Abstract
Chrysanthemum morifolium, is a well-known flowering plant worldwide, and has a high commercial, floricultural, and medicinal value. In this study, simple-sequence repeat (SSR) markers were generated from EST datasets and were applied to assess the genetic diversity among 32 cultivars. A total of 218 in silico SSR loci were identified from 7300 C. morifolium ESTs retrieved from GenBank. Of all SSR loci, 61.47% of them (134) were hexa-nucleotide repeats, followed by tri-nucleotide repeats (17.89%), di-nucleotide repeats (12.39%), tetra-nucleotide repeats (4.13%), and penta-nucleotide repeats (4.13%). In this study, 17 novel EST-SSR markers were verified. Along with 38 SSR markers reported previously, 55 C. morifolium SSR markers were selected for further genetic diversity analysis. PCR amplification of these EST-SSRs produced 1319 fragments, 1306 of which showed polymorphism. The average polymorphism information content of the SSR primer pairs was 0.972 (0.938–0.993), which showed high genetic diversity among C. morifolium cultivars. Based on SSR markers, 32 C. morifolium cultivars were separated into two main groups by partitioning of the clusters using the unweighted pair group method with arithmetic mean dendrogram, which was further supported by a principal coordinate analysis plot. Phylogenetic relationship among C. morifolium cultivars as revealed by SSR markers was highly consistent with the classification of medicinal C. morifolium populations according to their origin and ecological distribution. Our results demonstrated that SSR markers were highly reproducible and informative, and could be used to evaluate genetic diversity and relationships among medicinal C. morifolium cultivars.
Keywords: Chrysanthemum morifolium, EST-SSR, marker development, genetic diversity, phylogenetic relationship
Introduction
Chrysanthemum morifolium (Ramat.) Kitam is an important member of the family Asteraceae. A large number of C. morifolium cultivars are ornamental and medicinally important plants that are planted all over the world (Teixeira da Silva, 2003; Bhattacharya and da Silva, 2006). In addition to their esthetic value, some C. morifolium cultivars are used medicinally for their curative effects, particularly for treating common cold, headache, and dizziness (Chinese Pharmmacopoeia Editorial Committee, 2010). Based on growing regions and processing methods, the main domestic varieties of medicinal C. morifolium are divided into “Hang-ju,” “Bo-ju,” “Qi-ju,” “Gong-ju,” “Ji-ju,” “Chu-ju,” “Huai-ju,” and “Chuan-ju” (Shao et al., 2010; Zhao et al., 2013). In China, some regions have become important producing areas, such as Tongxiang (Zhejiang Province), Yancheng (Jiangsu Province); Wuzhi (Henan Province), Chuzhou, and Sexian (An'hui Province) (Shao et al., 2010; Wang T. et al., 2013; Chen et al., 2014).
For a long time, the genetic improvement of C. morifolium has been impeded because of its genome complexity, high level of heterozygosity, and the occurrence of both inbreeding depression and self-incompatibility (Anderson, 2006). Understanding genetic diversity is very important in plant breeding programs and the conservation of genetic resources. Molecular markers have potentials to reveal the genetic diversity among medicinal C. morifolium germplasms. Recently, a few studies have been reported on genetic diversity in cultivated chrysanthemums (Bhattacharya and da Silva, 2006; Xu et al., 2006; Shao et al., 2010; Zhang et al., 2011b), preliminary genetic linkage map construction, and QTL detection (Zhang et al., 2010, 2011a) using RAPD, AFLP, ISSR, and SRAP markers.
Simple-sequence repeats (SSRs), also known as microsatellites, are short tandem repeated motifs that may vary in the number of repeats at a given locus (Tautz, 1989). SSR markers have many advantages over other molecular markers, such as genetic co-dominance. They are multi-allelic, relatively abundant, widely dispersed across the genome, and easily and automatically scored (Powell et al., 1996). Over the past few years, SSR markers have been used in genetic diversity analysis (Dirlewanger et al., 2002; Hasnaoui et al., 2012; Shiferaw et al., 2012; Emanuelli et al., 2013; Ren et al., 2014), parentage assessment (Malysheva et al., 2003), species identification (Shirasawa et al., 2013), and mapping genetic linkage (Temnykh et al., 2000; Olmstead et al., 2008; Lu et al., 2012, 2013). In the genus Chrysanthemum, SSR markers have been reported for C. nankingense (Wang H. B. et al., 2013). In addition, SSR markers have been used to identify and classify Chinese traditional ornamental chrysanthemum cultivars (Zhang et al., 2014). Nevertheless, current genetic knowledge is very limited for Chinese traditional medicinal chrysanthemum varieties, which hinders genetic conservation and improvement of these endangered, but economically important Chinese medicinal herbs.
In this study, SSR markers were developed and were applied to investigate genetic diversity and phylogenetic relationships among medicinal C. morifolium cultivars, with the aim to provide new information that could be used to improve the utilization and conservation of C. morifolium genetic resources.
Materials and methods
Plant materials and DNA extraction
A total of 32 cultivars of C. morifolium were collected from the main distribution areas in China. The sampled germplasms and voucher specimens are shown in Table 1. These varieties were verified and confirmed using the specimens stored in the herbarium at the Institute of Botany, Chinese Academy of Sciences, Beijing, China (http://www.nhpe.org). Voucher samples were deposited in the Zhejiang Provincial Key Laboratory for Genetic Improvement and Quality Control of Medicinal Plants, Hangzhou Normal University, China.
Table 1.
List of medicinal Chrysanthemum morifolium samples included in this study.
