Abstract
The cellular response to stress is orchestrated by the expression of a family of proteins termed heat shock proteins (hsp) that are involved in the stabilization of basic cellular processes to preserve cell viability and homeostasis. The bulk of hsp function occurs within the cytosol and subcellular compartments. However, some hsp have also been found outside cells released by an active mechanism independent of cell death. Extracellular hsp act as signaling molecules directed at activating a systemic response to stress. The export of hsp requires the translocation from the cytosol into the extracellular milieu across the plasma membrane. We have proposed that membrane insertion is the initial step in this export process. We investigated the interaction of the major inducible hsp from mammalian (Hsp70) and bacterial (DnaK) species with liposomes. We found that mammalian Hsp70 displayed a high specificity for negatively charged phospholipids, such as phosphatidyl serine, whereas DnaK interacted with all lipids tested regardless of the charge. Both proteins were inserted into the lipid bilayer as demonstrated by resistance to acid or basic washes that was confirmed by partial protection from proteolytic cleavage. Several regions of mammalian Hsp70 were inserted into the membrane with a small portion of the N-terminus end exposed to the outer phase of the liposome. In contrast, the N-terminus end of DnaK was inserted into the membrane, exposing the C-terminus end outside the liposome. Mammalian Hsp70 was found to make high oligomeric complexes upon insertion into the membranes whereas DnaK only formed dimers within the lipid bilayer. These observations suggest that both Hsp70s interact with lipids, but mammalian Hsp70 displays a high degree of specificity and structure as compared with the bacterial form.
Keywords: Hsp70, DnaK, Lipids, Membranes, Heat shock proteins
Introduction
Organisms are continuously exposed to changing environments during natural conditions to which they adapt by modulating their gene expression profile. In some circumstances, organisms are confronted with extreme conditions or stresses that are likely to affect homeostasis. The cellular response to stress is orchestrated, at least in part, by the expression of a protein family named heat shock proteins (hsp). These proteins are involved in the stabilization of basic cellular processes during stress to preserve cell viability and a rapid return to homeostasis. In addition, they protect cells from subsequent insults in a time-dependent fashion that is known as stress tolerance (De Maio 1999). Some hsp are also present during normal physiological conditions in which they play a major role in protein folding as well as other cellular processes. Thus, they are also called molecular chaperones (De Maio 1999, Hartl and Hayer-Hartl 2009). Hsp have also been found outside cells released after cell death by necrosis or by an active mechanism independent of cell death (De Maio 2011, De Maio and Vazquez 2013). These extracellular hsp act as signaling molecules directed at activating a systemic response to stress that has been named the “stress observation system (SOS),” directed at avoiding the propagation of the insult (De Maio 2011).
The active release of hsp into the extracellular milieu has been controversial since these proteins do not contain any consensus signal that allows them to be exported via the classical RE-Golgi pathway. Indeed, the first observations of extracellular hsp were ignored for over 20 years (Tytell et al. 1986, Hightower and Guidon 1989). Currently, there is extensive evidence for the presence of extracellular hsp, in particular during various pathogenic conditions (De Maio 2011, Pockley et al. 2014). Hsp are likely exported by an alternative mechanism independent of the ER-Golgi pathway, termed “the non-classical secretory pathway” (Nickel and Seedorf 2008). The major element for this non-classical secretory pathway is that proteins need to pass from the cytosol into the extracellular milieu crossing the plasma membranes that appear thermodynamically unfavorable. This process has been proposed to be mediated by a membrane protein pore (Rabouille et al. 2012). In contrast, we have hypothesized that hsp are secreted by an active mechanism in which insertion into the plasma membrane is the first step, followed by their release associated with extracellular vesicles (ECV) (De Maio 2011). Indeed, hsp have been detected in a large number of ECV isolated from different sources (Thery et al. 2009, De Maio 2011). The caveat regarding this export mechanism is that hsp do not contain any hydrophobic domain that could predict partitioning within the lipid bilayer of the plasma membrane. However, a large number of reports have shown the presence of hsp within membranes. For example, the pioneering work of Multhoff’s group showed the presence of Hsp70 on the cell surface of transformed cells, which was not in association with membrane proteins but rather a true insertion into the plasma membrane (Multhoff and Hightower 1996; Multhoff 2007). This observation has been confirmed by others (De Maio 2011). Other hsp have also been detected within cellular membranes (Bausero et al. 2004, Belles et al. 1999, Roberts et al. 1999, Mills et al. 2010). Thus, Hsp70 presents an unorthodox capacity to get inserted into the lipid bilayer without any conventional transmembrane domains.
The question that emerges is what is the nature of the interaction between hsp and membranes? We have shown that Hsp70, both the inducible form (Hsp70 or HSPA1) and the constitutive form (Hsc70 or HSPA8), interact with artificial lipid membranes forming very stable ion conductance pathways (Arispe and De Maio 2000, Vega et al. 2008) that have been confirmed by others (Macazo and White 2014). In addition, the protein has been reportedly inserted into the membrane of liposomes (Armijo et al. 2014, McCalister et al. 2016) as well as inducing liposome aggregation (Arispe et al. 2002). However, the mechanism for membrane insertion has not been elucidated. In order to gain insight into this possible mechanism, we investigate the interaction of bacterial Hsp70 (DnaK) with liposomes. Bacterial and mammalian Hsp70 appear to play a very similar role (Mayer and Bukau 2005, Genevaux et al. 2007, Hartl and Hayer-Hartl 2009). DnaK shares a high degree of homology with mammalian Hsp70 but displays several differences that could provide us with important information regarding the molecular mechanism for membrane insertion. We found that DnaK is also capable of insertion into lipid membranes. However, we observed remarkable differences in the lipid interaction between mammalian and bacterial Hsp70.
