Abstract
Apoptosis is a biological process that removes damaged, excess, or infected cells through a genetically controlled mechanism. This process plays a crucial role in organismal development, immunity, and tissue homeostasis; and alterations in apoptosis contribute to human diseases including cancer and auto-immunity. In the past two decades, significant efforts have focused on understanding the function of the BCL-2 proteins, a complex family of pro-survival and pro-apoptotic alpha helical proteins that directly control the mitochondrial pathway of apoptosis. Diverse structural investigations of the BCL-2 family members have broadened our mechanistic understanding of their individual functions. However, an often over-looked aspect of the mitochondrial pathway of apoptosis is how the BCL-2 family specifically interacts with and targets the outer mitochondrial membrane (OMM) to initiate apoptosis. Structural information on the relationship between the BCL-2 family and the OMM is missing; likewise, biophysical mechanisms pertaining to how the OMM affects and effects apoptosis are lacking. In this mini-review, we will provide a current overview of the BCL-2 family members and discuss the latest structural insights of BAK/BAX activation and oligomerization in the context of the OMM and mitochondrial biology.
Keywords: Apoptosis, BAK, BAX, BCL-2 family, Lipids, Membrane, Mitochondria, MOMP, Structure
INTRODUCTION
Since the term “apoptosis” was introduced in 1972, significant efforts have focused on understanding its role in health and disease. Apoptosis, the most studied form of programmed cell death, is a biological process that triggers cells to commit suicide by activating a series of proteases referred to as caspases [1]. Once caspases are activated, proteolysis of key cellular components (e.g., cytoskeletal and nuclear proteins, DNA repair enzymes) pave the way for rapid detection by phagocytes, and this results in the clearance of dying cells with minimal damage to the surrounding tissue [2].
Two distinct molecular signaling mechanisms initiate apoptosis: (1) the “death receptor” or “extrinsic” pathway, and (2) the “mitochondrial pathway” or “intrinsic” pathway. The death receptor pathway is activated by a range of exogenous death-inducing ligands (e.g., tumor necrosis factor, TNF), which bind the death receptors (e.g., TNF receptor 1) and trigger caspase activation [2-3]. In contrast, the mitochondrial pathway of apoptosis is induced by various stress stimuli (e.g., DNA damage, growth-factor deprivation) and is often the target of chemotherapeutic interventions. This pathway is triggered by mitochondrial outer membrane permeabilization (MOMP), which releases pro-apoptotic factors (e.g., cytochrome c) from the mitochondrial intermembrane space (IMS) into the cytosol. The adaptor protein APAF-1 (apoptotic protease activating factor-1) binds cytosolic cytochrome c, undergoes oligomerization, and recruits pro-caspase 9 in a 2:1 (APAF-1:pro-caspase 9) complex forming the apoptosome. The apoptosome is the platform for caspase 9 activation, which in turn cleaves and activates the executioner caspases-3, -6, and -7, committing the cell to apoptosis [4-7].
MOMP initiates the mitochondrial pathway of apoptosis, and this event is mediated and regulated by the B cell lymphoma-2 (BCL-2) family of proteins at the outer mitochondrial membrane (OMM). Numerous structural investigations of BCL-2 family members have provided detailed molecular mechanisms of MOMP by defining how individual BCL-2 family members interact with one another (Table 1). However, many fundamental questions still remain unanswered, which restricts our understanding of how the BCL-2 proteins regulate MOMP and may be utilized as therapeutic targets. This mini-review focuses on the structural biology of pro-apoptotic BCL-2 proteins emphasizing recent structural data relating to BAK (BCL-2 antagonist killer 1) and BAX (BCL-2-associated x protein) activation, and their transitions from inactive monomers into high molecular weight homo-oligomers that mediate MOMP. In particular, what are the intra- and inter-molecular conformational changes associated with BAK/BAX activation leading to the initiation of MOMP; and what impact does mitochondrial network shape and composition contribute to pro-apoptotic BAK/BAX function.
Table 1.
Overview of BCL-2 family structures
| Pro-survival / Complex | PDB | Ref |
|---|---|---|
| hA1(Δc) + BOM-BH3 peptide | 2VM6 | 91 |
| mA1(Δc) + BMF-BH3 peptide | 2VOG | 18 |
| mA1(Δc) + BAK-BH3 peptide | 2VOH | 18 |
| mA1(Δc) + PUMA-BH3 peptide | 2VOF | 18 |
| mA1(Δc) + BID-BH3 peptide | 2VOI | 18 |
| hBCL-2(Δc) (isoform 1) | 1G5M | 92 |
| hBCL-2(Δc) (isoform 2) | 1GJH | 92 |
| mBCL-2(Δc) (Boo) | 2KUA | 93 |
| hBCL-2(Δc) + BAX-BH3 peptide | 2XA0 | 94 |
| hBCL-B(Δc) + BIM-BH3 peptide | 4B4S | 95 |
| hBCL-W(Δc) | 1MK3 | 96 |
| hBCL-W(Δc) | 1O0L | 97 |
| hBCL-W(Δc) + BID-BH3 peptide | 1ZY3 | 98 |
| hBCL-W(Δc) | 2Y6W | 99 |
| hBCL-W(Δc) + own BH3 domain | 4CIM | 19 |
| hBCL-xL(Δc) | 1MAZ | 11 |
| hBCL-xL(Δc) | 1LXL | 11 |
| rBCL-xL(Δc) | 1AF3 | 100 |
| hBCL-xL(Δc) + BAK-BH3 peptide | 1BXL | 13 |
| hBCL-xL(Δc) + BAD-BH3 peptide | 1G5J | 101 |
| mBCL-xL(Δc) | 1PQ0 | 102 |
| mBCL-xL(Δc) + mBID-BH3 peptide | 1PQ1 | 102 |
| hBCL-xL(Δc) dimer | 2B48 | 103 |
| hBCL-xL(Δc) + Beclin1-BH3 peptide | 2P1L | 104 |
| hBCL-xL(Δc) + Beclin1-BH3 peptide | 2PON | 105 |
| hBCL-xL(Δc) + BIM-BH3 peptide | 3FDL | 106 |
| hBCL-xL(Δc) + BAX-BH3 peptide | 3PL7 | 107 |
| hBCL-xL(Δc) + SOUL-BH3 peptide | 3R85 | 108 |
| hBCL-xL(Δc) + BIM-BH3 (BimSAHB) | 2YQ6 | 109 |
| hBCL-xL(Δc) + PUMA-BH3 peptide | 2M04 | 62 |
| hBCL-xL(Δc) + PUMA-BH3 peptide | 4HNJ | 62 |
| hBCL-xL(Δc) + own BH3 domain | 4CIN | 19 |
| hBCL-xL(Δc) + BID-BH3 peptide | 4QVE | 110 |
| hBCL-xL(Δc) hexamer | 4PPI | 111 |
| h-rMCL-1(Δnc) + hBIM-BH3 peptide | 2NL9 | 112 |
| mMCL-1(Δnc) + mNoxa-BH3 B peptide | 2JM6 | 112 |
| mMCL-1(Δnc) + mNoxa-BH3 A peptide | 2ROD | 113 |
| mMCL-1(Δnc) + PUMA-BH3 peptide | 2ROC | 113 |
| hMCL-1(Δnc) + BID-BH3 peptide | 2KBW | 114 |
| hMCL-1(Δnc) + BIM-BH3 peptide | 2PQK | 115 |
| hMCL-1(Δnc) + BAX-BH3 peptide | 3PK1 | 116 |
| hMCL-1(Δnc) + MCL-1-BH3 peptide | 3MK8 | 117 |
| hMCL-1(Δnc) + PUMA-BH3 peptide | 4BPI | 118 |
| hMCL-1(Δnc) | 2MHS | 119 |
| Pro-apoptotic / Complex | PDB | Ref |
|---|---|---|
| hBAX(fl) | 1F16 | 30 |
| hBAX(fl) + BIM_SAHB peptide | 2K7W | 41 |
| hBAX(Δc) dimer + BAX-BH3 peptide | 4BD6 | 40 |
| hBAX(Δc) dimer + BID-BH3 peptide | 4BD2 | 40 |
| hBAX(Δc) BIH-in-groove dimer (GFP) | 4BDU | 40 |
| hBAK(Δnc) | 2IMT | 29 |
| hBAK(Δnc) | 2YV6 | 120 |
| hBAK(Δnc) + BID-BH3 peptide | 2M5B | 43 |
| hBAK(Δnc) dimer | 4U2U | 44 |
| hSOUL(Δnc) | 3R8K | 108 |
X-ray crystal and NMR structures of the BCL-2 family with their PDB code are listed in black and red, respectively. Species are indicated by h: human, m: mouse, r: rat or h-r: human-rat chimeric protein. Δc, Δnc, and fl in parentheses denote delta c (lacking c-terminus), delta n & c (lacking n- & c-terminus), and full length, respectively.
Architectural Design of the BCL-2 Family
Eighteen members of the BCL-2 family have been identified and, based both on their structural and sequence homology, share conserved sequence regions known as the BCL-2 homology domains (BH1-BH4) (Fig1A). The BCL-2 family can further be divided into the pro-survival and pro-apoptotic subfamilies based on the composition of BH domains as well as the ability to activate or inhibit apoptosis. The pro-apoptotic subfamily has two sub-classes known as the ”BH3-only proteins” and the “effector” proteins [8-10].
