This study investigated whether diffusion tensor imaging tractography-based connectome can differentiate global developmental delay from speech delay in young children. Twelve children with pure speech delay, 14 children with global developmental delay, and 10 children with typical development underwent 3T DTI. Whole-brain connectome analysis was performed by using 116 cortical ROIs. Network metrics were measured at individual regions: strength, efficiency, cluster coefficient, and betweeness. Compared with typical development, global and local efficiency were significantly reduced in both global developmental delay and speech delay. Nodal strength of the cognitive network was reduced in global developmental delay, whereas the nodal strength of the language network was reduced in speech delay. This finding resulted in a high accuracy of >83% to discriminate global developmental delay from speech delay.
Abstract
BACKGROUND AND PURPOSE:
Pure speech delay is a common developmental disorder which, according to some estimates, affects 5%–8% of the population. Speech delay may not only be an isolated condition but also can be part of a broader condition such as global developmental delay. The present study investigated whether diffusion tensor imaging tractography-based connectome can differentiate global developmental delay from speech delay in young children.
MATERIALS AND METHODS:
Twelve children with pure speech delay (39.1 ± 20.9 months of age, 9 boys), 14 children with global developmental delay (39.3 ± 18.2 months of age, 12 boys), and 10 children with typical development (38.5 ± 20.5 months of age, 7 boys) underwent 3T DTI. For each subject, whole-brain connectome analysis was performed by using 116 cortical ROIs. The following network metrics were measured at individual regions: strength (number of the shortest paths), efficiency (measures of global and local integration), cluster coefficient (a measure of local aggregation), and betweeness (a measure of centrality).
RESULTS:
Compared with typical development, global and local efficiency were significantly reduced in both global developmental delay and speech delay (P < .0001). The nodal strength of the cognitive network is reduced in global developmental delay, whereas the nodal strength of the language network is reduced in speech delay. This finding resulted in a high accuracy of >83% ± 4% to discriminate global developmental delay from speech delay.
CONCLUSIONS:
The network abnormalities identified in the present study may underlie the neurocognitive and behavioral consequences commonly identified in children with global developmental delay and speech delay. Further validation studies in larger samples are required.
Global developmental delay (GD) is caused by a broad spectrum of etiologies that result in the impairment of multiple developmental domains such as language, motor function, cognition, social interaction, and activities of daily living.1 Its prevalence is estimated to be 1%–3% in children younger than 5 years of age.1 Children with isolated speech and language delay (SD) represent a distinct group with specific impairment in the receptive and/or expressive language domains in the context of otherwise intact neurocognitive and social functioning. SD in children is a common condition, which, according to some estimates, affects 5%–8% of the population.2,3
Even though speech and language are affected in both the GD and SD groups, the absence of additional abnormalities in other domains (ie, motor, daily living skills) characterizes the SD group. It is important to differentiate children with GD or SD into distinct subgroups as early as possible to provide accurate prognostic information and appropriate intervention.4 More important, direct developmental assessment by using psychometrics is often unreliable in young children, particularly those with developmental delay or impairment.5,6 Thus, new objective methods for potentially discriminating SD from GD in the first few years of life are needed to provide the most effective interventions in a timely manner.
Using noninvasive imaging approaches such as diffusion tensor imaging may provide critical clinical information and new insight into the neural basis of GD and SD. Conventional clinical MR imaging is typically unremarkable in most of these patients. Indeed, clinically used neuroimaging tools are of limited value in evaluating children with SD or GD except to rule out a lesional/structural etiology. Therefore, there is an urgent need to develop noninvasive neuroimaging approaches to improve our etiologic yield and understand the anatomic substrates of these disorders. With DTI tractography, it was found that a subset of children with GD showed poorly developed white matter tracts such as the arcuate fasciculus and inferior longitudinal fasciculus.7 In a subsequent tract-based morphometric study, it was further found that both diffusion and geometric properties of the arcuate fasciculus were abnormal in a subset of children with GD.8 In Angelman syndrome, a severe, syndromic form of GD, abnormalities were also found in multiple major cortical association tracts by using DTI tractography9 and Tract-Based Spatial Statistics (http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/TBSS).10 Overall, white matter abnormalities in GD and other related neurodevelopmental disorders have been replicated by many studies.11–13