Materials and methods
Liposome preparation and incorporation of DnaK or Hsp70
Liposomes were formed by resuspending the dried lipid film (400 μg, Avanti Polar Lipids, Alabaster, AL) in 50-mM Tris buffer pH 7.4 (120 μl), and vortexed every 5 min for 30 min. The preparation was extruded through a 100-nm membrane filter (15 passages). Thereafter, liposomes were incubated with recombinant DnaK (ADI-SPP-630, Escherichia coli K12 strain, Enzo life Sciences, Farmingdale, NY) or Hsp70 (HspA1A, ADI-ESP-555, Enzo Life Sciences) in 50-mM Tris buffer pH 7.4 for 30 min at 25 °C at a ratio of 400 μg lipids per 1 μg of protein or as indicated in the figure legend. DnaK/Hsp70-containing liposomes were centrifuged at 100,000×g for 45 min at 4 °C and washed once with a sodium carbonate (Na2CO3, pH 11.5) and centrifuged again. The final pellet after centrifugation (DnaK/Hsp70-liposomes) was solubilized in lithium dodecyl sulfate (LDS) sample buffer and boiled for 8 min. Material was resolved by LDS-polyacrylamide gel electrophoresis (PAGE) and visualized using Coomassie Brilliant Blue R-250 stain (ThermoFisher Scientific, Waltham, MA). In some experiments, samples were transferred onto a nitrocellulose membrane, and the presence of DnaK was detected by anti-DnaK monoclonal antibodies (SPA880, Enzo Life Sciences) and HRP-conjugated goat anti-mouse as secondary antibodies (Thermo Scientific, Rockford, IL). After incubation with the primary and secondary antibodies, the immuno-detection signal was visualized by chemiluminescence. All images were captured using the ChemiDoc MP Imaging System (Biorad, Hercules, CA) and analyzed using the ImageLab 5.2 software (Biorad).
Mass spectrometry analysis
Recombinant DnaK (2 μg) or recombinant Hsp70 (2 μg) were incubated with palmitoyloleoyl phosphatidylserine (POPS) liposomes (400 μg) in 50-mM Tris buffer, pH 7.4 for 30 min at 25 °C. Liposomes were centrifuged at 100,000×g for 45 min at 4 °C. Pellets were resuspended in Na2CO3 buffer (pH 11.5) and centrifuged again. The resulting proteoliposomes were incubated with 50 μg/ml proteinase K for 30 min at 37 °C, and the liposomes were pelleted at high-speed centrifugation and washed again. Pellets were solubilized and digested with trypsin. The resulting peptides were analyzed by HPLC coupled with tandem mass spectrometry (LC-MS/MS) using nano-spray ionization (TripleTOF 5600 hybrid mass spectrometer (AB SCIEX). Data were analyzed using MASCOT® (Matrix Science) and Protein Pilot 4.0 (AB SCIEX) for peptide identifications.
Results
DnaK interacts with lipid membranes
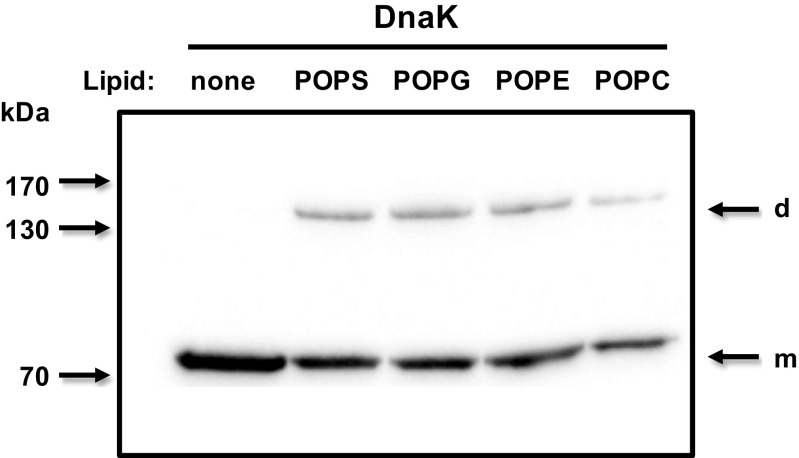
Recombinant pure DnaK was incubated with unilamellar liposomes (100 nm), each made of palmitoyloleoyl phosphatidylserine (POPS), palmitoyloleoyl phosphatidylcholine (POPC), palmitoyloleoyl phosphatidylethanolamine (POPE), or palmitoyloleoyl phosphatidylglycerol (POPG) at 25 °C for 30 min. Then the protein and liposome suspension was centrifuged at 100,000×g to separate incorporated and non-incorporated DnaK into the liposomes. The liposome pellet was washed with Na2CO3 buffer pH 11.5, and liposomes were solubilized in LDS sample buffer and liposome-incorporated proteins were separated by LDS-PAGE and detected by Western blotting. A similar incorporation of DnaK (50 %) was observed in POPS, POPE, and POPG liposomes and further reduced (30 %) in POPC liposomes (Fig. 1), indicating a capacity for membrane insertion but lacking the lipid selectivity observed for mammalian Hsp70 (Arispe et al. 2004, Schilling et al. 2009, Armijo et al. 2014). Monomers and dimers of DnaK were detected in samples corresponding to liposome preparations, but only the monomeric form was observed in the absence of liposomes (Fig. 1, see arrows).
Fig. 1.
