Abstract
Aeromonas is regarded as an important pathogen of freshwater animals but little is known about the genetics of its antimicrobial resistance in Chinese aquaculture. The aim of this study was to investigate the presence of integrons and characterize multidrug resistant Aeromonas spp. isolated from diseased farmed freshwater animals. These animal samples included fish, ornamental fish, shrimp, turtles, and amphibians which were collected from 64 farms in Guangdong province of South China. One hundred and twelve Aeromonas spp. isolates were examined for antimicrobial resistance phenotypes and the presence of class 1 integron sequences. Twenty-two (19.6%) of these isolates carried a class 1 integron comprising six different gene insertion cassettes including drfA12-orfF-aadA2, drfA12-orfF, aac(6′)-II-blaOXA-21-cat3, catB3, arr-3, and dfrA17. Among these, drfA12-orfF-aadA2 was the dominant gene cassette array (63.6%, 14/22) and this is the first report of aac(6′)-II-blaOXA-21-cat3 in an Aeromonas hydrophila isolate from a Chinese giant salamander (Andrias davidianus). All the integron-positive strains were resistant to more than five agents and 22 contained other resistance genes including blaCTX-M-3, blaTEM-1, aac(6′)-Ib-cr, and tetA. All integron-positive isolates also contained mutations in the quinolone resistance determining regions (QRDR). Our investigation demonstrates that freshwater animals can serve as a reservoir for pathogenic Aeromonas strains containing multiple drug-resistance integrons. This data suggests that surveillance for antimicrobial resistance of animal origin and a prudent and responsible use of antimicrobials in aquaculture is necessary in these farms.
Keywords: Aeromonas, multidrug resistance, class 1 integron, resistance genes, freshwater animal
Introduction
The genus Aeromonas is regarded as an important pathogen of freshwater animals. Motile aeromonads including A. hydrophila, A. veronii, A. caviae, and A. sobria are considered facultative pathogens and can infect fish as well as shrimp, reptiles, amphibians, and other aquatic species (Janda and Abbott, 2010). Aeromonas infections have been linked to major die-offs and fish kills around the globe over the past decade resulting in enormous economic losses. Furthermore, Aeromonas can cause disease in warm-blooded animals and can even infect humans via consumption of contaminated food or water (Janda and Abbott, 2010).
Treatment and prevention of diseases in animal husbandry including livestock and aquaculture have been relying on the extensive use of antimicrobials. This practice has been undoubtedly increased the frequency of antibiotic-resistant bacterial strains and this has been widely documented. Antimicrobial resistance in bacterial populations can result from clonal selection under antimicrobial selective pressure or through horizontal gene transfer. Mobile genetic elements such as plasmids, integrons, and transposons contribute to a wider dissemination of genetic resistance determinants among bacteria (Boerlin and Reid-Smith, 2008). Of these, the mobile integron encoded integrases can recombine gene cassettes and are primarily involved in the spread of antimicrobial resistance genes which contributed to multidrug-resistance (Mazel, 2006). The class 1 integron is the most common integron type and has been found in a wide range of gram-negative bacteria from various sources (Hall, 2012). Previous studies have found integron in Aeromonas spp. mainly belong to the class 1 and carried various antimicrobial resistance gene cassettes. The most often found resistance gene cassettes contained several aminoglycoside resistance gens aadA1, aadA2, and the trimethoprim resistance gene dfrA1 (Piotrowska and Popowska, 2015).
Motile Aeromonas species are important aquatic pathogens and are considered a public health concern. Several types of fluoroquinolones and sulfonamides have been approved for treatment of these infections and are frequently used in veterinary medicine [Ministry of Agriculture of the People's Republic of China (MOA), 2010; Su et al., 2011]. Consequently, resistance to antimicrobial has been widely spread in aquaculture environments. Integrons-inserted gene cassettes have been reported in widespread Aeromonas species isolated from diseased fish (Piotrowska and Popowska, 2014). However, only a few studies, to date, addressed the prevalence of class 1 integron in Chinese aquaculture (Feng et al., 2011; Su et al., 2011; Jiang et al., 2012).