| Original accession | Code | Voucher number | Longitude (E) | Latitude (N) | Location |
|---|---|---|---|---|---|
| Hangju “Dayangju” | Dyj-1 | CM1 | 120°32′ | 30°38′ | Tongxiang, Zhejiang province |
| Hangju “Dayangju” | Dyj-12 | CM12 | 120°43′ | 29°05′ | Pan'an, Zhejiang province |
| Hangju “Dayangju” | Dyj-21 | CM21 | 115°03′ | 31°18′ | Macheng, Hubei province |
| Hangju “Zaoxiaoyangju” | Zxyj-2 | CM2 | 120°32′ | 30°38′ | Tongxiang, Zhejiang province |
| Hangju “Zaoxiaoyangju” | Zxyj-13 | CM13 | 120°43′ | 29°05′ | Pan'an, Zhejiang province |
| Hangju “Xiaoyangju” | Xyj-3 | CM3 | 120°32′ | 30°38′ | Tongxiang, Zhejiang province |
| Hangju “Xiaoyangju” | Xyj-20 | CM20 | 115°03′ | 31°18′ | Macheng, Hubei province |
| Hangju “Yizhongdabaiju” | Yzdbj | CM4 | 120°32′ | 30°38′ | Tongxiang, Zhejiang province |
| Hangju “Xiaohuangju” | Xhj-5 | CM5 | 120°32′ | 30°38′ | Tongxiang, Zhejiang province |
| Hangju “Xiaohuangju” | Xhj-10 | CM10 | 120°43′ | 29°05′ | Pan'an, Zhejiang province |
| Hangju “No. 1 of Jinju” | Jj1 | CM6 | 120°32′ | 30°38′ | Tongxiang, Zhejiang province |
| Hangju “No. 2 of Jinju” | Jj2-7 | CM7 | 120°32′ | 30°38′ | Tongxiang, Zhejiang province |
| Hangju “No. 2 of Jinju” | Jj2-14 | CM14 | 120°43′ | 29°05′ | Pan'an, Zhejiang province |
| Hangju “No. 3 of Jinju” | Jj3 | CM8 | 120°32′ | 30°38′ | Tongxiang, Zhejiang province |
| Hangju “No. 4 of Jinju” | Jj4 | CM9 | 120°32′ | 30°38′ | Tongxiang, Zhejiang province |
| Hangju “Chidahuangju” | Cdhj | CM11 | 120°43′ | 29°05′ | Pan'an, Zhejiang province |
| Hangju “Dabaiju” | Dbj | CM15 | 120°25′ | 33°78′ | Sheyang, Jiangsu province |
| Hangju “Xiaobaiju” | Xbj | CM16 | 120°25′ | 33°78′ | Sheyang, Jiangsu province |
| Hangju “Changbanju” | Cbj | CM17 | 120°25′ | 33°78′ | Sheyang, Jiangsu province |
| Hangju “Hongxinju” | Hxj | CM18 | 120°25′ | 33°78′ | Sheyang, Jiangsu province |
| Hangju “Dahuangju” | Dhj | CM19 | 120°25′ | 33°78′ | Sheyang, Jiangsu province |
| Machengju | Mcj | CM22 | 115°03′ | 31°18′ | Macheng, Hubei province |
| Boju “Daboju” | Dboj | CM23 | 115°78′ | 33°85′ | Bozhou, Anhui province |
| Boju “Xiaoboju” | Xbj | CM24 | 115°78′ | 33°85′ | Bozhou, Anhui province |
| Gongju “Zaogongju” | Zgj | CM25 | 118°43′ | 29°87′ | Shexian, Anhui province |
| Gongju “Wangongju” | Wgj | CM26 | 118°43′ | 29°87′ | Shexian, Anhui province |
| Gongju “Huangyaoju” | Hyj | CM27 | 118°43′ | 29°87′ | Shexian, Anhui province |
| Chuju | Cj | CM28 | 118°32′ | 32°3′ | Chuzhou, Anhui province |
| Huaiju “Huaixiaohuangju” | Hxhj | CM29 | 113°38′ | 35°1′ | Wuzhi, Henan province |
| Huaiju “Huaidabaiju” | Hdbj | CM30 | 113°38′ | 35°1′ | Wuzhi, Henan province |
| Huaiju “Huaixiaobaiju” | Hxbj | CM31 | 113°38′ | 35°1′ | Wuzhi, Henan province |
| Huaiju “Huaizhenzhuju” | Hzzj | CM32 | 113°38′ | 35°1′ | Wuzhi, Henan province |
Fresh, young leaf tissues from 10 individuals of each cultivar were randomly collected for genomic DNA isolation. The genomic DNA was isolated as described previously (Feng et al., 2013). The integrity and quality of the DNA were evaluated by electrophoresis on 0.8% agarose gels, and the concentration of the genomic DNA samples was determined using a UV spectrometer.
SSR markers development
A total of 7300 C. morifolium EST sequences (total size 3.7 Mb) were retrieved from GenBank (http://www.ncbi.nlm.nih.gov/), with a mean length of 531 bp. These ESTs were analyzed to identify the perfect SSR loci using the MIcroSAtellite (MISA) software (http://pgrc.ipk-gatersleben.de/misa/), following the set to detect tandem repeats of hexa-, penta-, tetra-, tri-, and di-nucleotides, with a minimum number of 4, 4, 5, 7, and 10 tandem arrays of the core repeat, respectively. SSR loci embedded the ESTs with appropriate flanking sequences were selected for primer design using software Primer 3.0 (Untergasser et al., 2012). The parameters for designing the primers were set as follows: primer length with 20 ± 2 nucleotides, amplification product size of 100–300 bp, GC content of 40–60%, and optimum annealing temperature of at least 50°C.
SSR analysis
A total of 136 SSR primer pairs, targeting at 86 C. nankingense (Asteraceae) EST-SSRs (Wang H. B. et al., 2013) and 50 C. morifolium EST-SRRs identified in the present study, were synthesized by Shanghai Sangon Biological Engineering Technology and Service Co. Ltd (Shanghai, China). After a trial run of 136 pairs of SSR primers, 55 of them with clearly separated bands, stable amplification, and rich polymorphism were chosen for further analysis.
SSR amplification was performed using 20 μL of PCR mixture solution containing 2 μL 10 × PCR buffer (100 mM Tris-HCl, 100 mM (NH4)2SO4, 100 mM KCl, 1% Triton X-100, pH 8.8), 2 μL MgCl2 (20 mM), 0.4 μL dNTPs (10 mM), 1 μL of each primer (10 μM) (forward and reverse), 1 U Taq DNA polymerase (TaKaRa Bio., Kyoto, Japan), and 50 ng genomic DNA templates. PCR amplification was run using a GeneAmp PCR System 9700 (Applied Biosystems, Carlsbad, CA, USA) with the following program: 94°C for 5 min, followed by 35 cycles of 94°C for 40 s, then 40 s at the annealing temperature of each primer pair, 72°C for 1 min 30 s, and a final extension at 72°C for 10 min. PCR products were separated on 1.5% agarose gel, stained with ethidium bromide and photographed under UV light. Sanger sequencing was used to confirm SSRs in amplified genomic DNA fragments as described previously (Lu et al., 2013).
Data analysis
Only reproducible and consistent SSR fragments were scored as present (1) or absent (0) for each of the SSR markers. The polymorphism information content (PIC) of each pair of SSR primers was calculated using the formula:
Where n is the number of alleles (marker), qi is the ith allele frequency, and qj is the jth allele frequency (Botstein et al., 1980), The cluster analysis was conducted by NTSYS-pc version 2.10e software (Rohlf, 2000). A dendrogram was constructed using the unweighted pair group method with an arithmetic mean (UPGMA) based on similarity matrices calculated using the simple matching (SM) coefficient (Nei and Li, 1979). The data was also analyzed using principal coordinate analysis (PCoA) (Gower, 1966) to further demonstrate the multiple dimensional distributions of the chrysanthemum cultivars in a scatter-plot.
