3.Boolell M, et al. Sildenafil: an orally active type 5 cyclic GMP-specific phosphodiesterase inhibitor for the treatment of penile erectile dysfunction. Int. J. Impot. Res. 1996;8:47–52. [PubMed] [Google Scholar]
- 4.Chong CR, et al. Inhibition of angiogenesis by the antifungal drug itraconazole. ACS Chem. Biol. 2007;2:263–270. doi: 10.1021/cb600362d. [DOI] [PubMed] [Google Scholar]
- 5.Rudin CM, et al. Phase 2 study of pemetrexed and itraconazole as second-line therapy for metastatic nonsquamous non-small-cell lung cancer. J. Thorac. Oncol. 2013;8:619–623. doi: 10.1097/JTO.0b013e31828c3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nygaard HB, et al. A Phase Ib multiple ascending dose study of the safety, tolerability, and central nervous system availability of AZD0530 (saracatinib) in Alzheimer’s disease. Alzheimers Res. Ther. 2015;7:35. doi: 10.1186/s13195-015-0119-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaufman AC, et al. Fyn inhibition rescues established memory and synapse loss in Alzheimer mice. Annal. Neurol. 2015;77:953–971. doi: 10.1002/ana.24394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gray AL, et al. Essential medicines are still essential. Lancet. 2015;386:1601–1603. doi: 10.1016/S0140-6736(15)00514-0. [DOI] [PubMed] [Google Scholar]
- 9.Zheng W, et al. Phenotypic screens as a renewed approach for drug discovery. Drug Discov. Today. 2013;18:1067–1073. doi: 10.1016/j.drudis.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swinney DC, Anthony J. How were new medicines discovered? Nat. Rev. Drug Discov. 2011;10:507–519. doi: 10.1038/nrd3480. [DOI] [PubMed] [Google Scholar]
- 11.Sandercock AM, et al. Identification of anti-tumour biologics using primary tumour models, 3-D phenotypic screening and image-based multi-parametric profiling. Mol. Cancer. 2015;14:147. doi: 10.1186/s12943-015-0415-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun W, et al. Chemical signatures and new drug targets for gametocytocidal drug development. Sci. Rep. 2014;4:3743. doi: 10.1038/srep03743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Titus SA, et al. High-throughput multiplexed quantitation of protein aggregation and cytotoxicity in a Huntington’s disease model. Curr. Chem. Genom. 2012;6:79–86. doi: 10.2174/1875397301206010079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ebert AD, Svendsen CN. Human stem cells and drug screening: opportunities and challenges. Nat. Rev. Drug Discov. 2010;9:367–372. doi: 10.1038/nrd3000. [DOI] [PubMed] [Google Scholar]
- 15.Carlson C, et al. Phenotypic screening with human iPS cell-derived cardiomyocytes: HTS-compatible assays for interrogating cardiac hypertrophy. J. Biomol. Screen. 2013;18:1203–1211. doi: 10.1177/1087057113500812. [DOI] [PubMed] [Google Scholar]
- 16.Wainger BJ, et al. Intrinsic membrane hyperexcitability of amyotrophic lateral sclerosis patient-derived motor neurons. Cell Rep. 2014;7:1–11. doi: 10.1016/j.celrep.2014.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuan J, et al. Chemical genomic profiling for antimalarial therapies, response signatures, and molecular targets. Science. 2011;333:724–729. doi: 10.1126/science.1205216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mu J, et al. Plasmodium falciparum genome-wide scans for positive selection, recombination hot spots and resistance to antimalarial drugs. Nat. Genet. 2010;42:268–271. doi: 10.1038/ng.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen M, et al. Identification of therapeutic candidates for chronic lymphocytic leukemia from a library of approved drugs. PloS One. 2013;8:e75252. doi: 10.1371/journal.pone.0075252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu M, et al. delta-Tocopherol reduces lipid accumulation in Niemann-Pick type C1 and Wolman cholesterol storage disorders. J. Biol. Chem. 2012;287:39349–39360. doi: 10.1074/jbc.M112.357707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kouznetsova J, et al. Identification of 53 compounds that block Ebola virus-like particle entry via a repurposing screen of approved drugs. Emerging Microbes Infections. 2014;3:e84. doi: 10.1038/emi.2014.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palop JJ, et al. A network dysfunction perspective on neurodegenerative diseases. Nature. 2006;443:768–773. doi: 10.1038/nature05289. [DOI] [PubMed] [Google Scholar]
- 23.Barabasi AL, et al. Network medicine: a network-based approach to human disease. Nat. Rev. Genet. 2011;12:56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bolognesi ML. Polypharmacology in a single drug: multitarget drugs. Curr. Med. Chem. 2013;20:1639–1645. doi: 10.2174/0929867311320130004. [DOI] [PubMed] [Google Scholar]
