Abstract
MicroRNAs (miRNAs) have been found to be dysregulated in prostate cancer (PCa). In this study, we investigated if miR-1207-3p is capable of distinguishing between indolent and aggressive PCa and if it contributes to explaining the disproportionate aggressiveness of PCa in men of African ancestry (moAA). A total of 404 patients with primary adenocarcinoma of the prostate were recruited between 1988 and 2003 at the Moffitt Cancer Center, Tampa, FL, USA. Patient clinicopathological features and demographic characteristics such as race were identified. RNA samples from 404 postprostatectomy prostate tumor tissue samples were analyzed by real-time quantitative reverse transcription polymerase chain reaction for the mRNA expression of miR-1207-3p. miR-1207-3p expression in PCa that resulted in overall death or PCa-specific death is significantly higher than in PCa cases that did not. The same positive correlation holds true for other clinical characteristics such as biochemical recurrence, Gleason score, clinical stage, and prostate-specific antigen level. Furthermore, miR-1207-3p expression was significantly less in moAA in comparison to Caucasian men. We also evaluated whether miR-1207-3p is associated with clinical outcomes adjusted for age at diagnosis and tumor stage in the modeling. Using competing risk regression, the PCa patients with a high miR-1207-3p expression (≥ 6 vs 3) had a high risk to develop PCa recurrence (hazard rate = 2.5, P < .001) adjusting for age at diagnosis and tumor stage. In conclusion, miR-1207-3p is a promising novel prognostic biomarker for PCa. Furthermore, miR-1207-3p may also be important in explaining the disproportionate aggressiveness of PCa in moAA.
Introduction
Prostate cancer (PCa), with nearly 240,000 men diagnosed in the United States in 2014, is a major cause of cancer morbidity and mortality [1], [2], [3]. The annual morbidity continues to steadily grow as the rate of this cancer has increased by 14% over the last two decades [3], [4], [5]. One of the challenges faced in combating PCa is its considerable heterogeneity. PCa usually begins as being androgen dependent, and initially, most men are responsive to pharmacological and surgical androgen deprivation therapies [6]. However, in spite of these efforts, in a subset of patients, the cancer inevitably reoccurs and advances, with approximately 30% of men developing clinical recurrence [3]. Ultimately, it is this highly aggressive form of PCa that leads to death [6]. Therapeutic options for advanced PCa are currently limited and largely ineffective [7].
Furthermore, for reasons still unclear, men of African ancestry (moAA) have the worlds’ highest incidence of PCa [8]. moAA have approximately two-thirds increased incidence and two times the increased mortality rate compared with Caucasian men (CM) [9], [10]. Furthermore, moAA are more frequently diagnosed with worse clinicopathological features (prostate-specific antigen [PSA] levels, Gleason grades, advanced tumor stage, etc.) than CM [8], [11], [12]. To decrease the excess mortality, effective early detection and therapeutic strategies are required.
MicroRNAs (miRNAs) have potential as effective early detection and prognostic biomarkers in PCa [4]. miRNAs are small (18-22 nucleotides) nonprotein coding regulatory RNAs [13]. They are well conserved and endogenously synthesized and have been shown to regulate the expression of nearly 60% of human genes [13]. Many miRNAs have been reported to be implicated in cancer pathogenesis, functioning as either oncogenes or tumor suppressors according to the roles of their target genes [13], [14], [15]. Their expression is usually found to be aberrant in many cancers including PCa [1], [14], [16]. Studies of different cancers have demonstrated that miRNAs can be useful tumor markers. For example, specific oncogenic and tumor suppressor miRNAs have been detected in the serum and urine of patients with breast, ovarian, and bladder cancer [17], [18], [19], [20]. However, only few reports exist in literature describing the role of miRNAs in PCa aggressiveness and in PCa racial disparities.
miR-1207-3p is encoded by the PVT1 nonprotein coding gene located at the 8q24 PCa susceptibility locus [21]. In this study, we aimed to assess the role of miR-1207-3p in PCa aggressiveness because aggressive PCa is the life-threatening form of PCa [2]. At diagnosis, clinical and pathologic features, such as PSA level, pathologic Gleason grade, and tumor stage, are used to inform clinical decision making [4], [5], [22]. To determine if miR-1207-3p can be useful in PCa clinical decision making, we examined the expression of miR-1207-3p in prostate tumor tissue of patients by real-time quantitative reverse transcription polymerase chain reaction (qPCR). Then, we evaluated the possible prognostic implication of miR-1207-3p. Consequently, this study investigated the clinical significance of miR-1207-3p in PCa and determined if it contributes to explaining the disproportionate aggressiveness of PCa in moAA.