Results
SSR markers development and primer design
In total, 218 microsatellites were detected in 207 ESTs (Tables 2, 3). Among them, 10 (4.83%) ESTs contained more than one SSR loci (Table 2). Information about 218 SSR loci was showed in Supplementary Material. Of all detected SSR loci, hexa-nucleotide repeats were the most abundant with 134 loci, (61.47% of the total), followed by tri-nucleotide repeats with 39 loci (17.89% of the total), di-nucleotide repeats with 27 loci (12.39% of the total), tetra-nucleotide repeats with 9 loci (4.13% of the total), and penta-nucleotide repeats with 9 loci (4.13% of total) (Tables 2, 3). After removal of those ESTs with too short or inappropriate flanking sequences for primer design, 50 EST-SSRs were selected for primer design (Table 4).
Table 2.
Characterization of EST-SSRs in C. morifolium genome.
| Parameter | Value |
|---|---|
| Total number of ESTs searched | 7300 |
| Total size of examined sequences (bp): | 3,717,958 |
| Total number of ESTs with SSRs | 207 |
| Total number of ESTs with a single SSR | 197 |
| Total number of ESTs with more than 1 SSR | 10 |
| Repeat types | |
| Di-nucleotide | 27 (12.39%) |
| Tri-nucleotide | 39 (17.89%) |
| Tetra-nucleotide | 9 (4.13%) |
| Penta-nucleotide | 9 (4.13%) |
| Hexa-nucleotide | 134 (61.47%) |
| Total number of SSRs identified | 218 |
Table 3.
Distributions of microsatellite motifs observed in C. morifolium ESTs in the GenBank database.
| SSR motif | Number of repeat units in ESTs | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 3 | 4 | 5 | 6 | 7 | 8 | 9 | ≥10 | Total | |
| Di-nucleotide | – | – | – | – | – | – | – | 27 | 27 |
| (AC)n | – | – | – | – | – | – | – | 10 | 10 |
| (AG)n | – | – | – | – | – | – | – | 13 | 13 |
| (AT)n | – | – | – | – | – | – | – | 4 | 4 |
| Tri-nucleotide | – | – | – | – | 19 | 12 | 5 | 3 | 39 |
| (AAC)n | – | – | – | – | 4 | 4 | 3 | – | 11 |
| (AAG)n | – | – | – | – | 2 | – | – | 2 | |
| (AAT)n | – | – | – | – | 3 | 2 | – | 2 | 7 |
| (ACC)n | – | – | – | – | 8 | 2 | 1 | 1 | 12 |
| (ACT)n | – | – | – | – | 1 | – | 1 | ||
| (AGC)n | – | – | – | – | 1 | 1 | – | 2 | |
| (ATC)n | – | – | – | – | 3 | 1 | – | 4 | |
| Tetra-nucleotide | 6 | 2 | 1 | – | 9 | ||||
| Penta-nucleotide | 9 | – | 9 | ||||||
| Hexa-nucleotide | 110 | 17 | 6 | 1 | – | 134 | |||
| Total | 110 | 26 | 12 | 3 | 19 | 13 | 5 | 30 | 218 |
Table 4.
Polymorphism of 55 SSR primer pairs in medicinal Chrysanthemum morifolium samples.
| Primer pair (ID) | Primer Sequences (5′-3′) | Repeat motif | Tm | Size | No. of loci | No. of Polymorphic loci | Polymorphic loci % | PIC | References |
|---|---|---|---|---|---|---|---|---|---|
| CMeSSR001 | F:ATTCTGTCACTCAAACACCAC | (CATC)8 | 55.3 | 251 | 26 | 26 | 100.00 | 0.978 | This study |
| R:GGTTCAAACGAGCTAAATTACA | |||||||||
| CMeSSR002 | F:CTCACCATTTTCAGACCATTAT | (ACA)7 | 55.4 | 251 | 26 | 26 | 100.00 | 0.976 | This study |
| R:ACACATCTTGTACCTCTTGGTT | |||||||||
| CMeSSR003 | F:CTTTTTCACACACACTCAACAT | (TAA)7 | 54.5 | 256 | 26 | 26 | 100.00 | 0.976 | This study |
| R:TTGGAGACGTTGTTGTAAAGTA | |||||||||
| CMeSSR004 | F:AAAATGTTAGGTGCAGGATTAC | (AAT)7 | 54.3 | 248 | 27 | 27 | 100.00 | 0.977 | This study |
| R:AAAAACCGTTCCAGATTACAC | |||||||||
| CMeSSR005 | F:AAAACCTTCACTAGATCACACC | (CAC)7 | 56.3 | 252 | 29 | 29 | 100.00 | 0.978 | This study |
| R:TTTCAGTATCTTGGACCAGTCT | |||||||||
| CMeSSR006 | F:ATTCTCTTAATTAGCCAGCAAG | (CAC)7 | 54.3 | 247 | 17 | 17 | 100.00 | 0.958 | This study |
| R;GTGAATCGTAAATTCAGTTGG | |||||||||
| CMeSSR007 | F:GTCCTCCTTCAAAGCAAA | (CTA)7 | 51.3 | 157 | 28 | 28 | 100.00 | 0.976 | This study |
| R:GACGATTAATTATTGGGTAATA | |||||||||
| CMeSSR009 | F:AGTGATGATGAATTGAAAGAGC | (AAT)8 | 56.4 | 258 | 18 | 17 | 94.44 | 0.959 | This study |
| R:CTCTCAAGTGTTGAAGGAACTC | |||||||||
| CMeSSR010 | F:CATTTTCTTCATGGTACTCACA | (CAC)7 | 56.4 | 169 | 27 | 27 | 100.00 | 0.978 | This study |
| R:GTGAGGATGGAAATCTAGTAGG | |||||||||
| CMeSSR011 | F:AGGACAACTCAACTGTTAGGAG | (CCA)7 | 57.2 | 255 | 20 | 19 | 95.00 | 0.964 | This study |
| R:GTTTCTCAACCTCTTCTTCATC | |||||||||