- 25.Rastelli G, Pinzi L. Computational polypharmacology comes of age. Front. Pharmacol. 2015;6:157. doi: 10.3389/fphar.2015.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valencia A, et al. A novel QSAR model of Salmonella mutagenicity and its application in the safety assessment of drug impurities. Toxicol. Appl. Pharmacol. 2013;273:427–434. doi: 10.1016/j.taap.2013.09.015. [DOI] [PubMed] [Google Scholar]
- 27.Lamb J, et al. The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 28.Chen B, et al. Relating chemical structure to cellular response: an integrative analysis of gene expression, bioactivity, and structural data across 11,000 compounds. CPT Pharmacometrics Syst. Pharmacol. 2015;4:576–584. doi: 10.1002/psp4.12009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reutlinger M, et al. Multi-objective molecular de novo design by adaptive fragment prioritization. Angewandte Chemie. 2014;53:4244–4248. doi: 10.1002/anie.201310864. [DOI] [PubMed] [Google Scholar]
- 30.Rodrigues T, et al. Multidimensional de novo design reveals 5-HT2B receptor-selective ligands. Angewandte Chemie. 2015;54:1551–1555. doi: 10.1002/anie.201410201. [DOI] [PubMed] [Google Scholar]
- 31.Gujral TS, et al. Exploiting polypharmacology for drug target deconvolution. Proc. Natl. Acad. Sci. USA. 2014;111:5048–5053. doi: 10.1073/pnas.1403080111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rose PW, et al. The RCSB Protein Data Bank: views of structural biology for basic and applied research and education. Nucleic Acids Res. 2015;43:D345–D356. doi: 10.1093/nar/gku1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Bie S, et al. The role of electronic healthcare record databases in paediatric drug safety surveillance: a retrospective cohort study. Br. J. Clin. Pharmacol. 2015;80:304–314. doi: 10.1111/bcp.12610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Webster RM. Combination therapies in oncology. Nat. Rev. Drug Discov. 2016;15:81–82. doi: 10.1038/nrd.2016.3. [DOI] [PubMed] [Google Scholar]
- 35.Jia J, et al. Mechanisms of drug combinations: interaction and network perspectives. Nat. Rev. Drug Discov. 2009;8:111–128. doi: 10.1038/nrd2683. [DOI] [PubMed] [Google Scholar]
- 36.Knight ZA, et al. Targeting the cancer kinome through polypharmacology. Nat. Rev. Cancer. 2010;10:130–137. doi: 10.1038/nrc2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daud AI, et al. Phase I study of bosutinib, a Src/Abl tyrosine kinase inhibitor, administered to patients with advanced solid tumors. Clin. Cancer Res. 2012;18:1092–1100. doi: 10.1158/1078-0432.CCR-11-2378. [DOI] [PubMed] [Google Scholar]
- 38.Yu Z, et al. Metformin and temozolomide act synergistically to inhibit growth of glioma cells and glioma stem cells in vitro and in vivo. Oncotarget. 2015;6:32930–32943. doi: 10.18632/oncotarget.5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crystal AS, et al. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science. 2014;346:1480–1486. doi: 10.1126/science.1254721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ho DD. Time to hit Hiv, early and aard. N. Eng. J. Med. 1995;333:450–451. doi: 10.1056/NEJM199508173330710. [DOI] [PubMed] [Google Scholar]
- 41.Abramowicz M, et al. A 4-drug combination (Viekira Pak) for hepatitis C. JAMA. 2015;313:1857–1858. doi: 10.1001/jama.2015.4562. [DOI] [PubMed] [Google Scholar]
- 42.Ashley EA, et al. Spread of artemisinin resistance in Plasmodium falciparum malaria. N. Eng. J. Med. 2014;371:411–423. doi: 10.1056/NEJMoa1314981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mott BT, et al. High-throughput matrix screening identifies synergistic and antagonistic antimalarial drug combinations. Sci. Rep. 2015;5:13891. doi: 10.1038/srep13891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Planer JD, et al. Synergy testing of FDA-approved drugs identifies potent drug combinations against Trypanosoma cruzi. PLoS Negl. Trop. Dis. 2014;8:e2977. doi: 10.1371/journal.pntd.0002977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Polat ZA, et al. Miltefosine and polyhexamethylene biguanide: a new drug combination for the treatment of Acanthamoeba keratitis. Clin. Experiment. Ophthalmol. 2014;42:151–158. doi: 10.1111/ceo.12120. [DOI] [PubMed] [Google Scholar]
- 46.Matsuki E, et al. Successful treatment of adult onset Langerhans cell histiocytosis with multi-drug combination therapy. Int. Med. 2011;50:909–914. doi: 10.2169/internalmedicine.50.4808. [DOI] [PubMed] [Google Scholar]