DnaK incorporates into the membrane of liposomes made of different lipids. Recombinant DnaK (1 μg) was incubated with POPS, POPG, POPE, or POPC liposomes (400 μg) in 50-mM Tris buffer pH 7.4 for 30 min at 25 °C. At the end of the incubation period, the liposomes were centrifuged at 100,000×g for 40 min at 4 °C. Pellets were resuspended in Na2CO3 buffer (pH 11.5) and centrifuged again. Liposome pellets were then solubilized in LDS sample buffer, and liposome-incorporated proteins were separated by LDS-PAGE and analyzed by Western blotting using a monoclonal antibody against DnaK (SPA880, Enzo Life Sciences) and HRP-conjugated goat anti-mouse as secondary antibodies. Arrows indicate the location of monomeric (m) and dimeric (d) forms of DnaK, respectively
The interaction of DnaK with POPS liposomes depends on the incubation conditions
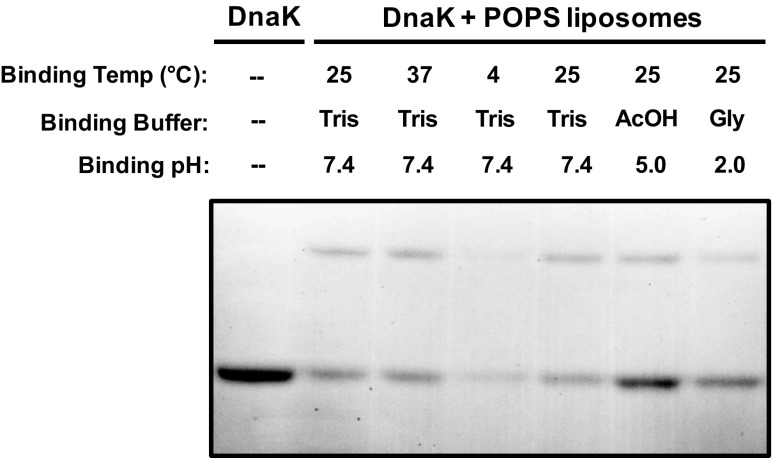
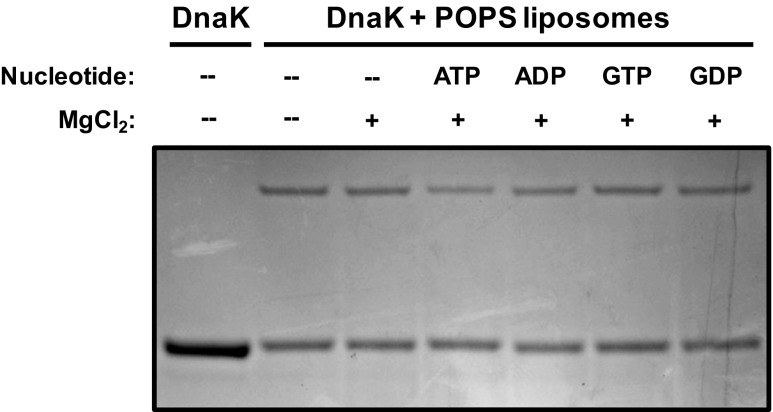
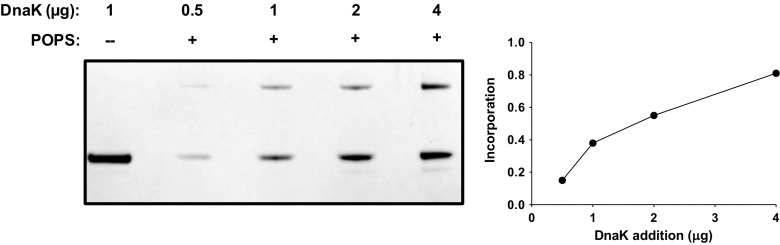
We wanted to address whether the interaction of DnaK with liposomes was modulated by the incubation conditions. For these experiments, only POPS liposomes were used. Recombinant DnaK was incubated with POPS liposomes (100 nm) at 4, 25, or 37 °C for 30 min, processed as described in “Materials and methods,” and the incorporation of DnaK into liposomes was detected by LDS-PAGE and Coomassie Brilliant Blue staining. There was no apparent difference in incorporation between incubations at 25 or 37 °C, but there was a decrease in incorporation in samples incubated at 4 °C (Fig. 2). DnaK was also incubated with POPS liposomes at pHs 2, 5 and 7.4 and analyzed as described above. There was an apparent increase in incorporation by decreasing the pH of the incubation buffer with maximal activity at pH = 5 (Fig. 2). Since the negative charge of the serine group is reduced at lower pHs, these observations suggest that DnaK does not have a high selectivity for negatively charged phospholipids, which is consistent with a similar interaction with POPC and POPE liposomes. We also investigated whether the incorporation of DnaK into POPS liposomes was dependent on the presence of nucleotides. DnaK was incubated with POPS liposomes in the absence or presence of MgCl2, or ATP/MgCl2, ADP/MgCl2, GTP/MgCl2, and GDP/MgCl2. The preparations were treated after the incubation as described above. The addition of MgCl2 without or in combination with nucleotides did not affect the incorporation of DnaK into POPS liposomes (Fig. 3). Finally, we investigated whether the incorporation of DnaK into POPS liposomes was dependent on the concentration of the protein in the incubation mixture. DnaK at concentrations of 0.5, 1, 2, and 4 μg/ml was incubated with POPS liposomes and the samples analyzed as described above. An increase in the incorporation of DnaK into liposomes was dependent on the initial concentration of the protein (Fig. 4).
Fig. 2.
The incorporation of DnaK into POPS liposomes is influenced by the incubation conditions. Recombinant DnaK (1 μg) was incubated with POPS liposomes (400 μg) in 50-mM Tris buffer pH 7.4 for 30 min at 4, 25, or 37 °C. DnaK was also incubated with POPS liposomes in glycine buffer pH 2 or acetate buffer pH 5 at 25 °C. At the end of the incubation period, the liposomes were centrifuged at 100,000×g for 40 min at 4 °C. Liposome pellets were then solubilized in LDS sample buffer, and liposome-incorporated proteins were separated by LDS-PAGE and detected by Coomassie Brilliant Blue staining
Fig. 3.
