Abstract
ICEApl1 was identified in the whole genome sequence of MIDG2331, a tetracycline-resistant (MIC = 8 mg/L) serovar 8 clinical isolate of Actinobacillus pleuropneumoniae, the causative agent of porcine pleuropneumonia. PCR amplification of virB4, one of the core genes involved in conjugation, was used to identify other A. pleuropneumoniae isolates potentially carrying ICEApl1. MICs for tetracycline were determined for virB4 positive isolates, and shotgun whole genome sequence analysis was used to confirm presence of the complete ICEApl1. The sequence of ICEApl1 is 56083 bp long and contains 67 genes including a Tn10 element encoding tetracycline resistance. Comparative sequence analysis was performed with similar integrative conjugative elements (ICEs) found in other members of the Pasteurellaceae. ICEApl1 is most similar to the 59393 bp ICEHin1056, from Haemophilus influenzae strain 1056. Although initially identified only in serovar 8 isolates of A. pleuropneumoniae (31 from the UK and 1 from Cyprus), conjugal transfer of ICEApl1 to representative isolates of other serovars was confirmed. All isolates carrying ICEApl1 had a MIC for tetracycline of 8 mg/L. This is, to our knowledge, the first description of an ICE in A. pleuropneumoniae, and the first report of a member of the ICEHin1056 subfamily in a non-human pathogen. ICEApl1 confers resistance to tetracycline, currently one of the more commonly used antibiotics for treatment and control of porcine pleuropneumonia.
Keywords: animal infections, antibiotic resistance, respiratory tract, conjugation, Pasteurellaceae
Introduction
Actinobacillus pleuropneumoniae is a major contributor to swine respiratory disease worldwide, causing considerable economic losses. Isolates can be differentiated into 15 established serovars, based on capsular polysaccharides, and a recently proposed serovar 16 identified on the basis of serology alone (Sárközi et al., 2015). There are geographical differences in the distribution of serovars. Within the UK, clinical isolates are predominantly serovar 8, with serovars 2, 6, 7, and 12 also represented (O’Neill et al., 2010).
There is growing concern regarding antimicrobial resistance in bacteria from food-producing animals (Michael et al., 2015). In Europe, tetracyclines are still the most commonly used antimicrobial for treatment of swine pleuropneumonia (European Medicines Agency, 2012). The genes tetB, tetH, tetL, and tetO, reported to mediate tetracycline resistance in A. pleuropneumoniae, are usually carried on small plasmids (Blanco et al., 2006, 2007). We recently sequenced the genome of MIDG2331, a serovar 8 UK clinical isolate of A. pleuropneumoniae (Bossé et al., 2016), and identified chromosomally encoded tetracycline resistance genes within a putative integrative conjugative element (ICE). Similar to genomic islands, ICE have the ability to integrate into bacterial chromosomes at specific sites, often in tRNA loci, via the action of an integrase (predominantly tyrosine recombinases) (Boyd et al., 2009; Wozniak and Waldor, 2010). However, ICE differ from genomic islands in that they are self-mobilizing, encoding all of the genes necessary for excision from the chromosome and conjugal transfer (Boyd et al., 2009). The core genes of ICE tend to group into functional modules, with syntenic regions responsible for maintenance, dissemination and regulation, which may be interspersed with accessory genes carried on transposons or other insertion elements (Burrus and Waldor, 2004; Wozniak and Waldor, 2010). The genes encoding the type 4 secretion system (T4SS), required for transport of DNA into recipient cells, include a ubiquitous ATPase encoded by virB4 or traU (Guglielmini et al., 2011).
ICEs are the most abundant conjugative elements identified in prokaryotes, and there is evidence of cross-clade transfer (Guglielmini et al., 2011). Within the Pasteurellaceae, ICE have been identified and characterized in Haemophilus influenzae and Haemophilus parainfluenzae (Juhas et al., 2007b), Pasteurella multocida (Michael et al., 2012), and Mannheimia haemolytica (Eidam et al., 2015). Here we report characterization of ICEApl1, to our knowledge the first ICE described in A. pleuropneumoniae.
Materials and Methods
Comparative Sequence Analysis
The full sequence of ICEApl1, identified within the genome of MIDG2331 (accession number LN908249) was analyzed using BLASTn and BLASTx1. A comparative alignment was generated for sequences most similar to ICEApl1 using Mauve version 2.3.12. Default parameters were used for all programs.