In this study, we investigated the distribution of integrons among aeromonads isolated from farmed diseased freshwater animals. We documented the presence of resistance genes and whether mutations of quinolone resistance determining regions (QRDR) were present among the integron-positive isolates. We evaluated the potential of multi-drug resistant aeromonads in these animals as a public health risk.
Materials and methods
Bacterial isolation and genetic identification
A total of 112 Aeromonas species isolated from diseased freshwater animal samples, including 67 fish from 37 farms, 7 ornamental fish from 6 farms, 12 shrimp from 6 farms, 21 turtles from 12 farms, and 5 amphibians from 3 farms in Guangdong province. All samples were collected at fisheries hospitals of the Pearl River Fisheries Research Institute from November 1995 to February 2012. All these freshwater animals were raised as food animals, pets or for other purposes. Gills, body surfaces, and livers were aseptically swabbed using sterile cotton buds and inoculated into Luria–Bertani broth for pre-enrichment at 30 ± 2°C for 18–24 h. The enriched cultures were streaked on Rimler–Shotts agar and incubated at 30 ± 2°C for 18–24 h. Yellow, oxidase-positive colonies were isolated and were presumptively considered as Aeromonas species. One colony per sample with typical Aeromonas morphology was selected and identified by API 20E biochemical tests (BioMérieux, France). For further identification, polymerase chain reaction (PCR) amplification of 16S rRNA gene and gyrB gene was performed as described in previous studies (Borrell et al., 1997; Yáñez et al., 2003). Taxonomic identification of the DNA sequences was performed using BLAST in GenBank (http://blast.ncbi.nlm.nih.gov/).
Antimicrobial susceptibility testing
All strains were evaluated for resistance to 14 antimicrobials by the disk diffusion method using commercially available disks (Oxoid, U.K.). These drugs included ampicillin (10 μg), cefotaxime (30 μg), sulfonamides (300 μg), trimethoprim/sulfamethoxazole (1.25/23.75 μg), rifampin (5 μg), nalidixic acid (30 μg), ciprofloxacin (5 μg), norfloxacin (10 μg), ofloxacin (5 μg), tetracycline (30 μg), doxycycline (30 μg), streptomycin (10 μg), amikacin (30 μg), and chloramphenicol (30 μg) discs. Escherichia coli ATCC 25922 was used for quality control. The results were evaluated as susceptible (S), intermediate (I), and resistant (R) based on the interpretative criteria from the [Clinical and Laboratory Standard Institute (CLSI), 2006a,b].
DNA extraction and PCR amplification of integrase genes
DNA prepared by the whole cell boiled lysate protocol was used for PCR templates. All isolates were screened for the presence of intI1, intI2, intI3. All the intI1-positive strains were also amplified for sul1 and qacEΔ1 fragments by PCR using primers described previously (Sandvang et al., 1997; Reyes et al., 2003). The PCR primers used in this study are presented in Supplementary Table 1.
Amplification and sequencing of gene cassette regions
Gene cassette arrays of intI1-positive strains were determined by PCR using primers described previously (Lévesque et al., 1995; Supplementary Table 1). PCR products were directly sequenced and the DNA sequences were analyzed using BLAST (http://blast.ncbi.nlm.nih.gov/).
PCR amplification of resistance genes and quinolone resistance determining regions (QRDR)
To characterize other resistance genes of the integron-positive isolates, extended spectrum beta-lactamase (ESBL) genes (blaTEM, blaCTX), tetracycline resistance genes (tetA, tetC, and tetE), and plasmid-mediated quinolone resistance (PMQR) genes [qnrA, qnrB, qnrS, aac(6′)-Ib-cr, and qepA] were also screened by PCR amplification as previously described (Liu et al., 2008). The sequencing primers are presented in Supplementary Table 1. Sequences were aligned with known sequences in the NCBI database using BLAST and are reported below as Genbank reference numbers.
The mutation within the QRDRs of gyrA and parC in integron-positive isolates were also determined as previously described (Giraud et al., 2004).
Results
Identification of Aeromonas spp. from different aquatic animals
Base on the biochemical and genetic characteristics, 112 Aeromonas isolates were identified up to the species level. They belonged to eight different species of Aeromonas (A. hydrophila, A. veronii, A. caviae, A. sobria, A. dhakensis, A. jandaei, A. trota, and A. media; Table 1). Over all, A. hydrophila was the most prevalent species, and were most frequently isolated from all kinds of animals analyzed, indicating that it is the predominant species in aquatic pathogens.
