Abstract
The American College of Medical Genetics and Genomics (ACMG) and Association of Molecular Pathology (AMP) recently published important new guidelines aiming to improve and standardize the pathogenicity classification of genomic variants. The Clinical Sequencing Exploratory Research (CSER) consortium evaluated the use of these guidelines across nine laboratories. One identified obstacle to consistent usage of the ACMG-AMP guidelines is the lack of a definition of cosegregation as criteria for pathogenicity classification. Cosegregation data differ from many other types of pathogenicity data in being quantitative. However, the ACMG-AMP guidelines do not define quantitative criteria for use of these data. Here, such quantitative criteria, in an easily implementable form, are proposed.
Introduction
The consideration of cosegregation of a genetic variant and disease is important data when evaluating the pathogenicity of a genomic variant. Thus, cosegregation is included as part of the recently published, important American College of Medical Genetics and Genomics (ACMG) and Association of Molecular Pathology (AMP) guidelines aiming to improve and standardize the pathogenicity classification of genomic variants.1 Such guidance is a crucial step in advancing a consistent implementation of genomic medicine.
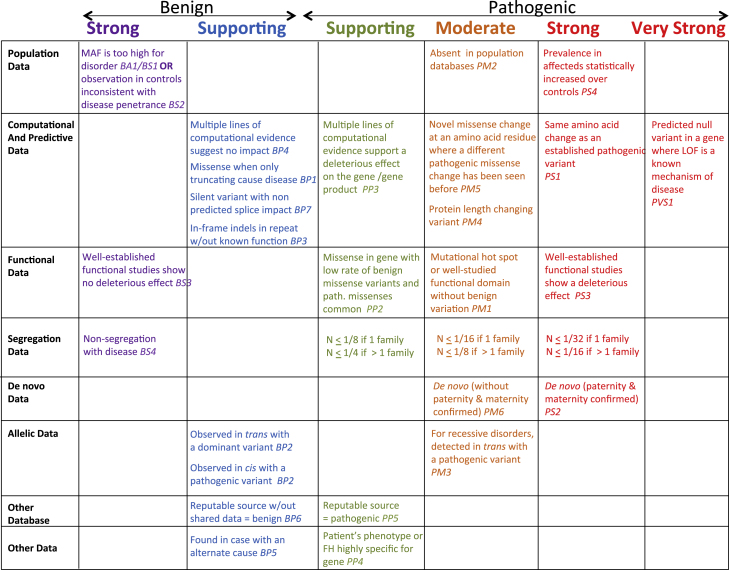
The ACMG-AMP pathogenicity classification guidelines offer a set of categories that can each be used to offer varying levels of support for classification of a variant as benign, likely benign, variant of uncertain significance, likely pathogenic, or pathogenic. These categories are summarized in the left column of Figure 1. In considering cosegregation evidence, non-segregation was considered strong evidence of a benign variant. Cosegregation with disease in multiple affected family members was considered supporting evidence of pathogenicity, and increased segregation data was considered moderate or strong evidence of pathogenicity. However, non-segregation, segregation, and increased segregation were not quantitatively defined. The AMCG-AMP guidelines cite the work of Thompson et al.2 and its extension by Bayrak-Toydemir et al.,3 but these authors do not propose specific evidence cutoffs for pathogenicity. Bayrak-Toydemir et al.3 do propose a so-called Bayes factor (BF) method that computes a likelihood ratio for quantitation of evidence, and they discuss the need for a threshold for calling a variant deleterious. In this issue of the American Journal of Human Genetics, the Clinical Sequencing Exploratory Research (CSER) consortium4 identifies the lack of quantitative guidelines for cosegregation as a source of discordance in the implementation of the ACMG-AMP guidelines across laboratories.5
Figure 1.
ACMG-AMP Pathogenicity Classification Guidelines Updated to Include Cosegregation
The summary table of evidence categories and levels from the ACMG-AMP pathogenicity classification guideline paper by Richards et al.1 is shown and is updated to include the proposed cosegregation thresholds as outlined in Table 1. The strongest evidence level supported by a given N is selected. Figure adapted with permission from Richards et al.1
The goal of this work is to propose a set of easily implemented, quantitative guidelines for the consideration of cosegregation of a variant and a disease in the classification of variant pathogenicity. These proposed guidelines support specific ACMG-AMP evidence levels. These guidelines are designed to be implementable by molecular pathologists and clinical geneticists without advanced statistical genetics training.