| CMeSSR012 | F:ATTCCCAACCTTCTTTAACC | (CA)11 | 54.1 | 254 | 29 | 29 | 100.00 | 0.979 | This study |
| R:AACTAAATCACCATCTCTTGCT | |||||||||
| CMeSSR013 | F:ATGAGAGGGAAATAGAAAGTGA | (GTAATA)3(TAA)4 | 56.4 | 219 | 26 | 26 | 100.00 | 0.973 | This study |
| R:TACTTGACGCTAACGGAGTAGT | |||||||||
| CMeSSR014 | F:CAAAACTTTCAACAGAGTCATC | (CAACAT)3(CAG)4 | 54.5 | 281 | 35 | 35 | 100.00 | 0.983 | This study |
| R:AGAAATAACGACTGGTCAGATT | |||||||||
| CMeSSR015 | F:TCTTGGTCAGCTTAATTACTCA | (TGG)7…(TGG)4(AGG)4 | 57.1 | 236 | 30 | 29 | 96.67 | 0.979 | This study |
| R:CATCACCTCCTCCTCCTC | |||||||||
| CMeSSR016 | F:GAATACTAAATGGGTGGAAGAA | (GGA)4(GGT)4 | 55.3 | 250 | 18 | 18 | 100.00 | 0.967 | This study |
| R:GCAAATAGATGTCCTTTAGGG | |||||||||
| CMeSSR017 | F:TCATGAAATCCGTGTATATGTC | (AC)5a(AC)6 | 54.5 | 229 | 30 | 30 | 100.00 | 0.980 | This study |
| R:ACCCTAATTCTCAAAATGAACC | |||||||||
| CMeSSR018 | F:ATCTACTATCCAAGCCATGAAC | (CAC)5…(GGT)10…(TGG)6 | 56.8 | 264 | 25 | 23 | 92.00 | 0.973 | This study |
| R:TATCCACCACCACCACCA | |||||||||
| gi298295865 | F:ACTCACTTGCCCCATTTGTC | (AACCCT)5 | 59.8 | 146 | 15 | 15 | 100.00 | 0.957 | Wang H. B. et al., 2013 |
| R:AGAGAAGCTCTCCAGGGACC | |||||||||
| gi298300528 | F:AGGGCATCGATAATCCATCA | (ATATC)4 | 56.8 | 135 | 12 | 12 | 100.00 | 0.938 | Wang H. B. et al., 2013 |
| R:AGATACGTGCCCATTTGAGG | |||||||||
| gi298295793 | F:ATAGAATTCCCCGACGACAA | (CCCTAT)4 | 56.8 | 111 | 15 | 14 | 93.33 | 0.948 | Wang H. B. et al., 2013 |
| R:GGCGGTTGAGATTGATAGGA | |||||||||
| gi298296818 | F:ATGTCCAGCTTGATGGGAAG | (GTG)7 | 58.8 | 210 | 24 | 23 | 95.83 | 0.977 | Wang H. B. et al., 2013 |
| R:GGCCCCTTGCAAATCCTC | |||||||||
| gi298298301 | F:CTTGACCGAAACACCGAAAT | (TTG)9 | 56.8 | 198 | 17 | 16 | 94.12 | 0.958 | Wang H. B. et al., 2013 |
| R:TGGCATCCTAGTTAGCAGCA | |||||||||
| gi298299323 | F:GCACATTTCCTTCATGGGTT | (ACA)9 | 57.8 | 264 | 33 | 33 | 100.00 | 0.982 | Wang H. B. et al., 2013 |
| R:TCCACGGTTTCAGATGATGA | |||||||||
| gi298297301 | F:TCAAACACCACCACCAACAC | (CATC)8 | 58.8 | 167 | 25 | 25 | 100.00 | 0.973 | Wang H. B. et al., 2013 |
| R:ATGTCACCAAGTCCTGGTCC | |||||||||
| 51 | F:CCCCCTCTTCTTCTTCAACC | (CCAA)4 | 57.8 | 202 | 22 | 22 | 100.00 | 0.974 | Wang H. B. et al., 2013 |
| R:CAATAGAAAGCGCGTGACAA | |||||||||
| 53 | F:TCGAAGACAATCAGCACCTG | (ATG)7 | 57.8 | 233 | 18 | 17 | 94.44 | 0.961 | Wang H. B. et al., 2013 |
| R:TAAGTGTTCTTCCAGCGCCT | |||||||||
| 64 | F:GGCGATGGATGATGATGATT | (TTC)9 | 56.8 | 267 | 22 | 22 | 100.00 | 0.991 | Wang H. B. et al., 2013 |
| R:GAAAGAGGTGGATCGGATGA | |||||||||
| 86 | F:AAACCACCAAACCCATCAAA | (TGG)8 | 54.7 | 223 | 25 | 25 | 100.00 | 0.990 | Wang H. B. et al., 2013 |
| R:AACTTTGCCAGCATCGACTT | |||||||||
| 135 | F:CATTCCTACCCATCCCTCCT | (GTGGAG)4 | 58.8 | 100 | 31 | 31 | 100.00 | 0.993 | Wang H. B. et al., 2013 |
| R:CGCATGAGTGAGCCTAATGA | |||||||||
| 204 | F:TGAGCTTCATCCGCTTCTTT | (TGA)8 | 55.8 | 262 | 10 | 10 | 100.00 | 0.985 | Wang H. B. et al., 2013 |
| R:TGGTCGTATTCCGTCCATTT | |||||||||
| 219 | F:AAAAGGTTGTGAGTGGGTCG | (GGGAAG)4…(TGAGGG)4 | 57.8 | 228 | 26 | 25 | 96.15 | 0.976 | Wang H. B. et al., 2013 |
| R:CCTCGGTCGATAAATCTCCA | |||||||||
| 221 | F:AACCATGAATCCAGACACCC | (TCA)7 | 57.8 | 181 | 20 | 20 | 100.00 | 0.964 | Wang H. B. et al., 2013 |
| R:ACCAAGCCAGTCGAGTTTTG | |||||||||
| 235 | F:GCCCCAATTTATTCACTCCA | (AAC)6 | 57.8 | 257 | 30 | 30 | 100.00 | 0.979 | Wang H. B. et al., 2013 |
| R:GCTCTTCCTCGTAAGCATCG | |||||||||
| 262 | F:TCTGCCAGCTTTGGGTAACT | (CTTTTT)4 | 57.8 | 260 | 13 | 13 | 100.00 | 0.946 | Wang H. B. et al., 2013 |
| R:GTGCGCCTGTATTGACTTGA | |||||||||
| 270 | F:AGGTGGAAAATACTGTGCGG | (ATAGTA)4 | 57.8 | 139 | 19 | 19 | 100.00 | 0.966 | Wang H. B. et al., 2013 |
| R:TGTTTCTGCACCTCAACAGC | |||||||||
| 285 | F:CCGGTGTTCGGTATAAATGG | (GTG)7 | 56.8 | 116 | 11 | 11 | 100.00 | 0.965 | Wang H. B. et al., 2013 |
| R:ACAATTCGCTTCGGCTCTAA | |||||||||
| 312 | F:GGCCCAAGTTTGAGACAAAA | (AAG)7 | 56.8 | 219 | 36 | 36 | 100.00 | 0.985 | Wang H. B. et al., 2013 |
| R:TCGGTATAAGTGCACCACGA | |||||||||
| 313 | F:GGCGTTCTCTTCCATTTCAA | (GAA)7 | 56.8 | 253 | 37 | 37 | 100.00 | 0.984 | Wang H. B. et al., 2013 |
