Abstract
Hearing impairment is the most common sensory deficit. It is frequently caused by the expression of an allele carrying a single dominant missense mutation. Herein, we show that a single intracochlear injection of an artificial microRNA carried in a viral vector can slow progression of hearing loss for up to 35 weeks in the Beethoven mouse, a murine model of non-syndromic human deafness caused by a dominant gain-of-function mutation in Tmc1 (transmembrane channel-like 1). This outcome is noteworthy because it demonstrates the feasibility of RNA-interference-mediated suppression of an endogenous deafness-causing allele to slow progression of hearing loss. Given that most autosomal-dominant non-syndromic hearing loss in humans is caused by this mechanism of action, microRNA-based therapeutics might be broadly applicable as a therapy for this type of deafness.
Introduction
Hearing impairment is the most common sensory deficit. It affects more than 360 million people worldwide and broadly impacts their quality of life (see Web Resources).1 Not only does it limit the ability to interpret speech sounds (leading to delayed language acquisition in infancy), but in adulthood hearing impairment can lead to economic disadvantage, social isolation, and stigmatization. Current treatment options focus on hearing aids and cochlear implants to bypass the biologic deficit by amplifying sounds (hearing aids) or by encoding them as electrical impulses that are transmitted to the auditory nerve through an implanted electrode array (cochlear implants). Although these two habilitation options are effective, they do not restore “normal” hearing. As life expectancy improves and populations grow, the hearing-impaired population will increase, making the development of therapeutics to restore or prevent hearing loss important to enhancing quality of life.2
Over the past decade, we have focused on RNA interference (RNAi) as a means of selectively suppressing mutant alleles in animal models of deafness.3, 4 Herein, we report on the use of an artificial microRNA (miRNA)-based approach to rescuing the progressive hearing-loss phenotype in the Beethoven (Bth) mutant mouse, a mouse model of human autosomal-dominant non-syndromic hearing loss (DFNA36 [MIM: 606705]). This mouse carries the semi-dominant Tmc1 c.1235T>A (p.Met412Lys) allele.5 The encoded protein, TMC1, is a transmembrane protein with six hydrophobic transmembrane domains (Figure 1A).7 TMC1 interacts with the tip-link proteins protocadherin 15 and cadherin 23 and, together with TMC2, is assumed to be a component of the mechanoelectrical transduction complex.8, 9 Five mutations have been reported in the human homolog, TMC1 (MIM: 606706], to cause autosomal-dominant non-syndromic hearing loss at the DFNA36 locus.10, 11, 12, 13, 14, 15 One TMC1 mutation, c.1253T>A (p.Met418Lys) (GenBank: NM_138691, NCBI build 36.3), is orthologous to the murine Bth mutation (Tmc1 c.1235T>A [p.Met412Lys]) and segregates in a large, 222 member Chinese family who suffers from progressive post-lingual sensorineural hearing loss (Figure 1B). In this kindred, age of onset varies from 5 to 25 years, potentially providing a window for therapeutic intervention to prevent the otherwise inevitable deterioration of hearing thresholds, which by 50 years of age are in the severe-to-profound range across all frequencies.15 This natural progression of hearing loss closely mimics the phenotype of the Bth-heterozygous mouse (Tmc1Bth/+).
Figure 1.
miRNA Design, Screening, and Viral-Vector Selection
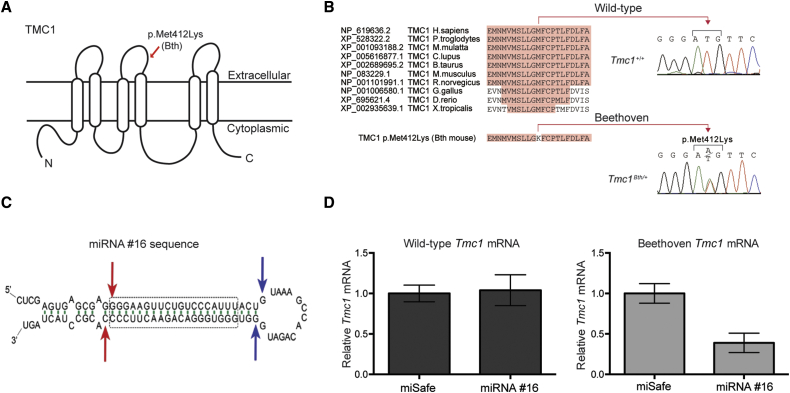
(A) Cartoon depiction of a possible configuration of TMC1 highlights the position of the Bth mutation.
(B) Multiple-sequence alignment shows conservation of Met412 in vertebrates and the Met412Lys change in the Tmc1Bth/+ mouse.
(C) siRNA sequence #16 embedded in an artificial miRNA scaffold. Of all miRNAs tested, #16 had the most specific and selective suppression of the mutant Tmc1 c.1235T>A allele. Blue and red arrows depict predicted Drosha and Dicer cleavage sites, respectively; the dashed box shows the core #16 sequence targeting the mutant Tmc1 variant.
(D) Real-time qPCR analysis of total RNA isolated from COS-7 cells cotransfected with constructs expressing both miRNA #16 and miSafe (a sequence specifically selected for its validated low off-targeting potential6) and either wild-type Tmc1 or mutant Tmc1 c.1235T>A. Relative mRNA expression levels were calculated with the ΔΔCt algorithm. Error bars represent the SD of three biological and nine technical replicates.
Herein, we report on the use of a single intracochlear injection of an artificial miRNA carried in an adeno-associated virus (AAV) vector to slow progression of hearing loss in the Tmc1Bth/+ mutant mouse. In some animals so treated and otherwise expected to be profoundly deaf by 35 weeks, hearing thresholds were approximately 40 dB better than those of untreated Tmc1Bth/+ control mice.
Material and Methods
Ethics Approval
The University of Iowa Institutional Biosafety Committee (rDNA Committee, rDNA approval notice 100024) and the University of Iowa Institutional Animal Care and Use Committee (protocol 0608169) approved all relevant procedures.
Mice
All procedures met NIH guidelines for the care and use of laboratory animals and were approved by the Institutional Animal Care and Use Committee at the University of Iowa. Mice were housed in a temperature-controlled environment on a 12 hr light-dark cycle. Food and water were provided ad libitum. Isogenic Bth-heterozygous mice (Tmc1Bth/+) maintained on a C3HeB/FeJ (C3H) background were obtained as a gift from Dr. Karen Steel.5 Inbred wild-type C3H mice were obtained from the Jackson Laboratory. Crossbred homozygous Tmc1Bth/Bth mice were caged with wild-type C3H mice for the generation of heterozygous Tmc1Bth/+ animals. Genotyping was done on DNA from tail-clip biopsies extracted by a phenol-chloroform procedure and amplified with forward (5′-CTAATCATACCAAGGAAACATATGGAC-3′) and reverse (5′-TAGACTCACCTTGTTGTTAATCTCATC-3′) primers in a 25 μl volume containing 150 ng DNA, 0.2 nM of each primer, and BioLase DNA polymerase (Bioline) for the generation of a 376 bp amplification product in Tmc1Bth/+ mice. Amplification conditions included an initial 2 min denaturation at 95°C followed by 35 step cycles of 30 s at 95°C, 30 s at 57°C, and 45 s at 72°C and a final elongation of 10 min at 72°C. PCR products were purified and sequenced on an automated sequencer (ABI PRISM model 3730XL, Applied Biosystems). For mechanotransduction experiments, two genotypes of Tmc-mutant mice were used (Tmc1+/−;Tmc2−/− and Tmc1Bth/-;Tmc2−/−) and maintained on a C57BL/6J background as previously described.16
RNAi Oligonucleotide Constructs
The RNAi oligonucleotides and vector plasmids were designed by the Viral Vector Core at the University of Iowa. Fifteen small interfering RNA (siRNA) sequences, either hand designed or selected with siSPOTR (siRNA Seed Probability of Off-Target Reduction) software, were chosen for walking through the target sequence one base at a time17 (Table S1). Forward and reverse oligonucleotide primers, which included an overlap in the common loop of the miRNA, were purchased from Integrated DNA Technologies and used for creating artificial miRNAs. The artificial-miRNA expression cassette was generated by PCR, purified, and digested as previously described.17, 18 Each artificial-miRNA expression cassette was cloned into a vector plasmid flanked by inverted terminal repeat sequences and containing the mouse U6 promoter (mU6) followed by a multiple-cloning site, cytomegalovirus (CMV)-promoter-driven eGFP, and a Pol II termination signal. miSafe, a sequence specifically selected for its validated low off-targeting potential, was used as a control.6
Virus Production
AAV vectors were prepared by the Viral Vector Core at the University of Iowa by a standard triple-transfection method in 293FT cells and subsequent purification in a cesium chloride gradient as previously described.19 For vector selection, single-stranded recombinant AAV serotypes (rAAV2/1 and rAAV2/9) that carried CMV-driven eGFP (rAAV2/1eGFP and rAAV2/9eGFP) were tested. Viral titers used in trans-round-window-membrane (RWM) injections were rAAV2/1 at 3.09 × 1013 vg/ml and rAAV2/9 at 1.59 × 1013 vg/ml. The selected therapeutic was single-stranded recombinant AAV serotype 2/9 (rAAV2/9) carrying a dual transgene cassette of mouse U6-driven miRNA #16, targeting the p.Met412Lys-encoding allele and downstream CMV-driven eGFP (rAAV2/9miTmc1k412.16eGFP [miTmc]). The control vector was mU6-driven miSafe and downstream CMV-driven eGFP (rAAV2/9miSafeGFP [miSafe]).6 Vector titers were determined by real-time PCR and were miTmc at 1.69 × 1012 and miSafe at 1.39 × 1013 DNase-resistant particles per mililiter. Virus aliquots were stored at −80°C before use.