Material and Methods
Although the Thompson-Bayrak-Toydemir BF method can achieve more precision when penetrance is incomplete and can be estimated, it is not easily computable by most laboratory personnel or clinicians. However, if the BF method is implemented computationally in a lab, the thresholds proposed here can be used for that method as well as the simpler method outlined here.
We calculate a simple probability that the observed variant-affected status data occur by chance, rather than due to cosegregation. We assume that the proband(s) have that variant and full penetrance and that the allele is rare enough that all occurrences in the observed pedigrees are identical by descent, rather than the same variant entering the pedigree from more than one ancestor. Under a dominant model, this probability is N = , where is the number of meioses of the variant of interest that are informative for cosegregation.
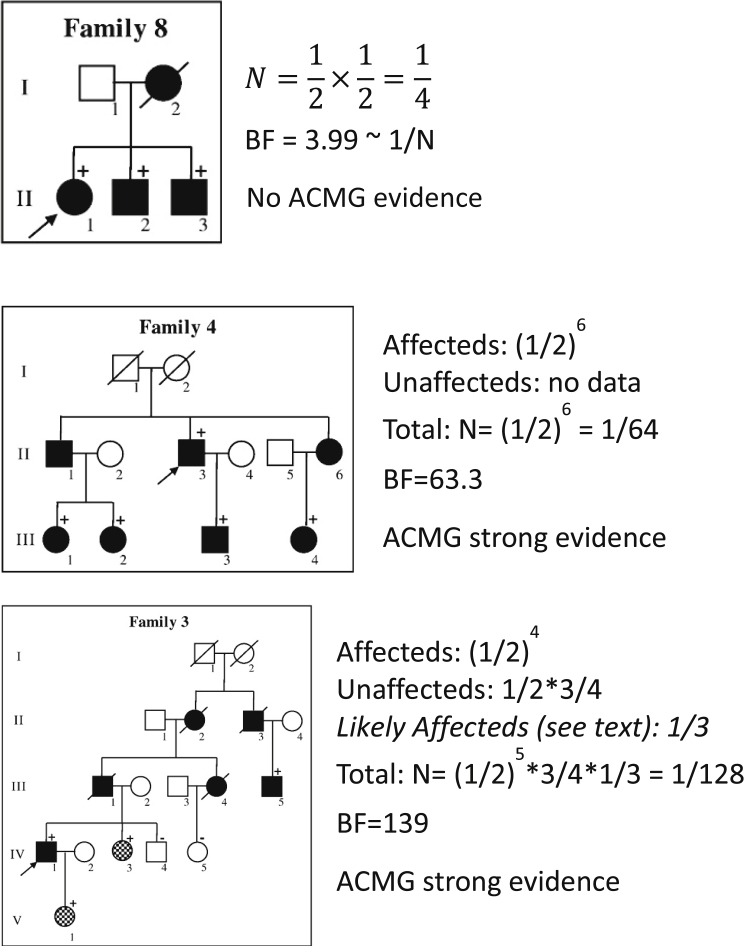
For example, if the only data are that an affected proband and one affected parent both carry the variant of interest for a dominant disorder, given that the proband carries the variant, the probability of the affected parent also carrying it is , because a single meioses informative for cosegregation is observed. It is important to note that absence of the variant of interest in an unaffected individual is cosegregation information. Informative meioses can be totaled across families. If some pedigree members are not phenotyped or genotyped, the probability of transmission of a variant can vary from 1/2. The probability for such individuals is multiplied by the probability for the other cosegregation events in the family to determine the final probability, given that these are independent events; this is demonstrated in Figure 2, family 3.
Figure 2.
Examples of Computation of Cosegregation Evidence
These examples demonstrate the computation of N proposed here and contrast it to BF as computed by Bayrak-Toydemir et al., considering pedigrees from their paper.3 Filled-in pedigree members are affected, and hashed are highly suspicious for the phenotype. A positive sign indicates that the individual harbors the variant of interest, and a negative sign indicates that the individual was genotyped and does not harbor the variant of interest. The proband is identified with an arrow. Pedigrees reprinted with permission from Bayrak-Toydemir et al.3
This method can be extended to uncertain phenotypes by weighting the probability of being affected (Figure 2, family 3). Similarly, this method can be extended to disorders with incomplete penetrance by considering the cosegregation in affected individuals only, although information is lost, or a penetrance estimate can be added to the calculation.