The goal of this study was to investigate a DTI tool by using connectome analysis to refine white matter abnormalities in children with GD and SD. This tool models the human brain as a network or a graph represented by a collection of nodes (ie, cortical and subcortical regions) and links (ie, axonal fiber counts between nodes), which may provide a powerful way of examining the structural connections in specific brain networks and how these connectivity strengths are associated with specific functional phenotypes. To quantify the degree of connectivity strength in clinical DTI data that typically samples the molecular displacement of water diffusion at a lower angular resolution, a novel DTI tractography method, referred to as “independent component analysis with ball-stick model” (ICA+BSM)14 was recently developed. This method combines 2 complementary approaches, ICA and BSM, to isolate multiple fiber bundles in a single voxel: The first is ICA, to approximate fiber orientations of multiple cylindric tensors existing in a local cluster, and the second is BSM, to refine the ICA-driven initial orientation of multiple cylindric tensors mixed in a single voxel of a local cluster. The major advantage of ICA+BSM is that it isolates independently attenuated diffusion profiles from “neighboring voxels” to optimize initial guesses of multiple tensor orientations existing in a single voxel. The method has an outstanding accuracy to detect different white matter pathways associated with primary motor and language functions by resolving the crossing-fiber problem in clinical DTI data.15,16
In this study, we used ICA+BSM tractography to determine whether abnormal connectivity patterns based on whole-brain connectome analysis can be used to improve the classification of young children with GD and SD. Several studies have noted that the volumes of the subcortical structures, including the hippocampus, correlated with intelligence quotient (IQ), which is a clinical measure defining the severity of GD.17–19 It seems likely that in children, poorly developed cortical/subcortical structures may exist and account for unrecognized distinctions between the subgroups of developmental delay (ie, GD versus pure SD). The present study presumes that the comprehensive evaluation by using whole-brain connectome analysis may allow us to clearly differentiate patients with GD and SD from healthy controls. We hypothesized that compared with healthy controls, patients with GD or pure SD will both have significantly reduced efficiency in both long- and short- range axonal connections in their whole-brain network and that while children with SD will show localized cortical connectivity abnormalities centered on the frontotemporal language network, children with GD will show broader corticosubcortical network abnormalities.
Materials and Methods
Subjects
Fourteen children with significant global developmental delay defined by impaired global cognition (IQ < 70) and adaptive behavioral functioning impaired in at least 2 developmental domains (gross/fine motor, speech/language, daily living skills, and socialization skills) were recruited for the GD group (39.3 ± 18.2 months of age, 12 boys). In addition, 12 children with isolated speech and language delay, defined by intact global cognition and measured language functioning in the impaired range (expressive and/or receptive language score of <70) and measured adaptive behavior measured within normal limits in daily living, socialization, and motor skills were recruited for the SD group (39.1 ± 20.9 months of age, 9 boys). Ten typically developing (TD) children, defined by measured global cognition, language, and adaptive behavior (communication, daily living, socialization, motor) skills within normal limits (standard score of ≥85) were recruited for healthy controls (38.5 ± 20.5 months of age, 7 boys). These children were recruited from the local area by an active community outreach effort.
Two-sample t tests showed that all 3 groups did not differ on age (P > .42) or sex (P > .37). For each group, we applied the following exclusion criteria: 1) history of seizures, 2) history of prematurity or a perinatal hypoxic-ischemic event, 3) focal deficits on clinical examination by a pediatric neurologist, 4) dysmorphic features suggestive of a clinical syndrome, 5) diagnosis of an autism spectrum disorder or attention deficit/hyperactivity disorder, 6) MR imaging findings interpreted as abnormal by a pediatric neuroradiologist, 7) comparative genomic hybridization microarray and/or Fragile X tests positive, 8) an inborn error of metabolism, 9) history of maltreatment, 10) being bilingual, and 11) being left-handed.
The present study was approved by institutional review board of the university, and written informed consent was obtained from all parents/guardians.
Data Acquisition
All MR imaging scans were obtained on a 3T Signa scanner (GE Healthcare, Milwaukee, Wisconsin) equipped with an 8-channel head coil and an array spatial sensitivity encoding technique. DTI was acquired with a multisection single-shot diffusion-weighted echo-planar imaging sequence at TR = 12,500 ms, TE = 88.7 ms, FOV = 24 cm, 128 × 128 acquisition matrix, contiguous 3-mm thickness to cover entire axial sections of the whole brain by using 55 isotropic gradient directions with b=1000 s/mm2, 1 b=0 acquisition, and NEX = 1. This DTI scan takes about 11 minutes. For anatomic reference, a 3D fast spoiled gradient-echo sequence was acquired for each participant at TR/TE/TI of 9.12/3.66/400 ms, section thickness of 1.2 mm, and planar resolution of 0.94 × 0.94 mm2, which takes approximately 3 minutes. Because the scans for children with GD and SD were clinical MR imaging studies, sedation was used as necessary by the sedation team. None of children with TD were sedated for the MR imaging. They were scanned while sleeping and were monitored for movement during scanning. If there was significant movement, the MR imaging was not used in the present study.
Data Processing
For each subject, an ICA+BSM tractography14 was applied for whole-brain tractography to avoid the intravoxel crossing-fiber problem and to isolate up to the orientations of 3 crossing-fiber bundles at every voxel. Before performing ICA+BSM tractography analysis for the structural connectivity, the National Institutes of Health TORTOISE package (https://science.nichd.nih.gov/confluence/display/nihpd/TORTOISE) was used to correct motion artifacts in the DTI data. Whole-brain streamline tractography was then performed by using ICA+BSM to reconstruct up to 3 crossing streamlines by applying 30 randomized seeding points at every voxel of fractional anisotropy of >0.20. The first eigenvectors of the stick components having a fractional ratio of >0.05 were considered as the reconstructed fiber orientations and were then used for the streamline tractography at step size = 0.2 voxel width, turning angle threshold = 60°, and maximal length = 250 mm.