Table 1.
Prevalence of Aeromonas spp. in different aquatic animals.
| Aeromonas spp. | Sources | ||||
|---|---|---|---|---|---|
| Fish | Ornamental fish | Shrimp | Turtles | Amphibians | |
| A. hydrophila (n = 62) | 34 | 5 | 4 | 16 | 3 |
| A. veronii (n = 23) | 17 | 2 | 3 | 1 | |
| A. caviae (n = 7) | 4 | 2 | 1 | ||
| A. sobria (n = 6) | 5 | 1 | |||
| A. dhakensis (n = 4) | 2 | 1 | 1 | ||
| A. trota (n = 4) | 1 | 3 | |||
| A. jandaei (n = 3) | 3 | ||||
| A. media (n = 1) | 1 | ||||
| Unidentified (n = 2) | 1 | 1 | |||
| Total (n = 112) | 67 | 7 | 12 | 21 | 5 |
Detection and characterization of integron and gene cassettes
Among 112 Aeromonas strains isolated from different farmed freshwater animals, 22 (19.6%) isolates were positive for intI1 while intI2 and intI3 were not detected in any of the isolates. All of integron-positive isolates also contained sul1 and qacEΔ1 which indicated that these isolates contained structurally complete integrons. Considering the different sources, A. hydrophila (81.8%, 18/22) was the most frequently isolated species carrying integrons, which were more prevalent in Aeromonas spp. isolated from fish and turtles (Table 2).
Table 2.
Characteristic of 22 integron-positive Aeromonas isolates.
| Strain | Aeromonas spp. | Year | Species | Farm | Class 1 integron gene cassettes | Resistance genes | QRDR mutation | Antimicrobial resistance profile | |
|---|---|---|---|---|---|---|---|---|---|
| GyrA | ParC | ||||||||
| A15 | A. hydrophila | 1998 | Fish (Channa argus) | FF6 | dfrA17 | tetA | Ile83 | Ser80 | AMP\S3\SXT\RD\NA\TE\C\S |
| A16 | A. hydrophila | 1998 | Fish (C. argus) | FF6 | dfrA17 | tetA | Ile83 | Ser80 | AMP\S3\SXT\RD\NA\TE\C\S |
| A36 | A. hydrophila | 2004 | Ornamental fish (Crypinus carpiod) | OF1 | drfA12-orfF-aadA2 | Ile83 | Ile80 | AMP\S3\SXT\RD\NA\OFL\TE\DO\C\S | |
| A25 | A. hydrophila | 2004 | Turtle (Chelydra serpentine) | TF2 | drfA12-orfF-aadA2 | blaCTX−M−3, blaTEM−1, aac(6′)-Ib-cr | Ile83 | Ile80 | AMP\CTX\S3\SXT\RD\NA\CIP\NOR\OFL\TE\C\AK\S |
| A21 | A. hydrophila | 2003 | Turtle (Mauremys mutica) | TF3 | drfA12-orfF-aadA2 | Ile83 | Ile80 | AMP\S3\SXT\RD\NA\NOR\OFL\TE\DO\C\S | |
| A27 | A. hydrophila | 2003 | Turtle (M. mutica) | TF3 | drfA12-orfF-aadA2 | Ile83 | Ile80 | AMP\S3\SXT\RD\NA\TE\DO\C\S | |
| A23 | A. hydrophila | 2003 | Turtle (M. mutica) | TF4 | drfA12-orfF-aadA2 | Ile83 | Ile80 | AMP\S3\SXT\RD\NA\NOR\OFL\TE\DO\C\S | |
| A26 | A. hydrophila | 2003 | Turtle (C. serpentine) | TF4 | drfA12-orfF-aadA2 | aac(6′)-Ib | Ile83 | Ile80 | AMP\S3\SXT\RD\NA\NOR\OFL\TE\ DO\C\S |