Our method is more intuitive to many, but under our assumptions of complete penetrance, a single causal allele, and no phenocopies, N = 1/BF as BF is defined by Thompson.2 Under these assumptions, the numerator of the BF equation is 1.0 (due to the complete penetrance) and the denominator is . Thus, N = 1/BF given simplifying assumptions.
Multiple families are jointly considered by adding the informative meioses across families to obtain m. Thus, if one observes four pedigrees for which the only data are that the affected child and affected parent share the same variant of interest in a dominant disorder, given that the proband in each family has the same variant, N = (1/2)4. Alternatively, when not all probabilities are 1/2, one multiplies the probability of the independent cosegregation events in each family together for a final probability.
Large pedigrees are less-often available for recessive disorders; however, similar calculations can be made. For example, for two affected siblings who carry the same two variants, the probability that the proband carries both variants of interest is 1 and N = 1/4; if three affected siblings share the same affected variant, N = 1/16. Information from unaffected pedigree members can also be considered. Cosegregation of X-linked disorders can be evaluated similarly, by setting the probability of the proband carrying the variant of interest as 1 and evaluating the probability of the observed cosegregation. For example, N = 1/2 when an affected male proband has either one affected brother with the variant of interest or, conversely, one unaffected brother without that variant.
Assigning ACMG-AMP Pathogenicity Evidence Level
The ACMG-AMP guideline paper2 suggests that increasing amounts of cosegregation evidence could lead to an evidence criterion of supporting evidence, moderate evidence, or strong evidence that a variant is pathogenic. Non-segregation is considered strong evidence that the variant is benign.
We propose the cutoffs summarized in Table 1 to define the supporting, moderate, and strong evidence levels. Inherent in our system is that including data for an individual very often changes the likelihood, N, by a factor of 1/2, so that these thresholds are multiples of 1/2. We propose that N be required to be smaller if all the segregation evidence comes from a single family, rather than two or more families, solely due to the concern that evidence from a single family can be due to physical linkage between the observed variant and an unobserved causal variant.
Table 1.
Proposed Cosegregation Evidence to Support Each ACMG-AMP1 Pathogenicity Evidence Level
| Single Family | >1 Family | |
|---|---|---|
| Strong evidence | ≤1/32 (≤0.03) | ≤1/16 (≤0.06) |
| Moderate evidence | ≤1/16 (≤0.06) | ≤1/8 (≤0.125) |
| Supporting evidence | ≤1/8 (≤0.125) | ≤1/4 (≤0.25) |
N, probability of observed cosegregation if not pathogenic, totaled over all families (or 1/BF). Note that the strongest evidence level supported by a given N is selected.
Given the well-accepted criteria for rejecting the null hypothesis of p ≤ 0.05 and the limitation of segregation data to factors of 1/2, N = 1/32 or 1/16 are reasonable levels for strong evidence of segregation. We propose that the 1/32 criteria be used if all cosegregation data come from one pedigree and 1/16 be used if at least two pedigrees have evidence supporting cosegregation (Table 1). We continue this dichotomy throughout all evidence levels.
Once the most restrictive threshold of N = 1/32 is met, further data need not be considered because the ACMG-AMP guidelines do not allow segregation data alone to lead to the determination of very strong evidence of pathogenicity.
Nonsegregation
In the case of a fully penetrant disorder, a single unaffected individual who has the variant of interest is evidence of nonsegregation. In our experience these negative data are rarely published, and evidence databases such as ClinVar6 do not often give nonsegregation details sufficient for a calculation. Nonsegregation can be difficult to access when penetrance is age dependent or incomplete and when the variant can enter the pedigree from more than one source. In that case, consideration of affected individuals only is conservative. However, for more common diseases the concern of phenocopies is relevant. For example, a proband with a pathogenic BRCA1 variant might not share this variant with a sister who also has breast cancer. Is this evidence that the variant is not pathogenic or does the sister have breast cancer from another cause or sporadic breast cancer? When inheritance is complex or when diseases are common and of heterogeneous etiology, the conclusion that nonsegregation supports a benign variant call should be made with caution.