Viral Inoculation
All mice were operated on at postnatal days 0–2 (P0–P2) under hypothermic anesthesia, for which animals were placed in a container with crushed ice for 3–5 min. trans-RWM injections were performed under an operating microscope. First, a post-auricular incision exposed the cochlea bulla, which was opened with fine forceps. Anatomic landmarks included the RWM and stapedial artery, which were identified before injections (Figure S1). Then, for the trans-RWM injection, either miTmc or miSafe mixed in a 10:1 ratio with 2.5% fast green dye was loaded into a borosilicate glass pipette (1.5 mm outer diameter [OD] × 0.86 mm inner diameter [ID], Harvard Apparatus) pulled with a Sutter P-97 micropipette puller to a final OD of ∼20 μm and affixed to an automated injection system pressured by compressed gas (Harvard Apparatus). Pipettes were manually controlled with a micropipette manipulator. A total volume of 0.5 μl was injected into the left ear of each mouse. After all procedures, mice were placed on a heating pad for recovery and rubbed with bedding before being returned to the mother. Recovery was closely monitored daily for at least 5 days post-operatively.
Auditory Testing
The hearing thresholds were recorded in the following groups: (1) wild-type littermates (C3HeB/FeJ inbred mice: n = 4 from 4 to 13 weeks; n = 5 from 26 to 35 weeks), (2) Tmc1Bth/+ non-injected mice (n = 11), (3) Tmc1Bth/++miSafe mice (n = 13), and (4) rescued Tmc1Bth/++miTmc mice (n = 10).
Auditory testing was conducted in a sound-attenuating room. Stimulus presentation and recording were controlled with MATLAB software (MathWorks) running on a PC connected to a 24-bit external sound card (Motu UltraLite mk3) as previously described.20 Stimuli were delivered via an ER-10B+ probe microphone (Etymotic Research) connected to two MF1 Multi-Field Magnetic Speakers (Tucker-Davis Technologies).
Calibration was performed with a 1.125 in microphone placed in a 0.028 cm3 cavity (length = 0.4 cm; diameter = 0.3 cm), as shown in Figure S2A. This method of calibration differs from that of many previous studies that used a standard 2 cm3 coupler (Figure S2B). For a given voltage drive to the loudspeaker, the sound pressure measured in the smaller cavity will be much larger than that measured in a 2 cm3 coupler. We based our calibration on the smaller cavity under the assumption that the actual pressure experienced at the mouse tympanic membrane is most accurately represented with a cavity that approximates the size of the mouse ear canal rather than the standard 2 cm3 coupler, which approximates a human ear canal. Measurements in our laboratory showed a 27 dB difference between pressures measured in the smaller (0.028 cm3) and larger (2 cm3) couplers. Therefore, when an auditory-brainstem-response (ABR) threshold is found, the reported sound pressure delivered by the loudspeaker will depend on the calibration cavity used. For identical thresholds, the values reported for the smaller coupler will be 27 dB higher (apparently worse) than the values reported for the larger coupler. Although the small-coupler values might reflect sound pressures delivered to the mouse eardrum more accurately, for ease of direct comparison to previous studies using C3HeB/FeJ mice, we report ABR thresholds in terms of equivalent 2 cm3 coupler values.21
ABR
Mice were anesthetized with intraperitoneal ketamine and xylazine at 100 and 6 mg per kg of body weight, respectively. ABR thresholds were obtained for both clicks and tone bursts. Clicks were square pulses 100 ms in duration. Tone bursts were 3 ms in length, including 1 ms onset and offset ramps (raised cosine shape) centered at 8, 16, and 32 kHz. The sweep length for each stimulus (both click and tone burst) was 29 ms. Between 500 and 1,000 sweeps were averaged at each stimulus level for obtaining the ABR waveforms. All recordings were conducted from both ears of all animals in a sound-attenuating room.
Distortion-Product Otoacoustic Emissions
Distortion-product otoacoustic emissions (DPOAEs) at 2f1 − f2 were measured with f2 frequencies from 4 to 32 kHz in half-octave steps (f2/f1 = 1.22). The levels of the primaries were fixed at 65 and 55 dB sound-pressure level (SPL) for f1 and f2, respectively. For each f2 frequency, ten 1 s stimulus presentations were averaged. DPOAE amplitudes and associated noise floors were calculated from fast-Fourier-transform (FFT) analysis of the averaged waveforms.
Immunohistochemistry and Histology
All injected and non-injected cochleae were harvested after animals were sacrificed by CO2 inhalation. Temporal bones were removed, perfused with 4% paraformaldehyde, and incubated for 1 hr. Cochleae were then rinsed in PBS and stored at 4°C in preparation for dissection and immunohistochemistry. Specimens were visualized with a dissecting microscope and dissected as previously described.22 Specimens were infiltrated with 0.3% Triton X-100 and blocked with 5% normal goat serum before tissues were incubated first in rabbit polyclonal Myosin-VIIA antibody (Proteus Biosciences) or mouse monoclonal antibody to GFP (Millipore) diluted at 1:1,000 in PBS for 1 hr and then in a 1:1,000 dilution of the secondary antibody, fluorescence-labeled anti-rabbit IgG Alexa Fluor 568 or goat anti-mouse IgG Alexa Flour 488 in a 1:2,000 dilution (Invitrogen, Molecular Probes) for 30 min. Filamentous actin was labeled by a 30 min incubation of phalloidin conjugated to Alexa Fluor 488 (Invitrogen, Molecular Probes). Specimens were mounted in ProLong Diamond mounting medium (Life Technologies).
z stack images of whole mounts were collected at 10×–40× on a Leica SP8 confocal microscope (Leica Microsystems). Maximum-intensity projections of z stacks were generated for each field of view, and composite images showing the whole cochlea were constructed in Adobe Photoshop CS6 in order to meet equal conditions and show the complete turns of the cochlea at high resolution. Distance from the apex was measured in 0.25 or 0.40 mm segments with imageJ (NIH Image). For the viral-vector-screening study, inner hair cells (IHCs) and outer hair cells (OHCs) with positive eGFP and overlapping MYO7A were counted with ImageJ Cell Counter. The total number of hair cells and GFP-positive hair cells were summed and converted to a percentage. For the miRNA study, IHCs and OHCs with positive MYO7A were counted with ImageJ Cell Counter; any segments that contained dissection-related damage were omitted from the analysis. IHC and OHC survival was quantified with 20×–40× images of whole-mount cochleae compiled into cochleograms at 35 weeks as previously described.23
Molecular Studies for In Vitro and In Vivo Expression Analyses
Previously reported cDNAs p.AcGFPmTmc1ex1WT and p.GEMT-easyTmc1ex1Bth were provided by Dr. Andrew Griffith. p.GEMT-easyTmc1ex1Bth was PCR amplified with forward (5′-GTCGACAGGATGCCACCCAAAAAAG-3′) and reverse (5′-ATGGATCCACTGGCCACCAGCAGC-3′) primers containing restriction sites SalI and BamH1. cDNA inserts were purified by 1% agarose gel electrophoresis and QIAquick Gel Extraction (QIAGEN) and subcloned and ligated into SalI- and BamH-digested p.AcGFP1-N2 vector (catalog no. 632483, Takara Clontech). Successful cloning was verified with Sanger sequencing (ABI PRISM model 3730XL, Applied Biosystems).
