Abstract
Multiple acyl-CoA dehydrogenase deficiencies (MADDs) are a heterogeneous group of metabolic disorders with combined respiratory-chain deficiency and a neuromuscular phenotype. Despite recent advances in understanding the genetic basis of MADD, a number of cases remain unexplained. Here, we report clinically relevant variants in FLAD1, which encodes FAD synthase (FADS), as the cause of MADD and respiratory-chain dysfunction in nine individuals recruited from metabolic centers in six countries. In most individuals, we identified biallelic frameshift variants in the molybdopterin binding (MPTb) domain, located upstream of the FADS domain. Inasmuch as FADS is essential for cellular supply of FAD cofactors, the finding of biallelic frameshift variants was unexpected. Using RNA sequencing analysis combined with protein mass spectrometry, we discovered FLAD1 isoforms, which only encode the FADS domain. The existence of these isoforms might explain why affected individuals with biallelic FLAD1 frameshift variants still harbor substantial FADS activity. Another group of individuals with a milder phenotype responsive to riboflavin were shown to have single amino acid changes in the FADS domain. When produced in E. coli, these mutant FADS proteins resulted in impaired but detectable FADS activity; for one of the variant proteins, the addition of FAD significantly improved protein stability, arguing for a chaperone-like action similar to what has been reported in other riboflavin-responsive inborn errors of metabolism. In conclusion, our studies identify FLAD1 variants as a cause of potentially treatable inborn errors of metabolism manifesting with MADD and shed light on the mechanisms by which FADS ensures cellular FAD homeostasis.
Introduction
Riboflavin, or vitamin B2, is the precursor of flavin adenine dinucleotide (FAD) and flavin mononucleotide (FMN), which are essential cofactors of numerous dehydrogenases involved in cellular metabolism and a number of other essential cellular pathways, such as antioxidant defense, protein folding, and apoptosis.1 These flavoenzymes are located in the cytosol or compartmented in the cellular organelles, where they ensure the functionality of mitochondrial electron transport, the metabolism of fatty acids, some amino acids, choline, and betaine, the synthesis of protoporphyrin, and the metabolism of vitamins B6, B9, and B12.2, 3, 4, 5, 6
Riboflavin cannot be synthesized in humans and must therefore be obtained as a nutrient via intestinal absorption. Riboflavin is transported into the bloodstream and taken up by target cells by the human riboflavin transporters (RFVTs). Three RFVTs (RFVT1, RFVT2, and RFVT3), each differing in tissue-specific presence, have been identified thus far and assigned to a new solute carrier family named SLC52.4 Within cells, riboflavin is converted into FMN by riboflavin kinase (RFK), and FMN is then converted into FAD by FAD synthase (FADS), previously known as FAD synthetase or FMN:ATP adenylyltransferase.7, 8 FADS is the product of FLAD1 (MIM: 610595), which according to the UCSC Genome Browser encodes two transcripts resulting in proteins with an intact FADS domain. Those two transcripts have been characterized in detail;7, 8, 9, 10, 11, 12 a longer isoform derives from a large exon 1 and has the potential to encode FADS1 with a predicted mitochondrial targeting sequence (MTS). In a shorter isoform, exon 1 is interrupted by an additional intron. This leads to usage of a downstream ATG start codon at position 98 of the longer transcript, resulting in FADS2.13 Both FADS proteins contain an N-terminal molybdopterin binding (MPTb) domain, which has been shown to have FAD hydrolase activity,10 and a C-terminal 3′-phosphoadenosine 5′-phosphosulfate (PAPS) reductase domain. This second domain is sufficient to catalyze FAD synthesis.11 Therefore, it is here named the FADS domain. All recombinant FADS produced up to now exhibit the ability to bind FAD, the product of their activity, tightly but not covalently.9 FAD release from FADS is likely to be tightly controlled and presumably requires well-defined conditions, including a correct redox state.9, 12 FAD (or as recently proposed, catalytically active FADS enzyme) binds to apoproteins and chaperones their folding into stable and functional flavoenzymes.14, 15, 16, 17, 18, 19 Accordingly, several studies have shown that a number of flavoproteins, and in particular mature mitochondrial acyl-CoA dehydrogenases, undergo fast degradation under depletion of riboflavin or flavin cofactors.20, 21, 22, 23 The biochemistry of riboflavin deficiency, with elevated multiple acylcarnitines and urine organic acids, resembles that seen in individuals who suffer from multiple acyl-CoA dehydrogenase deficiency (MADD [MIM: 231680]) or glutaric aciduria type 2 and have genetic defects in three genes encoding two enzymes, electron transfer flavoprotein (ETF) and electron transfer flavoprotein dehydrogenase (ETFDH). These two enzymes functionally link the acyl-CoA dehydrogenation reactions to ATP production by delivering their electrons to coenzyme Q10 and further on to complex III in the respiratory chain.24 Riboflavin-responsive forms of MADD leading to improvement in biochemical and clinical symptoms after high doses of riboflavin treatment were first explained by missense variants in ETFDH (MIM: 231675)25, 26, 27, 28, 29 and more recently by variants in the riboflavin transporter genes (SLC52A1 [MIM: 607883], SLC52A2 [MIM: 607882], and SLC52A3 [MIM: 613350]),30, 31, 32 as well as in the mitochondrial FAD transporter gene (SLC25A32 [MIM: 610815]).33
In this study, we describe individuals who are from seven unrelated families and have FLAD1 variants causing MADD and a disturbed riboflavin metabolism. The affected individuals presented with muscle symptoms and multiple-respiratory-chain deficiency. Depending on the FLAD1 genotype, some responded to riboflavin treatment.