Results
Examples of Calculating N and Evidence Level
Examples of computation of N for a dominant disorder are shown in Figure 2. These pedigrees were published by Bayrak-Toydemir et al.,3 and we use them here to contrast their computed BF with N computed here. We have ordered them by simplicity of computation, rather than the family number. Note that in each example, N is a reasonable approximation of 1/BF. Differences might be due to the consideration of minor-allele frequency (MAF) in each case, differences in assumed penetrance and phenocopy rates, and difference in weighting the highly suspicious for affected individuals in family 3.
In Figure 2, the parents of family 8 do not have genetic data and three affected siblings share the same variant. Because we assume that the variant of interest occurs in the proband, there are two additional meioses, to the siblings, to consider. Thus, N = (1/2)2 = 1/4. Note that it is not 1/8 because the probability that the proband has the variant is assumed to be 1. The ACMG-AMP evidence level for these data does not meet the 1/8 single family threshold for “pathogenic supporting” and is not used in variant classification.
In Figure 2, family 4, the assumption that a very rare variant does not enter the pedigree from two independent sources allows us to assume that the untyped relatives connecting those who carry the variant of interest are also carriers. The probability of these data given independent assortment of the variant and disorder and that the proband carries the variant is N = (1/2)6. These data yield an ACMG-AMP evidence level of pathogenic strong, encoded as PS.
Figure 2, family 3 adds data on unaffected individuals and challenges us to consider how to incorporate data on individuals that probably, but not definitely, have the disease. Again, we assume that the proband has the variant and that the variant is so rare that it only enters the pedigree once; thus, untyped relatives who must have passed the variant are assumed to have the variant. Considering definitely affected individuals, we observe four meioses, so that the affected individuals contribute a factor of (1/2)4 to the value of N. The probability that an individual is a noncarrier is 1 minus the probability that the individual is a carrier. So individual IV-4 contributes a factor of , and individual IV-5 contributes a factor of . Thus, for family 3, , without consideration of the individuals highly suspicious for disease. These individuals can be incorporated into the evidence with the BF approach. If the highly suspicious individuals are assumed to be affected, the segregation probability of 1/4 can be applied; it could also be reasonable to alter this based on the confidence that they are affected (e.g., use 1/3 or 1/2 instead of 1/4 to account for the reduced confidence in their diagnoses). Regardless of the handling of the highly suspicious individuals, these data yield an ACMG-AMP evidence level of pathogenic strong.
Discussion
Classification of the pathogenicity of variants will be an ongoing and important task. The repository ClinVar6 allows public reporting of pathogenicity classification, including the supporting evidence, of variant-disease pairs. However, inconsistency in the criteria different labs use to assess pathogenicity was identified as an obstacle to genomic medicine. The ACMG-AMP guidelines are an important effort to bring consistency to variant classification. Recent work from the CSER consortium identifies sources of variation in the implementation of ACMG-AMP guidelines and suggests clarifications and areas for improved guidance.5 One area of concern is a lack of standard criteria for cosegregation evidence to be used to support evidence levels. Given that cosegregation data are by definition quantifiable, we seek to provide such guidance.
As noted above, our use of the probability of the cosegregation data, given independent segregation of the variant and the disease, under simplifying assumptions, is equal to 1/BF, considering BF defined by Thompson et al.2 The BF can be used to incorporate incomplete penetrance, age-of-onset functions, and MAF and thus the possibility that the variant is not identical by descent in all pedigree members and uncertainty in diagnosis. Although some of these data will be estimated, they can add precision to the cosegregation evidence, and the BF can be inverted to evaluate the ACMG-AMP evidence level supported. The limitation of the BF computation is that it requires training and tools not required by the method suggested here. We note that, for disorders with incomplete penetrance, N can be computed considering only the affected individuals. However, this will lose information available in a calculation where penetrance-by-age functions are included.
We have made an effort to align our evidence levels with other data that ACMG-AMP identify as usable to support the supporting moderate or strong evidence of pathogenicity levels (Figure 1).1 A case-control study associating a variant with the phenotype is considered strong evidence. Given that this would customarily require a p value of 0.05 or less, our suggested criteria for segregation would yield similar evidence. A de novo “without maternity and paternity confirmed” would constitute moderate evidence under ACMG-AMP. The ACMG-AMP guidelines define supporting evidence of pathogenicity as “cosegregation with disease in multiple family members.” 1 This is aligned with our criterion of N = 1/8 in a single family.