The incorporation of DnaK into POPS liposomes is not affected by the presence of nucleotides. Recombinant DnaK (1 μg) was incubated with POPS liposomes (400 μg) in 50-mM Tris buffer pH 7.4 for 30 min at 25 °C in the absence or presence of MgCl2 (1 mM), ATP + MgCl2 (1 mM each), ADP + MgCl2 (1 mM each), GTP + MgCl2 (1 mM each), and GDP + MgCl2 (1 mM each). The liposomes were centrifuged at 100,000×g for 40 min at 4 °C. Pellets were resuspended in Na2CO3 buffer (pH 11.5) and centrifuged again. Liposome pellets were then solubilized in LDL sample buffer, and liposome-incorporated proteins were separated by LDS-PAGE and detected by Coomassie Brilliant Blue staining
Fig. 4.
The incorporation of DnaK into POPS liposomes is dependent on the concentration of DnaK in the incubation mixture. Increasing amounts of recombinant DnaK (0.5 to 4 μg) were incubated with POPS liposomes (400 μg) in 50-mM Tris buffer pH 7.4 for 30 min at 25 °C. The liposomes were centrifuged at 100,000×g for 40 min at 4 °C. Pellets were resuspended in Na2CO3 buffer (pH 11.5) and centrifuged again. Liposome pellets were then solubilized in LDL sample buffer, and liposome-incorporated proteins were separated by LDS-PAGE and detected by Coomassie Brilliant Blue staining
The incorporation of Hsp70 and DnaK into POPS liposomes is not identical
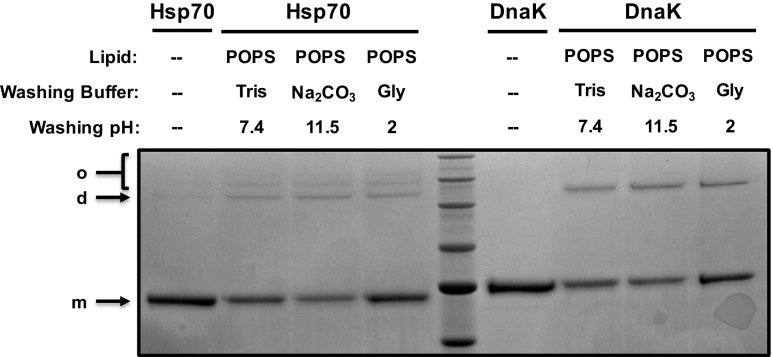
We have previously reported that human Hsp70 displayed a high affinity for POPS liposomes in contrast to liposomes made of non-negatively charged lipids (Armijo et al. 2014). We have found that DnaK did not display the same lipid specificity of Hsp70 since the protein interacted with all tested lipids. Therefore, it was interesting to perform a side-by-side comparison of Human Hsp70 (HspA1A) and bacterial DnaK. Both recombinant proteins were incubated with POPS liposomes at pH 7.4 for 30 min at 25 °C. Samples were processed as described in “Materials and methods,” and incorporation of the proteins into liposomes was detected by LDS-PAGE and Coomassie Brilliant Blue staining. The incorporation of both proteins in POPS liposomes were very similar, except that multioligomeric forms were observed in samples containing Hsp70 and only a dimeric form was observed for DnaK (Fig. 5, see arrows). In addition, the fact that the presence of the protein within the liposomes is not affected by washes at extreme pH (2 and 11.5) clearly demonstrates that the protein is inserted into the lipid membrane rather than being bound to the liposomes.
Fig. 5.
The incorporation of DnaK and Hsp70 into POPS liposomes results in different states of oligomerization. Recombinant DnaK (1 μg) or recombinant human Hsp70 (1 μg) were incubated with POPS liposomes (400 μg) in 50-mM Tris buffer pH 7.4 for 30 min at 25 °C. The liposomes were centrifuged at 100,000×g for 40 min at 4 °C. Pellets were resuspended in Tris buffer (pH 7.4), Na2CO3 buffer (pH 11.5), or glycine buffer (pH 2) and centrifuged again. Liposome pellets were then solubilized in LDS sample buffer, and liposome-incorporated proteins were separated by LDS-PAGE and detected by Coomassie Brilliant Blue staining. Arrows indicate the location of monomeric (m), dimeric (d) and oligomeric (o) forms of human Hsp70 and bacterial DnaK, respectively
The region inserted into POPS liposomes was different between DnaK and human Hsp70