Subjects and Methods
Subjects
Subjects (S1–S7) from seven unrelated families (F1–F7 in Figure 1A) recruited from medical centers located in Australia, Finland, Italy, Austria, Turkey, and France were enrolled in the current project. All studies were completed according to local approval by institutional review boards. Informed consent for participation in this study was obtained from the parents of all investigated subjects in agreement with the Declaration of Helsinki and approved by the ethical committees of the centers participating in this study, where biological samples were obtained.
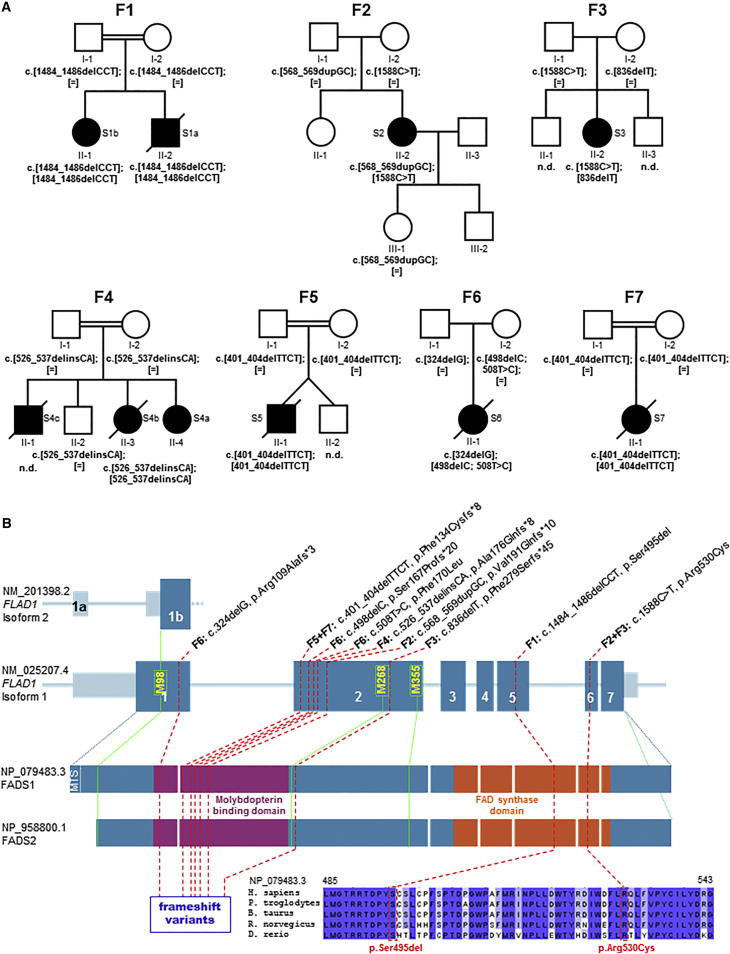
Figure 1.
FLAD1 Variants and Gene and Protein Structure
(A) Pedigrees of the investigated families (F1–F7) with recessively inherited FLAD1 variants. Affected individuals are indicated by closed symbols.
(B) Gene structure with exons and introns shows the localization of the investigated gene variations (homozygous variants are underlined). Met residues located upstream of the FADS domain are presented in yellow with their corresponding protein positions with respect to isoform 1 (GenBank: NM_025207.4). Isoforms 1 and 2 are reported in the UCSC Genome Browser as transcripts possessing an intact and active FADS domain. The protein structure highlights the MPTb domain in violet and the FADS domain in orange. Protein consequences of the identified FLAD1 mutations include the frameshift variants located in the MPTb domain and the two amino acid changes in a region of the FADS domain, which is highly conserved among eukaryotic species. Amino acid residues that are conserved across all species are highlighted in dark blue.