The authors of the ACMG guidelines raise the concern that cosegregation of a variant with disease in a family might be secondary to physical linkage between that observed variant and the actual pathogenic variant.1 This is often a concern in the identification of new disease—gene associations, where the gene has not previously been known to be associated with the disorder. As linkage regions are often very large in a single family, due to lack of recombination events, the concern over false positives due to physical linkage of the observed variant with the true pathogenic variant, which might even be in a neighboring gene, are reasonable. However, in the case of the full-sequence data of a gene known to be associated with the disorder, the likelihood of failing to identify the pathogenic change in favor of a marker in physical linkage is substantially reduced versus that of a partially sequenced linkage region with multiple possible associated loci. Further, the ACMG-AMP authors allow case-control association data as strong support of pathogenicity, and those results can also occur for a non-pathogenic variant in linkage disequilibrium with an unobserved pathogenic variant. Nonetheless, we do penalize the case where the variant only has disease cosegregation data from a single family by a somewhat arbitrary factor of 2.
Cosegregation cannot be considered very strong evidence of pathogenicity under the ACMG guidelines.1 Under the ACMG-AMP guidelines, the only evidence considered very strong evidence of pathogenicity is a nonsense mutation in a gene where nonsense mutations are a known cause of the disease. Even this is not considered strong enough stand-alone evidence to call the variant as pathogenic. Classifying a variant as pathogenic can occur when one very strong evidence of pathogenicity is combined with at least one strong support evidence level or two moderates, or a combination of lesser evidence levels. For this reason, it seems that the authors of the ACMG-AMP statement might reconsider whether there is a level of cosegregation which constitutes very strong evidence.
We propose here easily quantified criteria for cosegregation to support evidence levels defined by the ACMG-AMP variant classification guidelines1 in an effort to improve standardization of variant classification among clinical and other genomic laboratories. This is accompanied by a simplified calculation of N and also the ability to use these guidelines with fewer assumptions by calculation of a BF2, 3 and comparing 1/BF to the cutoffs proposed.
Acknowledgments
This work was supported by NIH grants U01 HG006507, U01 HG007307, U01HG008657, and R01 HG008359. The authors thank the journals Genetics in Medicine and Experimental and Molecular Pathology for permission to reprint and adapt the figures, Dr. Heidi L. Rehm for providing the original figure adapted for Figure 1, and Dr. Adam S. Gordon for helpful comments.
Published: May 26, 2016
References
- 1.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thompson D., Easton D.F., Goldgar D.E. A full-likelihood method for the evaluation of causality of sequence variants from family data. Am. J. Hum. Genet. 2003;73:652–655. doi: 10.1086/378100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bayrak-Toydemir P., McDonald J., Mao R., Phansalkar A., Gedge F., Robles J., Goldgar D., Lyon E. Likelihood ratios to assess genetic evidence for clinical significance of uncertain variants: hereditary hemorrhagic telangiectasia as a model. Exp. Mol. Pathol. 2008;85:45–49. doi: 10.1016/j.yexmp.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 4.Green R.C., Goddard K.A.B., Jarvik G.P., Amendola L.M., Appelbaum P.S., Berg J.S., Bernhardt B.A., Biesecker L.G., Biswas S., Blout C.L., Clinical Sequencing Exploratory Research Consortium Accelerating evidence-based practice of genomic medicine. Am. J. Hum. Genet. 2016;98:1051–1066. doi: 10.1016/j.ajhg.2016.04.011. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amendola L.M., Jarvik G.P., Leo M.C., McLaughlin H.M., Akkari Y., Amaral M.D., Berg J.S., Biswas S., Bowling K.M., Conlin L.K. Performance of ACMG/AMP variant interpretation guidelines among nine laboratories in the Clinical Sequencing Exploratory Research consortium. Am. J. Hum. Genet. 2016;98:1067–1076. doi: 10.1016/j.ajhg.2016.03.024. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Landrum M.J., Lee J.M., Benson M., Brown G., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Hoover J. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44(D1):D862–D868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]