For in vitro miRNA screening, the aforementioned miRNA expression plasmids were used. COS-7 cells, which do not contain native TMC1, were used in this study and grown in DMEM (Invitrogen, Thermo Fisher Scientific) with 10% fetal bovine serum at 37°C with 5% CO2.7 Prior to transfection, COS-7 cells were transferred and grown on a 24-well plate for 1 day. The transfection mix was made with Lipofectamine 2000 (Invitrogen, Thermo Fisher Scientific) according to the manufacturer’s protocol; miRNA expression plasmids were cotransfected with p.AcGFPmTmc1ex1Bth. RNA was extracted from cells with the use of TRIzol (Invitrogen, Thermo Fisher Scientific). Expression levels were assessed in triplicate by real-time PCR (StepOnePlus, Applied Biosystems) with intron-spanning Tmc1 forward (5′-GTTCGCCCAGCAAGATCCTGA-3′) and reverse (5′-GGATGGTAATCTTCCAGTTCAGCA-3′) primer sets and One Step SYBR PrimeScript RT-PCR Kit II (Clontech), and results were normalized to β-actin forward (5′-TGAGCGCAAGTACTCTGTGTGGAT-3′) and reverse (5′-ACTCATCGTACTCCTGCTTGCTGA-3′) primer sets. Controls included U6 miSafe and empty vector. The miRNA expression plasmids with greater than 50% suppression were selected and next cotransfected with p.AcGFPmTmc1ex1WT for the assessment of non-selective suppression of the wild-type allele.
For in vivo expression analysis, the left ear of Tmc1Bth/+ P0–P2 mice was injected with rAAV2/9 carrying miTmc; the right ear served as a non-injected control. Cochleae were harvested at P28. eGFP localization in hair cells was confirmed in the apical turns, which was dissected with a fluorescence dissecting micoscope. Cochlear tissue was incubated for 5 min in a collagenase solution at room temperature as previously described.24 To isolate populations of eGFP-positive hair cells, we isolated hair cells with a pulled glass micropipette as previously described24 and added a wash step prior to final collection (Movie S1). We changed glass pipettes after each isolation to minimize the introduction of contaminating RNAs into the lysis buffer from either the isolation buffer or previously isolated cells. GFP-positive hair cells were isolated and placed into tubes containing lysis buffer as previously described.25 Smart-seq2 protocol steps 1–13 were followed directly for sample processing from lysis through reverse transcription; however, the number of cycles in the PCR pre-amplification (step 14) was reduced to 7 for the purpose of minimizing amplification bias. The resulting product was used as the input for SYBR Green-based qRT-PCR. Allele-specific Bth (5′-AAACAGGGTGGGACAGAAC+T-3′) and wild-type Tmc1 (5′-AAACAGGGTGGGACAGAAC+A-3′) reverse primers were designed with Locked Nucleic Acid-modified nucleotides on the 3′ end, denoted as +T or +A. These reverse primers were paired with a single forward primer (5′-GCACAGGTGGAGGAGAACAT-3′) for the generation of a 219 bp PCR product. Allele specificity was confirmed with plasmid DNA containing Bth or Tmc1 sequence and optimized at an annealing temperature of 62.5°C in Power SYBR Green Master Mix (Applied Biosystems). qRT-PCR was carried out with a StepOnePlus Real-Time PCR System. Each sample was amplified with three primer sets in triplicate for a total of nine reactions per sample. Primer sets included primers specific to the wild-type Tmc1 allele, primers specific to the Bth Tmc1 allele, and β-actin primers. Melt curves and gel electrophoresis confirmed that PCR products were primer specific. Results were normalized to β-actin with the ΔΔCt algorithm.
Mechanotransduction
Inner ears of Tmc1Bth/−;Tmc2−/− mice and littermate control Tmc1+/−;Tmc2−/− mice were injected with 0.5 μl miTmc at P0–P1 by a trans-RWM approach. Cochleae were harvested at P8–P10, and the organs of Corti were bathed in standard artificial perilymph containing 137 mM NaCl, 0.7 mM NaH2PO4, 5.8 mM KCl, 1.3 mM CaCl2, 0.9 mM MgCl2, 10 mM HEPES, and 5.6 mM d-glucose. Vitamins (1:50) and amino acids (1:100) were added to the solution from concentrates (Invitrogen, Thermo Fisher Scientific), and NaOH was used to adjust the final pH to 7.40 (310 mOsm/kg). Recording pipettes (3–5 MΩ) were pulled from R6 capillary glass (King Precision Glass) and filled with intracellular solution containing 135 mM CsCl, 5 mM HEPES, 5 mM EGTA, 2.5 mM MgCl2, 2.5 mM Na2-ATP, and 0.1 mM CaCl2; CsOH was used to adjust the final pH to 7.40 (285 mOsm/kg). Whole-cell, tight-seal, voltage-clamp recordings were done at −84 mV at room temperature (22°C–24°C) with an Axopatch 200B amplifier (Molecular Devices). Hair bundles were deflected with stiff glass probes fabricated from capillary glass with a fire polisher (World Precision Instruments) for creating a rounded probe tip of ∼3–5 μm in diameter. Probes were mounted on a PICMA Chip piezo actuator (Physik Instrumente) and driven by a 400 mA ENV400 amplifier (Piezosystem). Sensory-transduction currents were recorded without knowledge of GFP expression from control and miTmc-treated hair cells. The data were filtered at 10 kHz with a low-pass Bessel filter and digitized at ≥20 kHz with a 16-bit acquisition board (Digidata 1440A, Molecular Devices) and pClamp 10 software (Molecular Devices). Data were stored for offline analysis with OriginPro 8 (OriginLab).
Statistical Analysis
Statistical analysis of ABR, DPOAE, and cell-counting data was completed in R with two-sample t tests for samples of equal variance. Samples with unequal variance were compared with Welch two-sample t tests. Sample variance was determined with F tests comparing two variances. All comparisons were made between the Tmc1Bth/++miTmc and Tmc1Bth/++miSafe cohorts.
Results
RNAi Suppresses the Tmc1 c.1235T>A Allele
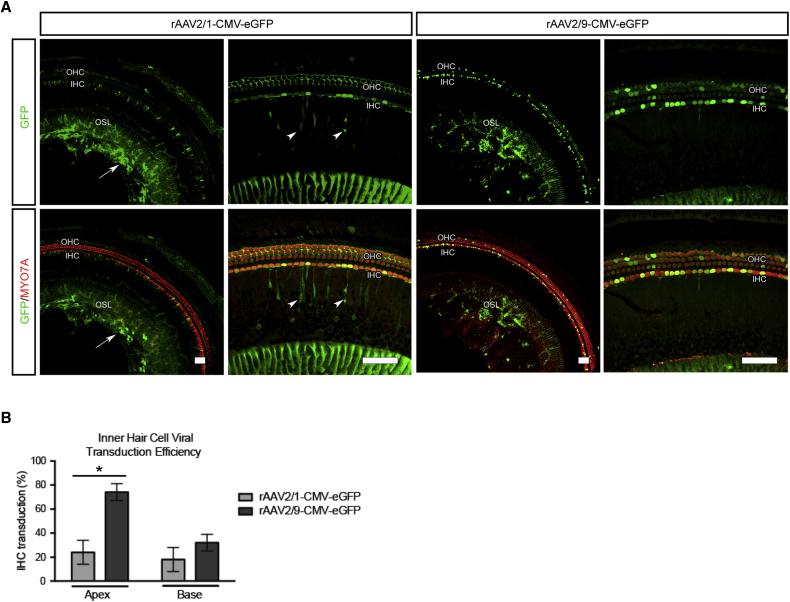
We sought to prevent progression of hearing loss in Tmc1Bth/+ mice by using an artificial miRNA to suppress the expression of the mutant Tmc1 c.1235T>A allele. Of 15 miRNA constructs evaluated (Table S1), one miRNA, #16, was chosen for further study because it robustly and selectively suppressed the Tmc1 c.1235A allele without affecting the wild-type allele (Figures 1C and 1D). We chose to use rAAV2/9 as the delivery vector after we compared the transduction efficiency of rAAV2/1-eGFP and rAAV2/9-eGFP injection into the cochleae of wild-type mice at P0–P2 by a trans-RWM approach. At 2 weeks, rAAV2/1-eGFP localized to a variety of cell types, including robust localization in supporting cells; rAAV2/9-eGFP, in comparison, predominantly localized to IHCs, which were transduced with 74% efficiency in the apical cochlear turn (Figures 2A and 2B). The corresponding transduction of OHCs was 7%. Because IHC dysfunction is primarily responsible for the deafness phenotype in Tmc1Bth/+ animals and OHC loss is subtle in the apical cochlear turns,5, 26 we considered the observed levels of hair cell transduction acceptable for this study.
Figure 2.