| R:GTTTTGGACCTTGCTTCTGC | |||||||||
| 320 | F:GGTCCTTCGTTTCATTTGGA | (TGG)7 | 57.8 | 235 | 21 | 21 | 100.00 | 0.973 | Wang H. B. et al., 2013 |
| R:CGGGGGTAGGAATAGAAAGC | |||||||||
| 327 | F:GAATGCAGCCTCAACAACAA | (TCAAAG)4 | 55.8 | 219 | 27 | 27 | 100.00 | 0.979 | Wang H. B. et al., 2013 |
| R:GAGCCGCCATTGTCATATTT | |||||||||
| 357 | F:ACCCAACCTGAACAAGATGC | (GGGTCA)4 | 58.8 | 252 | 24 | 24 | 100.00 | 0,990 | Wang H. B. et al., 2013 |
| R:ATACTGCTGCCACTGACCCT | |||||||||
| 581 | F:CCAATCCCAAACACTCCCTA | (CA)16 | 58.8 | 223 | 24 | 24 | 100.00 | 0.976 | Wang H. B. et al., 2013 |
| R:GCCGTTACCACTGCTCTTTC | |||||||||
| 984 | F:TCAAAACCCATCATCACCCT | (ACA)7 | 55.8 | 186 | 28 | 27 | 96.43 | 0.979 | Wang H. B. et al., 2013 |
| R:CGGCGTTTGTATCTTGGTTT | |||||||||
| 995 | F:TTGTTCCACGTGACGAGATT | (TGTTGG)4 | 56.9 | 244 | 16 | 16 | 100.00 | 0.961 | Wang H. B. et al., 2013 |
| R:CTCCCAAATGACCCATCATC | |||||||||
| 1036 | F:CTTTGGTAAGCGAAGGCTGT | (AATG)7 | 55.7 | 153 | 16 | 16 | 100.00 | 0.955 | Wang H. B. et al., 2013 |
| R:GCCATTTGTAAGCGGTTTGT | |||||||||
| 1187 | F:GAAAGCGATCATTGGGAAAA | (GAGAAG)4 | 54.7 | 112 | 19 | 19 | 100.00 | 0.963 | Wang H. B. et al., 2013 |
| R:TTACCCGTACATTCGGGATT | |||||||||
| 1424 | F:TAAAATCCATCCGTCCATCC | (CAA)7 | 57.8 | 278 | 26 | 26 | 100.00 | 0.975 | Wang H. B. et al., 2013 |
| R:CTTCCATATCTGCCAGTGGG | |||||||||
| 1428 | F:AACGCCCAAAACACCAACT | (AC)9 | 56.6 | 224 | 26 | 26 | 100.00 | 0.978 | Wang H. B. et al., 2013 |
| R:TAGAACCTTGTGCCCCCATA | |||||||||
| 1520 | F:AAATCACGGATCCCCTTCTT | (ATAGA)4 | 57.8 | 158 | 32 | 32 | 100.00 | 0.982 | Wang H. B. et al., 2013 |
| R:TTATCATCTTGGGGAGTGGC | |||||||||
| 1584 | F:CCTCCTCAAAACGACCATGT | (ACA)7 | 57.8 | 263 | 24 | 23 | 95.83 | 0.973 | Wang H. B. et al., 2013 |
| R:CGTCCCCATTACAATATCCG | |||||||||
| 1742 | F:AAGTGATAAGATGGGTGGCG | (TGG)7 | 57.8 | 166 | 28 | 28 | 100.00 | 0.974 | Wang H. B. et al., 2013 |
| R:GGTGGAGGCTCATTCAAATC | |||||||||
| 1762 | F:GCGTCAAATTACTGGTGGCT | (AAC)6 | 56.8 | 259 | 22 | 21 | 95.45 | 0.972 | Wang H. B. et al., 2013 |
| R:GTCTCATTTTCCGGCGATAA | |||||||||
| 1773 | F:CAAATGGGGTCGTTACGAAT | (CAT)6 | 55.8 | 215 | 17 | 17 | 100.00 | 0.965 | Wang H. B. et al., 2013 |
| R:AATCCCCGAATTCCCAATAG | |||||||||
| 1774 | F:TCACCACCACCACTGTCACT | (CACCGG)4 | 59.8 | 276 | 38 | 38 | 100.00 | 0.984 | Wang H. B. et al., 2013 |
| R:TGTGGGCTCTAGAGGTTTGG | |||||||||
| 1779 | F:AAAGTCCCCTTGCTTGTTCC | (AACC)5 | 57.8 | 148 | 25 | 25 | 100.00 | 0.975 | Wang H. B. et al., 2013 |
| R:CGACTCCATTTGATCCACCT | |||||||||
| Total | 1319 | 1306 | |||||||
| Average | 23.98 | 23.75 | 98.90% | 0.972 |
A total of 136 SSR primer pairs, including 50 C. morifolium EST-SSRs identified above and 86 C. nankingense EST-SSRs (Wang H. B. et al., 2013), were screened using three genomic DNA samples. Fifty-five of the primer pairs (40.44%) generated reproducible polymorphic DNA amplification products. The amplified bands with clear and expected size were sequenced. The corresponding repeat motifs were validated for 50 EST loci by Sanger sequencing. Finally, 17 novel C. morifolium EST-SSRs were developed successfully, and 38 C. nankingense EST-SSRs were confirmed with transferability for application in a related species. These 55 pairs of SSR primers were used for further genetic diversity analysis in C. morifolium cultivars (Table 4).
SSR analysis
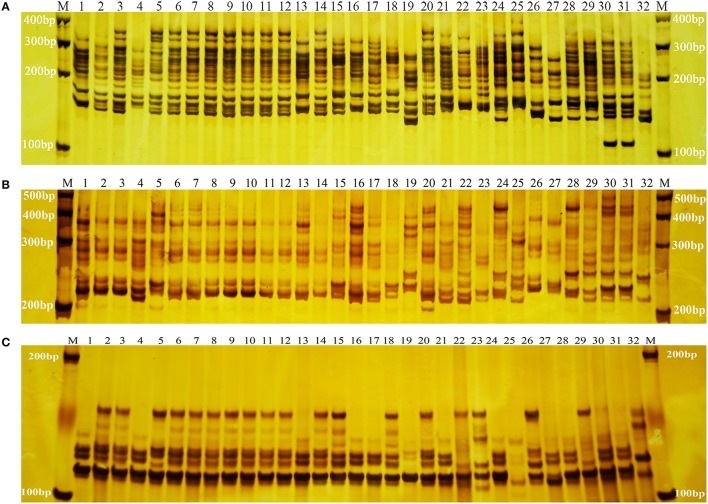
The 55 SSR primer pairs generated a total of 1319 fragments with an average of 23.98 fragments per primer pair and a range of 10 (primer pair ID. 204) to 38 (primer pair ID. 1774) (Table 4). A total of 1306 were polymorphic. The percentage of polymorphic bands across the primer pairs varied from 92.00 to 100.00% (Table 4), with an average 98.90%. Three representative profiles (primer pair ID. CMeSSR001, 219 and 285) are shown in (Figures 1A–C). The PIC value varied from 0.938 to 0.993 with an average of 0.972 (PIC > 0.5), which indicated that these loci contained a considerable amount of genetic information that could be used in genetic diversity studies on Chrysanthemum germplasms.