Detection of Other Isolates Containing ICEApl1
We screened 185 isolates of A. pleuropneumoniae (clinical isolates collected between 1995 and 2012 from the UK, Denmark, the Czech Republic, Cyprus, and Greece) for virB4 by PCR using primers virB4_for (CCTTCACGGTTAAAGAATCG AC)/virB4_rev (GCATCGTTTATTGGAAATGGAT). Primers were designed based on the virB4 gene in MIDG2331, amplifying the region from 1532511 to 1532894 in the genome sequence. Serovars 1 (1.2%), 2 (11.7%), 5 (2.4%), 6 (4.7%), 7 (10.6%), 8 (58.8%), 9/11 (2.4%), 10 (2.9%), and 12 (5.3%), were represented, and 84% of the isolates were from the UK. Genome sequence data was generated and assembled as previously described (Howell et al., 2013; Bossé et al., 2015) for 31 virB4 positive isolates. Sequences matching ICEApl1 were identified by BLASTn, assembled using Geneious 9.0.4, and deposited to Genbank (see Table 1 for accession numbers).
Table 1.
Clinical isolates of Actinobacillus pleuropneumoniae with ICEApl1.
| Isolate ID | Serovar | Location of isolationa | Year of isolation | Length of ICEApl1b | 5′ tRNAsc | Accession number for ICEApl1 sequence |
|---|---|---|---|---|---|---|
| MIDG2331 | 8 | Thirsk | 1995 | 56,083 bp | GLGLL | LN908249 (bases 1570419–1570505)d |
| MIDG2427 | 8 | Aberdeen | 1998 | 56,083 bp | GLGLL | KU551309 |
| MIDG2648 | 8 | Bury St Edmunds | 2005 | 54,898 bp | GLGL | KU551310 |
| MIDG2652 | 8 | Thirsk | 2005 | 56,083 bp | GLGLL | KU551311 |
| MIDG2654 | 8 | Winchester | 2005 | 56,070 bp | GLGLL | KU551312 |
| MIDG2657 | 8 | Winchester | 2005 | 56,083 bp | GLLGL | KU551313 |
| MIDG2663 | 8 | Thirsk | 2005 | 56,083 bp | GLLGL | KU551314 |
| MIDG2664 | 8 | Bury St Edmunds | 2005 | 56,083 bp | GLLGL | KU551315 |
| MIDG3200 | 8 | Thirsk | 2006 | 56,083 bp | GLGLL | KU551316 |
| MIDG3201 | 8 | Bury St Edmunds | 2006 | 56,083 bp | GLGLL | KU551317 |
| MIDG3221 | 8 | Langford | 2006 | 56,083 bp | GLGLL | KU551318 |
| MIDG3229 | 8 | Thirsk | 2007 | 56,083 bp | GLGLL | KU551320 |
| MIDG3232 | 8 | Thirsk | 2007 | 56,083 bp | GLGLL | KU551321 |
| MIDG3339 | 8 | Winchester | 2008 | 56,083 bp | GLGLL | KU551322 |
| MIDG3344 | 8 | Langford | 2005 | 56,083 bp | GLLGL | KU551323 |
| MIDG3346 | 8 | Thirsk | 2005 | 56,083 bp | GLLGL | KU551324 |
| MIDG3349 | 8 | Thirsk | 2006 | 56,083 bp | GLLGL | KU551325 |
| MIDG3357 | 8 | Shrewsbury | 2008 | 56,083 bp | GLGLL | KU551326 |
| MIDG3368 | 8 | Thirsk | 2008 | 56,083 bp | GLGLL | KU551327 |
| MIDG3370 | 8 | Thirsk | 2009 | 56,083 bp | GLLGL | KU551328 |
| MIDG3371 | 8 | Thirsk | 2009 | 56,083 bp | GLLGL | KU551329 |
| MIDG3372 | 8 | Thirsk | 2009 | 56,083 bp | GLLGL | KU551330 |
| MIDG3378 | 8 | Bury St Edmunds | 2009 | 56,047 bp | GLGLL | KU551331 |
| MIDG3381 | 8 | Thirsk | 2009 | 56,083 bp | GLGLL | KU551332 |
| MIDG3386 | 8 | Bury St Edmunds | 2009 | 56,083 bp | GLGLL | KU551333 |
| MIDG3388 | 8 | Thirsk | 2009 | 56,083 bp | GLGLL | KU551334 |
| MIDG3389 | 8 | Thirsk | 2009 | 56,083 bp | GLGLL | KU551335 |
| MIDG3395 | 8 | Thirsk | 2010 | 56, 011 bp | GLGLL | KU551336 |
| MIDG3401 | 8 | Bury St Edmunds | 2011 | 56,083 bp | GLGLL | KU551337 |
| MIDG3409 | 8 | Bury St Edmunds | 2011 | 56,083 bp | GLGLL | KU551338 |
| MIDG3458 | 8 | Cyprus | 2011 | 56,012 bp | GLGLL | KU551339 |
| MIDG3469 | 8 | Thirsk | 2012 | 56,083 bp | GLGLL | KU551340 |
aAll isolates except MIDG2427 and MIDG3458 were cultured from pigs submitted to the then Veterinary Laboratories Agency (now Animal and Plant Health Agency) regional laboratories in the UK, as indicated.