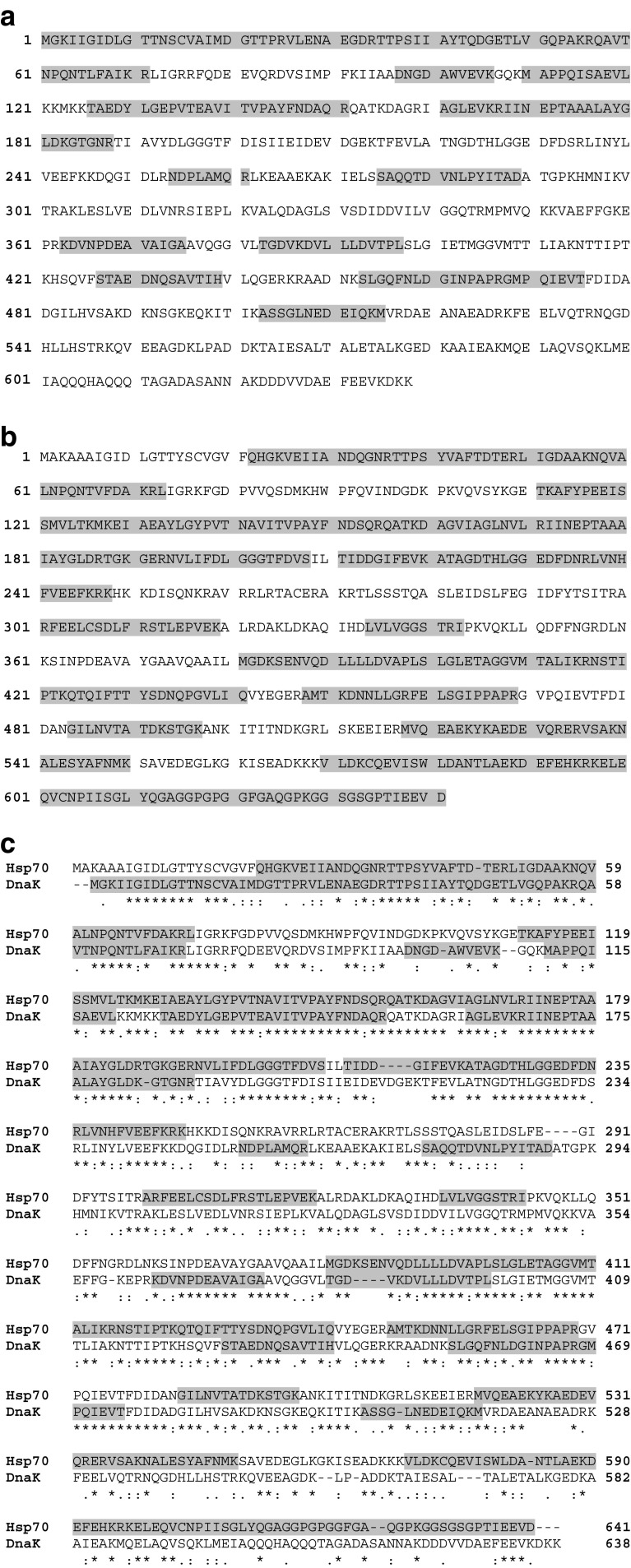
We next investigated the possible region of DnaK and Hsp70 (HspA1A) that is inserted into the lipid membrane. The proteins were incubated into liposomes and washed at pH 11.5 as described above. The resulting proteoliposomes were incubated with proteinase K for 30 min at 37 °C, and the liposomes were pelleted at high-speed centrifugation and washed again. The liposome pellet was analyzed by mass spectroscopy. Several peptides were detected for both DnaK and human Hsp70. These peptides match to regions of the protein that are protected from the proteinase treatment (grey areas on Fig. 6), which likely correspond to transmembrane regions or domains exposed to the lumen of the liposome. The N-terminus end of DnaK appeared as the region inserted into the liposomes with the C-terminus end exposed outside (Fig. 6a). In contrast, several regions of Hsp70 were observed inserted into the liposome with a small region of the N-terminus outside the liposome (Fig. 6b). These observations showed a disparity regarding lipid interaction between mammalian and bacterial Hsp70 (Fig. 6c).
Fig. 6.
The region inserted into POPS liposomes was different between DnaK and Hsp70. Recombinant DnaK (2 μg) or recombinant human Hsp70 (2 μg) were incubated with POPS liposomes (400 μg) in 50-mM Tris buffer pH 7.4 for 30 min at 25 °C. The liposomes were centrifuged at 100,000×g for 40 min at 4 °C. Pellets were resuspended in Na2CO3 buffer (pH 11.5) and centrifuged again. The resulting proteoliposomes were incubated with proteinase K (50 μg/ml) for 30 min at 37 °C, and the liposomes were centrifuged at 100,000×g for 40 min at 4 °C and washed again. The liposome pellet was analyzed by mass spectroscopy. DnaK (a) and Hsp70 (b) peptides that are protected from proteinase K digestion are highlighted in grey. Similarity between DnaK and Hsp70 peptide mapping is shown in the pairwise alignment of DnaK and Hsp70 sequences (c)
Discussion
The presence of extracellular hsp is a well-accepted phenomenon in spite of their controversial discovery over 20 years ago (De Maio 2011). The function of extracellular hsp is apparently different from their chaperone activity inside cells. They appear to act as signaling molecules directed at activating a systemic response to stress (De Maio 2011, Pockley et al. 2014), particularly in cells of the immune system (Asea et al. 2000, Gastpar et al. 2005, Vega et al. 2008). The mechanism for the export of extracellular hsp is not clear because they do not contain a consensus secretory signal necessary for their transport via the ER-Golgi compartment. Since hsp are localized in the cytosol and other subcellular compartments, they need to cross the plasma membrane to gain access to the extracellular milieu. We have proposed that one possible mechanism for the extracellular export of hsp requires the initial insertion into the plasma membrane, followed by their release associated with ECV (De Maio 2011). Indeed Hsp70 has been detected within the membrane of ECV (Gastpar et al. 2005, Vega et al. 2008). Moreover, several studies have demonstrated Hsp70 membrane interaction with artificial lipid membranes (Arispe et al. 2002, 2004, Vega et al. 2008, Schilling et al. 2009, Armijo et al. 2014, McCalister et al. 2016).