Materials and Methods
Patients and Samples
To evaluate miR-1207-3p expression in clinical samples, we used tissue specimens from 404 patients (389 CM and 15 moAA) with primary adenocarcinoma of the prostate from 1988 to 2003 at the Moffitt Cancer Center, Tampa, FL, USA. Institutional Review Board approvals were obtained for the study protocol at the University of South Florida and the Moffitt Cancer Center, Tampa, FL. Signed informed consent was obtained from all study participants. Clinicopathological and demographic information of the patients are summarized in Table 1, Table 2.
Table 1.
Correlation of miR-1207-3p PCa Tissue Expression with Clinical Characteristics of PCa in 404 Patients
| Variables | N | Mean ± SD | P | |
|---|---|---|---|---|
| Recurrence | Yes | 155 | 6.53 ± 3.75 | |
| No | 249 | 4.49 ± 3.54 | < .0001 | |
| Death | Yes | 129 | 7.05 ± 4.37 | |
| No | 275 | 4.44 ± 3.09 | < .0001 | |
| Death specific | Alive | 273 | 4.48 ± 3.08 | |
| Dead | 79 | 6.85 ± 4.60 | ||
| PCA specific | 50 | 7.38 ± 4.02 | < .0001 | |
| Race | Black | 15 | 3.00 ± 2.65 | |
| White | 389 | 5.36 ± 3.76 | .016 | |
| Gleason score | ≤ 6 | 167 | 4.81 ± 3.40 | < .0001 |
| 7 | 135 | 3.85 ± 2.52 | ||
| ≥ 8 | 29 | 6.33 ± 4.30 | ||
| Stage | 1-2 | 302 | 4.84 ± 3.34 | |
| 3-4 | 95 | 6.51 ± 4.44 | < .0001 | |
| PSA | ˂4 | 278 | 3.97 ± 3.79 | .002 |
| 4-10 | 168 | 4.03 ± 2.80 | ||
| ˃10 | 51 | 5.73 ± 3.58 | ||
| Marital status | Divorced | 22 | 3.80 ± 3.05 | .154 |
| Married | 349 | 5.37 ± 3.75 | ||
| Single | 22 | 5.47 ± 3.79 |
N, number; SD, standard deviation; P, P value. The level of significance was established using a P value less than .05.
Table 2.
PCa Tissue Expression of miR-1207-3p in moAA and CM
| Variables | N | Mean ± SD | P | |
|---|---|---|---|---|
| Race | Black | 15 | 3.00 ± 2.65 | |
| White | 389 | 5.36 ± 3.76 | .016 | |
The level of significance was established using a P value less than .05.
RNA Extraction and qPCR Analysis
Prostate tumor tissues were collected at prostatectomy from 404 patients with primary PCa. After histopathologic confirmation, PCa tissues were obtained from unstained slides with macrodissection. Total RNA was extracted from PCa tissues using superscript IV First-strand Synthesis system (Life Technology No. 18091200) and quantified using UV spectrophotometer NanoDrop 2000 (Thermo Scientific, USA). The cDNA templates were synthesized from RNA samples by SuperScript using random hexamers. RNA samples from the 404 patient samples were analyzed by qPCR to examine the expression of miR-1207-3p.
Gene expression was determined using power SYBR Green Real-Time PCR Master Mix (Life Technology No. 4367649) and 2.0 μl of cDNA template. qPCR was performed on an ABI7900 machine using the following amplification conditions: 10 minutes at 95°C followed by 45 cycles of 15 seconds at 95°C, 30 seconds at 55°C, and 30 seconds at 72°C. All assays were carried out in triplicate to control for technical variance. Cycle threshold values were determined using the SDS ver2.3 software (Bio-Rad). MicroRNA 1207-3p expression was normalized with U6 expression within each sample. Relative quantification of target gene expression was evaluated using the comparative cycle threshold method.