bThe length of each ICEApl1 sequence, calculated as the predicted circular form, is shown for each isolate. Size variation from that in MIDG2331 is indicated in bold. The sequence in MIDG2648 is missing 3 genes and has a truncated copy of the site-specific recombinase gene; whereas small deletions in the other sequences are all intergenic.
cOrder of tRNA genes upstream of ICEApl1: G, tRNA-Gly (GCC), L, tRNA-Leu (TAA). Variation from the order seen in MIDG2331 is indicated in bold.
dICEApl1 identified in MIDG2331 in previous study,4 all other ICEApl1 sequences were identified in this study.
Minimum Inhibitory Concentrations (MICs) for tetracycline were determined for isolates containing ICEApl1, according to the CLSI M37-A3 guidelines (Clinical and Laboratory Standards Institute [CLSI], 2008).
Conjugal Transfer of ICEApl1
MIDG2331ΔureC::nadV was used as the conjugal donor, with matings performed as previously described (Bossé et al., 2015). Plasmid-free, tetracycline-sensitive, nalidixic acid-resistant clinical isolates of serovars 6 (MIDG3376), 7 (MIDG2465), 8 (MIDG3217), and 12 (MIDG3347) were used as recipients. Transconjugants were selected on Brain Heart Infusion agar supplemented with 0.01% NAD, 5 mg/L tetracycline and 40 mg/L nalidixic acid. PCR was used to confirm the presence of the virB4 gene (as above), as well as serovar of, and the absence of nadV, in selected transconjugants using previously described primers (Bossé et al., 2014, 2015). Chromosomal insertion sites in transconjugants were determined by PCR using primers ICE5′_out1 (TGAGGGAGTAACAAGCAACACAG)/mfd3′_out (TTTACCGCTTGCCGATAATGCG) for the 5′ junction, and ICE3′_out1 (CAATGGAGAAAGAGAGTTGTTGGAC) /hybF5′_out (GACATCTCGTGCATAACCATTCC) for the 3′ junction, respectively. Amplicons were sequenced using internal primers ICE5′_out2 (GGAAGGTTCAATATCA CGACGG) or ICE3′_out2 (AGGCATACAGCAGCAACAAATC), as appropriate. For comparison, the region between mfd and hybF in the conjugal recipients (prior to conjugation) was amplified using mfd3′_out/hyb5′_out and sequenced in both directions.
Confirmation of the Circular Extrachromosomal form of the ICE by Nested PCR
DNA was extracted from MIDG2331ΔureC::nadV and selected transconjugants, and nested PCR was performed as previously described (Eidam et al., 2015), using primers ICE5′_out1/ICE3′_out1 followed by primers ICE5′_out2/ICE3′_out2. Amplicons were sequenced using primers ICE5′_out2 and ICE3′_out2.
Results and Discussion
Sequence of ICEApl1 and Comparative Analysis
ICEApl1, a 56083 bp element, is inserted into a tRNA-Leu (TAA) gene, a common insertion site for ICE in the family Pasteurellaceae (Dimopoulou et al., 2002; Michael et al., 2012; Eidam et al., 2015), in a tRNA cluster located between genes hybF and mfd. In the MIDG2331 genome (Bossé et al., 2016), this tRNA-Leu (TAA) gene is annotated as MIDG2331_01481, and is located between bases 1570419 and 1570505. Although all of the tRNA genes in the cluster, as well as hybF and mfd, are on the complement strand in the MIDG2331 genome, all further references to these genes, and the location of ICEApl1, will be with respect to the forward orientation. Insertion of ICEApl1 generated 66 bp imperfect direct repeats (DRs) at the left and right attachment sites, attL and attR (Figure 1A). The three bases that differ in the DRs reflect sequence variation in the tRNA-Leu (TAA) genes in A. pleuropneumoniae and H. influenzae (Figure 1B). The attI site in the closed circular form of ICEApl1 (confirmed by PCR; see below) is identical to that in ICEHin1056 (Figure 1C), both having 100% identity with the last 65 bases of the H. influenzae tRNA-Leu (TAA) gene followed by a T. These data suggest that insertion of ICEApl1 in MIDG2331 has generated an altered tRNA-Leu (TAA) gene, resulting from cross-over of the circular plasmid form of the ICE into the chromosome between the G at position 25 and the A at position 54 of MIDG2331_01481.
FIGURE 1.