- 47.Attarian S, et al. An exploratory randomised double-blind and placebo-controlled Phase 2 study of a combination of baclofen, naltrexone and sorbitol (PXT3003) in patients with Charcot-Marie-Tooth disease type 1A. Orphanet J. Rare Dis. 2014;9:199. doi: 10.1186/s13023-014-0199-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zilberberg MD, et al. Multi-drug resistance, inappropriate initial antibiotic therapy and mortality in Gram-negative severe sepsis and septic shock: a retrospective cohort study. Crit. Care. 2014;18:596. doi: 10.1186/s13054-014-0596-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhanel GG, et al. Ceftazidime-avibactam: a novel cephalosporin/beta-lactamase inhibitor combination. Drugs. 2013;73:159–177. doi: 10.1007/s40265-013-0013-7. [DOI] [PubMed] [Google Scholar]
- 50.Jin Y, et al. CYP2D6 genotype, antidepressant use, and tamoxifen metabolism during adjuvant breast cancer treatment. J. Natl. Cancer Inst. 2005;97:30–39. doi: 10.1093/jnci/dji005. [DOI] [PubMed] [Google Scholar]
- 51.Kelly CM, et al. Selective serotonin reuptake inhibitors and breast cancer mortality in women receiving tamoxifen: a population based cohort study. BMJ. 2010;340:c693. doi: 10.1136/bmj.c693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kiser JJ, et al. Drug–drug interactions during antiviral therapy for chronic hepatitis C. Nat. Rev. Gastroenterol. Hepatol. 2013;10:596–606. doi: 10.1038/nrgastro.2013.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ciesek S, et al. Second-wave protease inhibitors: choosing an heir. Clin. Liver Dis. 2011;15:597–609. doi: 10.1016/j.cld.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 54.Csermely P, et al. The efficiency of multi-target drugs: the network approach might help drug design. Trends Pharmacol. Sci. 2005;26:178–182. doi: 10.1016/j.tips.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 55.Frantz S. The trouble with making combination drugs. Nat. Rev. Drug Discov. 2006;5:881–882. doi: 10.1038/nrd2188. [DOI] [PubMed] [Google Scholar]
- 56.Jin GX, Wong STC. Toward better drug repositioning: prioritizing and integrating existing methods into efficient pipelines. Drug Discov. Today. 2014;19:637–644. doi: 10.1016/j.drudis.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nowak-Sliwinska P, et al. Optimization of drug combinations using feedback system control. Nat. Protocols. 2016;11:302–315. doi: 10.1038/nprot.2016.017. [DOI] [PubMed] [Google Scholar]
- 58.Barneh F, et al. Updates on drug-target network; facilitating polypharmacology and data integration by growth of DrugBank database. Brief. Bioinf. 2015 doi: 10.1093/bib/bbv094. pii: bbv094. [DOI] [PubMed] [Google Scholar]
- 59.Duke JD, et al. Literature based drug interaction prediction with clinical assessment using electronic medical records: novel myopathy associated drug interactions. PLoS Comput. Biol. 2012;8:e1002614. doi: 10.1371/journal.pcbi.1002614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu Y, et al. A novel algorithm for analyzing drug-drug interactions from MEDLINE literature. Sci. Rep. 2015;5:17357. doi: 10.1038/srep17357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vilar S, et al. Similarity-based modeling in large-scale prediction of drug–drug interactions. Nat. Protocols. 2014;9:2147–2163. doi: 10.1038/nprot.2014.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lounkine E, et al. Large-scale prediction and testing of drug activity on side-effect targets. Nature. 2012;486:361–367. doi: 10.1038/nature11159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Markowitz M, et al. A randomized open-label study of 3-versus 5-drug combination antiretroviral therapy in newly HIV-1-infected individuals. JAIDS. 2014;66:140–147. doi: 10.1097/QAI.0000000000000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chong CR, et al. A clinical drug library screen identifies astemizole as an antimalarial agent. Nat. Chem. Biol. 2006;2:415–416. doi: 10.1038/nchembio806. [DOI] [PubMed] [Google Scholar]
- 65.Huang R, et al. The NCGC pharmaceutical collection: a comprehensive resource of clinically approved drugs enabling repurposing and chemical genomics. Sci. Transl. Med. 2011;3:80ps16. doi: 10.1126/scitranslmed.3001862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Oprea TI, et al. Lead-like, drug-like or "Pub-like": how different are they? J. Comput. Aided Mol. Des. 2007;21:113–119. doi: 10.1007/s10822-007-9105-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang J, et al. Reversal of a full-length mutant huntingtin neuronal cell phenotype by chemical inhibitors of polyglutamine-mediated aggregation. BMC Neurosci. 2005;6:1. doi: 10.1186/1471-2202-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]