Statistical Analysis
Differences in two and three clinical outcome groups were compared using the two-tailed Student’s t test and analysis of variance test, respectively. For evaluating the association of miR-1207-3p expression with PCa death and PCa recurrence in CM, a competing risk approach was applied. miR-1207-3p expression was categorized into three groups: low (< 3), median (3-6), and high (≥ 6) based on the tertile cut points. Factors associated with PCa death were evaluated using competing-risk regression [23]. The cumulative incidence curves of PCa death were generated by the subgroups of miR-1207-3p expression. For evaluating miR-1207-3p expression associated with PCa survival, PCa death was the primary interest, and non-PCa death was treated as a competing risk. Survival time was defined as the time from date of PCa diagnosis to date of death. For evaluating association of miR-1207-3p expression with PCa recurrence, PCa recurrence was the primary interest, and nonrecurrence death was treated as a competing risk. Time to event was defined as time from the date of PCa diagnosis to PCa recurrence or nonrecurrence death, whichever comes first. Data were censored at the last follow-up date. The same approach was applied to evaluate the association of miR-1207-3p expression with PCa recurrence. The candidate factors taken into consideration in modeling included age at diagnosis, Gleason score, and tumor stage. PSA was excluded from the models because the PSA values were missing in 150 CM. The level of significance was established using a P value less than .05.
Results
Positive Correlation between miR-1207-3p Expression and Clinicopathological Features of PCa
To determine miR-1207-3p expression pattern in PCa and to investigate its ability to predict clinical progression (e.g., biochemical recurrence, PCa-specific death, etc.), miR-1207-3p expression pattern in PCa tissues was analyzed using tissues collected from 404 primary PCa patients at the Moffitt Cancer Center. Correlations between miR-1207-3p expression and known clinical and pathological parameters were determined. We observed that increased miR-1207-3p expression in PCa tissue is significantly positively associated with aggressive clinicopathological features. As shown in Table 1, miR-1207-3p overexpression is positively correlated with overall death and PCa-specific death in patients with PCa. miR-1207-3p expression in PCa tissues of patients that died from any cause (7.05 ± 4.37 vs 4.44.18 ± 3.09, P < .0001) or experienced PCa-specific death (7.38 ± 4.02 vs 4.48 ± 3.08, P < .0001) is significantly higher than expression in PCa tissues of patients that remained alive. Next, we investigated the expression of miR-1207-3p in PCa tissues of patients that had recurrent versus those that had nonrecurrent PCa to explore the involvement of miR-1207-3p in PCa progression. We observed that the expression of miR-1207-3p is significantly higher in recurrent PCa in comparison to nonrecurrent PCa (6.53 ± 3.75 vs 4.49 ± 3.54, P < .0001). This indicates that miR-1207-3p overexpression is associated with progression of PCa patients and may serve as a novel prognostic biomarker for PCa. Indeed, miR-1207-3p is significantly overexpressed in PCa tissues from patients with a Gleason score ≥ 8 (6.33 ± 4.30) in comparison to patients with a Gleason score ≤ 6 (4.81 ± 3.40) or Gleason score 7 (3.85 ± 2.52, P ≤ .0001), further confirming miR-1207-3p as a prognostic biomarker in PCa. In addition, miR-1207-3p expression’s positive correlation with advanced disease was also borne out for PCa stage (stage 1-2: 4.84 ± 3.34 vs stage 3-4: 6.51 ± 4.44, P < .0001) and PSA levels (PSA levels < 4: 3.97 ± 3.79, vs PSA levels 4-10: 4.03 ± 2.80 vs PSA levels > 10: 5.73 ± 3.58, P = .002). To our knowledge, this is the first observation of miR-1207-3p as a prognostic biomarker in PCa.
miR-1207-3p Is Differentially Expressed in PCa in moAA compared with CM
It is noteworthy that, in this cohort, moAA had a significantly decreased expression of miR-1207-3p in their PCa tissues in comparison to miR-1207-3p expression in the PCa tissue of CM. miR-1207-3p expression was significantly less in PCa tissues of moAA in comparison to the PCa tissues of CM (3.00 ± 2.65 vs 5.36 ± 3.76, P = .016) (Table 2). This suggests that miR-1207-3p may play a role in explaining the disproportionately increased aggressiveness of PCa in moAA.