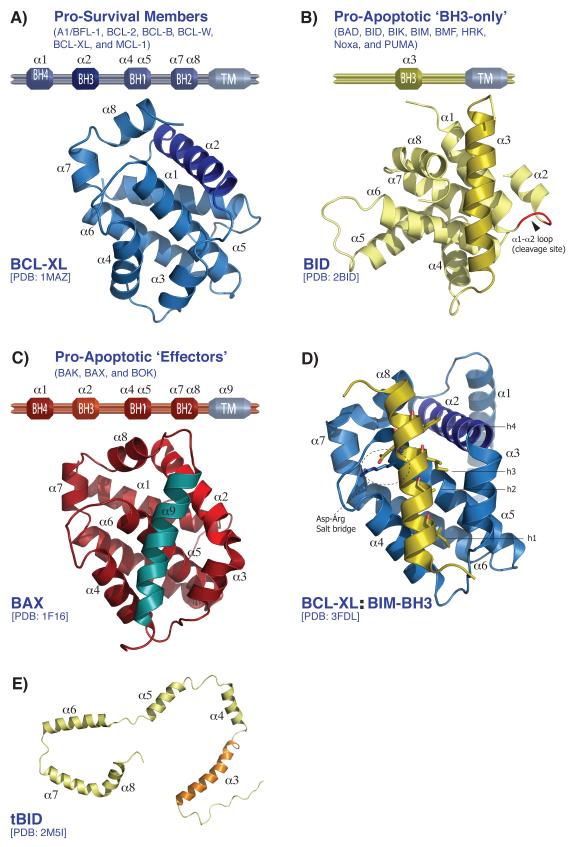
Figure 1. Structural overview of BCL-2 family.
(A-C) Domain architecture of the pro-survival members (A), the pro-apoptotic ‘BH3-only’ (B), and the pro-apoptotic ‘Effectors’ (C) in blue, yellow and red, respectively. The corresponding three-dimensional structures are depicted (BCL-XL, BID, and BAX). (D) Cartoon representation of BCL-XL bound to BIM-BH3 peptide showing the four hydrophobic residues (h1-h4) of the BIM-BH3 domain and the Asp-Arg salt bridge involved in the interaction with the BC-groove of BCL-XL. (E) The C-shape conformation of the NMR structure of tBID (pale yellow) presented with the BH3 domain (α3) colored orange. The tBID structure is determined in the presence of micelles and presumably represents an activated conformation of tBID, in which each α helix interacts with the micelles but no longer with each other, as shown in (B).
The Pro-Survival Subfamily
The pro-survival family contains six members: A1/BFL-1 (BCL-2-related gene A1), BCL-2, BCL-B, BCL-W (BCL-2-like 2 protein), BCL-XL (BCL-2-related protein long isoform), and MCL-1 (myeloid cell leukemia 1) all share four BH domains (BH1-BH4) with a transmembrane domain at the C-terminus (Fig1A) [8-9]. Human BCL-XL was the first published structure of the BCL-2 family (Fig1A) [11]. Since then, the three-dimensional structure of most members of the pro-apoptotic family has been determined (Table1). The overall structure of the pro-survival subfamily adopts a similar globular structure referred to as the “BCL-2 core” (Fig1A). The BCL-2 core is a conserved fold that is constructed by an eight α-helical bundle surrounding a central hydrophobic core helix [11]. The fold also generates a hydrophobic surface groove formed by α-helices 2, 3, 4, and 5 (BH1-BH3 domains) referred to as the BCL-2 family BH3 and C-terminus-binding groove (BC-groove). This canonical BC-groove serves as an important platform for interactions with the BH3 domain of different members of the BCL-2 family and enables homo- and heterodimerization within the family (e.g., BCL-2:BCL-2 homodimer and BID:BCL-2 heterodimer) [12-15]. Many three-dimensional structures of pro-survival members in complex with different BH3 domain peptides have revealed a conserved interaction mode between the BH3 domain and the canonical BC-groove (Table1). Structural insights demonstrate that the interaction is established through the insertion of four to six hydrophobic residues (h0-h5) on the amphipathic α-helix of the BH3 domain into corresponding binding pockets along the surface of the BC-groove. Furthermore, the interaction is enhanced through the formation of a salt bridge between the Asp residue on the BH3 domain and the conserved Arg residue on the BH1 domain present on all pro-survival protein members (Fig1D). However, certain subtle variations in the BH3 domain and the BC-groove determine the binding selectivity and affinity amongst the BCL-2 family members [16-19].
The Pro-Apoptotic Subfamily: BH3-only and Effector Proteins
The BH3-only subfamily members are expressed and/or activated in distinct cellular stress scenarios and are subdivided based on their ability to either exclusively interact with the pro-survival BCL-2 repertoire or both the pro-survival and pro-apoptotic effector subfamilies. BH3-only proteins that only bind to members of the pro-survival subfamily are classified as “sensitizer/de-repressor” BH3-only proteins. Members of this class of BH3-only proteins are: BAD (BCL-2 antagonist of cell death), BIK (BCL-2 interacting killer), BMF (BCL-2 modifying factor), HRK (Harakiri), and the latest member Soul [20].
The remaining BH3-only protein family members, BID (BCL-2-interacting domain death agonist), BIM (BCL-2-interacting mediator of cell death), Noxa, and PUMA (p53-upregulated modulator of apoptosis), are classified as “direct activators” [21,22]. The direct activators are able to interact and inhibit the pro-survival proteins, and directly activate BAK and BAX, allowing them to oligomerize, insert into the OMM and leading to MOMP. We will discuss the direct activation process in greater detail below.
The tri-partite balance of interactions between the pro-survival proteins, the proapoptotic proteins, and the BH3-only proteins is what ultimately regulates the decision to undergo MOMP and apoptosis. An example is the sequestration of stress-induced BIM by BCL-2 on the OMM. Following a subsequent cellular stress, PUMA is induced and competes with BIM for BCL-2 binding. When BIM is released, it interacts with BAX leading to BAX activation, BAX oligomerization, and MOMP [9,23].
The BH3-only protein subfamily members are primarily intrinsically disordered proteins and contain both a BH3 and C-terminal transmembrane domain. The BH3 domain becomes ordered upon binding to a globular BCL-2 family member and folds into the amphipathic α-helix. BID however, is structurally an exception: the overall tertiary fold is similar to the BCL-2 core with key residues of its BH3 domain buried, rendering it in a constitutively inactive state (Fig1B). Activation of BID is achieved by cleavage of the α1-α2 loop by multiple proteases including caspase-8 and granzymeB resulting in two fragments of BID: the N- and C-terminal fragments. The C-terminal fragment, often referred to as truncated BID (tBID), retains tight association with the N-terminal peptide. Upon contact with OMM, lipid vesicles, or detergents, tBID dissociates and interacts with the membrane and pro-survival BCL-2 member. Recent structural insights show that human tBID, in presence of membrane mimic micelles, takes a C-shape conformation with extensive interactions with micelles (Fig1E). Furthermore, the BH3-containing helix α3, which previously was believed to be exposed outside the membrane, may also be membrane associated [24-28].
The effector proteins in the pro-apoptotic subfamily consist of BAK, BAX and BOK (BCL-2-related ovarian killer). BAK and BAX are the BCL-2 family proteins that are responsible for the pro-apoptotic function at the OMM. After activation is triggered through the interaction with the BH3-domain of the direct activators BIM or BID, oligomerization of BAX and BAK occurs at the OMM leading to MOMP. The NMR and X-ray structures of non-activated BAK and BAX, respectively provided interesting structural insights [29,30] (Fig2). First, both protein structures show the conserved BCL-2 core with the hydrophobic BC-groove composed of helices α2-α5 and α8. Furthermore, another functionally important hydrophobic region is also buried: helix α1, where the BH4 domain resides. It contains certain hydrophobic and aromatic residues that make several contacts with helices α2, α5, and α6, thereby stabilizing the overall tertiary structure of the inactive form.
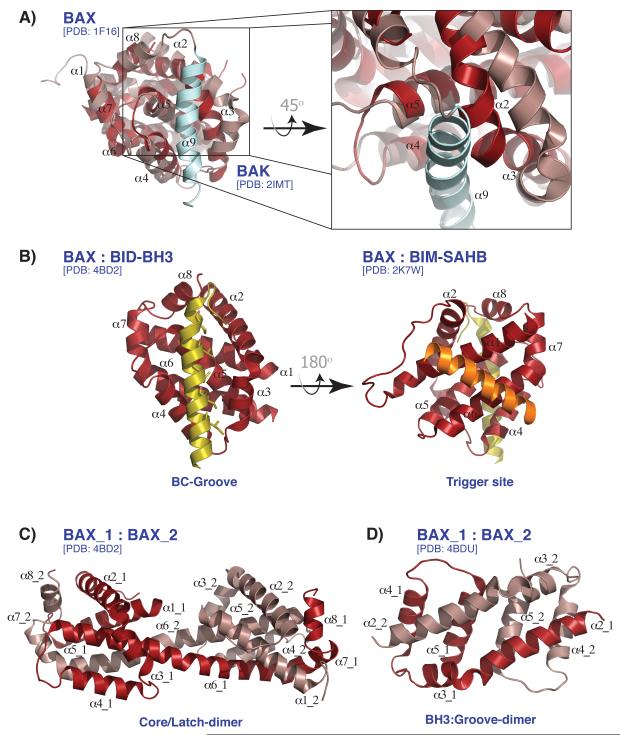
Figure 2. Activation, rearrangement, and dimerization of BAX.