Evaluation of the Efficiency and Specificity of AAV Transduction
(A) Comparative transduction efficiency and specificity of rAAV2/1 and rAAV2/9 carrying a CMV-eGFP expression construct delivered to wild-type murine cochlea at P0–P2 via a trans-RWM approach. rAAV2/1 transduces IHCs, OHCs, spiral ganglion cells (arrow), and inner sulcus cells (arrowhead) in the osseous spiral lamina (OSL). Overlapping MYO7A and eGFP localization represents positive hair cell transduction. Note that compared to rAAV2/1, rAAV2/9 shows specific IHC transduction. Scale bars represent 50 μm.
(B) The efficiency of viral transuction in IHCs was assessed in 400 μm segments in the apical and basal turns (rAAV2/1 [gray] and rAAV2/9 [black]). Note the high (∼74%) rAAV2/9 IHC transduction in the apical turn. Error bars represent the SD (n = 3). ∗p < 0.05, ∗∗p < 0.005.
The miRNA #16 expression cassette was cloned into rAAV2/9 as a dual transgene cassette of mouse U6-driven miRNA targeting the p.Met412Lys-encoding transcript coupled upstream of CMV-driven eGFP (rAAV2/9miTmck412.16eGFP, miTmc). A control vector was designed to carry a U6-driven specific sequence selected for its validated low off-targeting potential and CMV-driven eGFP (miSafe)6 (Figure 3A). trans-RWM inoculation surgery, whereby 0.5 μl of miTmc or miSafe was injected into only the left cochlea, was performed on Tmc1Bth/+ mice at P0–P2 (Figure S1).
Figure 3.
rAAV2/9-Mediated miTmc Suppresses Expression of Tmc1 c.1235T>A In Vivo
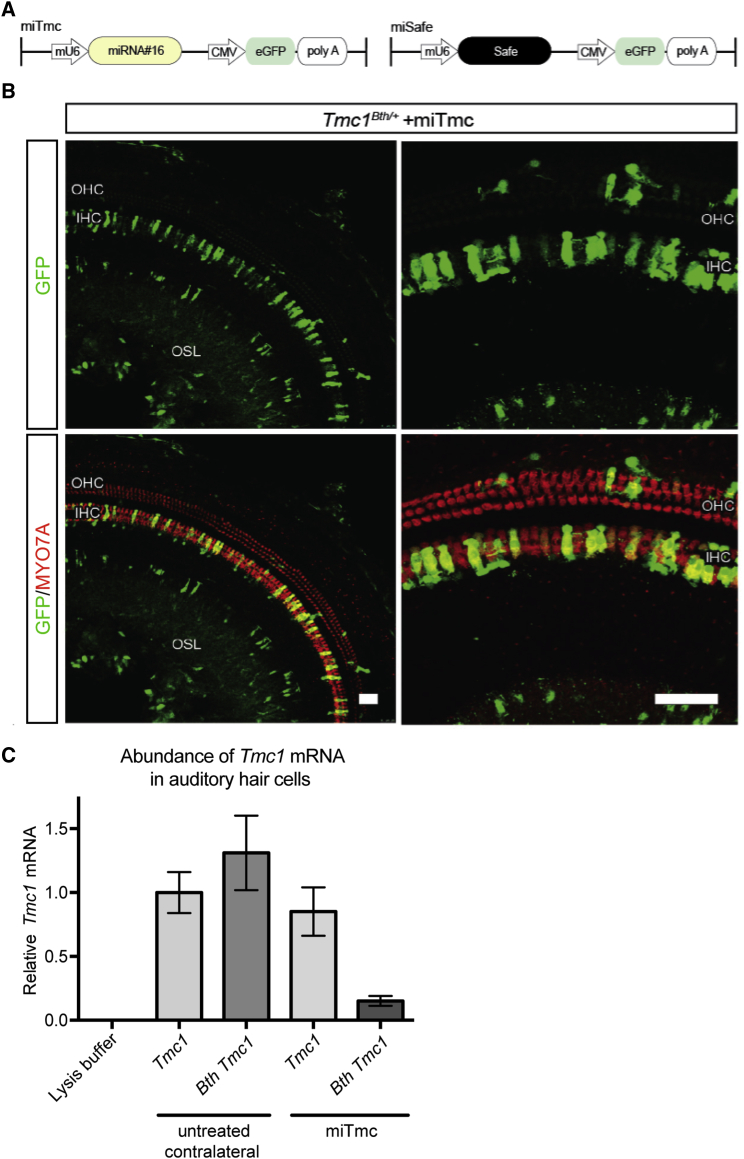
(A) Dual-promoter viral insert in which the U6 promoter drives miTmc miRNA expression and the CMV promoter drives eGFP expression. miSafe was specifically selected for its validated low off-targeting potential.24
(B) Two weeks after trans-RWM injection of miTmc at P0–P2, native eGFP localization was visible in transduced IHCs and OHCs in the organ of Corti.
(C) Expression of wild-type Tmc1 and Bth Tmc1 mRNA was measured by real-time qPCR using allele-specific primers. Allele-specific qPCR amplification was carried out on groups of individually isolated auditory hair cells (Movie S1). All samples were normalized to β-actin. Expression of wild-type Tmc1 mRNA measured in the untreated contralateral sample was set at a value of 1. mRNA abundance was calculated in relation to that of this untreated contralateral sample with wild-type Tmc1. Abundance of both wild-type Tmc1 and Bth Tmc1 were measured in samples containing 12 cells collected from either miTmc-treated ears or untreated contralateral ears from five 4-week-old Tmc1Bth/+ mice. Cells collected from untreated contralateral ears were GFP negative, whereas cells collected from miTmc-injected ears were GFP positive. mRNA abundance was calculated by the ΔΔCt method. The range indicated by the error bars represents the SD of ΔΔCt on the basis of the fold-difference calculation 2−ΔΔCt, where ΔΔCt + S and ΔΔCt − S.
We verified robust IHC eGFP localization in cochleae harvested from Tmc1Bth/+ P0–P2 miTmc-injected mice 2 weeks after surgery, consistent with the transduction pattern observed in ears injected with rAAV2/9-eGFP (Figure 3B; Figure S3). To assess in vivo allele-specific suppression in individual hair cells, 4 weeks after surgery we isolated GFP-positive hair cells from miTmc-injected cochleae and control hair cells from contralateral non-injected cochleae and completed real-time qPCR by using primers specific to the wild-type and Bth Tmc1 alleles (Movie S1). In non-injected ears, the relative expression of the Bth Tmc1 c.1235T>A allele and the wild-type allele was comparable (Figure 3C). In biological replicates from miTmc-injected ears, expression of the Bth Tmc1 c.1235T>A allele was suppressed by more than 88% in comparison to levels of Bth mRNA detected in the sample from the untreated contralateral ear (Figures 3C).
Mechanotransduction
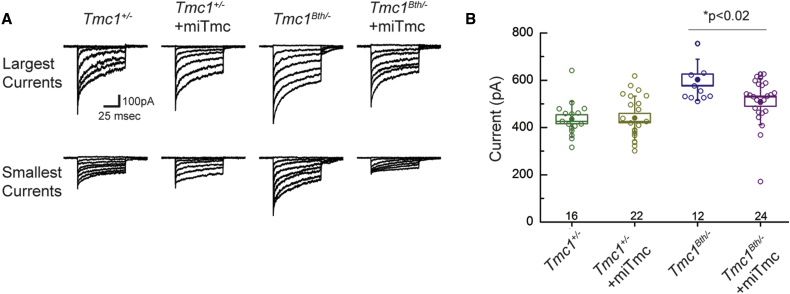
TMC1 and its closely related ortholog, TMC2, are assumed to be components of the mechanotransduction channel. In an earlier report, the mutant TMC1 p.Met412Lys variant was found to reduce calcium permeability and single-channel currents in IHCs.16 To test the effect of RNAi and allele-specific suppression, we therefore measured transduction currents at P8–P10 after exposing IHCs to miTmc. To ensure that observed currents were only due to expression of Tmc1, we recorded from mice on a Tmc2−/− background and compared Tmc1Bth/− mice (carrying only one mutant Tmc1 allele) to Tmc1+/− mice (carrying only one wild-type Tmc1 allele) before and after miTmc exposure. Tmc1Bth/− mice had larger currents than Tmc1+/− mice, consistent with data reported by Pan and colleagues.16 miTmc had no effect on currents observed in Tmc1+/− mice. In contrast, exposure to miTmc reduced current amplitudes in Tmc1Bth/− cells (p < 0.02), consistent with reduced expression of the mutant allele (Figures 4A and 4B).
Figure 4.
miTmc Lowers Mechanotransduction Currents in Tmc1Bth/− Mice
(A) Families of mechanotransduction currents were recorded from eight different IHCs (P8–P10) under four different conditions. Currents were evoked by step hair-bundle deflections that ranged in amplitude from −0.2 to 1 μm.