| A03 | A. hydrophila | 1996 | Fish (Carassius auratus) | FF1 | drfA12-orfF-aadA2 | Ile83 | Ile80 | AMP\S3\SXT\RD\NA\OFL\TE\DO\C\S | |
| A19 | A.veronii | 2001 | Fish (Siniperca chuatsi) | FF9 | drfA12-orfF-aadA2 | Ile83 | Ile80 | AMP\S3\SXT\NA\S | |
| A47 | A. hydrophila | 2005 | Fish (Tenualosa reevesii) | FF16 | drfA12-orfF-aadA2 | Ile83 | Ile80 | AMP\S3\SXT\RD\NA\NOR\OFL\TE\DO\C\S | |
| A72 | A. hydrophila | 2006 | Fish (Cirrhinus molitorella) | FF18 | drfA12-orfF-aadA2 | Ile83 | Ser80 | AMP\S3\SXT\NA\TE\S | |
| A78 | A. hydrophila | 2007 | Fish (Oreochromis niloticus) | FF22 | drfA12-orfF-aadA2 | Ile83 | Ile80 | AMP\S3\SXT\RD\NA\NOR\OFL\TE\DO\C\S | |
| A01 | A. sobria | 1995 | Shrimp (Macrobrachium rosenbergii) | SF1 | drfA12-orfF-aadA2 | Val83 | Ile80 | AMP\S3\SXT\RD\NA\TE\S | |
| A41 | A. veronii | 2004 | Ornamental fish (C. auratus) | OF2 | drfA12-orfF-aadA2, drfA17 | Val83 | Ile80 | AMP\S3\SXT\RD\NA\ NOR\OFL\TE\ DO\AK\S | |
| A42 | A. veronii | 2004 | Fish (Megalobrama terminalis) | FF11 | drfA12-orfF-aadA2, catB3, arr-3 | Ile83 | Ile80 | AMP\S3\SXT\RD\NA\S | |
| A24 | A. hydrophila | 2003 | Turtle (C. serpentine) | TF2 | drfA12-orfF-aadA2, catB3, arr-3 | aac(6′)-Ib | Ile83 | Ile80 | AMP\S3\SXT\RD\NA\CIP\NOR\OFL\TE\ DO\S |
| A33 | A. hydrophila | 2004 | Giant salamander (Andrias davidianus) | AF1 | aac(6′)-II-blaOXA−21-cat3 | Ile83 | Ile80 | AMP\S3\SXT\NA\TE\S | |
| A83 | A. hydrophila | 2007 | Turtle (Pelodiscus sinensis) | TF10 | drfA12-orfF, catB3 | Ile83 | Arg80 | AMP\S3\SXT\RD\NA\CIP\NOR\OFL\C\S | |
| A84 | A. hydrophila | 2007 | Turtle (Pelodiscus sinensis) | TF10 | drfA12-orfF, catB3 | aac(6′)-Ib-cr | Ile83 | Ile80 | AMP\S3\SXT\RD\NA\CIP\NOR\OFL\C\S |
| A22 | A. hydrophila | 2004 | Turtle (C. serpentine) | TF4 | No insert | blaTEM−1, aac(6′)-Ib-cr | Ile83 | Ile80 | AMP\CTX\S3\SXT\RD\NA\CIP\TE\AK\S |
| A38 | A. hydrophila | 2004 | Giant salamander (A. davidianus) | AF1 | ND | Ile83 | Ile80 | AMP\S3\SXT\RD\NA\CIP\NOR\OFL\TE\ DO\C\S | |
ND, not determined; FF, fish farm; OF, ornamental fish farm; TF, turtle farm; AF, amphibian farm; SF, shrimp farm; QRDR, quinolone resistance-determining regions; Ser, serine; Ile, iosleucine; Val, valine; Arg, arginine; AMP, ampicillin; CTX, cefotaxime; S3, sulfonamides; SXT, trimethoprim/sulfamethoxazole; RD, rifampin; NA, nalidixic acid; CIP, ciprofloxacin; NOR, norfloxacin; OFL, ofloxacin; TE, tetracycline; DO, doxycycline; S, streptomycin; AK, amikacin; C, chloramphenicol.