For the connectome analysis, 116 cortical regions (or nodes) of interest were generated by fitting a deformable template of the Automated Anatomical Labeling atlas (AAL, http://www.cyceron.fr/index.php/en/plateforme-en/freeware), resulting in 116 × 116 connectivity matrices in which the elements quantify the pair-wise connectivity scores (ie, streamline tract numbers connecting any 2 given cortical regions normalized by the corresponding tract mean lengths). The SPM8 Diffeomorphic Anatomical Registration Through Exponentiated Lie Algebra approach (http://www.fil.ion.ucl.ac.uk/spm/software/spm12) was used to obtain an optimal nonlinear deformation to warp the AAL template to individual subjects.
One-way ANOVA followed by the Benjamini and Hochberg procedure20 for multiple comparisons was applied to identify pairs of AAL regions showing significantly altered connectivity between 2 groups (TD versus GD, TD versus SD, and GD versus SD). For each of 3 between-group comparisons, the ANOVA was initially applied at each element of the upper triangular part of the 116 × 116 connectivity matrix to test the null hypothesis of equality in the mean value of connectivity score between groups (dependent variable: score; factor: group; covariate: age). Subsequently, the regions were combined to 6 bilateral anatomic regions (ie, frontal, temporal, parietal, occipital, cerebellum, and subcortical) to reduce the number of comparisons. The Benjamini and Hochberg procedure20 was used to adjust independent P values of these regions to control the false discovery rate for multiple comparisons (α = .05). In addition, the whole-brain false discovery rate connectome analysis by using the Network Based Statistic toolbox (https://sites.google.com/site/bctnet/comparison/nbs) was applied independently to determine statistical reproducibility.
For the subsequent analysis identifying specific regions with atypically altered connectivity patterns, single-subject connectivity matrices were first binarized by thresholding entire connectivity scores, whereby we only considered the existence/absence of fiber pathways (ie, the elements were considered as one if their scores were >5% of the maximal score and as zero otherwise). The proportional thresholding of 5% was heuristically selected to minimize the variances of the network metrics used in the TD group. This was performed to ensure that between-group differences reflect alterations in network organization rather than differences in absolute connectivity. The Brain Connectivity Toolbox (https://sites.google.com/site/bctnet) was then applied to the binary matrix to assess the following network metrics: nodal strength (the sum of links connected to the node measuring local connectivity at individual nodes), global efficiency (the average of the inverse of the shortest path lengths in the whole-brain measuring the ability of the whole-brain network for parallel information transfer), local efficiency (the inverse of the average shortest path connecting the given node with all other nodes measuring the efficiency of a given node in communicating with the rest of the nodes), cluster coefficient (the fraction of triangular links around a node measuring local aggregation at individual nodes), and betweeness (the number of all shortest paths at individual nodes measuring the importance of the node). In each of the metrics, 1-way ANOVA for the linear model (dependent variable: metric; factor: group; covariate: age) was used to assess the significance of between-group differences at each of 116 AAL regions.
Finally, a support vector machine approach21 was used to differentiate GD and SD by using each of the metrics, in which a grid search approach was adopted to optimize the radial basis function of each metric by using “training samples.” The optimized radial basis function was then used to classify the “testing samples.” The differentiation performance was validated by using conventional “hold-out” cross-validation—that is, in each trial, half of the entire sample in GD and SD was randomly selected as “training instances” and rest of the sample was used as “testing instances” to evaluate 3 performance measures of that trial (ie, accuracy, sensitivity, and specificity). A total of 10,000 trials were repeated in which the performance measures of each trial were averaged to assess overall performance. In addition, the permutation test was applied to evaluate the probability of getting accuracy values higher than the ones obtained during the cross-validation procedure by chance. We permuted the group labels 10,000 times without replacement, each time randomly assigning GD and SD labels to individual subjects, and we repeated the cross-validation procedure. The number of times the accuracy of the permuted labels was higher than that obtained for the real labels was reported in P values.