(B) Scatter plot shows maximal mechanotransduction currents recorded from 74 IHCs (open symbols) under four different conditions. Filled symbols indicate the mean ± SE (box) and SD (bars) for each condition. The number of cells for each condition is indicated at the bottom.
miTmc Treatment Prevents Hearing Loss
On the basis of the above results, we completed a longitudinal study to quantitate the effect of miTmc gene therapy on the hearing-loss phenotype in Tmc1Bth/+ mice (Figure 5A). To determine how auditory function was affected, we measured hearing thresholds as ABRs and DPOAEs in four groups of mice: (1) wild-type littermates, (2) Tmc1Bth/+ non-injected mice, (3) Tmc1Bth/++miSafe mice, and (4) Tmc1Bth/++miTmc mice. ABR thresholds were assessed as a response to both clicks and tone bursts at 8–32 kHz, whereas DPOAEs were measured at half-octave intervals across the same frequency range.
Figure 5.
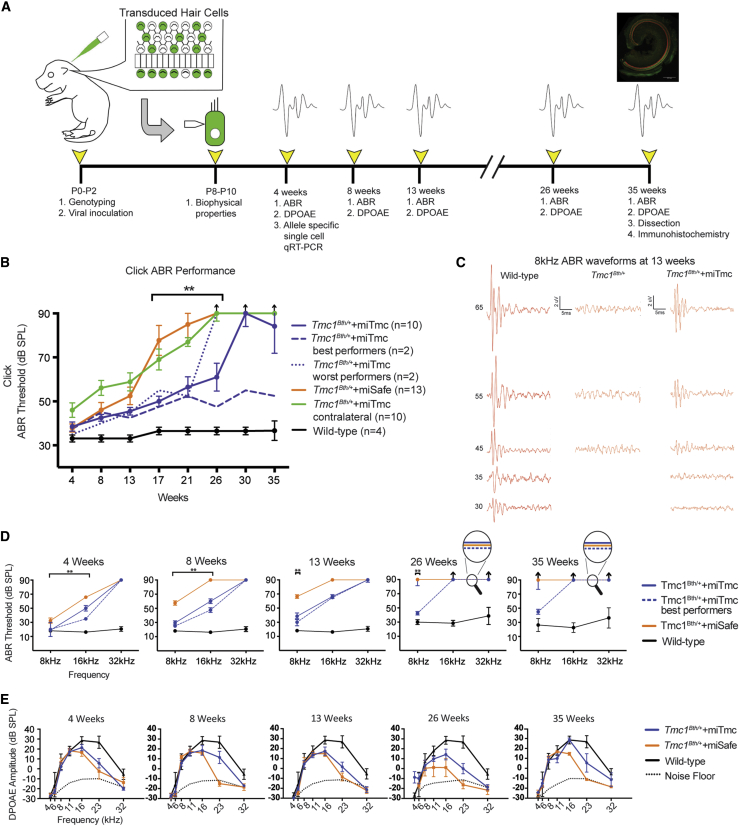
miTmc Gene Therapy Slows Progression of Hearing Loss in Tmc1Bth/+ Mice
(A) Experimental timeline catalogs the experimental procedures in Tmc1Bth/+ mice and controls from the time of artificial miRNA injection to the time of tissue collection.
(B) Click ABR thresholds in wild-type, Tmc1Bth/++miTmc contralateral, Tmc1Bth/++miSafe, and Tmc1Bth/++miTmc animals. The two best-performing and two worst-performing Tmc1Bth/++miTmc-treated animals are shown as dashed and dotted blue lines, respectively, to illustrate variability in performance within the treated cohort.
(C) Representative 8 kHz ABR traces recorded from the wild-type, non-injected Tmc1Bth/++miTmc contralateral, and Tmc1Bth/++miTmc 13-week-old mice.
(D and E) Tone-burst ABR thresholds (D) and DPOAE amplitudes and noise floors (E) in wild-type, Tmc1Bth/+, Tmc1Bth/++miSafe, and Tmc1Bth/++miTmc animals at 4, 8, 13, 26, and 35 weeks. The dotted black line indicates the average noise floor for each group of DPOAEs. Black arrows indicate no response at equipment limits. ∗p < 0.05, ∗∗p < 0.005. See also Figures S4 and S5.
Click ABRs cover a broad frequency and stimulus range (2–8 kHz and up to 90 dB SPL). We tested animals at 4 week intervals for 35 weeks and documented the expected deterioration of hearing thresholds in Tmc1Bth/+ mice, which by 17–21 weeks of age had hearing in the severe-to-profound range (Figure S4). The rate and degree of hearing loss were consistent with those reported by Noguchi and colleagues and were also seen in the untreated contralateral right ears of Tmc1Bth/++miTmc mice and in the injected left ears of Tmc1Bth/++miSafe mice, demonstrating that neither the viral inoculation procedure nor the vector itself added to the decline in auditory function26 (Figure 5B).
The progression of hearing loss was slower in the injected left ears of Tmc1Bth/++miTmc mice. In all Tmc1Bth/++miTmc mice, preservation of hearing was significant in comparison to hearing in controls for up to 21 weeks; however, by 30 weeks ABR thresholds in the Tmc1Bth/++miTmc mice increased (Figure 5B). In the two best-performing animals, hearing thresholds remained stable at 15–20 dB above the thresholds of wild-type C3HeB/FeJ littermate controls for the entire study (Figure 5B). In these two animals, hearing thresholds remained at least 40 dB better than in untreated Tmc1Bth/+ animals. In the two worst-performing animals, hearing protection was lost by 26 weeks.
Frequency-specific effects were measured with tone-burst ABRs (Figures 5C and 5D). Tmc1Bth/+ and Tmc1Bth/++miSafe mice both showed abnormal or absent hearing at 16 and 32 kHz by 4 weeks, consistent with earlier reports5, 27 (Figures 5D, 4 weeks). At 8 kHz, ABR responses were >70 dB by 13 weeks (Figure 5D, 13 weeks). The rate of progression was not increased in Tmc1Bth/++miSafe injected mice, supporting the nontraumatic nature of the trans-RWM injection at P0–P2.
In the left ears of Tmc1Bth/++miTmc mice, there was significant preservation of hearing at 8 and 16 kHz 4 weeks after treatment, although there was no protective effect at 32 kHz (Figures 5D, 4 weeks). By 13 weeks after injection, the protective effect at 16 kHz was lost; however, at 8 kHz a significant protective effect still remained (Figures 5C and 5D, 13 weeks). In the two best-performing animals, hearing thresholds at 8 kHz were preserved throughout the entire study period (dashed blue line); thresholds were stable and only mildly higher than those of wild-type littermate controls (solid black line) (Figure 5D, 4–35 weeks; Figure S5).
We also measured 8 kHz wave I amplitudes at 4 weeks as a measure of synaptic integrity. The treated left ears in Tmc1Bth/++miTmc mice had smaller amplitudes overall than did those of wild-type littermate controls. Wave I amplitudes were even smaller in Tmc1Bth/++miSafe mice and continued to dampen at 8 weeks. In the Tmc1Bth/++miTmc mice, the wave I amplitudes remained stable (Figures S6A and S6B).
DPOAEs are an objective measure of OHC function and were recorded at the same time points as the ABRs. Consistent with prior reports, we found DPOAE recordings to be comparable between Tmc1Bth/+ mice and wild-type controls, reflecting a high degree of OHC preservation in the cochlear apex26 (Figure 5E, 4–35 weeks). At ultra-high frequencies (22–32 kHz), we did identify a DPOAE decline in Tmc1Bth/+ mice (Figure 5E, 4–35 weeks). miTmc treatment did not affect DPOAEs in the first few weeks of the study, and at 4 weeks, we observed no differences between Tmc1Bth/++miTmc and Tmc1Bth/++miSafe mice. By 8 weeks, however, whereas Tmc1Bth/++miSafe mice showed a sharp decline in DPOAEs in the ultra-high-frequency range, DPOAEs in Tmc1Bth/++miTmc mice were maintained (Figures 5E, 8 weeks). This difference persisted, consistent with some preservation of OHCs in the basal turn of the cochlea in Tmc1Bth/++miTmc mice (Figure 5E, 4–35 weeks).
In summary, a single injection of miTmc significantly slowed progression of hearing loss in Tmc1Bth/+ mice for approximately 21 weeks. At the end of the 35-week study period, the two best-performing Tmc1Bth/++miTmc mice had hearing thresholds that were only 15–20 dB above thresholds for wild-type C3HeB/FeJ littermate controls. These thresholds were approximately 40 dB better than expected in the absence of treatment.
miTmc Improves Hair Cell Survival
The histological correlate of auditory function in Tmc1Bth/+ mice is hair cell survival. To quantitate the effect of miTmc on hair cell survival, we counted hair cells in 0.25 mm segments from cochlear whole mounts in four groups: (1) wild-type littermates, (2) Tmc1Bth/++miTmc mice, (3) Tmc1Bth/++miSafe mice, and (4) Tmc1Bth/+ non-injected mice.