(A) BAK and BAX structures superimposed shown in dark red and salmon, respectively. The transmembrane of BAX (α9, colored in light blue) sits in the BC-groove. Zoomed in, the superposition shows a narrow BAK BC-groove restricting the docking of the α9 helix. (B) BAX activation sites are shown. BAX bound to BID-BH3 peptide (yellow) in the BC-groove (left) and bound to BIM-SAHB peptide (orange) at the ‘trigger site’ of BAX on the opposite side of the BC-groove (right). (C) The ‘Core/Latch-dimer’ of BAX is shown with α6-α8 being interchanged between two BAX monomers. (D) The ‘BH3:Groove-dimer’ of BAX is shown with α2 of each monomer engaged with the BC-groove of the other.
In addition, BAK and BAX have a C-terminal transmembrane domain, termed α-helix 9, which is responsible for OMM insertion. BAK is found constitutively inserted within the OMM, whereas BAX is principally cytosolic and stably associates with the OMM only after activation by direct activator proteins [31]. Structural studies revealed that this difference in localization is governed by the structural position of α-helix 9. In BAX, α-helix 9 sits in the BC-groove in a ‘cis’ conformation, preventing it from inserting into the OMM (Fig2A) [30]. Disrupting this interaction between the α-helix 9 and the BC-groove, induces a conformational change in BAX leading to the release of α-helix 9 and its insertion into the OMM [32]. On the other hand, BAK α9 is exposed because its BC-groove is narrow and occluded by side chains that potentially restrict the docking of α-helix 9. Therefore, BAK skips the initial activation step and targets constitutively to the OMM [33,34].
BOK exhibits ~70-80% sequence homology to BAK and BAX and shares the conserved BH1-3 domains and a C-terminal transmembrane domain. Similar to BAK and BAX, BOK is widely expressed and induces cell death with the classical apoptotic features (e.g., release of cytochrome c, and activation of caspases). Despite these similarities the physiological role of BOK remained obscure, recent findings support a selective and distinguishing role for BOK in regulating the apoptotic response to endoplasmic reticulum (ER) or proteosomal stress [35].
Activation and Oligomerization Of the Effector Proteins
Understanding the biochemical and biophysical mechanisms by which BAK and BAX permeabilize the OMM to promote MOMP is considered the “holy grail” of apoptosis research [36]. Several structural studies have provided a glimpse into the model for BAK and BAX activation and oligomerization. In brief, BAK/BAX activation is a highly regulated multi-step process involving: 1) structural rearrangement exposing N- and C-termini, 2) insertion into the OMM, 3) dimerization, and 4) higher order oligomerization resulting to MOMP [37,38]. Of course, some of these steps are skipped for BAK, which constitutively resides within the OMM.
Interaction-Triggered Rearrangement
Two distinct activation sites on BAX are proposed to allow direct activator BH3-only proteins to interact and activate BAX (Fig2B, Fig3) [39,40]. Structural and biochemical studies in which (hydrocarbon stapled) BH3 peptides are utilized, demonstrated that the BIM BH3 domain can bind to an activator site proximal to the N-terminus of BAX. This first binding site, called the ‘trigger site’, is located opposite to the hydrophobic BC-groove and is defined by the two helices α1 and α6 forming a hydrophobic cleft [41,42]. Upon binding with the stapled BIM BH3 peptide, the unstructured loop between α-helices 1 and 2 is switched from a closed to an open position. The release of the C-terminal BAX α9 helix from the BC-groove follows and promotes OMM insertion. The exposure of the BH3 domain (α2) propagates the death signal through an auto-activating interaction with the trigger site of inactive BAX monomers while also allowing the BC-groove to interact with activator BH3 domains, which in turn induces further conformational changes [39]. While these mechanisms are convincing, whether the results obtained with minimal BH3 domain peptides reflect physiological control of BAX-dependent MOMP requires further investigation.
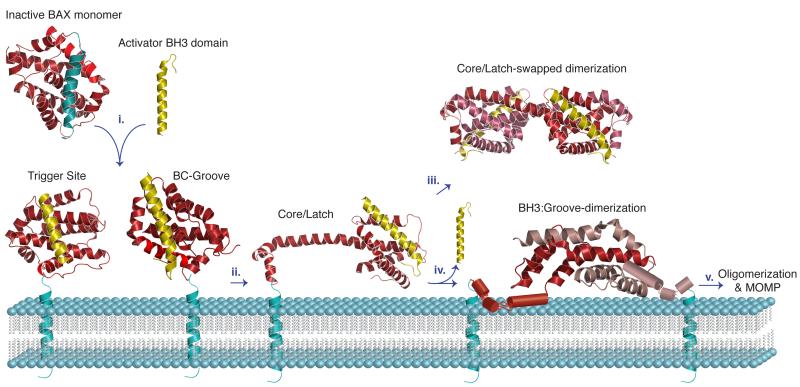
Figure 3. Model for the structural transitions of BAX activation, rearrangement, and dimerization leading to oligomerization and MOMP.
The BAX structures are shown in cartoon representation and color coded as in Figure 2. In step i. the inactivated BAX monomer interacts with the BH3 domain of an activator BH3-only protein via the BC-groove or the α1-α6 the trigger site. ii. BAX activation leads to major structural rearrangement in which the latch domain (α6-α8) detaches from the core. iii. In solution, two BAX monomers can engage in the core/latch swapped dimerization. iv. At the OMM, BAX engages in BH3:groove dimerization, after disassociation of the activator BH3 domain. The structure of the symmetrical α2-α5 core dimer of BAX revealed an extensive hydrophobic surface defined by residues from the α4 and α5 helices that stabilize the hydrophobic core of the inactivated BAX monomer. v. The formation of BAX oligomers remains unclear, although an interface involving helix α6 has been proposed. One of the emerging models suggests that the α2-α5 core dimers perturb the OMM through their hydrophobic α4-α5 surface, thereby inducing MOMP.
In addition, recent structural insights into the activation mechanism of BAK and BAX have revealed a second and common activation site (Fig2B, Fig3). In these studies a C-terminal deleted form of both proteins was utilized which mimics the full-length proteins with the tail inserted into the OMM. Crystal structures of both the truncated BAK and BAX in complex with the BH3 domain peptide of ‘direct activator’ BID and in presence of the detergent CHAPS revealed the second activation site at the canonical BC-groove of BAK and BAX. The NMR solution structure of BAK in complex with stapled BID BH3 was also determined, revealing the direct activation interface at the level of BAK monomer [40]. Furthermore, upon direct activation in the context of membranes or detergents, the N-terminal helix α1 and the BH3 domain of BAK and BAX are exposed permanently and transiently, respectively [40,43,44].
Translocation, Dimerization, and Oligomerization
As discussed above, BAK is constitutively located and inserted into the OMM via its transmembrane domain (α9). BAX however, is primarily cytosolic and recent data indicate that BAX retro-translocates back and forth from the cytoplasm to the OMM, possibly due to interactions with pro-survival protein members such as BCL-XL [45,46]. However, upon exposure of cells to apoptotic stimuli, BAX becomes activated, retro-translocation is halted, and translocates to the OMM.
Besides the interaction-triggered structural rearrangement of BAX upon activation, recent crystal structures of BAX bound to the BID BH3 domain have identified another structural revelation. It appears that a destabilizing cavity within BAX, close to its BH3 domain, is formed following interaction with the BID BH3 domain. This then might promote the extrusion of BAX BH3 domain (α2 helix). The crystal structure also shows that BAX is rearranged into a ‘core’ domain consisting of α2-α5 and a ‘latch’ domain comprised of helices α6-α8 [38,44]. Even though this particular BAX ‘core/latch’ dimer is believed to be off-pathway and does not play a role in oligomerization and is yet to be characterized physiologically, the disengagement of the core from the latch domain appears to be critical for BAX function (Fig2C, Fig3).
The crystal structure of the isolated core domain (α2-α5) revealed another homodimer in which the BH3 domain (α2) of each monomer engages the BC-groove (α3-α5) of the other (Fig2D, Fig3) [40]. This symmetric BAX homodimer seems to lie at the heart of the BH3:groove symmetric dimer [47,48]. BH3 peptide binding to BAK also produces N-terminal exposure and oligomerization and recent structural evidence confirms an analogous mechanism for activation and dimerization of BAK in response to certain BH3 peptides [44,49].
The BH3-in-groove symmetric dimer structure of BAX and BAK together with crosslinking and biophysical studies argues against the initial proposed head-to-tail model for oligomerization. An alternative model suggests a second interface which links the symmetric dimers to build the structure of the oligomers. In crosslinking studies an interface between α6 helices have been identified to be involved in the homo-oligomerization of dimers [48]. Until recently, the exact identity of the second interface and the precise structure of larger oligomers remained unknown. Recent reports using cysteine-scanning mutagenesis and hydrophilic labeling showed that the BAK α9 helix traverses the OMM and links itself to neighboring α9 segments, identifying an α9:α9 interface in BAK (and BAX) oligomers. This α9:α9 interaction might be the secondary interface that links the symmetric BH3-in-groove dimers together to form the oligomers [50].
Recent data question a longstanding model in which the α5/α6 helices are inserted as a hairpin into the OMM as a means of membrane association. In the new proposed in-plane model, it appears that two helices, α4 and α5, of both BAK and BAX collapse onto the membrane exposing multiple aromatic residues. Insertion of these residues between lipid head groups in the OMM might lead to increase in membrane tension and curvature which may promote stable bilayer breaks inducing formation of proteolipidic pores [44,51].