miR-1207-3p Expression Is Associated with PCa Recurrence and PCa-Specific Death in CM
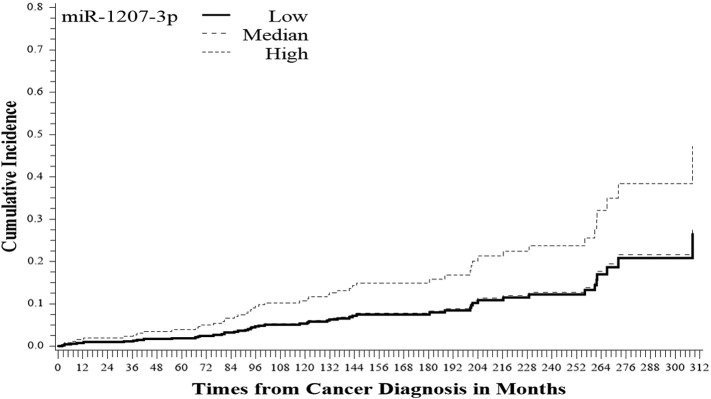
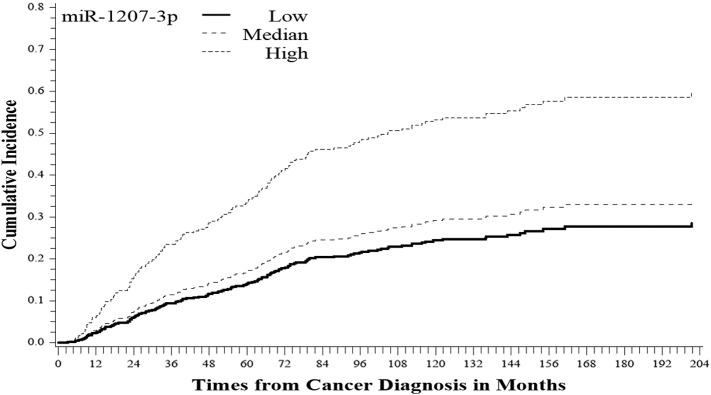
Among CM, there are 118, 131, and 140 patients with a low (< 3), median (3-6), and high (≥ 6) miR-1207-3p expression, respectively. In addition to miR-1207-3p expression, the candidate factors taken into consideration in modeling included age at diagnosis, Gleason score, and tumor stage. Age at diagnosis significantly correlated with PCa-specific death (hazard rate [HR] = 1.9, P < .01). As expected, stage is significantly correlated with recurrence (HR = 2.7, P < .01). Although Gleason score values for some patients were missing (missing n = 71 in CM), Gleason score was nevertheless highly correlated with tumor stage (Fisher’s exact P value < .001). We evaluated the association of miR-1207-3p with clinical outcomes adjusting for age at diagnosis and tumor stage in the modeling. Using competing risk regression, the CM with a high miR-1207-3p expression (≥ 6 vs 3) had a high risk of developing PCa recurrence (HR = 2.5, P < .001) adjusting for age at diagnosis and tumor stage. Also, miR-1207-3p expression had a marginally significant impact on PCa survival (HR = 1.8 P = .06) (Table 3, Figure 1, Figure 2).
Table 3.
miR-1207-3p Expressions Associated with PCa Death and Recurrence for CM
| Variables |
PCa Death |
PCa Recurrence |
|||
|---|---|---|---|---|---|
| Factor | Unadjusted |
Adjusted |
Unadjusted |
Adjusted |
|
| HR (95% CI)1 |
HR (95% CI)1 |
HR (95% CI)1 |
HR (95% CI)1 |
||
| N = 378 | N = 368 | ||||
| Age at diagnosis (in 10 years) | 1.9 (1.2-2.9)** | 1.9 (1.2-3.0)** | 1.1 (0.9-1.4) | 0.9 (0.8-1.2) | |
| Stage (3/4 vs 1/2) | 1.5 (0.8-2.6) | 1.3 (0.7-2.4) | 3.1 (2.2-4.3)** | 2.7 (1.9-3.8)** | |
| miR-1207-3p | < 3 | 1 | 1 | 1 | 1 |
| 3-6 | 1.0 (0.4-2.6) | 1.0 (0.4-2.7) | 1.2 (0.8-2.0) | 1.3 (0.8-2.1) | |
| ≥ 6 | 2.1 (0.9-4.6) | 1.8 (0.8-4.3) | 2.7 (1.8- 4.2)** | 2.5 (1.6-4.0)** | |
†P < .05.