The genetic content of the 22 integrons was determined through PCR amplification of the integron variable region. PCR products were obtained for 21 isolates, ranging from 750 to 2000 bp (Figure 1). One isolate was “empty” with no gene cassettes inserted between the conserved segments of the integron. The presence of six different gene cassette arrays were determined by sequencing and these were drfA12-orfF-aadA2 (KF442255 and KF442261), drfA12-orfF (KF442257), catB3 (KF442258 and KF442260), aac(6′)II-blaOXA−21-catB3 (KM009138), arr-3 (KF442259), and dfrA17 (KF442256). Additionally, we identified four isolates containing two to three integrons. Thirteen isolates contained an identical restriction fragment and carried the same gene cassette arrays (drfA12, orfF, and aadA2). This was the dominant gene cassette array found in this study.
Figure 1.
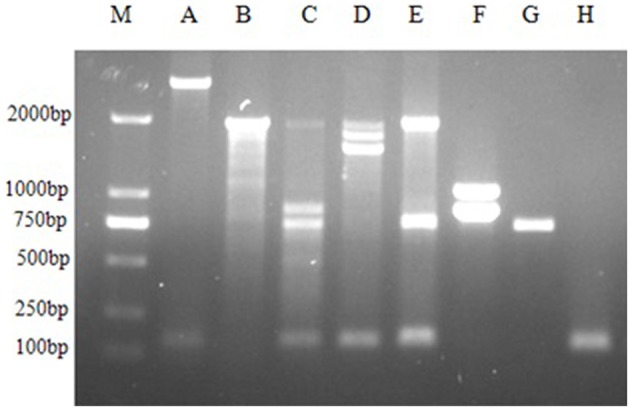
PCR amplification products of class 1 integron variable region M: DL2000 marker; A–H: eight groups of class 1 integrons.
Antimicrobial susceptibility of Aeromonas
The number of isolates resistant to antimicrobials was variable among the groups and 49.1% (55/112) were resistant to three or more different classes of antimicrobial agents. Resistance was most prevalent for ampicillin (85.7%), rifampin (57.1%), streptomycin (49.1%), and nalidixic acid (44.6%). Most of the isolates (≥90%) were susceptible to cefotaxime, amikacin, ciprofloxacin, norfloxacin, and doxycycline (Supplementary Table 2).
The isolates belonging to the eight identified Aeromonas spp. have varying levels of susceptibilities to the different antimicrobial agents. A. hydrophila and A. veronii isolates displayed greater levels of resistance as compared to other species (Supplementary Table 3). Considering the different sources, the Aeromonas isolates from turtles and amphibians were found to be more resistant to trimethoprim/sulfamethoxazole, quinolones, tetracyclines, and streptomycin (Supplementary Table 4).
With the exception of cefotaxime, the resistance rates of 13 antimicrobials of the integron-positive strains was significantly (P < 0.05) higher than the rate in integron-negative strains (Supplementary Table 2). All isolates carrying integrons were multi-drug resistant and were resistant to five or more commonly used antimicrobial agents. Resistance to more than 12 antimicrobial agents was observed in 12 isolates. The susceptibility pattern for the integron-containing strains is shown in Table 2.
Presence of resistance genes and analysis of QRDR mutations in integron-positive strains
Among the ESBL genes, blaCTX-M-3 was detected in one isolate in A. hydrophila from fish, whereas blaTEM-1 were detected in two isolates in A. hydrophila from fish and turtle, and one of these isolates harbored both genes. Among the tetracycline genes, only tetA was present in two isolates. Five integron-harboring isolates were positive for aac(6′)-Ib but only three of them were cr variants associated with ciprofloxacin resistance. Moreover, two isolates were associated with both beta-lactamase and PMQR genes.
The nucleotide sequences of gyrA and parC QRDR of 22 integron-positive strains were determined via PCR amplification and direct sequencing. With exception of two strains with a Ser-83-Val substitution, the other strains had mutations at codon 83 resulting in a Ser to Ile change in the GyrA QRDR. In the QRDR of ParC, the substitution Ser-80-Ile was observed in 18 of 22 integron-positive strains and one isolate had a Ser-83-Arg mutation (Table 2).