Genetic Analyses
For subjects S4a and S5, whole-exome sequencing (WES) was performed in genomic DNA by AROS Applied Biotechnology AS with Illumina’s TruSeq DNA Sample Prep Kit and Exome Enrichment Kit on the Illumina HiSeq 2000 platform (for S5)34 or with SureSelect Human All Exon 50Mb V5 Kit (Agilent) on a HiSeq 2500 system (Illumina) (for S4a).35, 36 The remaining affected individuals were investigated by sequencing of candidate genes associated with fatty-acid oxidation and/or mitochondrial disorders, including FLAD1 and other genes involved in riboflavin uptake or metabolism. For subjects S1a, S2, and S3, Sanger sequencing of the target genes listed in Table S1 was performed as described previously.37, 38 For subject S6, a customized gene panel39 (TruSeq Custom Amplicon, Illumina) containing 200 genes associated with mitochondrial disorders, including the genes listed in Table S1, was used for library preparation; then, samples were analyzed by a MiSeq system (Illumina). For subject S7, a panel of 17 genes (Table S1) was analyzed with a targeted resequencing approach by Ion AmpliSeq technology (Life Technologies) on a PGM analyzer (Life Technologies). All identified variants and their segregation in the affected families were validated by Sanger sequencing.
Skeletal-Muscle and Liver Homogenates and Fibroblast Cultures
Muscle biopsy specimens were obtained from seven subjects, and a post-mortem liver specimen was obtained from individual S5. Homogenization and preparation of mitochondrial fractions were performed according to standard procedures. Primary human dermal fibroblasts from subjects S1a, S2, and S4a and healthy control individuals were cultivated until confluent at 37°C and 5% (v/v) CO2 in RPMI-1640 media (Lonza) with 10% FCS, 2 mmol/L L-glutamine (Sigma-Aldrich), and 1% penicillin and streptomycin (Sigma-Aldrich).
Histological and Respiratory-Chain Analyses
Histological analyses and measurements of respiratory-chain function were performed via polarographic and spectrophotometric analyses as previously described.25, 40
SDS-PAGE and Immunoblotting
Liver tissue was homogenized with a potter-type homogenizer on ice in buffer (250 mmol/L sucrose, 50 mmol/L Tris-HCl [pH 7.4], 5 mmol/L MgCl2, EDTA-free protease inhibitor cocktail, and 1% Triton X-100), and frozen fibroblast pellets were lysed in a buffer containing 50 mmol/L Tris-HCL (pH 7.8), 5 mmol/L EDTA (pH 8.0), 1 mmol/L DTT, 10 μg/mL Aprotinin (Sigma-Aldrich), 1 mg/mL trypsin inhibitor (Roche) in 10 mL, and 1% Triton X-100. Aliquots of total liver protein (50–100 μg) were loaded on 14% SDS-PAGE gels, and 20 μg fibroblast protein lysate was loaded on Criterion GGX Stain-Free Precast Gels (Bio-Rad) and transferred to polyvinylidene difluoride membranes. Membranes were probed with human anti-FLAD1 antibody (fibroblasts: HPA028563, Sigma-Aldrich; liver: ab95312, Abcam) and anti-GAPDH (sc25778, Santa Cruz). Detected proteins were visualized with the ECL Plus Western Blotting Detection System (GE Healthcare).
Mass Spectrometry to Confirm the Identity of Anti-FADS Gel Bands
80 μg of total protein from control fibroblasts was extracted and separated by SDS-PAGE as described above. The bands of interest (∼60, 50, 37, 32, and 27 kDa) were excised and processed by an in-gel trypsin-digestion protocol as previously described.41 Selected reaction monitoring (SRM) was performed as previously described.42 Peptide selection, method development, and data analysis were performed with the software Skyline.43 Five FADS peptides passed selection criteria based on detectability and FADS uniqueness (checked in the whole human proteome, UniProt version 2012_06_19, containing 20,202 reviewed sequences), and these five FADS peptides were analyzed by liquid chromatography coupled with the TSQ Vantage Triple Quadrupole Mass Spectrometer (Thermo Fisher Scientific). Four or more transitions were detected for each peptide, and synthetic isotopically labeled tryptic peptides (SpikeTide from JPT Peptide Technologies) were added to each sample for quality assurance. The list of peptides and transitions can be found in Table S2.
Measurements of FAD Synthesis Rate and Flavin Content
For investigating different fractions of EDTA blood from subject S4a, plasma, mononuclear cells, and enriched erythrocytes purified by Ficoll gradient centrifugation, 50 μl of each was mixed with 375 μl of 10% trichloroacetic acid and 75 μl of water. After incubation on ice for 5 min and centrifugation at 13,000 × g for 5 min, the supernatant was extracted with equal amounts of diethyl ether twice, and 100 μl was applied to high-performance liquid chromatography (HPLC) separation on a reversed-phase column (Agilent, Eclipse Plus C18, 5 μm, 4.6 × 150 mm) for measuring riboflavin, FMN, and FAD.