More recently, a 3D model of active full length BAX dimer embedded in the membrane has been proposed [52,53]. Based on double electron-electron resonance (DEER) spectroscopy using spin-labeled BAX variants inserted into large unilamellar vesicles (LUVs), the model describes the relative structural arrangement of the full length oligomeric BAX at the membrane. The BAX dimer adopts a clamp-like conformation at the membrane with the opening of the hairpin of helices α5 and α6 as suggested being the central core in the mechanism of membrane permeabilization [51]. Furthermore, the 3D model shows that the α2-α5 core of one BAX molecule forms a stable interaction interface with the α2-α5 core of another BAX molecule, in agreement with the crystal structure of the GFP-fused α2-α5 core dimer of BAX [40]. We believe that while there is conflicting evidence, we speculate that monomers insert the membrane prior to the oligomerization based on recent Mode1/2 data and heterodimerization data (anti- & BAX).
BCL-2 Family Interactions With p53
Apart from BH3-only proteins, BAX and BAK activation can also be initiated by other stimuli, such as low or high pH, hydrogen peroxide, and mild heat [54-56]. Another intriguing stimulus is the non-BCL-2 family protein, p53 [57]. In addition to direct transcriptional regulation of apoptosis, this tumor suppressor protein has also been shown to regulate the BCL-2 family proteins by direct interaction with BAX, BAK, BCL-2, and BCL-XL allowing for MOMP and apoptosis to occur. Furthermore, the p53 protein possesses both sensitizer/de-repressor and direct activator BH3-only protein functions and can directly control MOMP and apoptosis [58-62]. The molecular mechanism of direct BAX activation by p53 follows a different path leading to the structural rearrangement of BAX. Very recent structural findings show that both the cis isomer of p53 proline 47 (Pro47) and its ability to isomerize are required for BAX activation. The structural model of p53 in complex with BAX suggests that Pro47 isomerization destabilizes the C-terminus of BAX (region α6-α9) triggering BAX rearrangement for activation. Other data also show that the prolyl isomerase Pin1 (peptidyl-prolyl cis-trans isomerase NIMA-interacting 1) enhances p53-dependent BAX activation by catalyzing the cis-trans interconversion of p53 Pro47 [63].
The Pro-Apoptotic Landscape of the Outer Mitochondrial Membrane
The classical textbook view of mitochondria is that they are small bean-shaped organelles scattered throughout the cytosol. Mitochondria, however, are dynamic organelles that vary their shape from spherical to elongated through homeostatic cycles of fusion and fission [64]. In addition, several reports indicate that the BCL-2 family regulates mitochondrial shape, but direct mechanistic contributions to the decision to undergo MOMP are scarce. For quite some time, it has been assumed that stress-induced apoptosis proceeds via collaborative efforts between the BCL-2 family and mitochondria. However, whether all changes in mitochondrial shape are a consequence of apoptosis or contribute an important role in the cellular decision to undergo MOMP and apoptosis requires further investigation.
While progress has been made in unraveling the molecular mechanism for the transition from inactive to active BAK or BAX monomers and construction of large oligomers, how these oligomers form a pore into the OMM remain unanswered. Several reports have suggested that at least nine to twelve BAX molecules are required to release cytochrome c [65,66], and upwards to twenty BAX molecules to release other (maybe larger) proteins from the mitochondria [67]. Various experimental studies utilizing isolated mitochondria and liposomal-based systems have demonstrated that the pores are composed of both proteinacious and lipidic parts [68-70].
Previously published observations reveal that BAK/BAX-mediated apoptosis is regulated by at least three factors: (i) a stress-specific combination of pro-apoptotic BH3-only proteins, (ii) an actively maintained and regulated lipid composition of the OMM via an array of lipid metabolic pathways, and more recently (iii) a specific mitochondrial shape and size that contributes to BAX activation [9,71,72]. Recent studies have shown that the mitochondrial shape and size has an impact on cell death by contributing to BAX activation, MOMP, and apoptosis. The diameter of the OMM cooperates with BAX helix α9 to establish stable BAX-membrane interactions to promote MOMP and apoptosis [72]. These findings are supported by previous observations showing that DRP1, a large GTPase of the dynamin superfamily involved in mitochondrial fission, is able to directly remodel the OMM by triggering membrane tethering and hemifusion and thereby promoting BAX oligomerization [73].
In addition, biophysical data show that BAX-derived helical peptides induce pore formation and that these pores are at least partially framed by a lipid monolayer. Furthermore, the data suggests that the formation of such lipidic pores is a major mechanism for α-pore-forming proteins, such as BAX. This mechanism involves the spontaneously binding of amphipathic pore-forming peptides to the water-lipid chain interface of the lipid bilayer. This results to the increase of interfacial area, which stretches the hydrocarbon core of the bilayer, causing an elastic strain in the lipid bilayer and ultimately forming the pores. This model implies and speculates that the curvature properties of the OMM influence the pore formation [69].
Besides the landscape of mitochondria, there is increasing evidence supporting a key role of the lipid milieu in BAX-induced MOMP [9,68]. In particular, mitochondrial cholesterol has potential to emerge as an important regulator of MOMP in response to apoptotic stimuli (e.g., hypoxia, TNF, or BAX) [76]. The presence of cholesterol in the bilayer hindered BAX-mediated MOMP due to the combination of reduced membrane dynamics and decreased ability of BAX to insert or oligomerize into the OMM. This inhibition does not require direct interaction with BAX but functions on the membrane environment to prevent BAX integration [77-79].
An important component of the inner mitochondrial membrane (IMM) is cardiolipin, a negatively charged phospholipid. Cardiolipin is almost exclusively found in the IMM, where it constitutes 20% of the total lipid composition. However, small amounts of cardiolipin have been found in the OMM [78]. Liposome and OMM vesicle studies demonstrated the requirement of cardiolipin in the bilayer for BAX-induced permeabilization of mitochondrial LUVs. Interestingly, LUVs composed of lipids that resemble the endoplasmic reticulum (ER) resist BAX-mediated permeabilization unless cardiolipin is added [68,79,80]. Additional roles for cardiolipin in the OMM include the BID translocation to liposomes, to the OMM, and to mitochondrial contact sites [81-83]. Furthermore, cardiolipin is involved in the recruitment and activation of caspase 8 to the OMM providing a docking site for the interaction and activation of tBID [84]. Besides BID and BAX, cardiolipin also seems to bind to a truncated form of BAK leading to an increased sensitivity to BID-induced BAK activation and membrane permeabilization [85]. Finally, experimental data also provide another role for cardiolipin in which it induces unique membrane curvature at the OMM contact sites that stress the mitochondrial membrane [82]. It is believed that the interaction with this stress-related membrane curvature is responsible for the C-shape conformation of tBID.
Another key component in the lipid control of BAK/BAX-dependent MOMP is the sphingolipid family of ceramides. Studies have shown that ceramide is able to form pores in planar membranes as well as in isolated mitochondria and that the pore-forming activity of ceramide is regulated by several members of the pro-survival BCL-2 subfamily [86-88]. In contrast, numerous studies demonstrate that BAX synergizes with ceramide leading to enhanced permeabilization of planar membranes and isolated mitochondria [89]. Interestingly, recent data suggest that small molecule inhibitors against the pro-survival BCL-2 proteins engage ceramide accumulation in the OMM, and this depends on BAK function [90].
Ceramides can be reversibly metabolized into a variety of lipid species that influence cellular sensitivity to apoptosis. For example, ceramides can be converted into sphingosine-1-phosphate (S1P), which is further metabolized into the fatty aldehyde, hexadecenal. Experimental data show that S1P and hexadecenal promote BAK and BAX activation, respectively [71]. Purified mitochondria deficient in S1P metabolism were resistant to MOMP induced by BID and BIM; yet could be re-sensitized to BAK/BAX dependent MOMP after the addition of S1P and hexadecenal, respectively. Furthermore, data shows hexadecenal to bind directly to BAX promoting BAX activation, oligomerization, and MOMP in mitochondria and LUVs [71]. These data reveal that apoptotic pathways regulate mitochondrial shape and composition in a BCL-2 family dependent manner, and that apoptosis proceeds when the same mitochondrial components cooperate with BAK/BAX to induce MOMP.
Future Perspectives
Detailed structural studies complemented with elegant biochemical model systems have enriched our understanding of the BCL-2 family and provided fascinating insights into how mitochondrial biology controls BCL-2 family function and cell death commitment. Three-dimensional structures of the BCL-2 family proteins with and without their cognate protein partners have revealed key molecular insights into the mechanisms of cell death, along with providing a foundation into the development of small molecule regulators of BAK/BAX-dependent apoptosis. Indeed, we are just beginning to appreciate BAK/BAX oligomerization and MOMP at the structural level (Fig3), and significant efforts should be focused on understanding how mitochondrial membranes and lipids directly impact upon both the pro-survival and pro-apoptotic members of the BCL-2 family. Likewise, a broader repertoire of membrane biology biochemical techniques and structural approaches should be applied to the BCL-2 family to unveil deeper molecular details on this intriguing family of proteins that function in the deadly landscape of the outer mitochondrial membrane.
ACKNOWLEDGEMENTS
This work was supported by: NIH grant CA157740, the JJR Foundation, the William A. Spivak Fund, the Fridolin Charitable Trust, an American Cancer Society Research Scholar Award, and an Irma T. Hirschl/Monique Weill-Caulier Trust Research Award. This work was also supported in part by two research grants (5-FY11-74 and 1-FY13-416) from the March of Dimes Foundation, and the Developmental Research Pilot Project Program within the Department of Oncological Sciences at Mount Sinai. Finally, we would like to acknowledge Doug Green for his outstanding mentorship, scientific contributions, and sincere friendship.