Hazard rate (95% confidence interval) analyzed using competing risk regression.
P < 0.01.
Figure 1.
Cumulative incidence of PCa death by miR-1207-3p expression level (P = .062).
Figure 2.
Cumulative incidence of PCa recurrence by miR-1207-3p expression level (P < .001).
Discussion
Approximately 30% of PCa patients develop clinical recurrence, and the survival period of this phase of PCa is limited and extremely variable [3]. The challenge lies in the identification of those patients most at risk for relapse. Currently, factors such as serum PSA level, Gleason score, and tumor stage are used for diagnosis, prognostication, and treatment decision making [4], [24]. However, none of these alone or in combination are adequate indicators for accurate clinical decision making [3]. It is likely that heterogeneity of PCa at the molecular level may account for the fact that even those patients displaying similar PSA levels, tumor stage, and Gleason score can still have different clinical outcomes [3]. Furthermore, current measures for predicting/detecting recurrence such as increased serum PSA levels may actually lead to false-positive diagnosis [25]. Moreover, increased PSA following radical prostatectomy can be caused by the presence of residual benign prostate tissue [3]. These issues with the use of PSA are considered so problematic that the US Preventive Services Task Force has advised against the widespread use of PSA based on their opinion that the service has no real clear benefits and in actuality may cause more harm than good [26]. Consequently, further understanding of the molecular abnormalities underlying PCa progression is critical to the discovery of good molecular biomarkers for diagnosis, prognostication, and clinical decision making.
Understanding the molecular mechanisms of PCa progression is essential for the discovery of robust prognostic markers which are capable of helping to identify those patients that are at the greatest risk of relapse. This will, in turn, enable us to optimize management strategies to control PCa progression. In recent studies, several putative biomarkers have been suggested which, thus far, have only achieved limited success. Among them are prostate cancer antigen 3, prostate-specific membrane antigen, and altered expression of apoptotic regulators Bcl-2 and Bax [7], [24], [27]. These proposed prognostic biomarkers either have not been validated in multiple studies/large scale studies or have failed to reliably and accurately estimate the clinical outcome due to the heterogeneity of PCa [7]. Considering these current limitations, it remains a priority in PCa research to discover novel biomarkers to add to our “tool box” to improve the clinical decision-making process. Identification of biomarkers which can efficiently predict recurrence after radical prostatectomy would be of great clinical significance. Besides, a biomarker that can predict distinct outcomes regardless of similar clinical characteristics would be invaluable. To contribute to this effort, in the present study, we evaluated the expression of miR-1207-3p in the PCa tissues of patients with both aggressive and nonaggressive PCa.
The importance of miRNAs in cancer biology has become apparent in many different cancers, including PCa [14], [15], [16]. Typically, they regulate biological processes such as proliferation, migration, apoptosis, and differentiation by negatively regulating gene expression at the posttranscriptional level via binding to the 3′ untranslated region of their mRNA target [4], [14]. There are several advantages to the discovery of microRNA prognostic biomarkers [22]. First, they have been shown to be able to distinguish differentiation states of several malignancies including breast, lung, and colon cancers [13], [22]. Hence, their aberrant expression can be exploited as a prognostic tool to distinguish between less aggressive and more aggressive forms of the same cancer type. Consequently, investigations of miRNA differential expression in cancers are proving to be very beneficial. Second, they are relatively stable in tissues and upon their release into a variety of biofluids such as blood and urine [28], [29]. Repeated measurements of miRNA levels in biofluids are very feasible, are minimally invasive, and can provide critical information on a disease’s molecular signature status over time through the course of treatment and recurrence [2]. Real-time qPCR assays are able to detect specific RNAs indicative of disease status from a simple noninvasive sampling without the need for a biopsy or surgically resected tissue [2], [22]. Consequently, we assessed the role of miR-1207-3p expression in PCa patients with a focus on a potential role in prognostication.