Consistent with reported data, in Tmc1Bth/+ mice IHC loss was more pronounced than OHC loss and occurred in a base-to-apex gradient.26 By 35 weeks, we found complete IHC loss in the basal turn and 40%–50% IHC loss in the apical turn (Figures 6A and 6B, Tmc1Bth/+). As expected, there was no IHC or OHC loss in 35-week-old wild-type control animals (Figures 6A and 6B, wild-type).
Figure 6.
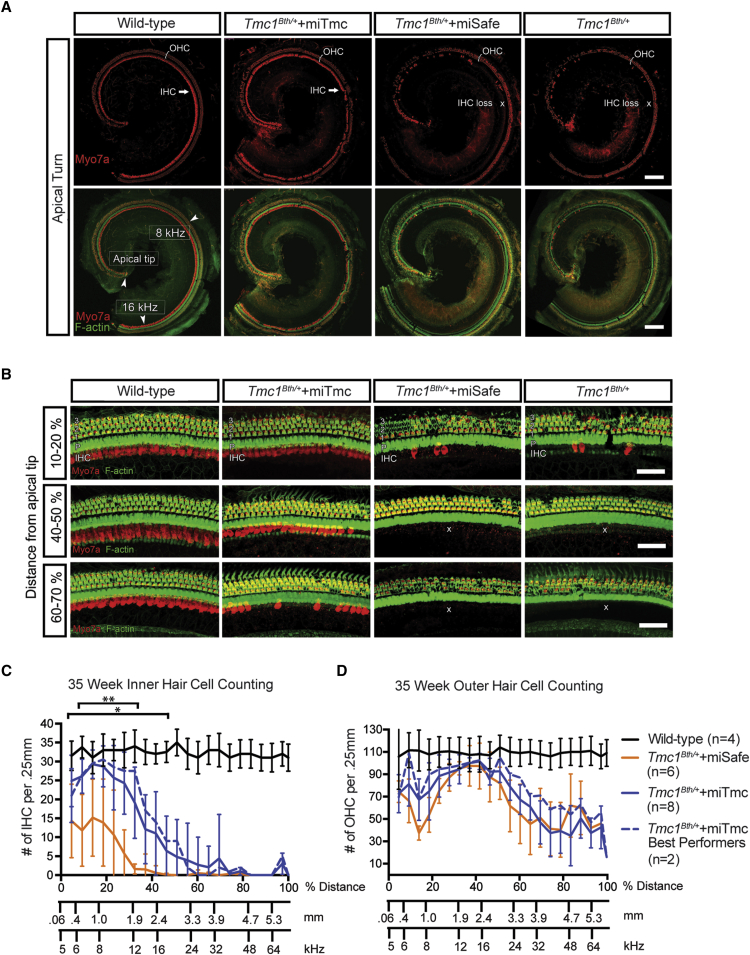
miTmc Gene Therapy Improves Hair Cell Survival
Wild-type, Tmc1Bth/+, Tmc1Bth/++miSafe, and Tmc1Bth/++miTmc animals sacrificed 35 weeks after treatment. Ears were fixed, dissected, and stained as cochlear whole mounts.
(A) 10× images of representative whole-mount apical turns from wild-type, Tmc1Bth/++miTmc, Tmc1Bth/++miSafe, and Tmc1Bth/+ animals. Samples were stained with MYO7A (red) and phalloidin (green) for labeling hair cells and filamentous actin, respectively. Arrowheads show the apical tip and 8 and 16 kHz regions along the apical turn of the cochlea. Note IHC preservation in the Tmc1Bth/++miTmc animals. The white cross shows the area devoid of IHCs. Scale bars represent 150 μm.
(B) 40× magnification at the indicated position in relation to the cochlear apex. The three rows of OHCs (1–3), pillar cells (P), and IHCs are shown. Areas with dark hallows illustrate OHC or IHC loss. The white cross shows the area devoid of IHCs. Scale bars represent 50 μm.
(C and D) IHC (C) and OHC (D) survival was quantified with 20×–40× images of whole-mount cochlea compiled into cochleograms at 35 weeks. Hair cells were counted in 0.25 mm segments and plotted against the distance (%) from the apex. Tmc1Bth/++miSafe, Tmc1Bth/++miTmc, and Tmc1Bth/++miTmc best performers (n = 2) are shown. ∗p < 0.05, ∗∗p < 0.005.
Hair cell survival in all Tmc1Bth/++miSafe-treated left ears was indistinguishable from hair survival in Tmc1Bth/+ mice (Figures 6A and 6B, Tmc1Bth/++miSafe and Tmc1Bth/+). In contrast, Tmc1Bth/++miTmc mice showed markedly improved hair cell survival (Figures 6A and 6B, Tmc1Bth/++miTmc). IHC counts in Tmc1Bth/++miTmc mice were greatest in the apical region of the cochleae (10%–20%), and visible gaps occurring in the mid-modiolar region (40%–50%) showed inter-animal variability. In the lower mid-modiolar to basal regions of the cochleae (60%–70%), the pattern of IHC loss was indistinguishable between Tmc1Bth/++miTmc mice and Tmc1Bth/+ and Tmc1Bth/++miSafe mice. When we compared the entire Tmc1Bth/++miTmc cohort to the two best performers, we observed only a slight difference in mean IHC survival in the apical to mid-modiolar regions of the cochleae (10%–50%; Figure 6C). Compared to Tmc1Bth/++miSafe animals, Tmc1Bth/++miTmc mice showed minimal improvement in OHC survival in the apical 10% of the cochleae and in the lower mid-modiolar to basal regions of the cochlea, although these differences were not significant. OHC survival was otherwise comparable between Tmc1Bth/++miTmc and Tmc1Bth/+ animals (Figure 6D).
We also examined stereocilia-bundle morphology in surviving hair cells in both the best-performing and worst-performing Tmc1Bth/++miTmc mice alongside Tmc1Bth/+ and wild-type controls at 35 weeks (Figure S7). In the Tmc1Bth/++miTmc mice, stereocilia bundles appeared to be thinner but nevertheless well organized and comparable to those of wild-type controls in the apical regions; in the basal region, any remaining stereocilia bundles were sparse and distorted. This apex-to-base gradient of stereocilia-bundle degeneration is consistent with earlier observations in Tmc1Bth/+ mice.5, 27 In Tmc1Bth/+ mice, fewer surviving hair cells remained, and in those cells, the stereocilia bundles were distorted or absent (Figure S7, Tmc1Bth/+). We observed no difference between the two best performers and the entire cohort treated with Tmc1Bth/++miTmc (Figure S7).
Discussion
This report demonstrates that using RNAi to suppress expression of an endogenous deafness-causing allele can slow progression of hearing loss. We selectively suppressed the Tmc1 c.1235T>A (p.Met412Lys) dominant gain-of-function allele in Tmc1Bth/+ mice at an early developmental stage. We were able to prevent profound hearing loss from developing for over 35 weeks (the duration of the study) in some animals otherwise destined to have severe to profound levels of deafness across all frequencies by 17–21 weeks. Treatment with miTmc also appeared to have a particularly striking protective effect on hair cell survival. Compared to Tmc1Bth/++miSafe and Tmc1Bth/+ non-injected mice, Tmc1Bth/++miTmc mice showed significantly enhanced IHC survival in the apical turn of the cochlea. Although the reasons for enhanced IHC survival remain to be determined, these observations are consistent with both the pattern of rAAV2/9 transduction and audiometric data.
In the majority of Tmc1Bth/++miTmc mice, a single injection of miTmc maintained hearing acuity for ∼26 weeks. Thereafter, the protective effect of miTmc on hearing was variable, a result consistent with well-documented differences in the efficiency and longevity of viral transduction.22, 28, 29 For example, Akil et al. used trans-RWM injection of AAV1-Vglut3 to restore hearing in neonatal Vglut3−/− mice and observed variable degrees of hearing loss 7 weeks after treatment.28 Kim et al. used in utero rAAV2/1-MsrB3 (methionine sulfoxide reductase B3) to rescue MsrB3−/− mice and showed hearing deterioration after 4 weeks.30
There are also reports of successful restoration of normal inner-ear morphology but failed restoration of auditory function after inner-ear gene therapy employing AAV vectors. Chien et al. attempted to restore hearing in deaf whirler (whirlin−/−) mice with trans-RWM injections of AAV8-Whirlin, and although they demonstrated restored morphology of the stereocilia, hearing sensitivity was not rescued.31 Similarly, attempts using AAV1-Gjb2 to restore hearing in cCX26 knockout mice by Yu et al. demonstrated restoration of gap-junction function in supporting cells without successful hearing restoration.32 Explanations for this dichotomy have included (1) injection methods, (2) injection timing, (3) promoter types, (4) vector serotypes, and (5) transduction efficiency.