The mechanism of Hsp70 insertion into membranes is still unclear. This protein does not contain a stretch of hydrophobic amino acids that could predict membrane insertion. We have proposed that the insertion of Hsp70 into membranes is composed of several steps, including recognition (binding) of the phospholipid polar region, induction of a conformational change, insertion into the plasma membrane, and oligomerization (Armijo et al. 2014). To gain insight into this hypothesis, we investigated the interaction of bacterial Hsp70 (DnaK) with lipids. DnaK has substantial homology with mammalian Hsp70 but enough differences to allow us to make predictions about the insertion process. We observed that DnaK was indeed incorporated into liposomes in a concentration-dependent manner. We and others have previously shown that Hsp70 displays a high degree of selectivity for negatively charged lipids, such as POPS (Arispe et al. 2004, Armijo et al. 2014), POPG (Nylandsted et al. 2004, Armijo et al. 2014) and glycolipids (Harada et al. 2007, Gehrmann et al. 2008). However, we did not observe the same selectivity for DnaK that was found to interact with the bulk of lipids found in natural membranes (POPS, POPC, POPE, and POPG). DnaK membrane insertion was demonstrated by resistance to stringent washes at low pH (2) and high pH (11.5) buffers and proteolysis/mass spectroscopic analysis. Finally, the incorporation of DnaK into liposomes was not dependent on the presence of nucleotides as opposed to prior reports about human Hsp70 (McCalister et al. 2016).
We detected that the N-terminus end of DnaK was the region that was inserted into the membrane with the C-terminus end exposed outside the liposome as determined by limited proteolysis and mass spectroscopic analysis of the inserted proteins within liposomes (Fig. 6a). This observation is in contrast with the insertion of mammalian Hsp70 into membranes that display a more extensive region imbedded within the liposome, with the N-terminus end exposed outside the liposome (Fig. 6b) and the C-terminus end of mammalian Hsp70 inserted into the membrane, as previously reported (Armijo et al. 2014). In contrast, the C-terminus end of DnaK is located outside the liposome. The orientation of Hsp70 within the membrane of liposomes may resemble the translocation of the protein from the cytosol into the plasma membrane. In this case, the lumen of the liposome is equivalent to the extracellular milieu and the external medium is comparable to the cytosol. Prior studies have shown that the C-terminus end of Hsp70 is likely exposed to the cell exterior. Indeed, the C-terminus end of Hsp70 inserted into the liposomes was found to be protected, at least in part, from proteolytic cleavage by exogenous proteases. Moreover, the epitope (TDK) recognized by the cmHsp70.1 antibody that has been used to detect cell surface Hsp70 (Botzeler et al. 1998) was found protected from the protease treatment in Hsp70 proteoliposomes, suggesting that this region is likely exposed to the lumen of the liposome, inferring the same conformation within the plasma membrane.
We have proposed that Hsp70 undergoes a conformational change during or upon membrane insertion resulting in a multimeric oligomerization (Armijo et al. 2014). Our results showed disparity between DnaK and Hsp70, particularly at the levels of membrane oligomerization. Prior studies have shown that a small degree of DnaK dimerization occurs in solution in a nucleotide-dependent manner. These dimers were important for the interaction with DnaJ (Sarbeng et al. 2015). We only detected DnaK dimers after insertion into the plasma membrane, and their presence was not affected by the presence of nucleotides. It is possible that the lack of detection of DnaK dimers in solution was due to their disruption during the conditions of LDS-PAGE. In contrast, dimers within the lipid bilayer may be preserved during LDS-PAGE, perhaps due to a conformational change of the protein within the membrane. In contrast to DnaK, we observed high molecular weight discreet oligomers upon insertion of mammalian Hsp70 into membranes as shown in this study and prior observations (Armijo et al. 2014). The capacity of forming multimeric subunits within the lipid bilayer is a requirement for the conductance of ions across the membrane as the one observed for Hsp70. Therefore, it is not surprising that we never detected ion channel activity when DnaK was exposed to artificial lipid bilayers (unpublished observation). Since the C-terminus end is the least conserved region between bacterial and mammalian Hsp70, it is possible that the oligomerization domain resides within this region. It is also possible that oligomerization only occurred when the C-terminus end is inserted into the lipid bilayer, and this region is not within the membrane in the case of DnaK as mentioned above.
The detection of DnaK on membranes has been reported previously. Bukau et al. (1993) showed the presence of DnaK on the membrane of E. coli, as detected by electron microscopy, an observation that has been forgotten for many years. The presence of DnaK in membranes has also been reported in Fusobacterium nucleatum (Skar et al. 2003) and Bacillus subtilis (Seydlova et al. 2012). The latter was observed after ethanol stress suggesting that changes in membrane fluidity may play a role in its insertion (Horváth et al. 2008). Indeed, Hsp70 membrane insertion was enhanced by a decrease of membrane fluidity (Armijo et al. 2014), resembling the presence of Hsp70 in lipid rafts of cells (Broquet et al. 2003; Hunter-Lavin et al. 2004; Chen et al. 2005; Wang et al. 2006; Vega et al. 2008), which are membrane domains enriched in saturated lipids, such as sphingolipids (Lingwood and Simons 2010).
In summary, DnaK is added to the list of proteins that could spontaneously translocate into membranes without displaying an evident hydrophobic domain. Since so much information is available about the structure and function of DnaK (Genevaux et al. 2007, Kityk et al. 2015), it may be possible to formulate hypotheses that could be tested experimentally in order to elucidate the unusual ability of the proteins to get embedded into the lipid bilayer. In addition, the biological significance of DnaK and other Hsp70 membrane insertion remains to be revealed. However, it is possible that the mechanism of membrane insertion and extracellular release may be different between DnaK and Hsp70.
Acknowledgments
This work was supported by the National Institutes of Health, grant numbers GM R01 09845 and GM R25 083275.