Analysis of ICEApl1 insertion site. The imperfect direct repeats (DRs) (A) flanking ICEApl1 share sequence identity with the last 65 bases of tRNA-Leu genes (B) from Actinobacillus pleuropneumoniae (e.g., MIDG2331_01482 and _01484) and Haemophilus influenzae (e.g., accession number LK008335), which differ at 3 bases (in light blue for bases normally found in A. pleuropneumoniae and red for bases normally found in H. influenzae). The attI sequences (C) in the closed circular forms of ICEApl1 and ICEHin1056 are identical, and match the end of the H. influenzae tRNA-Leu (TAA) gene sequence, with an additional T which is also present in the DRs (A). Insertion of ICEApl1 in the A. pleuropneumoniae tRNA-Leu (TAA) gene MIDG2331_01481 has resulted in an altered sequence, indicating cross-over of the circular ICEApl1 into the chromosome occurred between the G at position 25 and the A at position 54.
Comparative sequence analysis revealed that ICEApl1 is related to the ICEHin1056 subfamily of elements (Figure 2) found in H. influenzae and H. parainfluenzae (Mohd-Zain et al., 2004; Juhas et al., 2007b). ICEApl1 encodes 67 genes that share extensive sequence homology and gene order with ICEHin1056 and other members of this subfamily. The first 14687 bp of ICEApl1 shares 99% identity with the region of ICEHin1056 reported to contain replication and stabilization genes (Juhas et al., 2007a,b, 2013). The 8933 bp Tn10 element in ICEApl1, although in the same location and orientation as that in ICEHin1056, more closely resembles that in ICEHpaT3T1 (99% identity, but inverted), with tetracycline resistance genes tetR, tetB, tetC, and tetD; and gltS encoding glutamate permease (Juhas et al., 2007b). The Tn10 element in ICEHin1056 has a further IS5 insertion (encoding chloramphenicol resistance) within it that is not seen in ICEApl1 (Juhas et al., 2007b). The 20466 bp following the Tn10 insertion shares 99% identity with the region containing genes encoding components of the type IV secretion system (required for conjugal transfer) in ICEHin1056 (Juhas et al., 2007a,b, 2013). This region is well conserved in all of the members of the ICEHin1056 family (Juhas et al., 2007a,b, 2013). The gene order in ICEApl1 remains syntenic with that of ICEHin1056 up to traC, where in ICEHin1056 there is a Tn3 insertion (encoding beta-lactamase resistance) that is not found in ICEApl1. From traC in ICEApl1, the nucleotide sequence and gene order more closely resemble those in ICEHin2866 up to the site-specific tyrosine recombinase gene, which is the final gene in ICEApl1 on the attR side. This 8.5 kb region shares 98% identity with sequences in ICEHin2866, and includes accessory genes encoding a type I restriction enzyme M subunit, and a transposon gamma-delta resolvase, as well as four hypothetical genes of unknown function (Juhas et al., 2007b).
FIGURE 2.
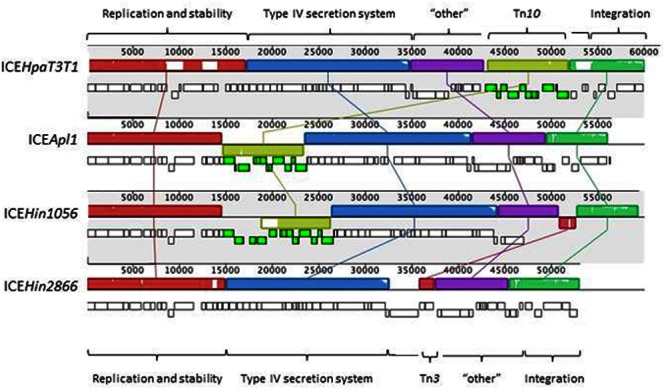
Mauve alignments of ICEApl1 and closely related members of the ICEHin1056 subfamily of elements. The orientation and relative size of genes in each ICE are indicated by the small rectangular blocks (lower blocks are on the complementary strand), with those colored green indicating genes of Tn10 elements present in all except ICEHin2866. Note the extra genes in the Tn10 insertion in ICEHin1056 are due to the presence of a further IS5 insertion not seen in the other ICE. Regions containing contiguous genes of related function are indicated by colored blocks above the genes, and are connected by lines to matching blocks in each ICE sequence. The labels at the top and bottom of the figure indicate functional sets of genes defined by Juhas et al. (2007b).
Surprisingly, the ICEHin1056 subfamily of conjugative elements has previously only been reported in H. influenzae and H. parainfluenzae, two human species of Haemophilus, where they appear to be evolving by descent (Dimopoulou et al., 2007; Juhas et al., 2007b). To our knowledge, this is the first report of a member of the ICEHin1056 subfamily in a Pasteurellaceae species that infects livestock. There have been ICE reported for bovine isolates of P. multocida (ICEPmu1) and M. haemolytica (ICEMh1), as well as an uncharacterized putative ICE in Histophilus somni strain 2336, which are related and appear to have evolved from a common ancestor, but are part of a different subfamily than ICEHin1056 elements (Juhas et al., 2007b; Michael et al., 2012; Eidam et al., 2015). The identity of the attI sites in both ICEApl1 and ICEHin1056 with the last 65 bases of the H. influenzae tRNA-Leu (TAA) gene would suggest more recent acquisition of an ICEHin1056 element in A. pleuropneumoniae.