Figure 1.
SSR amplification profiles of primer pairs CMeSSR001 (A), 219 (B), and 285 (C). Lane M: DNA molecular standards with length (bp) on left and right. Lanes 1–32: genotypes of the 32 Chrysanthemum morifolium samples (CM1–CM32) in Table 1.
Genetic diversity and relationships among genotypes
A total of 1319 loci were accounted to calculate the genetic diversity among the 32 Chrysanthemum cultivars. Binary data matrices produced by SSRs were used to estimate the genetic similarity of the genotyped Chrysanthemum samples. The pairwise similarity coefficient among the 32 cultivars ranged from a maximum of 0.809 (between Huaiju “Hdbj” and Huaiju “Hxbj”) to a minimum of 0.533 (between Huangju “Jj3” and Gongju “Wgj”).
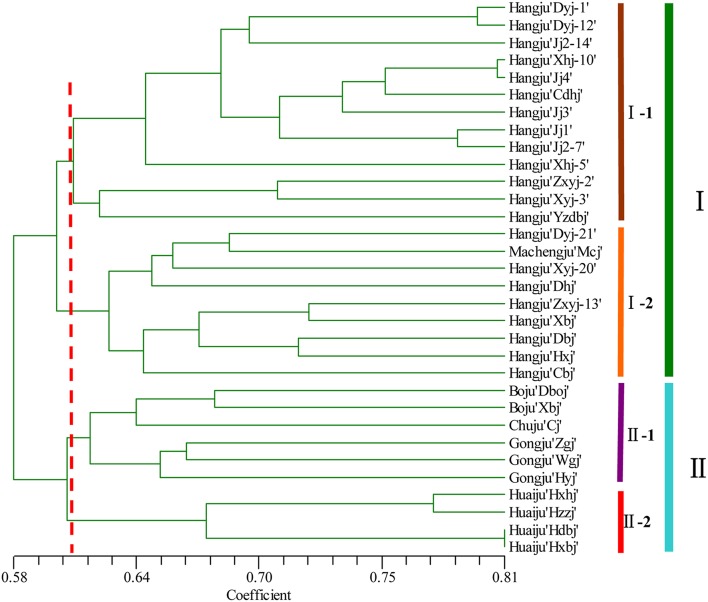
A dendrogram using UPGMA analysis was constructed based on the corresponding genetic similarity coefficient among the tested 32 C. morifolium populations (Figure 2). In this study, all the C. morifolium samples could be grouped into two main clusters, with a similarity index of 0.584. Cluster I consisted of 22 cultivars, including all the “Hangju” and “Machengju” samples. This cluster was further subdivided into three subgroups. Subgroup “I-1” consisted of 13 samples, all of which belong to Hangju cultivars. Machengju “Mcj” and eight Hangju cultivars were assigned to subgroup “I-2.” Group II comprised of 10 cultivars, which belonged to “Boju,” “Huaiju,” “Chuju,” and “Gongju.” Among them, the “Boju,” “Chuju,” and “Gongju” cultivars were classified into subgroup II-1, while four “Huaiju” cultivars constituted subgroup II-2.
Figure 2.
Relationships among Chrysanthemum morifolium varieties based on the genetic similarities between DNA fingerprinting patterns from SSR markers used in the UPGMA dendrogram.
Principal coordinate analysis
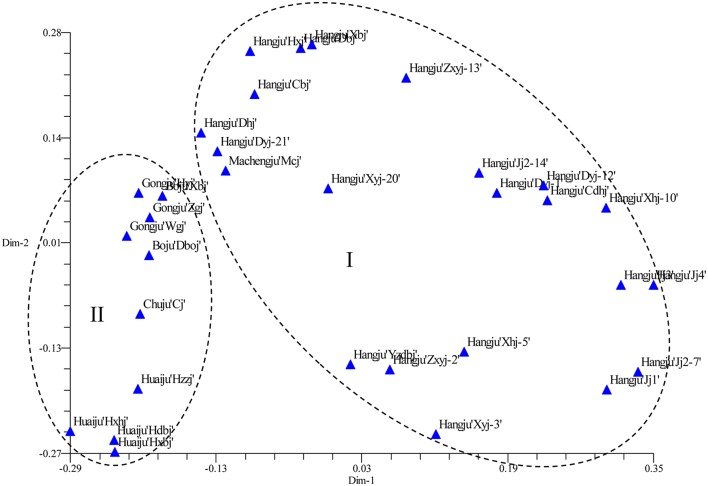
The SSR data were subjected to PCoA in order to obtain an alternative view of the phylogenetic relationships among the cultivars (Figure 3). In the two-dimensional PCoA plot, C. morifolium cultivars were divided into two groups (Figure 3), which was similar to the pattern as shown by the UPGMA dendrogram. The first two principal axes explained 10.60 and 6.70% of the total molecular variation observed, respectively.
Figure 3.
Two-dimensional projection of the PCoA of 32 Chrysanthemum morifolium samples based on SSR markers along the first two principal axes.
Discussion
Compared with anonymous markers, SSR markers, as a type of co-dominance markers, may yield more accurate estimates of genetic diversity. SSRs have been used successfully to determine genetic diversity among many plants (Dirlewanger et al., 2002; Hasnaoui et al., 2012; Shiferaw et al., 2012; Emanuelli et al., 2013; Ren et al., 2014). SSRs were previously identified in C. nankingense and C. nankingense SSRs were proved to be useful for genetic analysis in the genus Chrysanthemum and its related genera (Wang H. B. et al., 2013). In the present study, we found that 44% (38 out 86) of C. nankingense SSRs were also proved to be useful for genetic diversity study among medicinal C. morifolium cultivars (Table 4).
A previous study used 20 SSR markers for identification and classification of Chinese traditional ornamental Chrysanthemum cultivars (Zhang et al., 2014). However, few studies have explored development and application of SSR markers for genetic diversity among medicinal C. morifolium cultivars. The diversity and genetic relationship among 29 C. morifolium populations were investigated using the types of dominant molecular markers (Shao et al., 2010). The present study report discovery of novel SSRs in C. morifolium.