Discussion
Characterization of integrons in Aeromonas
In aquaculture, disease diagnoses are often presumptive and therapeutic measures are usually administered in the absence of reliable antimicrobial resistance data. There are few antimicrobial agents that have been approved for Chinese aquaculture [Ministry of Agriculture of the People's Republic of China (MOA), 2010], so impudent and abusive use of antimicrobials is common. The emergence and dissemination of antimicrobial resistance among Aeromonas spp. from aquaculture has attracted much attention. As such, Aeromonas strains have developed resistance to various antimicrobial agents as we and others have found. Our data indicates that these resistance patterns in Aeromonas can be derived from integrons encoding genes conferring resistance to trimethoprim-sulfamethoxazole, streptomycin, and other agents. Multi-drug resistance mediated by integrons in Aeromonas may complicate future antibiotic therapies used in aquaculture as well as stimulate resistance gene transfer.
Integrons with gene cassettes found primarily in gram-negative bacteria belonging to the family Enterobacteriaceae. These are often located on conjugative plasmids or transposons which can facilitate lateral transfer between pathogens (Mazel, 2006). Recently, several studies reported the presence of integrons on similar inserted-gene cassettes in fish-pathogenic Aeromonas isolates with a frequency of 30–50% (Nawaz et al., 2010; Ndi and Barton, 2011; Lukkana et al., 2012). In the present study, 19.6% of Aeromonas isolates from farmed freshwater animals carried the class 1 integron. Taken together, the frequency of class 1 integrons in Aeromonas isolates from farmed freshwater animals in South China is less than those from other freshwater animals. This is most likely due to different sources of isolates and the particular background of antimicrobial usage.
The addA2 gene responsible for resistance to streptomycin and the dfrA12 gene encoding resistance to trimethoprim were identified in most of the integron-borne Aeromonas identified in this study. Both the addA and dfr genes are the most frequently found resistance genes in the variable region of integrons among different species of bacteria. A similar pattern has been reported in Aeromonas from aquaculture sources (Kadlec et al., 2011; Lukkana et al., 2012), environmental samples (Moura et al., 2012) as well as clinical isolates (Lee et al., 2008). The predominance of the addA and dfr genes suggests that these genes cassettes are more stable when integrated than other gene cassettes. Additionally, both the selection and dispersion of addA and dfr genes in integrons could be related to the extensive use of streptomycin and trimethoprim-sulfamethoxazole in the control of animal diseases.
We identified a new cassette combination aac(6′)-II-blaOXA-21-cat3 in A. hydrophila isolated from a Chinese giant salamander (Andrias davidianus). This combination had previously only been found in A. hydrophila from clinical isolates in Taiwan (GenBank DQ993182.1). The Chinese giant salamander is both high medicinal value and evolutionary significance. The breeding of this animal is important in scientific research and has an economic impact as well. However, a variety of highly infectious, lethal diseases have hindered development of a breeding industry for giant salamander. The high value of these animals has motivated farmers to increase investments in antimicrobials and the same situation also appears in turtle farming and ornamental fish farming (Verner-Jeffreys et al., 2009; Kadlec et al., 2011). In this study, we also demonstrated that Aeromonas isolates from turtles, the Chinese giant salamander and ornamental fish were multi-drug resistant indicating that a serious problem of antimicrobial resistance may be emerging in high-value animal farming.
Detection of other resistance genes in integron-positive isolates
We found 22 isolates containing class 1 integrons that were resistant to five or more antimicrobial agents. We then screened for genes encoding 10 other resistance determinants and four [aac(6′)-Ib-cr, tetA, blaCTX-M-3, blaTEM-1] were detected. Cephalosporins are effective human antimicrobials but are seldom used in aquaculture. Unexpectedly, we acquired one cefotaxime-resistant strain among 22 integron-positive Aeromonas isolates which carried blaCTX-M-3. Two isolates carrying blaTEM-1 were also associated with aac(6′)-Ib-cr.
The occurrence of PMQR and ESBL determinants in Enterobacteriaceae from aquatic animals has been previously demonstrated (Jiang et al., 2012). We found three A. hydrophila isolates from turtles (Chelydra serpentine and Pelodiscus sinensis) that harbored aac(6′)-Ib-cr, blaTEM-1, and/or blaCTX-M-3. Farmed animals with high economic values are grown over long culture cycles and farmers are seeking more effective antimicrobial agents such as ciprofloxacin, ceftriaxone, and amikacin. With the intensive use of these antimicrobial in aquaculture, the incidence of drug abuse might also post a potential risk to public health.