ABBREVIATIONS
- OMM
outer mitochondrial membrane
- TNF
tumor necrosis factor
- MOMP
mitochondrial outer membrane permeabilization
- IMS
intermembrane space
- APAF-1
apoptotic protease activating factor-1
- BCL-2
B cell lymphoma-2
- BAK
BCL-2 antagonist killer 1
- BAX
BCL-2-associated x protein
- BH
BCL-2 homology
- A1/BFL-1
BCL-2-related gene A1
- BCL-W
BCL-2-like 2 protein
- BCL-XL
BCL-2-related protein long isoform
- MCL-1
myeloid cell leukemia 1
- BC-groove
BH3 and C-terminus-binding groove
- BAD
BCL-2 antagonist of cell death
- BIK
BCL-2 interacting killer
- BMF
BCL-2 modifying factor
- HRK
Harakiri
- BID
BCL-2-interacting domain death agonist
- BIM
BCL-2-interacting mediator of cell death
- PUMA
p53-upregulated modulator of apoptosis
- tBID
truncated BID
- BOK
BCL-2-related ovarian killer
- NMR
nuclear magnetic resonance
- ER
endoplasmic reticulum
- DEER
double electron-electron resonance
- LUV
large unilamellar vesicles
- Pro
proline
- Pin1
peptidyl-prolyl cis-trans isomerase NIMA-interacting 1
- DRP1
dynamin-related protein 1
- IMM
inner mitochondrial membrane
- S1P
sphingosine-1-phosphate
Footnotes
AUTHOR CONTRIBUTIONS
JEC conceived and supervised the manuscript; MPALV and JEC wrote the manuscript; MPALV made the figures and graphical abstract and made the manuscript revisions.
REFERENCES
- 1).Kerr JF, Wyllie AH, Currie AR. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer. 1972;26:239–57. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2).Hotchkiss RS, Strasser A, McDunn JE, Swanson PE. Cell death in disease: mechanisms and emerging therapeutic concepts. N Engl J Med. 2009;361:1670–83. doi: 10.1056/NEJMra0901217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3).Riedl SJ, Galvesen GS. The apoptosome: signaling platform of cell death. Nat Rev Mol Cell Biol. 2007;8:405–13. doi: 10.1038/nrm2153. [DOI] [PubMed] [Google Scholar]
- 4).Tait SW, Green DR. Mitochondria and cell death: outer membrane permeabilization and beyond. Nat Rev Mol Cell Biol. 2010;11:621–32. doi: 10.1038/nrm2952. [DOI] [PubMed] [Google Scholar]
- 5).Yuan S, Akey CW. Apoptosome structure, assembly, and procaspase activation. Structure. 2013;21:501–15. doi: 10.1016/j.str.2013.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6).Czabotar PE, Lessene G, Strasser A, Adams JM. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat Rev Mol Cell Biol. 2014;15:49–63. doi: 10.1038/nrm3722. [DOI] [PubMed] [Google Scholar]
- 7).Hu Q, Wu D, Chen W, Yan Z, Yan C, He T, Liang Q, Shi Y. Molecular determinants of caspase-9 activation by the Apaf-1 apoptosome. Proc Natl Acad Sci U S A. 2014;111:16254–61. doi: 10.1073/pnas.1418000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8).Chipuk JE, Moldoveanu T, Llambi F, Parsons MJ, Green DR. The BCL-2 family reunion. Mol Cell. 2010;37:299–310. doi: 10.1016/j.molcel.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9).Renault TT, Chipuk JE. Death upon a kiss: mitochondrial outer membrane composition and organelle communication govern sensitivity to BAK/BAX-dependent apoptosis. Chem Biol. 2014;21:114–23. doi: 10.1016/j.chembiol.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10).Rech de Laval V, Deleage G, Aouacheria A, Combet C. BCL2DB: database of BCL-2 family members and BH3-only proteins. Database (Oxford) 2014 doi: 10.1093/database/bau013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11).Muchmore SW, Sattler M, Liang H, Meadows RP, Harlan JE, Yoon HS, Nettesheim D, Chang BS, Thompson CB, Wong SL, Ng SL, Fesik SW. X-ray and NMR structure of human BCL-XL, an inhibitor of programmed cell death. Nature. 1996;381:335–41. doi: 10.1038/381335a0. [DOI] [PubMed] [Google Scholar]
- 12).Yin XM, Oltvai ZN, Korsmeyer SJ. BH1 and BH2 domains of BCL-2 are required for inhibition of apoptosis and heterodimerization with BAX. Nature. 1994;369:321–3. doi: 10.1038/369321a0. [DOI] [PubMed] [Google Scholar]
- 13).Sattler M, Liang H, Nettesheim D, Meadows RP, Harlan JE, Eberstadt M, Yoon HS, Shuker SB, Chang BS, Minn AJ, Thompson CB, Fesik SW. Structure of BCL-CL-BAK peptide complex: recognition between regulators of apoptosis. Science. 1997;275:983–6. doi: 10.1126/science.275.5302.983. [DOI] [PubMed] [Google Scholar]
- 14).Diaz JL, Oltersdorf T, Horne W, McConnell M, Wilson G, Weeks S, Garcia T, Fritz LC. A common binding site mediates heterodimerization and homodimerization of BCL-2 family members. J Biol Chem. 1997;17:11350–5. doi: 10.1074/jbc.272.17.11350. [DOI] [PubMed] [Google Scholar]
- 15).Wang K, Gross A, Waksman G, Korsmeyer SJ. Mutagenesis of the BH3 domain of BAX identifies residues critical for dimerization and killing. Mol Cell Biol. 1998;18:6083–9. doi: 10.1128/mcb.18.10.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16).Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, Colman PM, Day CL, Adams JM, Huang DC. Differential targeting of prosurvival BCL-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell. 2005;17:393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 17).Kuwana T, Bouchier-Hayes L, Chipuk JE, Bonzon C, Sullivan BA, Green DR, Newmeyer DD. BH3 domains of BH3-only proteins differentially regulate BAX-mediated mitochondrial membrane permeabilization both directly and indirectly. Mol Cell. 2005;17:525–35. doi: 10.1016/j.molcel.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 18).Smits C, Czabotar PE, Hinds MG, Day CL. Structural plasticity underpins promiscuous binding of the prosurvival protein A1. Structure. 2008;16:818–29. doi: 10.1016/j.str.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 19).Lee EF, Dewson G, Evangelista M, Pettikiriachchi A, Gold GJ, Zhu H, Colman PM, Fairlie WD. The functional difference between pro-survival and pro-apoptotic B cell lymphoma 2 (BCL-2) proteins depend on structural differences in their BCL-2 homology 3 (BH3) domains. J Biol Chem. 2014;289:36001–17. doi: 10.1074/jbc.M114.610758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20).Doerflinger M, Glab JA, Puthalakath H. BH3-only proteins: a 20-year stock-take. FEBS. 2015;282:1006–16. doi: 10.1111/febs.13190. [DOI] [PubMed] [Google Scholar]
- 21).Ren D, Tu HC, Kim H, Wang GX, Bean GR, Takeuchi O, Jeffers JR, Zambetti GP, Hsieh JJ, Cheng EH. BID, BIM, and PUMA are essential for activation of the BAX- and BAK-dependent cell death program. Science. 2010;330:1390–3. doi: 10.1126/science.1190217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22).Chen HC, Kanai M, Inoue-Yamauchi A, Tu HC, Huang Y, Ren D, Kim H, Takeda S, Reyna DE, Chan PM, Ganesan YT, Liao CP, Gavathiotis E, Hsieh JJ, Cheng EH. An interconnected hierarchical model of cell death regulation by the BCL-2 family. Nat Cell Biol. 2015;17:1270–81. doi: 10.1038/ncb3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23).Chipuk JE, Fisher JC, Dillon CP, Kriwacki RW, Kuwana T, Green DR. Mechanism of apoptosis induction by inhibition of the anti-apoptotic BCL-2 proteins. Proc Natl Acad Sci U S A. 2008;105:20327–32. doi: 10.1073/pnas.0808036105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24).McDonnel JM, Fushman D, Milliman CL, Korsmeyer SJ, Cowburn D. Solution structure of the proapoptotic molecule BID: a structural basis for apoptotic agonists and antagonists. Cell. 1999;96:625–34. doi: 10.1016/s0092-8674(00)80573-5. [DOI] [PubMed] [Google Scholar]
- 25).Chou JJ, Li H, Salvesen GS, Yuan J, Wagner G. Solution structure of BID, an intracellular amplifier of apoptotic signaling. Cell. 1999;96:615–24. doi: 10.1016/s0092-8674(00)80572-3. [DOI] [PubMed] [Google Scholar]
- 26).Li H, Zhu H, Xu CJ, Yuan J. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell. 1998;94:491–501. doi: 10.1016/s0092-8674(00)81590-1. [DOI] [PubMed] [Google Scholar]
- 27).Luo X, Budihardjo I, Zou H, Slaughter C, Wang X. BID, a BCL-2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell. 1998;94:481–90. doi: 10.1016/s0092-8674(00)81589-5. [DOI] [PubMed] [Google Scholar]
- 28).Wang Y, Tjandra N. Structural insights of tBid, the Caspase-8-activated Bid, and its BH3 domain. J Biol Chem. 2013;288:35840–35851. doi: 10.1074/jbc.M113.503680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29).Moldoveanu T, Liu Q, Tocilj A, Watson M, Shore G, Gehring K. The X-ray structure of a BAK homodimer reveals an inhibitory zinc binding site. Mol Cell. 2006;24:677–88. doi: 10.1016/j.molcel.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 30).Suzuki M, Youle RJ, Tjandra N. Structure of BAX: coregulation of dimer formation and intracellular localization. Cell. 2000;103:645–54. doi: 10.1016/s0092-8674(00)00167-7. [DOI] [PubMed] [Google Scholar]