Distribution of ICEApl1 in A. pleuropneumoniae Isolates
PCR analysis revealed the presence of a virB4 amplicon in 32/185 A. pleuropneumoniae isolates, including MIDG2331. All 32 (31 from the UK, 1 from Cyprus) were serovar 8 and had an MIC for tetracycline of 8 mg/L, i.e., above the CLSI breakpoint of ≥ 2 mg/L for A. pleuropneumoniae (Clinical and Laboratory Standards Institute [CLSI], 2008). ICEApl1 sequences were detected in the whole genomes of the 32 isolates, and comparative analysis revealed that, other than minor nucleotide differences in some, all of the sequences were complete except the element from MIDG2648, which was lacking three genes (encoding a putative DNA-binding protein and two hypothetical proteins) in the accessory gene region, and has a truncated copy of the tyrosine recombinase gene (Table 1).
Conjugal Transfer of ICEApl1 and Detection of Circular Intermediate Form
As ICEApl1 appeared to be present only in serovar 8 isolates of A. pleuropneumoniae, it was possible that other serovars blocked conjugal entry of the ICE, either due to restriction modification systems or CRISPR mediated restriction (Elhai et al., 1997; Garneau et al., 2010). We therefore tested the ability to conjugally transfer ICEApl1 to clinical isolates of A. pleuropneumoniae representing serovars 6, 7, 8, and 12 (MIDG3376, MIDG2465, MIDG3217, and MIDG3347, respectively) that are commonly found in the UK (O’Neill et al., 2010). All tested isolates produced transconjugants, as initially confirmed by PCR (data not shown). The frequencies of conjugation were similar for the serovar 7, 8, and 12 recipients (between 10-4 and 10-5), but much lower (5 × 10-8) for the serovar 6 isolate tested. Similar frequencies have been reported for ICEHin1056 elements in H. influenzae, with strain related differences also noted (Juhas et al., 2007b). More serovar 6 isolates would need to be tested in order to determine if the difference in frequency of conjugation for ICEApl1 is serovar-specific. The presence of a circular intermediate form of ICEApl1 was confirmed in the donor strain and in transconjugants by nested PCR. Sequenced amplicons confirmed a single copy of the 66 bp attI (Figure 1C) at the closed junction of the circular intermediates.
Sequencing PCR products generated at both the attL and attR ends in the transconjugants confirmed insertion of ICEApl1 in the same tRNA cluster between mfd and hybF as in the donor strain (MIDG2331ΔureC::nadV). However, in all transconjugants tested, the sequences between mfd and hybF contained only the altered tRNA-Leu (TAA) gene in which the ICE inserted, flanked by tRNA-Gly (GCC) genes at either end of the cluster (Figure 3B). This is in contrast to the cluster in the donor strain where there are 5 tRNA genes on the attL side, and a tRNA-Gly (GCC) gene on the attR side (Figure 3C). Sequencing across the tRNA cluster in the recipient strains prior to conjugation revealed that, although a different order of tRNA genes was present in MIDG3376 compared to the other strains (Figure 3A), all contained 3 copies each of tRNA-Gly (GCC) and tRNA-Leu (TAA) genes. These results indicate that in all transconjugants tested, a deletion of 3 tRNA genes was associated with ICEApl1 insertion. However, examination of the insertion sites in the serovar 8 clinical isolates with endogenous ICEApl1 (Table 1) revealed conservation of the 6 tRNA genes normally found in this cluster – i.e., 3 copies each of tRNA-Gly (GCC) and tRNA-Leu (TAA) – with the order of genes showing one of 3 patterns (see Figure 3C and Table 1 for details). In MIDG2648, the truncated element present in this isolate is located in a different copy of the tRNA-Leu (TAA) gene than seen in isolates with intact ICEApl1 (Figure 3C), with 4 tRNA genes on the attL side, and a tRNA-Leu (TAA) followed by tRNA-Gly (GCC) gene on the attR side. It would appear that, although the majority of clinical isolates show integration in the same tRNA-Leu (TAA) gene (i.e., the final copy in the tRNA cluster in the forward orientation), ICEApl1 has the ability to integrate into different copies of this gene, as has been reported for ICEHin1056 in H. influenzae (Dimopoulou et al., 2002). In Pseudomonas knackmussii, ICEclc was found to insert into different copies of the tRNA-Gly (GCC) gene, with double integration in some transconjugants (Sentchilo et al., 2009). In that study, excision and reintegration was associated with generation of a heterogenous population in which ICEclc was found to move from its original insertion site to alternate tRNA-Gly genes, but only those