In conclusion, our investigation demonstrated that freshwater animals can serve as a reservoir for Aeromonas species containing integron-mediated multiple drug-resistant phenotypes. The association of multi-drug resistance with integrons increases the risk of co-selection and persistence of other resistance determinants under the selective pressure imposed by the use of antimicrobial agents. Therefore, prudent and responsible use of antimicrobials is a mandatory requirement to promote the healthy development of Chinese aquaculture.
Author contributions
YD and LJ conceived the study, designed the trial, and supervised conduct of the trial. AT and LL supervised the data collection. YD and YW drafted the manuscript. RZ contributed substantially to the revision. LJ takes responsibility for the article as a whole.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was funded by Young Scientists Fund in National Natural Science Foundation of China (Grant No. 31302228) and Special Fund for Agro-scientific Research in the Public Interest (Grant No. 201203085).
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00935
References
- Boerlin P., Reid-Smith R. J. (2008). Antimicrobial resistance: its emergence and transmission. Anim. Health Res. Rev. 9, 115–126. 10.1017/S146625230800159X [DOI] [PubMed] [Google Scholar]
- Borrell N., Acinas S. G., Figueras M. J., Martínez-Murcia A. J. (1997). Identification of Aeromonas clinical isolates by restriction fragment length polymorphism of PCR-amplified 16S rRNA genes. J. Clin. Microbiol. 35, 1671–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical Laboratory Standard Institute (CLSI) (2006a). Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria. Wayne, PA: CLSI. Approved Guideline – M45-A. [Google Scholar]
- Clinical Laboratory Standard Institute (CLSI) (2006b). Methods for Antimicrobial Disk Susceptibility Testing of Bacteria Isolated from Aquatic Animals, Wayne, PA: CLSI. Approved Standards M42-A. [Google Scholar]
- Feng J. L., Yan H., Chowdhury N., Neogi S. B., Yamasaki S., Shi L., et al. (2011). Identification and characterization of integron-associated antibiotic resistant Laribacter hongkongensis isolated from aquatic products in China. Int. J. Food Microbiol. 144, 337–341. 10.1016/j.ijfoodmicro.2010.10.014 [DOI] [PubMed] [Google Scholar]
- Giraud E., Blanc G., Bouju-Albert A., Weill F. X., Donnay-Moreno C. (2004). Mechanisms of quinolone resistance and clonal relationship among Aeromonas salmonicida strains isolated from reared fish with furunculosis. J. Med. Microbiol. 53, 895–901. 10.1099/jmm.0.45579-0 [DOI] [PubMed] [Google Scholar]
- Hall R. M. (2012). Integrons and gene cassettes: hotspots of diversity in bacterial genomes. Ann. N.Y. Acad. Sci. 1267, 71–78. 10.1111/j.1749-6632.2012.06588.x [DOI] [PubMed] [Google Scholar]
- Janda J. M., Abbott S. L. (2010). The genus Aeromonas: taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 23, 35–73. 10.1128/CMR.00039-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H. X., Tang D., Liu Y. H., Zhang X. H., Zeng Z. L., Xu L., et al. (2012). Prevalence and characteristics of β-lactamase and plasmid-mediated quinolone resistance genes in Escherichia coli isolated from farmed fish in China. J. Antimicrob. Chemother. 67, 2350–2353. 10.1093/jac/dks250 [DOI] [PubMed] [Google Scholar]
- Kadlec K., Von Czapiewski E., Kaspar H., Wallmann J., Michael G. B., Steinacker U., et al. (2011). Molecular basis of sulfonamide and trimethoprim resistance in fish-pathogenic Aeromonas isolates. Appl. Environ. Microbiol. 77, 7147–7150. 10.1128/AEM.00560-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. F., Peng C. F., Lin Y. H., Lin S. R., Chen Y. H. (2008). Molecular diversity of class 1 integrons in human isolates of Aeromonas spp. from southern Taiwan. Jpn. J. Infect. Dis. 61, 343–349. [PubMed] [Google Scholar]
- Lévesque C., Piché L., Larose C., Roy P. H. (1995). PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39, 185–191. 10.1128/AAC.39.1.185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. H., Deng Y. T., Zeng Z. L., Gao J. H., Chen L., Arakawa Y., et al. (2008). Coprevalence of plasmid-mediated quinolone resistance determinants QepA, Qnr, and AAC(6′)-Ib-cr among 16S rRNA methylase RmtB-producing Escherichia coli isolates from pigs. Antimicrob. Agents Chemother. 52, 2992–2993. 10.1128/AAC.01686-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukkana M., Wongtavatchai J., Chuanchuen R. (2012). Class 1 integrons in Aeromonas hydrophila isolates from farmed Nile tilapia (Oreochromis nilotica). J. Vet. Med. Sci. 74, 435–440. 10.1292/jvms.11-0441 [DOI] [PubMed] [Google Scholar]
- Mazel D. (2006). Integrons: agents of bacterial evolution. Nat. Rev. Microbiol. 4, 608–620. 10.1038/nrmicro1462 [DOI] [PubMed] [Google Scholar]
- Ministry of Agriculture of the People's Republic of China (MOA) (2010). Announcement No.1435 of the Ministry of Agriculture. Available online at: www.moa.gov.cn/govpublic/SYJ/201008/t20100823_1622639.html (Online in Chinese.)