Our results demonstrate that it may be possible to predict clinical behavior of PCa based upon miR-1207-3p expression in primary PCa. To our knowledge, this is the first study to evaluate the expression of miR-1207-3p in any human disease. PCa is a heterogeneous disease, with the exact mechanisms for its development and progression unclear [30]. However, our data show that the expression of miR-1207-3p in PCa tissues displays a significant and distinctly different pattern between indolent and aggressive PCa. Overexpression of miR-1207-3p is clearly positively related to PCa progression and worse prognosis. Therefore, our data support the potential use of miR-1207-3p as a prognostic biomarker in PCa. We anticipate that this beneficial use will advance into the clinical setting soon.
Despite progress being made in the field, significant PCa racial disparity persists [26], [31], [32]. And the aggressiveness of the disease may be due to specific biological factors [31], [32]. Here, we present data that suggest that miR-1207-3p could possibly contribute to the disproportionate aggressiveness of miR-1207-3p in moAA. The discovery of a miRNA biomarker that is capable of distinguishing between indolent and aggressive PCa and displays differential expression in men of different ancestry would be of great significance for the development and optimization of personalized treatment approaches [33]. Moreover, as moAA have a two-fold higher chance of getting PCa and aggressive PCa, it is of great importance to enhance our understanding of the molecular mechanisms which may explain the disproportionately increased aggressiveness of PCa in moAA [8], [31], [32]. Differential expression of miRNAs such as miR-26a has been reported in PCa [9]. We also previously reported that miR-21 is associated with recurrence, especially in obese cases [16]. However, the importance of miRNAs in PCa in moAA is still unclear. In this regard, we have discovered that miR-1207-3p is differentially expressed in primary tumors of moAA tissues compared with primary tumors of CM. To our knowledge, this is the first report of a microRNA that directly correlates with aggressive PCa but has divergent expression between CM and moAA. These data suggest that it may be possible to predict clinical behavior of PCa based upon miRNA-1207-3p expression in primary PCa. We anticipate that this beneficial use will advance into the clinical setting.
Although the overall sample size of the present study is relatively large, a limitation is the small sample size of the moAA subgroup. Although challenging because of issues related to recruitment, a larger study enriched with a substantial proportion of moAA is warranted. In addition, the present study did not examine miR-1207-3p expression in nontumor versus tumor tissues. Therefore, the current data do not provide information regarding whether miR-1207-3p is differentially expressed in normal prostate versus PCa tissue. Likewise, because miRNAs exert functions via regulating the expression levels of their target genes, it is of great importance to identify the target genes of miR-1207-3p in PCa and to investigate the involvement of miR-1207-3p target genes in PCa. Further studies are needed to address these issues.
Conclusion
Our study identifies miR-1207-3p as a novel prognostic biomarker of aggressive PCa that is differentially expressed between CM and moAA. As such, an individual PCa patient’s miR-1207-3p PCa tissue expression level has the potential to provide a novel and clinically relevant basis for making decisions in PCa. It is also possible that miR-1207-3p–based therapeutic strategies may have efficacy in PCa but must be applied in a personalized fashion. Further studies are required to establish this.
Human and Animal Rights
Signed informed consent was obtained from all study participants.
Conflict of Interest
The authors declare no potential conflicts of interests. Olorunseun O. Ogunwobi and Jong Park had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Author contributions
Jong Y. Park led data acquisition. Dibash K. Das, Jong Y. Park, and Olorunseun O. Ogunwobi contributed to data analysis. Dibash K. Das, Joseph R. Osborne, Jong Y. Park, and Olorunseun O. Ogunwobi contributed to data interpretation and manuscript writing. Hui-Yi Lin contributed to biostatistical analysis and manuscript writing.
Acknowledgements
Work in Dr. Ogunwobi’s laboratory is supported by the NIMHD/NIH grant to Hunter College: 8 G12 MD007599. This research was also supported in part by the National Cancer Institute grant R01 CA128813 (J. Y. P.).
Footnotes
Work in Dr. Ogunwobi’s laboratory is supported by the NIMHD/NIH grant to Hunter College: 8 G12 MD007599. This research was also supported in part by the National Cancer Institute grant R01 CA128813 (J. Y. P.).
Contributor Information
Jong Y. Park, Email: jong.park@moffitt.org.
Olorunseun O. Ogunwobi, Email: ogunwobi@genectr.hunter.cuny.edu.