References
- Arispe N, De Maio A. ATP and ADP modulate a cation channel formed by Hsc70 in acidic phospholipid membranes. J Biol Chem. 2000;275:30839–30843. doi: 10.1074/jbc.M005226200. [DOI] [PubMed] [Google Scholar]
- Arispe N, Doh M, De Maio A. Lipid interaction differentiates the constitutive and stress-induced heat shock proteins Hsc70 and Hsp70. Cell Stress Chaperones. 2002;7:330–338. doi: 10.1379/1466-1268(2002)007<0330:LIDTCA>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arispe N, Doh M, Simakova O, Kurganov B, De Maio A. Hsc70 and Hsp70 interact with phosphatidylserine on the surface of PC12 cells resulting in a decrease of viability. FASEB J. 2004;18:1636–1645. doi: 10.1096/fj.04-2088com. [DOI] [PubMed] [Google Scholar]
- Armijo G, Okerblom J, Cauvi DM, Lopez V, Schlamadinger DE, Kim J, Arispe N, De Maio A. Interaction of heat shock protein 70 with membranes depends on the lipid environment. Cell Stress Chaperones. 2014;19:877–886. doi: 10.1007/s12192-014-0511-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asea A, Kraeft SK, Kurt-Jones EA, Stevenson MA, Chen LB, Finberg RW, Koo GC, Calderwood SK. HSP70 stimulates cytokine production through a CD14-dependant pathway, demonstrating its dual role as a chaperone and cytokine. Nat Med. 2000;6:435–442. doi: 10.1038/74697. [DOI] [PubMed] [Google Scholar]
- Bausero MA, Page DT, Osinaga E, Asea A. Surface expression of Hsp25 and Hsp72 differentially regulates tumor growth and metastasis. Tumour Biol. 2004;25:243–251. doi: 10.1159/000081387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belles C, Kuhl A, Nosheny R, Carding SR. Plasma membrane expression of heat shock protein 60 in vivo in response to infection. Infect Immun. 1999;67:4191–4200. doi: 10.1128/iai.67.8.4191-4200.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botzeler C, Li G, Issels RD, Multhoff G. Definition of extracellular localized epitopes of Hsp70 involved in an NK immune response. Cell Stress Chaperones. 1998;3:6–11. doi: 10.1379/1466-1268(1998)003<0006:DOELEO>2.3.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broquet AH, Thomas G, Masliah J, Trugnan G, Bachelet M. Expression of the molecular chaperone Hsp70 in detergent-resistant microdomains correlates with its membrane delivery and release. J Biol Chem. 2003;278:21601–21606. doi: 10.1074/jbc.M302326200. [DOI] [PubMed] [Google Scholar]
- Bukau B, Reilly P, McCarty J, Walker GC. Immunogold localization of the DnaK heat shock protein in Escherichia coli cells. J Gen Microbiol. 1993;139:95–99. doi: 10.1099/00221287-139-1-95. [DOI] [PubMed] [Google Scholar]
- Chen S, Bawa D, Besshoh S, Gurd JW, Brown IR. Association of heat shock proteins and neuronal membrane components with lipid rafts from the rat brain. J Neurosci Res. 2005;81:522–529. doi: 10.1002/jnr.20575. [DOI] [PubMed] [Google Scholar]
- De Maio A. Heat shock proteins: facts, thoughts, and dreams. Shock. 1999;11:1–12. doi: 10.1097/00024382-199901000-00001. [DOI] [PubMed] [Google Scholar]
- De Maio A. Extracellular heat shock proteins, cellular export vesicles, and the Stress Observation System: a form of communication during injury, infection, and cell damage. Cell Stress Chaperones. 2011;16:235–249. doi: 10.1007/s12192-010-0236-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Maio A, Vazquez D. Extracellular heat shock proteins: a new location, a new function. Shock. 2013;40:239–246. doi: 10.1097/SHK.0b013e3182a185ab. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gastpar R, Gehrmann M, Bausero MA, Asea A, Gross C, Schroeder JA, Multhoff G. Heat shock protein 70 surface-positive tumor exosomes stimulate migratory and cytolytic activity of natural killer cells. Cancer Res. 2005;65:5238–5247. doi: 10.1158/0008-5472.CAN-04-3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrmann M, Liegisch G, Schmitz G, Anderson R, Steinem C, De Maio A, Pockley G, Multhoff G. Tumor-specific Hsp70 plasma membrane localization is enabled by the glycosphingolipid Gb3. PLoS One. 2008;3 doi: 10.1371/journal.pone.0001925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genevaux P, Georgopoulos C, Kelley WL. The Hsp70 chaperone of Escherichia coli: a paradigm for the repartition of chaperone functions. Mol Microbiol. 2007;66:840–857. doi: 10.1111/j.1365-2958.2007.05961.x. [DOI] [PubMed] [Google Scholar]
- Harada H, Murakami T, Tea SS, Takeuchi A, Koga T, Okada S, Suico MA, Shuto T, Kai H. Heat shock suppresses human NK cell cytotoxicity via regulation of perforin. Int J Hyperth. 2007;23:657–665. doi: 10.1080/02656730701822087. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M. Converging concepts of protein folding in vitro and in vivo. Nat Struct Mol Biol. 2009;16:574–581. doi: 10.1038/nsmb.1591. [DOI] [PubMed] [Google Scholar]
- Hightower LE, Guidon PT., Jr Selective release from cultured mammalian cells of heat-shock (stress) proteins that resemble glia-axon transfer proteins. J Cell Physiol. 1989;138:257–266. doi: 10.1002/jcp.1041380206. [DOI] [PubMed] [Google Scholar]