Results
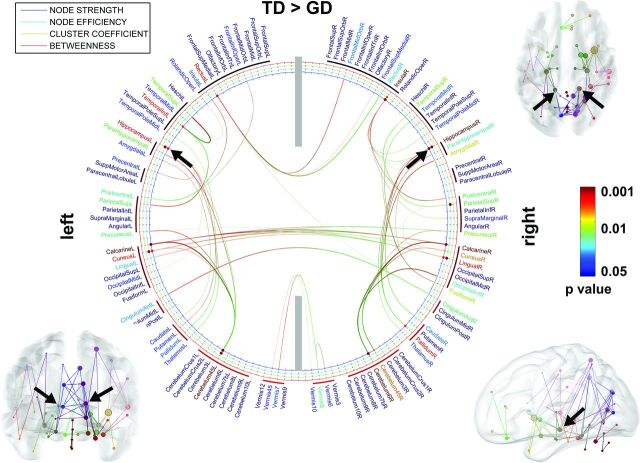
In the comparison between the TD and GD groups (Fig 1), it was observed that the TD group showed increased pair-wise connectivity (P < .05), leading to significantly increased node strength and/or efficiency in the bilateral hippocampi (F > 14.44, P < .001), bilateral caudate/putamen/globus pallidus/thalamus (F > 6.52, P < .02), left rectus (F = 7.82, P = .01), left inferior temporal (F > 7.82, P < .01), left postcentral (F > 6.52, P < .02), left insular (F > 6.52, P < .02), left superior parietal (F = 6.52, P = .02), left calcarine (F = 15.24, P < .001), left cuneus (F >13.21, P < .003), left cerebellum 4, 5 (F > 13.89, P < .002), right superior parietal (F > 6.51, P < .02), right insular (F > 13.93, P < .003), right calcarine (F = 14.82, P < .001), right cuneus (F = 7.92, P = .008), right lingual (F = 14.01, P < .005), right fusiform (F = 6.49, P = .02), and right cerebellum 4, 5, 6 (F > 7.81, P < .01). Compared with other regions such as the calcarine, cuneus, and cerebellum, the hippocampus (marked by black arrows) has more short paths to adjacent neighbors, including the parahippocampus, caudate, putamen, thalamus, and pallidum, suggesting its higher modular connectivity to detect sparse axonal connections in the GD group.
Fig 1.
ROIs showing significantly altered network metrics in the group comparison of TD > GD. In the 2D connectogram, the color of anatomic label scales the P value of group difference in the AAL template. Similarly, the color of each circle represents the P value of individual metrics. The 3D connectogram shows individual pair-wise pathways having significant group differences in nodal strength (ie, the greater radius of the sphere, the greater the group difference). In both 2D and 3D connectograms, block arrows indicate the hippocampal network whose nodal properties are significantly reduced in GD compared with TD.
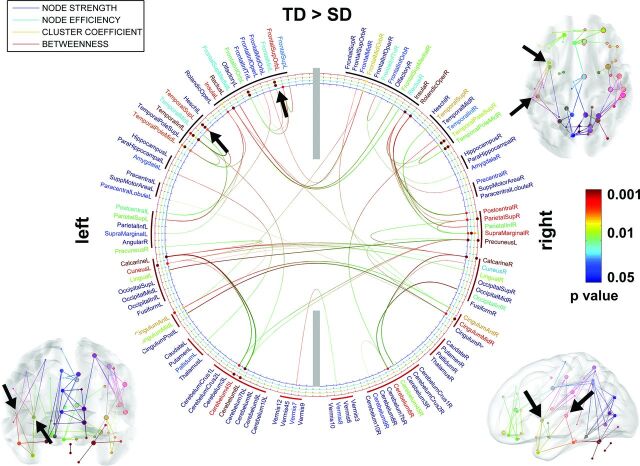
Figure 2 presents the comparison of the TD and SD groups. Compared with the SD group, the TD group shows increased pair-wise connectivity (P < .05), resulting in significantly increased node strength, efficiency, and/or clustering coefficient in the bilateral superior temporal (F >14.91, P < .001), bilateral midtemporal pole (F = 6.32, P < .01), bilateral insular (F > 6.35, P < .01), bilateral midfrontal (F > 6.29, P < .01), bilateral calcarine (F > 14.89, P < .001), bilateral inferior orbitofrontal (F > 6.26, P < .01), bilateral inferior temporal (F > 5.01, P < .03), left rectus (F > 6.20, P < .01), left superior temporal (F > 11.23, P < .005), left cuneus (F > 11.14, P < .005), left anterior cingulum (F > 10.13, P < .008), left amygdala (F = 5.73, P = .02), left pallidum (F = 5.71, P = .02), left cerebellum 4, 5(F > 11.21, P < .005), right midorbitofrontal (F > 10.62, P < .006), right inferior frontal triangularis (F > 5.12, P < .03), right superior temporal pole (F > 15.24, P < .001), right rolandic operculum (F > 5.62, P = .02), right postcentral (F = 10.67, P = .006), right superior parietal (F > 6.28, P < .01), right supramarginal (F > 5.81, P < .02), right precuneus (F > 15.11, P < .001), right calcarine (F > 15.10, P < .001), right cuneus (F = 5.89, P = .02), right inferior occipital (F = 5.85, P = .02), right anterior cingulum (F = 10.66, P = .006), right midcingulum (F > 11.01, P < .005), and right cerebellum 4, 5, 6 (F > 5.78, P < .02). A frontotemporal language network having increased axonal connections at the left midfrontal and left superior temporal gyrus was apparently distinctive in this comparison (indicated by black arrows).