- 31).Wolter KG, Hsu YT, Smith CL, Nechushtan A, Xi XG. Movement of BAX from the cytosol to mitochondria during apoptosis. J Cell Biol. 1997;139:1281–92. doi: 10.1083/jcb.139.5.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32).Brock SE, Li C, Wattenberg BW. The BAX carboxy-terminal hydrophobic helix does not determine organelle-specific targeting but is essential for maintaining BAX in an inactive state and for stable mitochondrial membrane insertion. Apoptosis. 2010;15:14–27. doi: 10.1007/s10495-009-0410-2. [DOI] [PubMed] [Google Scholar]
- 33).Setoguchi K, Otera H, Mihara K. Cytosolic factor- and TOM-independent import of C-tail-anchored mitochondrial outer membrane proteins. EMBO J. 2006;25:5635–47. doi: 10.1038/sj.emboj.7601438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34).Leshchiner ES, Braun CR, Bird GH, Walensky LD. Direct activation of full-length proapoptotic BAK. Proc Natl Acad Sci U S A. 2013;110:986–95. doi: 10.1073/pnas.1214313110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35).Carpio MA, Michaud M, Zhou W, Fisher JK, Walensky LD, Katz SG. BCL-2 family member BOK promotes apoptosis in response to endoplasmic reticulum stress. Proc Natl Acad Sci U S A. 2015;112:7201–6. doi: 10.1073/pnas.1421063112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36).Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 37).Westphal D, Dewson G, Czabotar PE, Kluck RM. Molecular biology of BAX and BAK activation and action. Biochim Biophys Acta. 2011;1813:521–31. doi: 10.1016/j.bbamcr.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 38).Lovell JF, Billen LP, Bindner S, Shama-Din A, Fradin C, Leber B, Andrews DW. Membrane binding by tBID initiates an ordered series of events culminating in membrane permeabilization by BAX. Cell. 2008;135:1074–84. doi: 10.1016/j.cell.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 39).Gavathiotis E, Reyna DE, Davis ML, Bird GH, Walensky LD. BH3-triggered structural reorganization drives the activation of proapoptotic BAX. Mol Cell. 2010;40:481–92. doi: 10.1016/j.molcel.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40).Czabotar PE, Westphal D, Dewson G, Ma S, Hockings C, Fairlie WD, Lee EF, Yao S, Robin AY, Smith BJ, Huang DC, Kluck RM, Adams JM, Colman PM. BAX crystal structures reveal how BH3 domains activate BAX and nucleate its oligomerization to induce apoptosis. Cell. 2013;152:519–31. doi: 10.1016/j.cell.2012.12.031. [DOI] [PubMed] [Google Scholar]
- 41).Gavathiotis E, Suzuki M, Davis ML, Pitter K, Bird GH, Katz SG, Tu HC, Kim H, Cheng EH, Tjandra N, Walensky LD. BAX activation is initiated at a novel interaction site. Nature. 2008;255:1076–81. doi: 10.1038/nature07396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42).Walensky LD, Pitter K, Morash J, Oh KJ, Barbuto S, Fisher J, Smith E, Verdine GL, Korsmeyer SJ. A stapled BID BH3 helix directly binds and activates BAX. Mol Cell. 2006;24:199–210. doi: 10.1016/j.molcel.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 43).Moldoveanu T, Grace CR, Llambi F, Nourse A, Fitzgerald P, Gehring K, Kriwacki RW, Green DR. BID-induced structural changes in BAK promotes apoptosis. Nat Struct Mol Biol. 2013;20:589–97. doi: 10.1038/nsmb.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44).Brouwer JM, Westphal D, Dewson G, Robin AY, Uren RT, Bartolo R, Thompson GV, Colman PM, Kluck RM, Czabotar PE. BAK core and latch domains separate during activation, and freed core domains from symmetric homodimers. Mol Cell. 2014;55:938–46. doi: 10.1016/j.molcel.2014.07.016. [DOI] [PubMed] [Google Scholar]
- 45).Edlich F, Banerjee S, Suzuki M, Cleland MM, Arnoult D, Wang C, Neutzner A, Tjandra N, Youle RJ. BCL-XL retrotranslocates BAX from the mitochondria into the cytosol. Cell. 2011;145:104–16. doi: 10.1016/j.cell.2011.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46).Schellenberg B, Wang P, Keeble JA, Rodriguez-Enriquez R, Walker S, Owens TW, Foster F, Tanianis-Hughes J, Brennan K, Streuli CH, Gilmore AP. BAX exists in a dynamic equilibrium between the cytosol and mitochondria to control apoptotic priming. Mol Cell. 2013;49:959–71. doi: 10.1016/j.molcel.2012.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47).Dewson G, Ma S, Frederick P, Hockings C, Tan I, Kratina T, Kluck RM. BAX dimerizes via a symmetric BH3:groove interface during apoptosis. Cell Death Differ. 2012;19:661–70. doi: 10.1038/cdd.2011.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48).Dewson G, Kratina T, Czabotar P, Day CL, Adams JM, Kluck RM. Bak activation for apoptosis involves oligomerization of dimers via their alpha6 helices. Mol Cell Sci. 2009;36:696–703. doi: 10.1016/j.molcel.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 49).Oh KJ, Singh P, Lee K, Foss K, Lee S, Park M, Lee S, Aluvila S, Park M, Singh P, Kim RX, Symersky J, Walters DE. Conformational changes in BAK, a pore-forming proapoptotic BCL-2 family member, upon membrane insertion and direct evidence for the existence of BH3-BH3 contact surface in BAK homo-oligomers. J Biol Chem. 2010;285:28924–37. doi: 10.1074/jbc.M110.135293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50).Iyer S, Bell F, Westphal D, Anwari K, Gulbis J, Smith BJ, Dewson G, Kluck RM. BAK apoptotic pores involve a flexible C-terminal region and juxtaposition of the C-terminal transmembrane domains. Cell Death Differ. 2015:1–11. doi: 10.1038/cdd.2015.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51).Westphal D, Dewson G, Menard M, Frederick P, Iyer S, Bartolo R, Gibson L, Czabotar PE, Smith BJ, Adams JM, Kluck RM. Apoptotic pore formation is associated with in-plane insertion of BAK or BAX central helices into the mitochondrial outer membrane. Proc Natl Acad Sci U S A. 2014;111:4076–85. doi: 10.1073/pnas.1415142111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52).Bleicken S, Jeschke G, Stegmueller C, Salvador-Gallego R, Garcia-Saez AJ, Bordignon E. Structural model of active BAX at the membrane. Mol Cell. 2014;56:496–505. doi: 10.1016/j.molcel.2014.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53).Sung TC, Li CY, Lai YC, Hung CL, Shih O, Yeh YQ, Jeng US, Chiang YW. Solution structure of apoptotic BAX oligomer: Oligomerization likely precedes membrane insertion. Structure. 2015;23:1878–88. doi: 10.1016/j.str.2015.07.013. [DOI] [PubMed] [Google Scholar]
- 54).Cartron PF, Oliver L, Mayat E, Meflah K, Vallette FM. Impact of pH on Bax alpha conformation, oligomerization and mitochondrial integration. FEBS Lett. 2004;587:41–46. doi: 10.1016/j.febslet.2004.10.080. [DOI] [PubMed] [Google Scholar]
- 55).Nie C, Tian C, Zhao L, Petit PX, Mehrpour M, Chen Q. Cysteine 62 of Bax is critical for its conformational activation and its proapoptotic activity in response to H2O2-induced apoptosis. J Biol Chem. 2008;283:15359–15369. doi: 10.1074/jbc.M800847200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56).Pagliari LJ, Kuwana T, Bonzon C, Newmeyer DD, Tu S, Beere HM, Green DR. The multidomain propapoptotic molecules Bax and Bak are directly activated by heat. Proc Natl Acad Sci U S A. 2005;102:17975–80. doi: 10.1073/pnas.0506712102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57).Elkholi R, Chipuk JE. How do I kill thee? Let me count the ways: p53 regulates PARP-1 dependent necrosis. Bioessays. 2014;36:46–51. doi: 10.1002/bies.201300117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58).Mihara M, Erster S, Zaika A, Petrenko O, Chittenden T, Pancoska P, Moll UM. p53 has a direct apoptogenic role at the mitochondria. Mol Cell. 2003;11:577–90. doi: 10.1016/s1097-2765(03)00050-9. [DOI] [PubMed] [Google Scholar]
- 59).Leu JI, Dumont P, Hafey M, Murphy ME, George DL. Mitochondrial p53 activates Bak and causes disruption of a Bak-Mcl1 complex. Nat Cell Biol. 2004;6:443–50. doi: 10.1038/ncb1123. [DOI] [PubMed] [Google Scholar]
- 60).Chipuk JE, Kuwana T, Bouchier-Hayes L, Droin NM, Newmeyer DD, Schuler M, Green DR. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science. 2004;303:1010–1014. doi: 10.1126/science.1092734. [DOI] [PubMed] [Google Scholar]