- Horváth I, Multhoff G, Sonnleitner A, Vígh L. Membrane-associated stress proteins: more than simply chaperones. Biochim Biophys Acta. 2008;1778:1653–1664. doi: 10.1016/j.bbamem.2008.02.012. [DOI] [PubMed] [Google Scholar]
- Hunter-Lavin C, Davies EL, Bacelar MM, Marshall MJ, Andrew SM, Williams JH. Hsp70 release from peripheral blood mononuclear cells. Biochem Biophys Res Commun. 2004;324:511–517. doi: 10.1016/j.bbrc.2004.09.075. [DOI] [PubMed] [Google Scholar]
- Kityk R, Vogel M, Schlecht R, Bukau B, Mayer MP. Pathways of allosteric regulation in Hsp70 chaperones. Nat Commun. 2015;6:8308. doi: 10.1038/ncomms9308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- Macazo FC, White RJ. Monitoring charge flux to quantify unusual ligand-induced ion channel activity for use in biological nanopore-based sensors. Anal Chem. 2014;86:5519–5525. doi: 10.1021/ac500832a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCalister C, Kdeiss B, Nikolaidis N. Biochemical characterization of the interaction between HspA1A and phospholipids. Cell Stress Chaperones. 2016;21:41–53. doi: 10.1007/s12192-015-0636-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills DR, Haskell MD, Callanan HM, Flanagan D, Brilliant K, Yang D, Hixson D. Monoclonal antibody to novel cell surface epitope on Hsc70 promotes morphogenesis of bile ducts in newborn rat liver. Cell Stress Chaperones. 2010;15:39–53. doi: 10.1007/s12192-009-0120-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Multhoff G. Heat shock protein (Hsp70): membrane location, export and immunological relevance. Methods. 2007;43:229–237. doi: 10.1016/j.ymeth.2007.06.006. [DOI] [PubMed] [Google Scholar]
- Multhoff G, Hightower LE. Cell surface expression of heat shock proteins and the immune response. Cell Stress Chaperones. 1996;1:167–176. doi: 10.1379/1466-1268(1996)001<0167:CSEOHS>2.3.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickel W, Seedorf M. Unconventional mechanisms of protein transport to the cell surface of eukaryotic cells. Annu Rev Cell Dev Biol. 2008;24:287–308. doi: 10.1146/annurev.cellbio.24.110707.175320. [DOI] [PubMed] [Google Scholar]
- Nylandsted J, Gyrd-Hansen M, Danielewicz A, Fehrenbacher N, Lademann U, Høyer-Hansen M, Weber E, Multhoff G, Rohde M, Jäättelä M. Heat shock protein 70 promotes cell survival by inhibiting lysosomal membrane permeabilization. J Exp Med. 2004;200:425–435. doi: 10.1084/jem.20040531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pockley AG, Henderson B, Multhoff G. Extracellular cell stress proteins as biomarkers of human disease. Biochem Soc Trans. 2014;42:1744–1751. doi: 10.1042/BST20140205. [DOI] [PubMed] [Google Scholar]
- Rabouille C, Malhotra V, Nickel W. Diversity in unconventional protein secretion. J Cell Sci. 2012;125:5251–5255. doi: 10.1242/jcs.103630. [DOI] [PubMed] [Google Scholar]
- Roberts J, Menoret A, Cohen N. Cell surface expression of the endoplasmic reticular heat shock protein gp96 is phylogenetically conserved. J Immunol. 1999;163:4133–4139. [PubMed] [Google Scholar]
- Sarbeng EB, Liu Q, Tian X, Yang J, Li H, Wong JL, Zhou L, Liu Q. A functional DnaK dimer is essential for the efficient interaction with Hsp40 heat shock protein. J Biol Chem. 2015;290:8849–8862. doi: 10.1074/jbc.M114.596288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling D, Gehrmann M, Steinem C, De Maio A, Pockley AG, Abend M, Molls M, Multhoff G. Binding of heat shock protein 70 to extracellular phosphatidylserine promotes killing of normoxic and hypoxic tumor cells. FASEB J. 2009;23:2467–2477. doi: 10.1096/fj.08-125229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydlova G, Halada P, Fiser R, Toman O, Ulrych A, Svobodova J. DnaK and GroEL chaperones are recruited to the Bacillus subtilis membrane after short-term ethanol stress. J Appl Microbiol. 2012;112:765–774. doi: 10.1111/j.1365-2672.2012.05238.x. [DOI] [PubMed] [Google Scholar]
- Skar CK, Kruger PG, Bakken V. Characterisation and subcellular localisation of the GroEL-like and DnaK-like proteins isolated from Fusobacterium nucleatum ATCC 10953. Anaerobe. 2003;9:305–312. doi: 10.1016/j.anaerobe.2003.08.004. [DOI] [PubMed] [Google Scholar]
- Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev. 2009;9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- Tytell M, Greenberg SG, Lasek RJ. Heat shock-like protein is transferred from glia to axon. Brain Res. 1986;363:161–164. doi: 10.1016/0006-8993(86)90671-2. [DOI] [PubMed] [Google Scholar]
- Vega VL, Rodríguez-Silva M, Frey T, Gehrmann M, Diaz JC, Steinem C, Multhoff G, Arispe N, De Maio A. Hsp70 translocates into the plasma membrane after stress and is released into the extracellular environment in a membrane-associated form that activates macrophages. J Immunol. 2008;180:4299–4307. doi: 10.4049/jimmunol.180.6.4299. [DOI] [PubMed] [Google Scholar]
- Wang R, Kovalchin JT, Muhlenkamp P, Chandawarkar RY. Exogenous heat shock protein 70 binds macrophage lipid raft microdomain and stimulates phagocytosis, processing, and MHC-II presentation of antigens. Blood. 2006;107:1636–1642. doi: 10.1182/blood-2005-06-2559. [DOI] [PubMed] [Google Scholar]