Fig 2.
ROIs showing significantly altered network metrics in the group comparison of TD > SD. In the 2D connectogram, the color of anatomic label scales the P value of group difference in the AAL template. Similarly, the color of each circle represents the P value of individual metrics. The 3D connectogram shows individual pair-wise pathways having significant group differences in nodal strength (ie, the greater the radius of the sphere, the greater the group difference). In both 2D and 3D connectograms, block arrows indicate the frontotemporal language network in which nodal properties are significantly reduced in SD compared with TD.
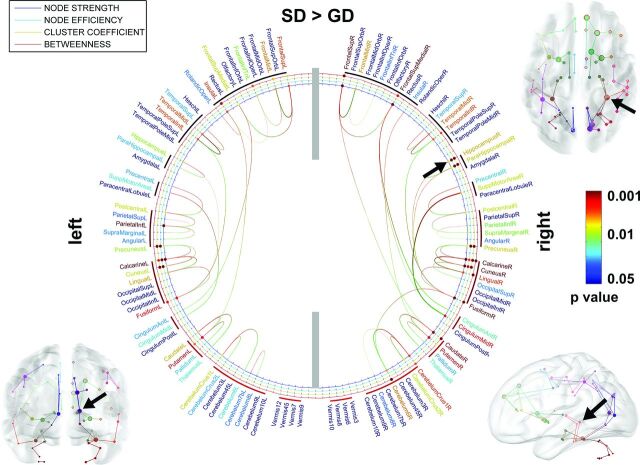
In the comparison of the SD and GD groups (Fig 3), we found that compared with the GD group, the SD group had increased node strength, efficiency, and/or cluster coefficient in both hemispheres, hippocampus (F > 6.26, P < .02), parahippocampal (F = 6.25, P < .02), superior frontal (F > 10.23, P < .004), midfrontal (F > 6.29, P < .02), inferior frontal triangularis (F > 6.26, P < .02), superior medial frontal (F > 6.28, P < .02), insular (F > 6.38, P < .02), superior temporal (F > 5.01, P < .03), midtemporal (F > 6.26, P < .02), inferior temporal (F > 6.29, P < .02), caudate/putamen/pallidum/thalamus (F > 6.89, P < .01), anterior and midcingulum (F > 6.91, P < .01), precentral/inferior parietal/supramarginal/angular (F > 6.25, P < .02), precuneus (F > 14.99, P < .001), calcarine and cuneus (F > 15.21, P < .001), lingual (F > 9.67, P < .005), fusiform (F = 9.69, P = .005), cerebellum crus 1, 2 (F > 6.27, P < .02), and cerebellum 6 and 8 (F > 6.28, P < .02). Sparser local connections are apparent in the bilateral hippocampal networks of the GD group but are more severe at the right hippocampus as indicated by black arrows.
Fig 3.
ROIs showing significantly altered network metrics in the group comparison of SD > GD. In the 2D connectogram, the color of anatomic label scales the P value of the group difference in the AAL template. Similarly, the color of each circle represents the P value of individual metrics. The 3D connectogram shows individual pair-wise pathways having significant group differences in nodal strength (ie, the greater the radius of the sphere, the greater the group difference). In both 2D and 3D connectograms, block arrows indicate the right hippocampus whose nodal properties are significantly reduced in GD compared with SD.
No significant differences were observed at P < .05 for other group contrasts such as GD > TD, SD > TD, and GD > SD.
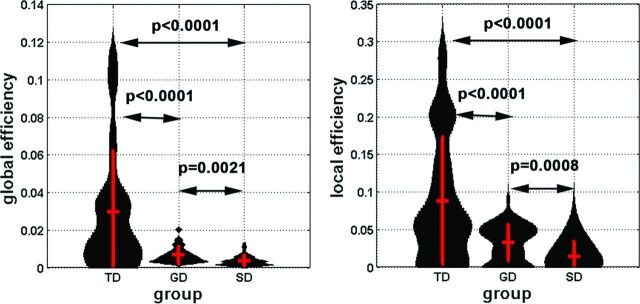
In Figs 1 and 2, we found that compared with the TD group, both the SD and GD groups showed significantly reduced inter-/intrahemispheric connections in the calcarine gyrus, lingual gyrus, rectal gyrus, superior frontal gyrus, and cerebellum, resulting in significantly impaired axonal efficiency (both global and local efficiency) in long- and short-range whole-brain connections (P < .001, Fig 4). The Network Based Statistic toolbox could replicate our findings at a small number of permutations (≤500), which reflects the lower power of the nonparametric permutation test.
Fig 4.