- 61).Chipuk JE, Bouchier-Hayes L, Kuwana T, Newmeyer DD, Green DR. PUMA couples the nuclear and cytoplasmic proapototic function of p53. Science. 2005;309:1732–1735. doi: 10.1126/science.1114297. [DOI] [PubMed] [Google Scholar]
- 62).Follis AV, Chipuk JE, Fisher JC, Yun ML, Grace CR, Nourse A, Baran K, Ou L, Min L, White SW, Green DR, Kriwacki RW. PUMA binding induces partial unfolding within BCL-XL to disrupt p53 binding and promote apoptosis. Nat Chem Biol. 2013;9:163–8. doi: 10.1038/nchembio.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63).Follis AV, Llambi F, Merritt P, Chipuk JE, Green DR, Kriwacki RW. Pin1-Induced proline isomerization in cytosolic p53 mediates BAX activation and apoptosis. Mol Cell. 2015;59:1–8. doi: 10.1016/j.molcel.2015.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64).Autret A, Martin SJ. Emerging role for members of the BCL-2 family in mitochondrial morphogenesis. Mol Cell. 2009;36:355–63. doi: 10.1016/j.molcel.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 65).Saito M, Korsmeyer SJ, Schlesinger PH. BAX-dependent transport of cytochrome c reconstituted in pure liposomes. Nat Cell Biol. 2000;2:553–5. doi: 10.1038/35019596. [DOI] [PubMed] [Google Scholar]
- 66).Martinez-Caballero S, Dejean LM, Kinnally MS, Oh KJ, Mannella CA, Kinnally KW. Assembly of the mitochondrial apoptosis-induced channel, MAC. J Biol Chem. 2009;284:12235–12245. doi: 10.1074/jbc.M806610200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67).Antonsson B, Montessuit S, Sanchez B, Martinou JC. BAX is present as a high molecular weight oligomer/complex in the mitochondrial membrane of apoptotic cells. J Biol Chem. 2001;276:11615–23. doi: 10.1074/jbc.M010810200. [DOI] [PubMed] [Google Scholar]
- 68).Kuwana T, Mackey MR, Perkins G, Ellisman MH, Latterich M, Schneiter R, Green DR, Newmeyer DD. BID, BAX, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell. 2002;111:331–42. doi: 10.1016/s0092-8674(02)01036-x. [DOI] [PubMed] [Google Scholar]
- 69).Qian S, Wang W, Yang L, Hyang HW. Structure of transmembrane pore induced by BAX-derived peptide: evidence for lipidic pores. Proc Natl Acad Sci U S A. 2008;105:17379–83. doi: 10.1073/pnas.0807764105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70).Schafer B, Quispe J, Choudhary V, Chipuk JE, Ajero TG, Du H, Schneiter R, Kuwana T. Mitochondrial outer membrane proteins assist BID in BAX-mediated lipidic pore formation. Mol Biol Cell. 2009;20:2276–85. doi: 10.1091/mbc.E08-10-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71).Chipuk JE, McStay GP, Bharti A, Kuwana T, Clarke CJ, Siskind LJ, Obeid LM, Green DR. Sphingolipid metabolism cooperates with BAK and BAX to promote the mitochondrial pathway of apoptosis. Cell. 2012;148:988–1000. doi: 10.1016/j.cell.2012.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72).Renault TT, Floros KV, Elkholi R, Corrigan KA, Kushnareva Y, Wieder SY, Lindtner C, Serasinghe MN, Asciolla JJ, Buettner C, Newmeyer DD, Chipuk JE. Mitochondrial shape governs BAX-induced membrane permeabilization and apoptosis. Mol Cell. 2015;57:69–82. doi: 10.1016/j.molcel.2014.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73).Montessuit S, Somasekharan SP, Terrones O, Lucken-Ardjomande S, Herzig S, Schwarzenbacher R, Manstein DJ, Bossy-Wetzel E, Basanez G, Meda P, Martinou JC. Membrane remodeling induced by the dynamin-related protein Drp1 stimulates BAX oligomerization. Cell. 2010;142:889–901. doi: 10.1016/j.cell.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74).Garcia-Ruiz C, Mari M, Collel A, Morales A, Caballero F, Montero J, Terrones O, Basanez G, Fernandez-Checa JC. Mitochondrial cholesterol in health and disease. Histol Histolpathol. 2009:117–132. doi: 10.14670/HH-24.117. [DOI] [PubMed] [Google Scholar]
- 75).Christenson E, Merlin S, Saito M, Schlesinger P. Cholesterol effects on BAX pore activation. J Mol Biol. 2008;381:1168–83. doi: 10.1016/j.jmb.2008.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76).Lucken-Ardjomande S, Montessuit S, Martinou JC. BAX activation and stress-induced apoptosis delayed by the accumulation of cholesterol in mitochondrial membranes. Cell Death Differ. 2008;15:484–93. doi: 10.1038/sj.cdd.4402280. [DOI] [PubMed] [Google Scholar]
- 77).Montero J, Montserrat M, Collel A, Morales A, Basanez G, Garcia-Ruiz C, Fernandez-Checa JC. Cholesterol and peroxidized cardiolipin in mitochondrial membrane properties, permeabilization and cell death. Biochim Biophys Acta. 2010;1797:1217–1224. doi: 10.1016/j.bbabio.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78).De Kroon AI, Dolis D, Mayer A, Lill R, de Kruijff B. Phospholipid composition of highly purified mitochondrial outer membranes of rat liver and Neurospora crassa: Is cardiolipin present in the mitochondrial outer membrane? Biochim Biophys Acta. 1997;1325:108–116. doi: 10.1016/s0005-2736(96)00240-4. [DOI] [PubMed] [Google Scholar]
- 79).Gonzalvez F, Pariselli F, Dupaigne P, Budihardjo I, Lutter M, Antonsson B, Diolez P, Manon S, Martinou JC, Goubern M, Wang X, Bernard S, Petit PX. tBID interaction with cardiolipin primarily orchestrates mitochondrial dysfunctions and subsequently activates BAX and BAK. Cell Death Differ. 2005;12:614–26. doi: 10.1038/sj.cdd.4401571. [DOI] [PubMed] [Google Scholar]
- 80).Lucken-Ardjomande S, Montessuit S, Martinou JC. Contributions to BAX insertion and oligomerization of lipids of the mitochondrial outer membrane. Cell Death Differ. 2008;15:929–37. doi: 10.1038/cdd.2008.9. [DOI] [PubMed] [Google Scholar]
- 81).Garcia-Saez AJ, Ries J, Orzaez M, Perez-Paya E, Schwille P. Membrane promotes tBID interaction with BCL-XL. Nat Struct Mol Biol. 2009;16:1178–1185. doi: 10.1038/nsmb.1671. [DOI] [PubMed] [Google Scholar]
- 82).Lutter M, Fang M, Luo X, Nishjima M, Xie X, Wang X. Cardiolipin provides specificity for targeting of tBID to mitochondria. Nat Cell Biol. 2000;2:754–761. doi: 10.1038/35036395. [DOI] [PubMed] [Google Scholar]
- 83).Lutter M, Perkins GA, Wang X. The pro-apoptotic BCL-2 family member tBid localizes to mitochondrial contact sites. BMC Cell Biol. 2001;2:22. doi: 10.1186/1471-2121-2-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84).Gonzalvez F, Schug ZT, Houtkooper RH, MacKenzie ED, Brooks DG, Wanders RJA, Petit PX, Vaz FM, Gottlieb E. Cardiolipin provides an essential activating platform for caspase-8 on mitochondria. J Cell Biol. 2008;183:681–696. doi: 10.1083/jcb.200803129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85).Landeta O, Landajuela A, Gil D, Taneva S, Di Primo C, Sot B, Valle M, Frolov VA, Basanez G. Reconstitution of proapoptotic BAK function in liposomes reveals a dual role for mitochondrial lipids in the BAK-drive membrane permeabilization process. J Biol Chem. 2011;286:8123–8230. doi: 10.1074/jbc.M110.165852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86).Obeid LM, Linardic CM, Karolak LA, Hannun YA. Programmed cell death induced by ceramide. Science. 1993;259:1769–71. doi: 10.1126/science.8456305. [DOI] [PubMed] [Google Scholar]
- 87).Siskind LJ, Feinstein L, Yu T, David JS, Jones D, Choi J, Zuckerman JE, Tan W, Hill RB, Hardwick JM, Colombini M. Anti-apoptotic BCL-2 family proteins disassemble ceramide channels. J Biol Chem. 2008;283:6622–6630. doi: 10.1074/jbc.M706115200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88).Perera MN, Lin SH, Peterson YK, Bielawska A, Szulc ZM, Bittman R, Colombini M. BAX and BCL-XL exert their regulation on different sites of the ceramide channel. Biochem J. 2012;445:81–91. doi: 10.1042/BJ20112103. [DOI] [PubMed] [Google Scholar]
- 89).Ganesan V, Perera MN, Colombini D, Datsakovskiy D, Chadka K, Colombini M. Ceramide and activated BAX act synergistically to permeabilize the mitochondrial outer membrane. Apoptosis. 2010;15:553–562. doi: 10.1007/s10495-009-0449-0. [DOI] [PubMed] [Google Scholar]
- 90).Beverly LJ, Howell LA, Hernandez-Corbacho M, Casson K, Chipuk JE, Siskind LJ. BAK activation is necessary and sufficient to drive ceramide synthase-dependent ceramide accumulation following inhibition of BCL2-like proteins. Biochem J. 2013;452:111–9. doi: 10.1042/BJ20130147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91).Herman MD, Nyman T, Welin M, Lehtiö L, Flodin S, Trésaugues L, Kotenyova T, Flores A, Nordlund P. Completing the family portrait of the anti-apoptotic Bcl-2 proteins: crystal structure of human Bfl-1 in complex with Bim. FEBS Lett. 2008;582:3590–4. doi: 10.1016/j.febslet.2008.09.028. [DOI] [PubMed] [Google Scholar]