with the GCC anticodon (Sentchilo et al., 2009). Similarly, following in vitro conjugal transfer of ICEKp1 into a recipient strain of Klebsiella pneumoniae, integration was found at any of four tRNA-Asn genes, with insertion in multiple copies in some transconjugants (Lin et al., 2008). Furthermore, in some transconjugants, there was evidence of deletions between copies of the tRNA-Asn genes associated with integration of ICEKp1, which may have been due to recombination between multiple insertion sites (Lin et al., 2008). We did not detect multiple insertions of ICEApl1 in our transconjugants, as only one PCR product was generated using the outward facing primers designed to amplify the closed junction of the circular ICE. These primers would also have generated a secondary product in the presence of tandem insertions, given the proximity of the copies of the tRNA-Leu (TAA) genes in A. pleuropneumoniae. It is also possible that recombination may occur between the DRs found in alternate copies of the target tRNA, with or without the presence of an integrated ICE, resulting in deletion of the intervening sequences. However, given the results of Lin et al. (2008), it is likely that conditions during in vitro conjugation favor integration of multiple copies of ICE leading to deletions, whereas this does not appear to be common amongst clinical isolates. Little is known regarding the signals that govern initiation of horizontal transfer of ICE between pathogens in a host animal environment.
FIGURE 3.

Schematic representation of the tRNA cluster located between mfd and hybF in A. pleuropneumoniae isolates. Note that all sequences are shown in the forward orientation for simplicity. In the MIDG2331 genome, these sequences are on the complement strand. (A) The tRNA genes in isolates used as conjugal recipients are in the same order in (i) MIDG2465 (serovar 7), MIDG3217 (serovar 8), and MIDG3347 (serovar 12); and a different order in (ii) MIDG3376 (serovar 6). Note that all of the tRNA-Leu (TAA) genes have the sequence shown in Figure 1B. (B) Following conjugation, ICEApl1 integration resulted in loss of 3 tRNA genes, with all of the transconjugants (MIDG2465::ICEApl1, MIDG3217::ICEApl1, MIDG3347::ICEApl1, and MIDG3376::ICEApl1) showing identical sequences flanking the insertion (ICEApl1 shown boxed in blue). Note that the tRNA-Leu (TAA) gene shown in red has the altered bases of the attL DRs in Figure 1A. (C) In the 32 isolates with endogenous ICEApl1, the tRNA genes are found in three different orders in (i) MIDG2331 and 21 other isolates; (ii) 9 other isolates; and (iii) MIDG2648. Note the truncated element in MIDG2648 is shown as ICEApl1∗ (in purple text, boxed in purple). See Table 1 for details of specific isolates. Again, the tRNA-Leu (TAA) gene shown in red has the altered bases of the attL DRs in Figure 1A.
Conclusion
Identification of ICEApl1 in only serovar 8 clinical isolates of A. pleuropneumoniae may simply be a reflection of this being the most common in the UK (O’Neill et al., 2010), and thus in our collection. It may also indicate a tendency for ICE to be inherited by vertical transmission rather than horizontal transfer. The similar in vitro conjugation frequencies of ICEApl1 into isolates of serovars 7, 8, and 12 suggests there are no restriction endonuclease or CRISPR barriers to transfer between these serovars. Futhermore, the variation in order of tRNA genes flanking ICEApl1 suggests that horizontal transfer may have occurred independently into different isolates, as a similar variation in tRNA gene order was also seen in clinical isolates lacking ICEApl1. As reported for other ICE, ICEApl1 has the ability to integrate into different copies of the target tRNA gene, in this case tRNA-Leu (TAA). Although we did not detect multiple insertions following in vitro conjugal transfer of ICEApl1, it is likely that recombination between tandem insertions was responsible for the deletions detected in transconjugants.
To our knowledge, this is the first description of an ICE identified in A. pleuropneumoniae, and the first report of a member of the ICEHin1056 subfamily found in a non-human pathogen. The presence of ICEApl1 in isolates of A. pleuropneumoniae confers resistance to tetracycline, which is commonly used for treatment and control of porcine pleuropneumonia (European Medicines Agency, 2012). Although currently only found in serovar 8 isolates, the ability to transfer to other serovars was confirmed in vitro, and has implications for the spread of antimicrobial resistance in this important pig pathogen.