Global and local efficiency of the whole-brain network was obtained from individual subjects and is presented in the violin plots. Group mean and 1 SD are represented by red vertical lines. The black curve of each violin indicates the probabilistic attenuation function of the measure. To avoid the effect of arbitrary thresholding, we calculated the values of efficiencies at 3 discrete thresholds (5, 7, 10) of pair-wise connectivity scores of individual subjects to minimize the potential confounding across subjects.
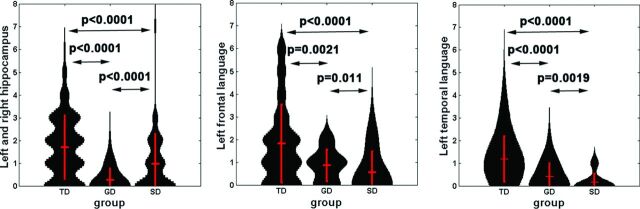
The subsequent support vector machine analysis by using leave-one-out cross-validation revealed that the nodal strengths of 3 regions, bilateral hippocampi, left frontal language (mid-/superior frontal gyrus and insular), and left temporal language (superior temporal gyrus), have significant group differences between SD and GD (P < .01, Fig 5) and achieved a high accuracy of >83% ± 4% to discriminate GD from SD (Table). The other 3 measures, including nodal efficiency, clustering coefficient, and betweeness, had relatively lower statistical significance compared with the nodal strength.
Fig 5.
Violin plots show the probability attenuation functions of nodal strengths (black) measured from bilateral hippocampus (left), left frontal language region (mid-/superior frontal gyrus/insular, middle), and left temporal language region (superior temporal gyrus, right) of each group. To estimate the probability attenuation function of individual groups, we calculated the values of nodal strength by applying 3 discrete thresholds (5, 7, 10) to the single connectivity matrix. Vertical red lines show mean ± 1 SD of each function.
Results of differentiation between GD and SD groups using SVM with nodal strengtha
| Network | Accuracy | Sensitivity | Specificity | P Value |
|---|---|---|---|---|
| Hippocampal | 89 (4) | 96 (5) | 74 (15) | .02 |
| Frontal language | 83 (4) | 93 (6) | 71 (16) | .04 |
| Temporal language | 88 (5) | 94 (5) | 77 (14) | .02 |
Note:—SVM indicates support vector machine.
The mean (SD) of accuracy, sensitivity, and specificity were reported in percentages. The P value indicates the probability of the permutation in that the accuracy of the permuted label is higher than the one obtained for the real label.
Discussion
In the present study, we found that global and local efficiency were significantly reduced in GD and SD. However, the nodal strengths of cognitive/language networks are differentially reduced between children with SD and those with GD. The GD group showed abnormal connectivity centered around the bilateral hippocampal network, whereas the left frontotemporal network was abnormal in the SD group. These abnormalities may represent the neurocognitive and behavioral features commonly identified in these children and allow subjects with SD to be distinguished from those with GD on the basis of objective parameters at a very young age when differentiation between these 2 conditions is usually difficult in the clinical setting. Furthermore, the present approach may encourage translation of advanced DTI techniques (ICA+BSM tractography effective for short-acquisition-time DTI) to clinical practice in the pediatric population, in which currently available approaches are suboptimal for whole-brain connectome analysis.
The anatomic basis of IQ, a measure defining the severity of GD, has been previously studied by neuroimaging techniques. On the basis of a review of 37 functional neuroimaging studies, Jung and Haier17 proposed a parietal-frontal integration theory of intelligence. However, other studies have noted that the volume of subcortical structures such as the hippocampus and cerebellum correlate with IQ.18,19 Such a cortical-versus-subcortical (ie, hippocampal and cerebellar) dichotomy has long been established for neurocognitive conditions such as aphasia and dementia in adults.22–24 Results of the present study are consistent with the notion that both cortical and subcortical connectivity abnormalities reported in the above studies may account for unrecognized distinctions within the GD and SD groups. Thus, the present study provides preliminary evidence to support the existence of cortical/subcortical subgroups of GD and SD. Future studies with both task-based functional imaging and meta-analysis are required to further validate this notion with a larger sample size.
It has also been observed that whole-brain, gray matter, and white matter volumes correlate with IQ.25 In particular, volumes of different white matter tracts, a measure proportional to some of the network metrics used in the present study, were found to have high heritability.26 Given such high heritability of tract volumes for IQ, it seems likely that a focused effort to identify the genetic variants responsible for low IQ in GD, by using connectivity measures such as endophenotypes, is likely to be successful. In fact, such an effort could identify mutations in 2 axon guidance genes (EN2 and MID1) in patients with GD.27 Our future studies will expand on this theme by using the network abnormalities as endophenotypes to identify the underlying genetic mechanisms driving the white matter abnormalities. By combining connectome and genetic techniques (eg, whole exome sequencing), we may be able to more comprehensively define the origin of abnormal cognitive/language networks in children with GD and SD.