References
- 1.Larne O, Ostling P, Haflidadottir BS, Hagman Z, Aakula A, Kohonen P, Kallioniemi O, Edsjo A, Bjartell A, Lilja H. miR-183 in prostate cancer cells positively regulates synthesis and serum levels of prostate-specific antigen. Eur Urol. 2015;68(4):581–588. doi: 10.1016/j.eururo.2014.12.025. [DOI] [PubMed] [Google Scholar]
- 2.Huang X, Yuan T, Liang M, Du M, Xia S, Dittmar R, Wang D, See W, Costello BA, Quevedo F. Exosomal miR-1290 and miR-375 as prognostic markers in castration-resistant prostate cancer. Eur Urol. 2015;67(1):33–41. doi: 10.1016/j.eururo.2014.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahmad I, Singh LB, Yang ZH, Kalna G, Fleming J, Fisher G, Cooper C, Cuzick J, Berney DM, Moller H. Mir143 expression inversely correlates with nuclear ERK5 immunoreactivity in clinical prostate cancer. Br J Cancer. 2013;108(1):149–154. doi: 10.1038/bjc.2012.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun X, Yang Z, Zhang Y, He J, Wang F, Su P, Han J, Song Z, Fei Y. Prognostic implications of tissue and serum levels of microRNA-128 in human prostate cancer. Int J Clin Exp Pathol. 2015;8(7):8394–8401. [PMC free article] [PubMed] [Google Scholar]
- 5.Parnes HL, House MG, Tangrea JA. Prostate cancer prevention: strategies for agent development. Curr Opin Oncol. 2013;25(3):242–251. doi: 10.1097/CCO.0b013e32835fc8d4. [DOI] [PubMed] [Google Scholar]
- 6.Nouri M, Ratther E, Stylianou N, Nelson CC, Hollier BG, Williams ED. Androgen-targeted therapy-induced epithelial mesenchymal plasticity and neuroendocrine transdifferentiation in prostate cancer: an opportunity for intervention. Front Oncol. 2014;4:370. doi: 10.3389/fonc.2014.00370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karatas OF, Guzel E, Suer I, Ekici ID, Caskurlu T, Creighton CJ, Ittmann M, Ozen M. miR-1 and miR-133b are differentially expressed in patients with recurrent prostate cancer. PloS one. 2014;9(6):e98675. doi: 10.1371/journal.pone.0098675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koochekpour S, Willard SS, Shourideh M, Ali S, Liu C, Azabdaftari G, Saleem M, Attwood K. Establishment and characterization of a highly tumorigenic African American prostate cancer cell line, E006AA-hT. Int J Biol Sci. 2014;10(8):834–845. doi: 10.7150/ijbs.9406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Theodore SC, Rhim JS, Turner T, Yates C. MiRNA 26a expression in a novel panel of African American prostate cancer cell lines. Ethn Dis. 2010;20(1 Suppl. 1):S1–S96. [100] [PMC free article] [PubMed] [Google Scholar]
- 10.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62(1):10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 11.Hoffman RM, Gilliland FD, Eley JW, Harlan LC, Stephenson RA, Stanford JL, Albertson PC, Hamilton AS, Hunt WC, Potosky AL. Racial and ethnic differences in advanced-stage prostate cancer: the Prostate Cancer Outcomes Study. J Natl Cancer Inst. 2001;93(5):388–395. doi: 10.1093/jnci/93.5.388. [DOI] [PubMed] [Google Scholar]
- 12.Chu KC, Miller BA, Springfield SA. Measures of racial/ethnic health disparities in cancer mortality rates and the influence of socioeconomic status. J Natl Med Assoc. 2007;99(10):1092–1100. [102–4] [PMC free article] [PubMed] [Google Scholar]
- 13.Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2012;4(3):143–159. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 15.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101(9):2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amankwah EK, Anegbe E, Park H, Pow-Sang J, Hakam A, Park JY. miR-21, miR-221 and miR-222 expression and prostate cancer recurrence among obese and non-obese cases. Asian J Androl. 2013;15(2):226–230. doi: 10.1038/aja.2012.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105(30):10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu W, Qin W, Atasoy U, Sauter ER. Circulating microRNAs in breast cancer and healthy subjects. BMC Res Notes. 2009;2:89. doi: 10.1186/1756-0500-2-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Resnick KE, Alder H, Hagan JP, Richardson DL, Croce CM, Cohn DE. The detection of differentially expressed microRNAs from the serum of ovarian cancer patients using a novel real-time PCR platform. Gynecol Oncol. 2009;112(1):55–59. doi: 10.1016/j.ygyno.2008.08.036. [DOI] [PubMed] [Google Scholar]