Author Contributions
JB, PL, AR, BW, DM, and AT conceived the study; JB, YL, RFC, RRC, MH, and JR produced the data; JB, YL, RFC, and RRC analyzed the data; JB and PL wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The BRaDP1T Consortium comprises: DM, AT, Sarah E. Peters, Lucy A. Weinert, Jinhong (Tracy) Wang, Shi-Lu Luan, RRC (University of Cambridge; present address for RRC is: Department of Molecular Biology and Biotechnology, University of Sheffield, Firth Court, Western Bank, Sheffield, S10 2TN, UK), AR, Gareth A. Maglennon, Jessica Beddow (Royal Veterinary College); BW, Jon Cuccui, Vanessa S. Terra (London School of Hygiene and Tropical Medicine); and PL, JB, YL (Imperial College London). The authors wish to thank Susanna Williamson and Chris Teale from the APHA for their advice and input.
Funding. This work was supported by a Longer and Larger (LoLa) grant from the Biotechnology and Biological Sciences Research Council (BBSRC grant numbers BB/G020744/1, BB/G019177/1, BB/G019274/1, and BB/G018553/1), the UK Department for Environment, Food and Rural Affairs, and Zoetis (formerly Pfizer Animal Health) awarded to the Bacterial Respiratory Diseases of Pigs-1 Technology (BRaDP1T) Consortium. MTGH was supported by the Wellcome Trust (grant number 098051). JR was funded from the former AHVLA’s Research and Development Internal Investment Fund (grant number RD0030c).
References
- Blanco M., Gutiérrez-Martín C. B., Rodríguez-Ferri E. F., Roberts M. C., Navas J. (2006). Distribution of tetracycline resistance genes in Actinobacillus pleuropneumoniae isolates from Spain. Antimicrob. Agents Chemother. 50 702–708. 10.1128/AAC.50.2.702-708.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco M., Kadlec K., Gutiérrez-Martín C. B., de la Fuente A. J. M., Schwarz S., Navas J. (2007). Nucleotide sequence and transfer properties of two novel types of Actinobacillus pleuropneumoniae plasmids carrying the tetracycline resistance gene tet(H). J. Antimicrob. Chemother. 60 864–867. 10.1093/jac/dkm293 [DOI] [PubMed] [Google Scholar]
- Bossé J. T., Chaudhuri R. R., Li Y., Leanse L. G., Fernandez Crespo R., Coupland P., et al. (2016). Complete genome sequence of MIDG2331, a genetically tractable serovar 8 clinical isolate of Actinobacillus pleuropneumoniae. Genome Announc. 4:e1667–15. 10.1128/genomeA.01667-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossé J. T., Li Y., Angen Ø., Weinert L. A., Chaudhuri R. R., Holden M. T., et al. (2014). A multiplex PCR to unequivocally differentiate A. pleuropneumoniae serovars 1-3, 5-8, 10 and 12. J. Clin. Microbiol. 52 2380–2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossé J. T., Li Y., Walker S., Atherton T., Fernandez Crespo R., Williamson S. M., et al. (2015). Identification of dfrA14 in two distinct plasmids conferring trimethoprim resistance in Actinobacillus pleuropneumoniae. J. Antimicrob. Chemother. 70 2217–2222. 10.1093/jac/dkv121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd E. F., Almagro-Moreno S., Parent M. A. (2009). Genomic islands are dynamic, ancient integrative elements in bacterial evolution. Trends Microbiol. 17 47–53. 10.1016/j.tim.2008.11.003 [DOI] [PubMed] [Google Scholar]
- Burrus V., Waldor M. K. (2004). Shaping bacterial genomes with integrative and conjugative elements. Res. Microbiol. 155 376–386. 10.1016/j.resmic.2004.01.012 [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute [CLSI] (2008). Performance Standards for (Antimicrobial) Disk and Dilution Susceptibility Tests for Bacteria Isolated From Animals - Approved Standard M31-A3 3rd Edn Wayne, PA: CLSI. [Google Scholar]
- Dimopoulou I. D., Kartali S. I., Harding R. M., Peto T. E. A., Crook D. W. (2007). Diversity of antibiotic resistance integrative and conjugative elements among haemophili. J. Med. Microbiol. 56 838–846. 10.1099/jmm.0.47125-0 [DOI] [PubMed] [Google Scholar]
- Dimopoulou I. D., Russell J. E., Mohd-Zain Z., Herbert R., Crook D. W. (2002). Site-specific recombination with the chromosomal tRNALeu gene by the large conjugative Haemophilus resistance plasmid. Antimicrob. Agents Chemother. 46 1602–1603. 10.1128/AAC.46.5.1602-1603.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eidam C., Poehlein A., Leimbach A., Michael G. B., Kadlec K., Liesegang H., et al. (2015). Analysis and comparative genomics of ICEMh1, a novel integrative and conjugative element (ICE) of Mannheimia haemolytica. J. Antimicrob. Chemother. 70 93–97. 10.1093/jac/dku361 [DOI] [PubMed] [Google Scholar]