The present study was limited by a small sample size and low spatial resolution to parcellate a small number of discrete regions in the whole brain. Due to the small sample size, most false discovery rate–corrected ANOVA P values reported in this study were statistically significant (ie, P < .05) only at the level of cortical lobar and subcortical regions. Further research needs to evaluate potential associations between axonal connectivity and network property at higher spatial resolutions and larger sample sizes to improve the statistical power of between-group comparison and also verify the reproducibility.28,29 Although the above limitations exist, our preliminary results suggest that the abnormalities of network properties reported at the bilateral hippocampi and the left frontal-temporal language network may underlie the presence of sparse connections in both cognitive and language systems. Most important, our findings also reveal differential associations between distinct structural connectivities and specific behavioral problems that are suggestive of distinct neural substrates in children with GD and SD.
Despite the group-level differences found in this study, more studies with larger samples sizes are required before connectome data can be used in individual diagnosis. Especially, current neuropsychological tests are less reliable in younger children than in older children though they are still primarily used as the clinical standard. The impact of young age may not completely invalidate the tests but may increase the noise level in group classification. This possibility could, in turn, potentially inflate the statistical significance of the group differences reported in this study. Future studies that can evaluate these children with follow-up neuropsychological assessment (when they are more reliable) will be able to validate the results of the present study. Furthermore, a combinatorial model integrating all the abnormalities found in this study, including temporal pole (semantic memory), calcarine/fusiform/cuneus (visual perception), putamen/caudate (motor skill), and insular (social emotion) can be used as the starting basis to make individual diagnosis feasible.
Conclusions
By combining ICA+BSM tractography with whole-brain connectome analysis to differentiate subjects with GD and SD from healthy controls, the present study found that nodal strengths of cognitive/language networks are differentially reduced between children with SD and those with GD. The results of the present study promise a new, refined imaging tool to better examine the subgroups of developmental disorders at a very young age and evaluate their anatomic substrates in vivo.
ABBREVIATIONS:
- AAL
Automated Anatomical Labeling
- GD
global developmental delay
- ICA+BSM
independent component analysis with ball-stick model
- IQ
intelligence quotient
- SD
speech delay
- TD
typical development
Footnotes
Disclosures: Jeong-Won Jeong—RELATED: Grant: National Institutes of Health National Institute of Neurological Disorders and Stroke 1R01NS089659 (Principal Investigator).* Senthil Sundaram—UNRELATED: Grants/Grants Pending: National Institutes of Health,* Comments: supported by a National Institute of Child Health and Human Development grant 1R01HD059817 (2009–2014), “Diffusion Tensor Imaging Biomarker in Developmental Delay.” *Money paid to the institution.
This work was supported by grant R01-NS089659 to J.-W.J. from the National Institute of Neurological Disorders and Stroke.
References
- 1. Shevell MI, Ashwal S, Donley D, et al. ; Quality Standards Subcommittee of the American Academy of Neurology, Practice Committee of the Child Neurology Society. Practice parameter: evaluation of the child with global developmental delay—report of the Quality Standards Subcommittee of the American Academy of Neurology and the Practice Committee of the Child Neurology Society. Neurology 2003;60:367–80 10.1212/01.WNL.0000031431.81555.16 [DOI] [PubMed] [Google Scholar]
- 2. Burden V, Stott CM, Forge J, et al. The Cambridge Language and Speech Project (CLASP), I: detection of language difficulties at 36 to 39 months. Dev Med Child Neurol 1996;38:613–31 [DOI] [PubMed] [Google Scholar]
- 3. Nelson HD, Nygren P, Walker M, et al. Screening for speech and language delay in preschool children: systematic evidence review for the US Preventive Services Task Force. Pediatrics 2006;117:e298–319 10.1542/peds.2005-1467 [DOI] [PubMed] [Google Scholar]
- 4. Bashir AS, Scavuzzo A. Children with language disorders: natural history and academic success. J Learn Disabil 1992;25:53–65; discussion 66–70 10.1177/002221949202500109 [DOI] [PubMed] [Google Scholar]
- 5. Downing JE, Perino DM. Functional versus standardized assessment procedures: implications for educational programming. Ment Retard 1992;30:289–95 [PubMed] [Google Scholar]
- 6. Sattler JM. Assessment of Children: Cognitive Applications. 4th ed. Vol 1. San Diego: Jerome M. Sattler; 2001 [Google Scholar]