The SSR markers selected in this study yielded reproducible polymorphic bands in 32 C. morifolium cultivars and showed that they provide a powerful and reliable molecular tool for analyzing genetic diversity and relationships among C. morifolium cultivars. In this study, 98.90% of the bands generated by the SSR assay were polymorphic, which was higher than the polymorphic proportions of 53.85% detected by SSR among celery cultivars (Fu et al., 2014), 97.40% among grass pea from different regions (Shiferaw et al., 2012), and 97.50% among melon accessions (Kacar et al., 2012). Molecular markers with higher PIC values have a greater ability to identify cultivars. A locus with a PIC greater than 0.5 is considered to be highly diverse, as a previous study reported (Botstein et al., 1980). The PIC values of the SSR markers used in the Chrysanthemum cultivars analysis ranged between 0.938 and 0.993, with an average of 0.972, which indicated that the highly informative SSR markers could be employed in genetic diversity studies of medicinal Chrysanthemum cultivars.
Evaluation of genetic diversity and relationship among plant populations is the foundation of selective breeding programs. Using SSR markers, our study found considerable diversity among Chrysanthemum cultivars, which could be used in breeding programs for Chrysanthemum improvement. According to their origin and ecological distribution, 32 C. morifolium cultivars were classified into six sources: Hangju, Machengju, Chuju, Boju, Gongju, and Huaiju (Table 1). A dendrogram constructed with SSR data using the UPGMA method indicated that the C. morifolium cultivars were grouped into two main groups. All the Hangju cultivars were clustered in the first group along with Machengju “Mcj” (Group I), which means that the genetic relationship between Hangju cultivars and Machengju “Mcj” is very close, consistant with a previous study (Shao et al., 2010). The main growing regions for Boju, Gongju and Chuju are Bozhou (115°78′, 33°85′), Shexian (118°43′, 29°87′), and Chuzhou (118°32′, 32°3′) in Anhui Province, China. In theory, the genetic relationships between the cultivars of these three C. morifolium populations may be closer than between other C. morifolium populations (Hangju, Machengju and Huaiju). In our study, all the Boju, Chuju and Gongju cultivars were grouped together within subgroup II-1, which confirmed the inference above (Figures 2, 3). The Huaiju cultivars collected from Wuzhi, Henan Province (113°38′, 35°1′), were grouped into subgroup II-2. Geographically, Henan Province is adjacent to Anhui Province, which may explain why the four Huaiju cultivars have a close relationship with other three C. morifolium populations (Chuju, Boju, and Gongju) (Figure 2). The results of the present study showed that cluster analysis using SSR markers mainly supported the classification of medical C. morifolium accessions according to their origin and ecological distribution.
Increased urbanization has meant that C. morifolium' cultivation has greatly declined and some populations are rare. Therefore, it is imperative to undertake effective measures to protect C. morifolium germplasms. Our study found that there was a high level of genetic diversity between Chrysanthemum populations. As the study by Shao et al. (2010) suggested, a priority for in situ conservation should be to rescue and conserve the core populations.
In conclusion, our study demonstrates that SSR technology is a powerful tool for evaluating genetic diversity and relationships among the medical C. morifolium cultivars. SSR analysis showed that medicinal C. morifolium populations could be classified according to their origin and ecological distribution. In future studies, more medicinal C. morifolium cultivars will be included to verify whether these findings are true for more closely related taxa.
Author contributions
Conceived and designed the study: SF, HW. Collected plant samples XS, ZW. Performed the experiments: SF, RH, MJ. Analyzed the data: SF, RH, JL, YJ. Wrote the manuscript: SF, HW.
Funding
This study was supported in part by the Zhejiang Provincial Natural Science Foundation of China (LQ13H280006); the National science and technology support Programme project (2011BAI04B02); Zhejiang Provincial New Agricultural Varieties Breeding Of Traditional Chinese Medicinal Materials Major R&D Projects (2012C12912).
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fgene.2016.00113
References
- Anderson N. O. (2006). Chrysanthemum. Flower Breeding and Genetics. Issues, Challenges and Opportunities for the 21st Century. Heidelberg: Springer. [Google Scholar]
- Bhattacharya A., da Silva J. A. T. (2006). Molecular systematics in Chrysanthemum x grandiflorum (Ramat.) Kitamura. Sci. Horticult. 109, 379–384. 10.1016/j.scienta.2006.06.004 [DOI] [Google Scholar]
- Botstein D., White R. L., Skolnick M., Davis R. W. (1980). Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 32, 314–331. [PMC free article] [PubMed] [Google Scholar]
- Chen C.-W., Yan H., Han B.-X. (2014). Rapid identification of three varieties of Chrysanthemum with near infrared spectroscopy. Rev. Brasil. Farmacog. 24, 33–37. 10.1590/0102-695X20142413387 [DOI] [Google Scholar]
- Dirlewanger E., Cosson P., Tavaud M., Aranzana M., Poizat C., Zanetto A., et al. (2002). Development of microsatellite markers in peach [Prunus persica (L.) Batsch] and their use in genetic diversity analysis in peach and sweet cherry (Prunus avium L.). Theor. Appl. Genet. 105, 127–138. 10.1007/s00122-002-0867-7 [DOI] [PubMed] [Google Scholar]
- Chinese Pharmmacopoeia Editorial Committee (2010). Pharmmacopoeia of the People's Republic of China. Beijing: Chemical Industry Press. [Google Scholar]
- Emanuelli F., Lorenzi S., Grzeskowiak L., Catalano V., Stefanini M., Troggio M., et al. (2013). Genetic diversity and population structure assessed by SSR and SNP markers in a large germplasm collection of grape. BMC Plant Biol. 13:39. 10.1186/1471-2229-13-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Zhao H., Lu J., Liu J., Shen B., Wang H. (2013). Preliminary genetic linkage maps of Chinese herb Dendrobium nobile and D. moniliforme. J. Genet. 92, 205–212. 10.1007/s12041-013-0246-y [DOI] [PubMed] [Google Scholar]
- Fu N., Wang P. Y., Liu X. D., Shen H. L. (2014). Use of EST-SSR markers for evaluating genetic diversity and fingerprinting celery (Apium graveolens L.) cultivars. Molecules 19, 1939–1955. 10.3390/molecules19021939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gower J. C. (1966). Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika 53, 325–338. 10.1093/biomet/53.3-4.325 [DOI] [Google Scholar]