- Elhai J., Vepritskiy A., Muro-Pastor A. M., Flores E., Wolk C. P. (1997). Reduction of conjugal transfer efficiency by three restriction activities of Anabaena sp. strain PCC 7120. J. Bacteriol. 179 1998–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Medicines Agency (2012). Sales of Veterinary Antimicrobial Agents in 19 EU/EEA Countries – Second ESVAC Report. London: European Medicines Agency; 1–74. [Google Scholar]
- Garneau J. E., Dupuis M.- È., Villion M., Romero D. A., Barrangou R., Boyaval P., et al. (2010). The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468 67–71. 10.1038/nature09523 [DOI] [PubMed] [Google Scholar]
- Guglielmini J., Quintais L., Garcillán-Barcia M. P., de la Cruz F., Rocha E. P. C. (2011). The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet. 7:e1002222 10.1371/journal.pgen.1002222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell K. J., Weinert L. A., Luan S.-L., Peters S. E., Chaudhuri R. R., Harris D., et al. (2013). Gene content and diversity of the loci encoding biosynthesis of capsular polysaccharides of the 15 serovar reference strains of Haemophilus parasuis. J. Bacteriol. 195 4264–4273. 10.1128/JB.00471-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhas M., Crook D. W., Dimopoulou I. D., Lunter G., Harding R. M., Ferguson D. J. P., et al. (2007a). Novel type IV secretion system involved in propagation of genomic islands. J. Bacteriol. 189 761–771. 10.1128/JB.01327-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhas M., Dimopoulou I., Robinson E., Elamin A., Harding R., Hood D., et al. (2013). Identification of another module involved in the horizontal transfer of the Haemophilus genomic island ICEHin1056. Plasmid 70 277–283. 10.1016/j.plasmid.2013.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhas M., Power P. M., Harding R. M., Ferguson D. J. P., Dimopoulou I. D., Elamin A. R. E., et al. (2007b). Sequence and functional analyses of Haemophilus spp. genomic islands. Genome Biol. 8:R237 10.1186/gb-2007-8-11-r237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T.-L., Lee C.-Z., Hsieh P.-F., Tsai S.-F., Wang J.-T. (2008). Characterization of integrative and conjugative element ICEKp1-associated genomic heterogeneity in a Klebsiella pneumoniae strain isolated from a primary liver abscess. J. Bacteriol. 190 515–526. 10.1128/JB.01219-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael G. B., Freitag C., Wendlandt S., Eidam C., Feßler A. T., Lopes G. V., et al. (2015). Emerging issues in antimicrobial resistance of bacteria from food-producing animals. Future Microbiol. 10 427–443. 10.2217/fmb.14.93 [DOI] [PubMed] [Google Scholar]
- Michael G. B., Kadlec K., Sweeney M. T., Brzuszkiewicz E., Liesegang H., Daniel R., et al. (2012). ICEPmu1, an integrative onjugative element (ICE) of Pasteurella multocida: analysis of the regions that comprise 12 antimicrobial resistance genes. J. Antimicrob. Chemother. 67 84–90. 10.1093/jac/dkr406 [DOI] [PubMed] [Google Scholar]
- Mohd-Zain Z., Turner S. L., Cerdeño-Tárraga A. M., Lilley A. K., Inzana T. J., Duncan A. J., et al. (2004). Transferable antibiotic resistance elements in Haemophilus influenzae share a common evolutionary origin with a diverse family of syntenic genomic islands. J. Bacteriol. 186 8114–8122. 10.1128/JB.186.23.8114-8122.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill C., Jones S. C. P., Bossé J. T., Watson C. M., Williamson S. M., Rycroft A. N., et al. (2010). Prevalence of Actinobacillus pleuropneumoniae serovars in England and Wales. Vet. Rec. 167 661–662. 10.1136/vr.c5106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sárközi R., Makrai L., Fodor L. (2015). Identification of a proposed new serovar of Actinobacillus pleuropneumoniae: serovar 16. Acta Vet. Hung. 63 444–450. 10.1556/004.2015.041 [DOI] [PubMed] [Google Scholar]
- Sentchilo V., Czechowska K., Pradervand N., Minoia M., Miyazaki R., van der Meer J. R. (2009). Intracellular excision and reintegration dynamics of the ICEclc genomic island of Pseudomonas knackmussii sp. strain B13. Mol. Microbiol. 72 1293–1306. 10.1111/j.1365-2958.2009.06726.x [DOI] [PubMed] [Google Scholar]
- Wozniak R. A. F., Waldor M. K. (2010). Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat. Rev. Microbiol. 8 552–563. 10.1038/nrmicro2382 [DOI] [PubMed] [Google Scholar]