- Moura A., Pereira C., Henriques I., Correia A. (2012). Novel gene cassettes and integrons in antibiotic-resistant bacteria isolated from urban wastewaters. Res. Microbiol. 163, 92–100. 10.1016/j.resmic.2011.10.010 [DOI] [PubMed] [Google Scholar]
- Nawaz M., Khan S. A., Khan A. A., Sung K., Tran Q., Kerdahi K., et al. (2010). Detection and characterization of virulence genes and integrons in Aeromonas veronii isolated from catfish. Food Microbiol. 27, 327–331. 10.1016/j.fm.2009.11.007 [DOI] [PubMed] [Google Scholar]
- Ndi O. L., Barton M. D. (2011). Incidence of class 1 integron and other antibiotic resistance determinants in Aeromonas spp. from rainbow trout farms in Australia. J. Fish Dis. 34, 589–599. 10.1111/j.1365-2761.2011.01272.x [DOI] [PubMed] [Google Scholar]
- Piotrowska M., Popowska M. (2014). The prevalence of antibiotic resistance genes among Aeromonas species in aquatic environments. Ann. Microbiol. 64, 14 10.1007/s13213-014-0911-2 [DOI] [Google Scholar]
- Piotrowska M., Popowska M. (2015). Insight into the mobilome of Aeromonas strains. Front. Microbiol. 6:494. 10.3389/fmicb.2015.00494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes A., Bello H., Domínguez M., Mella S., Zemelman R., González G. (2003). Prevalence and types of class 1 integrons in aminoglycoside-resistant Enterobacteriaceae from several Chilean hospitals. J. Antimicrob. Chemother. 51, 317–321. 10.1093/jac/dkg083 [DOI] [PubMed] [Google Scholar]
- Sandvang D., Aarestrup F. M., Jensen L. B. (1997). Characterisation of integrons and antibiotic resistance genes in Danish multiresistant Salmonella enterica Typhimurium DT104. FEMS Microbiol. Lett. 157, 177–181. 10.1111/j.1574-6968.1997.tb12770.x [DOI] [PubMed] [Google Scholar]
- Su H. C., Ying G. G., Tao R., Zhang R. Q., Fogarty L. R., Kolpin D. W. (2011). Occurrence of antibiotic resistance and characterization of resistance genes and integrons in Enterobacteriaceae isolated from integrated fish farms in South China. J. Environ. Monit. 13, 3229–3236. 10.1039/c1em10634a [DOI] [PubMed] [Google Scholar]
- Verner-Jeffreys D. W., Welch T. J., Schwarz T., Pond M. J., Woodward M. J., Haig S. J., et al. (2009). High prevalence of multidrug-tolerant bacteria and associated antimicrobial resistance genes isolated from ornamental fish and their carriage water. PLoS ONE 4:e8388. 10.1371/journal.pone.0008388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yáñez M. A., Catalán V., Apráiz D., Figueras M. J., Martínez-Murcia A. J. (2003). Phylogenetic analysis of members of the genus Aeromonas based on gyrB gene sequences. Int. J. Syst. Evol. Microbiol. 53, 875–883. 10.1099/ijs.0.02443-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