- Hasnaoui N., Buonamici A., Sebastiani F., Mars M., Zhang D., Vendramin G. G. (2012). Molecular genetic diversity of Punica granatum L. (pomegranate) as revealed by microsatellite DNA markers (SSR). Gene 493, 105–112. 10.1016/j.gene.2011.11.012 [DOI] [PubMed] [Google Scholar]
- Kacar Y. A., Simsek O., Solmaz I., Sari N., Mendi Y. Y. (2012). Genetic diversity among melon accessions (Cucumis melo) from Turkey based on SSR markers. Genet Mol. Res. 11, 4622–4631. 10.4238/2012.November.29.2 [DOI] [PubMed] [Google Scholar]
- Lu J. J., Kang J. Y., Feng S. G., Zhao H. Y., Liu J. J., Wang H. Z. (2013). Transferability of SSR markers derived from Dendrobium nobile expressed sequence tags (ESTs) and their utilization in Dendrobium phylogeny analysis. Sci. Horticult. 158, 8–15. 10.1016/j.scienta.2013.04.011 [DOI] [Google Scholar]
- Lu J. J., Wang S., Zhao H. Y., Liu J. J., Wang H. Z. (2012). Genetic linkage map of EST-SSR and SRAP markers in the endangered Chinese endemic herb Dendrobium (Orchidaceae). Genet. Mol. Res. 11, 4654–4667. 10.4238/2012.December.21.1 [DOI] [PubMed] [Google Scholar]
- Malysheva L., Sjakste T., Matzk F., Röder M., Ganal M. (2003). Molecular cytogenetic analysis of wheat–barley hybrids using genomic in situ hybridization and barley microsatellite markers. Genome 46, 314–322. 10.1139/g02-117 [DOI] [PubMed] [Google Scholar]
- Nei M., Li W. H. (1979). Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. U.S.A. 76, 5269–5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmstead J. W., Sebolt A. M., Cabrera A., Sooriyapathirana S. S., Hammar S., Iriarte G., et al. (2008). Construction of an intra-specific sweet cherry (Prunus avium L.) genetic linkage map and synteny analysis with the Prunus reference map. Tree Genet. Genomes 4, 897–910. 10.1007/s11295-008-0161-1 [DOI] [Google Scholar]
- Powell W., Machray G. C., Provan J. (1996). Polymorphism revealed by simple sequence repeats. Trends Plant Sci. 1, 215–222. 10.1016/1360-1385(96)86898-1 [DOI] [Google Scholar]
- Ren X., Jiang H., Yan Z., Chen Y., Zhou X., Huang L., et al. (2014). Genetic diversity and population structure of the major peanut (Arachis hypogaea L.) cultivars grown in China by SSR markers. PLoS ONE 9:e88091. 10.1371/journal.pone.0088091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohlf F. J. (2000). NTSYS-PC: Numerical Taxonomy And Multivariate Analysis System, version 2.00. Setauket, NY: Exeter Software. [Google Scholar]
- Shao Q. S., Guo Q. S., Deng Y. M., Guo H. P. (2010). A comparative analysis of genetic diversity in medicinal Chrysanthemum morifolium based on morphology, ISSR and SRAP markers. Biochem. Syst. Ecol. 38, 1160–1169. 10.1016/j.bse.2010.11.002 [DOI] [Google Scholar]
- Shiferaw E., Pe M. E., Porceddu E., Ponnaiah M. (2012). Exploring the genetic diversity of Ethiopian grass pea (Lathyrus sativus L.) using EST-SSR markers. Mol. Breed 30, 789–797. 10.1007/s11032-011-9662-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasawa K., Ishii K., Kim C., Ban T., Suzuki M., Ito T., et al. (2013). Development of Capsicum EST-SSR markers for species identification and in silico mapping onto the tomato genome sequence. Mol. Breed. 31, 101–110. 10.1007/s11032-012-9774-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D. (1989). Hypervariability of simple sequences as a general source for polymorphic DNA markers. Nucleic Acids Res. 17, 6463–6471. 10.1093/nar/17.16.6463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira da Silva J. A. (2003). Chrysanthemum: advances in tissue culture, cryopreservation, postharvest technology, genetics and transgenic biotechnology. Biotechnol. Adv. 21, 715–766. 10.1016/S0734-9750(03)00117-4 [DOI] [PubMed] [Google Scholar]
- Temnykh S., Park W. D., Ayres N., Cartinhour S., Hauck N., Lipovich L., et al. (2000). Mapping and genome organization of microsatellite sequences in rice (Oryza sativa L.). Theor. Appl. Genet. 100, 697–712. 10.1007/s001220051342 [DOI] [Google Scholar]
- Untergasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B. C., Remm M., et al. (2012). Primer3 - new capabilities and interfaces. Nucleic Acids Res. 40, e115. 10.1093/nar/gks596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. B., Jiang J. F., Chen S. M., Qi X. Y., Peng H., Li P. R., et al. (2013). Next-generation sequencing of the Chrysanthemum nankingense (Asteraceae) transcriptome permits large-scale unigene assembly and SSR marker discovery. PLoS ONE 8:62293. 10.1371/journal.pone.0062293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Zhu Z. B., Guo Q. S., Mao P. F. (2013). Variation in major flavonoids glycosides and caffeoylquinic acids during florescence of three Chrysanthemum morifolium Ramat cv. ‘Hangju’ genotypes. Biochem. Syst. Ecol. 47, 74–79. 10.1016/j.bse.2012.11.004 [DOI] [Google Scholar]
- Xu W. B., Guo Q. S., Wang C. L. (2006). RAPD analysis for genetic diversity of Chrysanthemum morifolium. Zhongguo Zhong Yao Za Zhi 31, 18–21. 10.3321/j.issn:1001-5302.2006.01.004 [DOI] [PubMed] [Google Scholar]
- Zhang F., Chen S. M., Chen F. D., Fang W. M., Chen Y., Li F. T. (2011a). SRAP-based mapping and QTL detection for inflorescence-related traits in chrysanthemum (Dendranthema morifolium). Mol. Breed. 27, 11–23. 10.1007/s11032-010-9409-1 [DOI] [Google Scholar]
- Zhang F., Chen S. M., Chen F. D., Fang W. M., Deng Y. M., Chang Q. S., et al. (2011b). Genetic analysis and associated SRAP markers for flowering traits of chrysanthemum (Chrysanthemum morifolium). Euphytica 177, 15–24. 10.1007/s10681-010-0239-3 [DOI] [Google Scholar]
- Zhang F., Chen S. M., Chen F. D., Fang W. M., Li F. T. (2010). A preliminary genetic linkage map of chrysanthemum (Chrysanthemum morifolium) cultivars using RAPD, ISSR and AFLP markers. Sci. Horticult. 125, 422–428. 10.1016/j.scienta.2010.03.028 [DOI] [Google Scholar]
- Zhang Y., Dai S. L., Hong Y., Song X. B. (2014). Application of genomic SSR locus polymorphisms on the identification and classification of Chrysanthemum cultivars in China. PLoS ONE 9:104856. 10.1371/journal.pone.0104856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Zhao J., He L., Sun Y., Cai H. (2013). Molecular structure and the second introns variation of gene F3'H of two medicinal Chrysanthemum morifolium populations. Biochem. Syst. Ecol. 51, 251–258. 10.1016/j.bse.2013.09.012 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.