- 7. Sundaram SK, Sivaswamy L, Makki MI, et al. Absence of arcuate fasciculus in children with global developmental delay of unknown etiology: a diffusion tensor imaging study. J Pediatr 2008;152:250–55 10.1016/j.jpeds.2007.06.037 [DOI] [PubMed] [Google Scholar]
- 8. Jeong JW, Sundram SK, Kumar A, et al. Aberrant diffusion and geometric properties in left arcuate fasciculus of developmentally delayed children: a diffusion tensor imaging study. AJNR Am J Neuroradiol 2011;32:323–30 10.3174/ajnr.A2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wilson BJ, Sundaram SK, Huq AH, et al. Abnormal language pathway in children with Angelman syndrome. Pediatr Neurol 2011;44:350–56 10.1016/j.pediatrneurol.2010.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tiwari VN, Jeong JW, Wilson BJ, et al. Relationship between aberrant brain connectivity and clinical features in Angelman syndrome: a new method using tract based spatial statistics of DTI color-coded orientation maps. Neuroimage 2012;59:349–55 10.1016/j.neuroimage.2011.07.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nagae LM, Zarnow DM, Blaskey L, et al. Elevated mean diffusivity in the left hemisphere superior longitudinal fasciculus in autism spectrum disorders increases with more profound language impairment. AJNR Am J Neuroradiol 2012;33:1720–25 10.3174/ajnr.A3037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Peters SU, Kaufmann WE, Bacino CA, et al. Alterations in white matter pathways in Angelman syndrome. Dev Med Child Neurol 2011;53:361–67 10.1111/j.1469-8749.2010.03838.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Catani M, Mesulam MM, Jakobsen E, et al. A novel frontal pathway underlies verbal fluency in primary progressive aphasia. Brain 2013;136(pt 8):2619–28 10.1093/brain/awt163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jeong JW, Asano E, Yeh FC, et al. Independent component analysis tractography combined with ball and stick model to isolate intra-voxel crossing fibers of the corticospinal tracts in clinical diffusion MRI. Mag Reson Med 2013;70:441–53 10.1002/mrm.24487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jeong JW, Asano E, Brown EC, et al. Automatic detection of primary motor areas using diffusion MRI tractography: comparison with functional MRI and electrical stimulation mapping. Epilepsia 2013;54:1381–90 10.1111/epi.12199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jeong JW, Asano E, Juhász C, et al. Localization of specific language pathways using diffusion-weighted imaging tractography for presurgical planning of children with intractable epilepsy. Epilepsia 2015;56:49–57 10.1111/epi.12863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jung RE, Haier RJ. The Parieto-Frontal Integration Theory (P-FIT) of intelligence: converging neuroimaging evidence. Behav Brain Sci 2007;30:135–54; discussion 154–87 10.1017/S0140525X07001185 [DOI] [PubMed] [Google Scholar]
- 18. Haier RJ, Karama S, Leyba L, et al. MRI assessment of cortical thickness and functional activity changes in adolescent girls following three months of practice on a visual-spatial task. BMC Res Notes 2009;2:174 10.1186/1756-0500-2-174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Frangou S, Chitins X, Williams SC. Mapping IQ and gray matter density in healthy young people. Neuroimage 2004;23:800–05 10.1016/j.neuroimage.2004.05.027 [DOI] [PubMed] [Google Scholar]
- 20. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57:289–300 [Google Scholar]
- 21. Fan RE, Chang KW, Hsieh CJ, et al. LIBLINEAR: a library for large linear classification. J Mach Learn Res 2008;9:1871–74 [Google Scholar]
- 22. Cappa SF, Cavallotti G, Guidotti M, et al. Subcortical aphasia: two clinical-CT scan correlation studies. Cortex 1983;19:227–41 10.1016/S0010-9452(83)80016-1 [DOI] [PubMed] [Google Scholar]
- 23. Huber SJ, Shuttleworth EC, Paulson GW, et al. Cortical vs subcortical dementia: neuropsychological differences. Arch Neurol 1986;43:392–94 10.1001/archneur.1986.00520040072023 [DOI] [PubMed] [Google Scholar]
- 24. Kirk A, Kertesz A. Cortical and subcortical aphasias compared. Aphasiology 1994;8:65–82 10.1080/02687039408248641 [DOI] [Google Scholar]
- 25. Posthuma D, De Geus EJ, Baaré WF, et al. The association between brain volume and intelligence is of genetic origin. Nat Neurosci 2002;5:83–84 10.1038/nn0202-83 [DOI] [PubMed] [Google Scholar]
- 26. Hulshoff Pol HE, Schnack HG, Posthuma D, et al. Genetic contributions to human brain morphology and intelligence. J Neurosci 2006;26:10235–42 10.1523/JNEUROSCI.1312-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sundaram S, Huq AH, Hsia T, et al. Exome sequencing and diffusion tensor imaging in developmental disabilities. Pediatr Res 2014;75:443–47 10.1038/pr.2013.234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zalesky A, Fornito A, Harding IH, et al. Whole-brain networks: does the choice of nodes matter? Neuroimage 2010;50:970–83 10.1016/j.neuroimage.2009.12.027 [DOI] [PubMed] [Google Scholar]
- 29. Ge B, Tian Y, Hu X, et al. Construction of multi-scale consistent brain networks: methods and applications. PLoS One 2015;10:e0118175 10.1371/journal.pone.0118175 [DOI] [PMC free article] [PubMed] [Google Scholar]