- 92).Petros AM, Medek A, Nettesheim DG, Kim DH, Yoon HS, Swift K, Matayoshi ED, Olterdorf T, Fesik SW. Solution structure of the antiapoptotic protein bcl-2. Proc Natl Acad Sci U S A. 2001;98:3012–17. doi: 10.1073/pnas.041619798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93).Rautureau GJ, Day CL, Hinds MG. The structure of Boo/Diva reveals a divergent Bcl-2 protein. Proteins. 2010;78:2181–6. doi: 10.1002/prot.22728. [DOI] [PubMed] [Google Scholar]
- 94).Ku B, Liang C, Jung JU, Oh BH. Evidence that inhibition of BAX activation by BCL-2 involves its tight and preferential interaction with the BH3 domain of BAX. Cell Res. 2011;21:627–41. doi: 10.1038/cr.2010.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95).Rautureau GJ, Yabal M, Yang H, Huang DC, Kvansakul M, Hinds MG. The restricted binding repertoire of Bcl-B leaves Bim as the universal BH3-only prosurvival Bcl-2 protein antagonist. Cell Death Dis. 2012;3:e443. doi: 10.1038/cddis.2012.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96).Denisov AY, Madiraju MS, Chen G, Khadir A, Beauparlant P, Attardo G, Shore GC, Gehring K. Solution structure of human BCL-w: modulation of ligand binding by the C-terminal helix. J Biol Chem. 2003;287:21124–8. doi: 10.1074/jbc.M301798200. [DOI] [PubMed] [Google Scholar]
- 97).Hinds MG, Lackmann M, Skea GL, Harrison PJ, Huang DC, Day CL. The structure of Bcl-w reveals a role for the C-terminal residues in modulating biological activity. EMBO J. 2003;22:1497–507. doi: 10.1093/emboj/cdg144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98).Denisov AY, Chen G, Sprules T, Moldoveanu T, Beauparlant P, Gehring K. Structural model of the BCL-w-BID peptide complex and its interactions with phospholipid micelles. Biochemistry. 2006;45:2250–6. doi: 10.1021/bi052332s. [DOI] [PubMed] [Google Scholar]
- 99).Lee EF, Dewson G, Smith BJ, Evangelista M, Pettikiriarachchi A, Dogovski C, Perugini MA, Colman PM, Fairlie WD. Crystal structure of a BCL-W domain-swapped dimer: implications for the functions of BCL-2 family proteins. Structure. 2011;19:1467–76. doi: 10.1016/j.str.2011.07.015. [DOI] [PubMed] [Google Scholar]
- 100).Aritomi M, Kunishima N, Inohora N, Ishibashi Y, Ohta S, Morikawa K. Crystal structure of rat Bcl-xL. Implications for the function of the Bcl-2 protein family. J Biol Chem. 1997;272:27886–92. doi: 10.1074/jbc.272.44.27886. [DOI] [PubMed] [Google Scholar]
- 101).Petros AM, Nettesheim DG, Wang Y, Olejniczak ET, Meadows RP, Mack J, Swift K, Matayoshi ED, Zhang H, Thompson CB, Fesik SW. Rationale for Bcl-xL/Bad peptide complex formation from structure, mutagenesis, and biophysical studies. Protein Sci. 2000;9:2528–34. doi: 10.1110/ps.9.12.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102).Liu X, Dai S, Zhu Y, Marrack P, Kappler JW. The structure of a Bcl-xL/Bim fragment complex: implications for Bim function. Immunity. 2003;19:341–52. doi: 10.1016/s1074-7613(03)00234-6. [DOI] [PubMed] [Google Scholar]
- 103).O’Neill JW, Manion MK, Maguire B, Hockenberry DM. BCL-XL dimerization by three-dimensional domain swapping. J Mol Biol. 2006;356:367–81. doi: 10.1016/j.jmb.2005.11.032. [DOI] [PubMed] [Google Scholar]
- 104).Oberstein A, Jeffrey PD, Shi Y. Crystal structure of the Bcl-XL-Beclin 1 peptide comples: Beclin 1 is a novel BH3-only protein. J Biol Chem. 2007;282:13123–32. doi: 10.1074/jbc.M700492200. [DOI] [PubMed] [Google Scholar]
- 105).Feng W, Huang S, Wu H, Zhang M. Molecular basis fo Bcl-xL’s target recognition versatility revealed by the structure of Bcl-xL in complex with the BH3 domain of Beclin-1. J Mol Biol. 2007;372:223–35. doi: 10.1016/j.jmb.2007.06.069. [DOI] [PubMed] [Google Scholar]
- 106).Lee EF, Sadowsky JD, Smith BJ, Czabotar PE, Peterson-Kaufman KJ, Colman PM, Gellman SH, Fairlie WD. High-resolution structural characterization of a helical alpha/beta-peptide foldamer bound to the anti-apoptotic protein Bcl-xL. Angew Chem Int Ed Engl. 2009;48:4318–22. doi: 10.1002/anie.200805761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107).Czabotar PE, Lee EF, Thompson GV, Wardak AZ, Fairlie WD, Colman PM. Mutation to Bax beyond the BH3 domain disrupts interactions with pro-survival proteins and promotes apoptosis. J Biol Chem. 2011;286:7123–31. doi: 10.1074/jbc.M110.161281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108).Ambrosi E, Capaldi S, Bovi M, Saccomani G, Perduca M, Monaco HL. Structural changes in the BH3 domain of SOUL protein upon interaction with the anti-apoptotic protein Bcl-xL. Biochem J. 2011;438:291–301. doi: 10.1042/BJ20110257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109).Okamoto T, Zobel K, Fedorova A, Quan C, Yang H, Fairbrother WJ, Huang DC, Smith BJ, Deshayes K, Czabotar PE. Stabilizing the pro-apoptotic BimBH3 helix (BimSAHB) does not necessarily enhance affinity or biological activity. ACS Chem Biol. 2013;8:297–302. doi: 10.1021/cb3005403. [DOI] [PubMed] [Google Scholar]
- 110).Rajan S, Choi M, Baek K, Yoon HS. Bh3 induced conformational changes in Bcl-Xl revealed by crystal structure and comparative analysis. Proteins. 2015 doi: 10.1002/prot.24816. [DOI] [PubMed] [Google Scholar]
- 111).Rajan S, Choi M, Nguyen QT, Ye H, Liu W, Toh HT, Kang C, Kamariah N, Li C, Huang H, White C, Baek K, Grüber G, Yoon HS. Structural transitions in Bcl-xL and its potential association with mitochondrial calcium ion transport. Sci Rep. 2015;5:10609. doi: 10.1038/srep10609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112).Czabotar PE, Lee EF, van Delft MF, Day CL, Smith BJ, Huang DC, Fairlie WD, Hinds MG, Colman PM. Structural insights into the degradation of Mcl-1 induced by BH3 domains. Proc Natl Acad Sci U S A. 2007;104:6217–22. doi: 10.1073/pnas.0701297104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113).Day CL, Smits C, Fan FC, Lee EF, Fairlie WD, Hinds MG. Structure of the BH3 domains from the p53-inducible BH3-only proteins Noxa and Puma in complex with Mcl-1. J Mol Biol. 2008;380:958–71. doi: 10.1016/j.jmb.2008.05.071. [DOI] [PubMed] [Google Scholar]
- 114).Liu Q, Moldoveanu T, Sprules T, Matta-Camacho E, Mansur-Azzam N, Gehring K. Apoptotic regulation by MCL-1 through heterodimerization. J Biol Chem. 2010;285:19615–24. doi: 10.1074/jbc.M110.105452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115).Fire E, Gullá SV, Grant RA, Keating AE. Mcl-1-Bim complexes accommodate surprising point mutations via minor structural changes. Protein Sci. 2010;19:507–19. doi: 10.1002/pro.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116).Czabotar PE, Lee EF, Thompson GV, Wardak AZ, Fairlie WD, Colman PM. Mutation to Bax beyond the BH3 domain disrupts interactions with pro-survival proteins and promotes apoptosis. J Biol Chem. 2011;286:7123–31. doi: 10.1074/jbc.M110.161281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117).Stewart ML, Fire E, Keating AE, Walensky LD. The MCL-1 BH3 helix is an exclusive MCL-1 inhibitor and apoptosis sensitizer. Nat Chem Biol. 2010;6:595–601. doi: 10.1038/nchembio.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118).Smith BJ, Lee EF, Checco JW, Evangelista M, Gellman SH, Fairlie WD. Structure-guided rational design of α/β-peptide foldamers with high affinity for BCL-2 family prosurvival proteins. Chembiochem. 2013;14:1564–72. doi: 10.1002/cbic.201300351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119).Liu G, Poppe L, Aoki K, Yamane H, Lewis J, Szyperski T. High-quality NMR structure of human anti-apoptotic protein domain Mcl-1 (171-327) for cancer drug design. PLoS One. 2014;9:e96521. doi: 10.1371/journal.pone.0096521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120).Wang H, Takemato C, Akasaka R, Uchikubo-Kamo T, Kishishita S, Murayama K, Tereda T, Chen L, Liu ZJ, Wang BC, Sugano S, Tanaka A, Inoue M, Kigawa T. Novel dimerization mode of the human Bcl-2 family protein Bak, a mitochondrial apoptosis regulator. J Struct Biol. 2009;166:32–7. doi: 10.1016/j.jsb.2008.12.003. [DOI] [PubMed] [Google Scholar]