The low rate of viral transduction appears to be the most common conclusion for unsuccessful restoration of hearing sensitivity and could be the reason for the variability observed in our study as well. Indeed, although we achieved high viral transduction in the apical turn and observed hearing preservation at 8 kHz, transduction in the lower turns was very low, and at 32 kHz, the progression of hearing loss was not affected. These observations, however, do not explain the hearing deterioration we documented in the majority of treated mice after 26 weeks.
The longevity of rAAV therapeutics is believed to maintain stable transgene expression for a year or longer, as noted in studies of human gene therapy with AAV2-hRPE65v2 in Leber’s congenital amaurosis.33 One explanation for the time-limited hearing preservation we observed could be the natural progression of OHC loss that occurs in Tmc1Bth/+ mice in a base-to-apex gradient overtime, although this loss is minimal in the apical region at 20 weeks.26 An alternate explanation could be the variability of RNAi in Tmc1Bth/++miTmc mice at the molecular level, which leads to early loss of the RNAi-mediated effect. Side effects of RNAi include miRNA off-target effects, saturation of miRNA endogenous machinery, and immune stimulation via siRNA.34 A combination of these side effects could possibly lead to molecular changes at the hair cell level, although we did not observe structural damage, significant IHC loss, or signs of inflammation even in treated animals that were profoundly deaf at the end of the study, suggesting that miTmc gene therapy did not cause oto-toxicity at the cellular level. Further studies will be needed to improve OHC transduction and to investigate any long-term consequences of RNAi-mediated gene therapy at the molecular level.
Missense mutations underlie 85% of all human autosomal-dominant non-syndromic hearing loss, raising the possibility that an RNAi-based therapeutic strategy could be broadly applicable to this type of hearing loss.35 Targeting autosomal-dominant non-syndromic hearing loss would be attractive because this type of loss is postlingual and progressive, thus providing a large window of opportunity for surgical intervention with a miRNA-based gene-silencing strategy. Prior to any clinical trial, however, several challenges must be addressed. First, although this and other studies attest to the value of mouse models for testing different forms of gene therapy directed at hearing preservation or restoration,28, 36 because functional maturation of the cochlea occurs during the second postnatal week in mice and during the third trimester in humans, direct translation of these studies across species is not possible. Additional studies in rodents must focus on the age at injection, which remains a potential challenge given that transduction efficiency of inner-ear structures appears to be inversely related to the postnatal time of exposure. Target tissue must also be addressed because although HCs express one-third of all genes known to be associated with deafness37—which makes them excellent transduction candidates—efforts to effectively and selectively target other cells must be established.
Route of injection and vector choice must be optimized. Because of the small volume of the inner ear, direct-injection techniques will limit the total number of viral-genome-containing particles per mililiter that can be delivered, suggesting that alternative approaches should be explored to permit higher total injection loads. These experiments will require testing a variety of vectors to define the injection route, injection time, and inner-ear cell specificity for each. Finally, the possibility that therapeutic intervention after hearing loss begins could improve some genetic types of deafness if irreversible damage has not occurred should be explored.
Many of these questions will be addressed with murine models of hearing loss, but ultimately proof-of-concept studies in non-human primates will be needed to confirm vector trophism for specific types of inner-ear cells in the absence of off-target effects. Clinical trials will then be possible. Their design might require the recruitment of persons with gene-specific or even mutation-specific types of hearing loss, a selection process that will be facilitated through genetic databases that many laboratories are maintaining on persons who receive comprehensive genetic testing as part of their clinical diagnostic evaluation for hearing impairment.
We believe that preventing hearing loss in an organ destined to fail presents fewer challenges than restoring function in a cochlea that has already undergone substantial degenerative changes. Thus, although using gene therapy to replace lost hair cells or regrow inner-ear architecture is likely to be exceedingly difficult, its use for replacing defective genes, suppressing a deafness-causing allele, or correcting abnormal splicing before cochlear damage is rampant might be more feasible. Because substantial challenges remain in using gene therapy to restore the functional integrity of this unique and complex organ, we believe that efforts to prevent autosomal-dominant non-syndromic hearing loss warrant a concerted effort by the research community as an example of “low-hanging fruit.”
Acknowledgments
We thank Dr. Karen Steel for providing the Tmc1Bth/+ mutant mouse and Drs. Kiyoto Kurima and Andrew Griffith for the generous gift of the p.AcGFPmTmc1ex1WT and p.GEMT-easyTmc1ex1Bth constructs. We thank Maria Scheel and Susan Stamnes (University of Iowa Viral Vector Core) for assistance and providing the viral vectors. We thank and acknowledge Drs. Hela Azaiez, Yuzhou Zhang, Yukihide Maeda, Samuel P. Gubbels, Michael S. Hildebrand, A. Eliot Shearer, Abraham M. Sheffield, Mark A. Behlke, and Scott D. Rose for earlier work on using RNA interference to suppress hearing loss. We thank Drs. Beverly Davidson and Andrew Griffith for their valuable critiques of the paper. The current work was supported in part by NIH National Institute on Deafness and Other Communication Disorders grants RO1 DC003544 (to R.J.H.S.) and T32 DC000040-17 (to S.B.S.), a Resident Research Award from the American Academy of Otolaryngology – Head and Neck Surgery Foundation (to S.B.S.), and the Bertarelli Program in Translational Neuroscience and Neuroengineering (J.R.H.).
Published: May 26, 2016
Footnotes
Supplemental Data include seven figures, one table, and one movie and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.03.028.
Web Resources
OMIM, http://omim.org/
World Health Organization, http://www.who.int/pbd/deafness/estimates/en/
Supplemental Data
This movie demonstrates the methodology used for rapid isolation of single auditory hair cells.