- 20.Yamada Y, Enokida H, Kojima S, Kawakami K, Chiyomaru T, Tatarano S, Yoshino H, Kawahara K, Nishiyama K, Seki N. MiR-96 and miR-183 detection in urine serve as potential tumor markers of urothelial carcinoma: correlation with stage and grade, and comparison with urinary cytology. Cancer Sci. 2011;102(3):522–529. doi: 10.1111/j.1349-7006.2010.01816.x. [DOI] [PubMed] [Google Scholar]
- 21.Huppi K, Volfovsky N, Runfola T, Jones TL, Mackiewicz M, Martin SE, Mushinski JF, Stephens R, Caplen NJ. The identification of microRNAs in a genomically unstable region of human chromosome 8q24. Mol Cancer Res. 2008;6(2):212–221. doi: 10.1158/1541-7786.MCR-07-0105. [DOI] [PubMed] [Google Scholar]
- 22.Cannistraci A, Di Pace AL, De Maria R, Bonci D. MicroRNA as new tools for prostate cancer risk assessment and therapeutic intervention: results from clinical data set and patients' samples. Biomed Res Int. 2014;2014:146170. doi: 10.1155/2014/146170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng SC, Fine JP, Wei LJ. Prediction of cumulative incidence function under the proportional hazards model. Biometrics. 1998;54(1):219–228. [PubMed] [Google Scholar]
- 24.Roberts MJ, Chow CW, Schirra HJ, Richards R, Buck M, Selth LA, Doi SA, Samaratunga H, Perry-Keene J, Payton D. Diagnostic performance of expression of PCA3, Hepsin and miR biomarkers inejaculate in combination with serum PSA for the detection of prostate cancer. Prostate. 2015;75(5):539–549. doi: 10.1002/pros.22942. [DOI] [PubMed] [Google Scholar]
- 25.Sita-Lumsden A, Dart DA, Waxman J, Bevan CL. Circulating microRNAs as potential new biomarkers for prostate cancer. Br J Cancer. 2013;108(10):1925–1930. doi: 10.1038/bjc.2013.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Powell IJ, Bock CH, Ruterbusch JJ, Sakr W. Evidence supports a faster growth rate and/or earlier transformation to clinically significant prostate cancer in black than in white American men, and influences racial progression and mortality disparity. J Urol. 2010;183(5):1792–1796. doi: 10.1016/j.juro.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bubendorf L, Sauter G, Moch H, Jordan P, Blochlinger A, Gasser TC, Mihatsch MJ. Prognostic significance of Bcl-2 in clinically localized prostate cancer. Am J Pathol. 1996;148(5):1557–1565. [PMC free article] [PubMed] [Google Scholar]
- 28.Ronnau CG, Verhaegh GW, Luna-Velez MV, Schalken JA. Noncoding RNAs as novel biomarkers in prostate cancer. Biomed Res Int. 2014;2014:591703. doi: 10.1155/2014/591703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mall C, Rocke DM, Durbin-Johnson B, Weiss RH. Stability of miRNA in human urine supports its biomarker potential. Biomark Med. 2013;7(4):623–631. doi: 10.2217/bmm.13.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X, Wu J. Prognostic role of microRNA-145 in prostate cancer: a systems review and meta-analysis. Prostate Int. 2015;3(3):71–74. doi: 10.1016/j.prnil.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Presley CJ, Raldow AC, Cramer LD, Soulos PR, Long JB, Yu JB, Makarov DV, Gross CP. A new approach to understanding racial disparities in prostate cancer treatment. J Geriatr Oncol. 2013;4(1):1–8. doi: 10.1016/j.jgo.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peters N, Armstrong K. Racial differences in prostate cancer treatment outcomes: a systematic review. Cancer Nurs. 2005;28(2):108–118. doi: 10.1097/00002820-200503000-00004. [DOI] [PubMed] [Google Scholar]
- 33.Mata DA, Katchi FM, Ramasamy R. Precision medicine and men's health. Am J Mens Health. 2015 doi: 10.1177/1557988315595693. [DOI] [PMC free article] [PubMed] [Google Scholar]