References
- 1.Smith R.J., Bale J.F., Jr., White K.R. Sensorineural hearing loss in children. Lancet. 2005;365:879–890. doi: 10.1016/S0140-6736(05)71047-3. [DOI] [PubMed] [Google Scholar]
- 2.Géléoc G.S., Holt J.R. Sound strategies for hearing restoration. Science. 2014;344:1241062. doi: 10.1126/science.1241062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maeda Y., Fukushima K., Nishizaki K., Smith R.J. In vitro and in vivo suppression of GJB2 expression by RNA interference. Hum. Mol. Genet. 2005;14:1641–1650. doi: 10.1093/hmg/ddi172. [DOI] [PubMed] [Google Scholar]
- 4.Maeda Y., Fukushima K., Kawasaki A., Nishizaki K., Smith R.J. Cochlear expression of a dominant-negative GJB2R75W construct delivered through the round window membrane in mice. Neurosci. Res. 2007;58:250–254. doi: 10.1016/j.neures.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 5.Vreugde S., Erven A., Kros C.J., Marcotti W., Fuchs H., Kurima K., Wilcox E.R., Friedman T.B., Griffith A.J., Balling R. Beethoven, a mouse model for dominant, progressive hearing loss DFNA36. Nat. Genet. 2002;30:257–258. doi: 10.1038/ng848. [DOI] [PubMed] [Google Scholar]
- 6.Boudreau R.L., Spengler R.M., Davidson B.L. Rational design of therapeutic siRNAs: minimizing off-targeting potential to improve the safety of RNAi therapy for Huntington’s disease. Mol. Ther. 2011;19:2169–2177. doi: 10.1038/mt.2011.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Labay V., Weichert R.M., Makishima T., Griffith A.J. Topology of transmembrane channel-like gene 1 protein. Biochemistry. 2010;49:8592–8598. doi: 10.1021/bi1004377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawashima Y., Géléoc G.S., Kurima K., Labay V., Lelli A., Asai Y., Makishima T., Wu D.K., Della Santina C.C., Holt J.R., Griffith A.J. Mechanotransduction in mouse inner ear hair cells requires transmembrane channel-like genes. J. Clin. Invest. 2011;121:4796–4809. doi: 10.1172/JCI60405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kurima K., Ebrahim S., Pan B., Sedlacek M., Sengupta P., Millis B.A., Cui R., Nakanishi H., Fujikawa T., Kawashima Y. TMC1 and TMC2 Localize at the Site of Mechanotransduction in Mammalian Inner Ear Hair Cell Stereocilia. Cell Rep. 2015;12:1606–1617. doi: 10.1016/j.celrep.2015.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bakhchane A., Charoute H., Nahili H., Roky R., Rouba H., Charif M., Lenaers G., Barakat A. A novel mutation in the TMC1 gene causes non-syndromic hearing loss in a Moroccan family. Gene. 2015;574:28–33. doi: 10.1016/j.gene.2015.07.075. [DOI] [PubMed] [Google Scholar]
- 11.Hilgert N., Monahan K., Kurima K., Li C., Friedman R.A., Griffith A.J., Van Camp G. Amino acid 572 in TMC1: hot spot or critical functional residue for dominant mutations causing hearing impairment. J. Hum. Genet. 2009;54:188–190. doi: 10.1038/jhg.2009.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kitajiri S., Makishima T., Friedman T.B., Griffith A.J. A novel mutation at the DFNA36 hearing loss locus reveals a critical function and potential genotype-phenotype correlation for amino acid-572 of TMC1. Clin. Genet. 2007;71:148–152. doi: 10.1111/j.1399-0004.2007.00739.x. [DOI] [PubMed] [Google Scholar]
- 13.Kurima K., Peters L.M., Yang Y., Riazuddin S., Ahmed Z.M., Naz S., Arnaud D., Drury S., Mo J., Makishima T. Dominant and recessive deafness caused by mutations of a novel gene, TMC1, required for cochlear hair-cell function. Nat. Genet. 2002;30:277–284. doi: 10.1038/ng842. [DOI] [PubMed] [Google Scholar]
- 14.Yang T., Kahrizi K., Bazazzadeghan N., Meyer N., Najmabadi H., Smith R.J. A novel mutation adjacent to the Bth mouse mutation in the TMC1 gene makes this mouse an excellent model of human deafness at the DFNA36 locus. Clin. Genet. 2010;77:395–398. doi: 10.1111/j.1399-0004.2009.01338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao Y., Wang D., Zong L., Zhao F., Guan L., Zhang P., Shi W., Lan L., Wang H., Li Q. A novel DFNA36 mutation in TMC1 orthologous to the Beethoven (Bth) mouse associated with autosomal dominant hearing loss in a Chinese family. PLoS ONE. 2014;9:e97064. doi: 10.1371/journal.pone.0097064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pan B., Géléoc G.S., Asai Y., Horwitz G.C., Kurima K., Ishikawa K., Kawashima Y., Griffith A.J., Holt J.R. TMC1 and TMC2 are components of the mechanotransduction channel in hair cells of the mammalian inner ear. Neuron. 2013;79:504–515. doi: 10.1016/j.neuron.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boudreau R.L., Davidson B.L. Generation of hairpin-based RNAi vectors for biological and therapeutic application. Methods Enzymol. 2012;507:275–296. doi: 10.1016/B978-0-12-386509-0.00014-4. [DOI] [PubMed] [Google Scholar]
- 18.McBride J.L., Boudreau R.L., Harper S.Q., Staber P.D., Monteys A.M., Martins I., Gilmore B.L., Burstein H., Peluso R.W., Polisky B. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc. Natl. Acad. Sci. USA. 2008;105:5868–5873. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang G.S., Schmidt M., Yan Z., Lindbloom J.D., Harding T.C., Donahue B.A., Engelhardt J.F., Kotin R., Davidson B.L. Virus-mediated transduction of murine retina with adeno-associated virus: effects of viral capsid and genome size. J. Virol. 2002;76:7651–7660. doi: 10.1128/JVI.76.15.7651-7660.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soken H., Robinson B.K., Goodman S.S., Abbas P.J., Hansen M.R., Kopelovich J.C. Mouse cochleostomy: a minimally invasive dorsal approach for modeling cochlear implantation. Laryngoscope. 2013;123:E109–E115. doi: 10.1002/lary.24174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng Q.Y., Johnson K.R., Erway L.C. Assessment of hearing in 80 inbred strains of mice by ABR threshold analyses. Hear. Res. 1999;130:94–107. doi: 10.1016/s0378-5955(99)00003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shibata S.B., Di Pasquale G., Cortez S.R., Chiorini J.A., Raphael Y. Gene transfer using bovine adeno-associated virus in the guinea pig cochlea. Gene Ther. 2009;16:990–997. doi: 10.1038/gt.2009.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Viberg A., Canlon B. The guide to plotting a cochleogram. Hear. Res. 2004;197:1–10. doi: 10.1016/j.heares.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 24.Liu H., Pecka J.L., Zhang Q., Soukup G.A., Beisel K.W., He D.Z. Characterization of transcriptomes of cochlear inner and outer hair cells. J. Neurosci. 2014;34:11085–11095. doi: 10.1523/JNEUROSCI.1690-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Picelli S., Faridani O.R., Björklund A.K., Winberg G., Sagasser S., Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 26.Noguchi Y., Kurima K., Makishima T., de Angelis M.H., Fuchs H., Frolenkov G., Kitamura K., Griffith A.J. Multiple quantitative trait loci modify cochlear hair cell degeneration in the Beethoven (Tmc1Bth) mouse model of progressive hearing loss DFNA36. Genetics. 2006;173:2111–2119. doi: 10.1534/genetics.106.057372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marcotti W., Erven A., Johnson S.L., Steel K.P., Kros C.J. Tmc1 is necessary for normal functional maturation and survival of inner and outer hair cells in the mouse cochlea. J. Physiol. 2006;574:677–698. doi: 10.1113/jphysiol.2005.095661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Akil O., Seal R.P., Burke K., Wang C., Alemi A., During M., Edwards R.H., Lustig L.R. Restoration of hearing in the VGLUT3 knockout mouse using virally mediated gene therapy. Neuron. 2012;75:283–293. doi: 10.1016/j.neuron.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Askew C., Rochat C., Pan B., Asai Y., Ahmed H., Child E., Schneider B.L., Aebischer P., Holt J.R. Tmc gene therapy restores auditory function in deaf mice. Sci. Transl. Med. 2015;7:295ra108. doi: 10.1126/scitranslmed.aab1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim M.A., Cho H.J., Bae S.H., Lee B., Oh S.K., Kwon T.J., Ryoo Z.Y., Kim H.Y., Cho J.H., Kim U.K., Lee K.Y. Methionine Sulfoxide Reductase B3-Targeted In Utero Gene Therapy Rescues Hearing Function in a Mouse Model of Congenital Sensorineural Hearing Loss. Antioxid. Redox Signal. 2016;24:590–602. doi: 10.1089/ars.2015.6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chien W.W., Isgrig K., Roy S., Belyantseva I.A., Drummond M.C., May L.A., Fitzgerald T.S., Friedman T.B., Cunningham L.L. Gene Therapy Restores Hair Cell Stereocilia Morphology in Inner Ears of Deaf Whirler Mice. Mol. Ther. 2016;24:17–25. doi: 10.1038/mt.2015.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu Q., Wang Y., Chang Q., Wang J., Gong S., Li H., Lin X. Virally expressed connexin26 restores gap junction function in the cochlea of conditional Gjb2 knockout mice. Gene Ther. 2014;21:71–80. doi: 10.1038/gt.2013.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Testa F., Maguire A.M., Rossi S., Pierce E.A., Melillo P., Marshall K., Banfi S., Surace E.M., Sun J., Acerra C. Three-year follow-up after unilateral subretinal delivery of adeno-associated virus in patients with Leber congenital Amaurosis type 2. Ophthalmology. 2013;120:1283–1291. doi: 10.1016/j.ophtha.2012.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jackson A.L., Linsley P.S. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat. Rev. Drug Discov. 2010;9:57–67. doi: 10.1038/nrd3010. [DOI] [PubMed] [Google Scholar]
- 35.Sloan-Heggen C.M., Bierer A.O., Shearer A.E., Kolbe D.L., Nishimura C.J., Frees K.L., Ephraim S.S., Shibata S.B., Booth K.T., Campbell C.A. Comprehensive genetic testing in the clinical evaluation of 1119 patients with hearing loss. Hum Genet. 2016;135:441–450. doi: 10.1007/s00439-016-1648-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lentz J.J., Jodelka F.M., Hinrich A.J., McCaffrey K.E., Farris H.E., Spalitta M.J., Bazan N.G., Duelli D.M., Rigo F., Hastings M.L. Rescue of hearing and vestibular function by antisense oligonucleotides in a mouse model of human deafness. Nat. Med. 2013;19:345–350. doi: 10.1038/nm.3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scheffer D.I., Shen J., Corey D.P., Chen Z.Y. Gene Expression by Mouse Inner Ear Hair Cells during Development. J. Neurosci. 2015;35:6366–6380. doi: 10.1523/JNEUROSCI.5126-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This movie demonstrates the methodology used for rapid isolation of single auditory hair cells.