Flavin content in muscle homogenate from subject S5 was measured by HPLC with a reverse-phase C18 column and fluorescence detection (excitation and emission wavelengths of 470 and 530 nm, respectively) according to the previously described method.44 200 μl of muscle homogenate was mixed with 500 μl methanol and centrifuged, and the supernatant was injected for HPLC analysis (mobile phase: 50 mmol/L NaH2PO4 and 13% acetonitrile [pH 2.9]; flow rate = 1.2 mL/min).
For measurements in fibroblast cells, the frozen cells were re-suspended in 150 μl lysis buffer (50 mmol/L Tris-HCl [pH 7.5], 1% Triton X-100, 5 mmol/L β-mercaptoethanol [2-ME], 1 mmol/L NaF, 0.1 mmol/L PMSF, and 1× protease-inhibitor cocktail) and passed through a 26G needle. After incubation for 30 min on ice, the cell suspension was centrifuged at 13,000 × g for 10 min at 4°C, and the supernatant was recovered as cell lysate. Riboflavin, FMN, and FAD were measured in neutralized perchloric acid extracts of cell lysates (0.2 mg) by HPLC as previously described.45, 46, 47, 48 Separation was achieved with a Gilson HPLC system, including a model 306 pump and a model 307 pump equipped with a FP-2020 Plus Jasco Fluorescence Detector and uniPoint software. Quantitative determination of riboflavin, FMN, and FAD was carried out with a calibration curve made in each experiment with standard solutions diluted in the extraction solution. The rate of FAD synthesis was measured at 37°C in 500 μl of 50 mM Tris-HCl (pH 7.5) in the presence of 0.08 mg cell lysate, 1 μmol/L FMN, 5 mmol/L ATP, and 5 mmol/L MgCl2. At the appropriate time, 100 μl aliquots were taken, extracted with perchloric acid, and neutralized. Riboflavin, FMN, and FAD were analyzed with HPLC (see above).
qRT-PCR and Transcriptome Analysis
Total RNA was isolated according to the TRIzol Reagent protocol (Ambion) or with the RNeasy Plus Mini Kit (QIAGEN). The quality of the RNA was determined with the Agilent 2100 BioAnalyzer (RNA 6000 Nano Kit, Agilent). cDNA was synthesized from subject S2’s RNA with the iScript cDNA Synthesis Kit (Bio-Rad), and the c.1588C>T genotype was assessed by sequencing.
qRT-PCR analysis (iQ SYBR Green Supermix, iCycler, Bio-Rad) of subject S4a’s fibroblast RNA (TRI Reagent, Molecular Research Center), treated with DNase (TURBO DNase, Thermo Fisher), was performed with oligo-dT-amplified cDNA (Maxima Reverse Transcriptase, Thermo Fisher) with a set of primers spanning all coding regions of FLAD1 (Figure S1). Threshold cycles (Ct) were normalized (ΔCt) to housekeeping genes HPRT (GenBank: NM_000194.2; MIM: 308000) and RPL27 (GenBank: NM_000988.3; MIM: 607526), and the expression of PCR products was compared to that of control fibroblasts (ΔΔCt).
For transcriptome analysis, quantitative library preparation and enrichment were performed as described in Haack et al.49 RNA libraries were assessed for quality and quantity with the Agilent 2100 BioAnalyzer and the Quant-iT PicoGreen dsDNA Assay Kit (Life Technologies) and sequenced as 100 bp paired-end runs on an Illumina HiSeq 2500 platform. Sashimi plots were used for quantitative visualization of splice junctions of their alignment data in the Integrative Genomics Viewer.50
Lentiviral Gene Expression
The wild-type (WT) sequence of isoform 2 (GenBank: NM_201398.2) was obtained as a plasmid from DNASU (HsCD00039838). Shorter FLAD1 transcripts coding for FADS proteins starting at methionine 268 (Met268) or methionine 355 (Met355) were amplified from a human control cDNA and cloned into the pLenti6.3/V5-TOPO TA Cloning Kit (Invitrogen) for production in human fibroblasts.36 The following primers were used: 5′-CGCCATGAAGGGACTATTCC-3′ (forward Met268), 5′-CGCCATGCCCAACGCTGTGG-3′ (forward Met355), and 5′-TCATGTGCGGGAGTTCCGCT-3′ (reverse).
Biophysical Studies of Recombinant 6His-p.Ser495del and 6His-p.Arg530Cys FADS2
Site-Directed Mutagenesis
The coding region for human FADS isoform 2 (FADS2) was amplified as previously described.7 The Ser495 deletion and the p.Arg530Cys substitution were obtained via overlap-extension PCR12 with the following primers: 5′-ATCTGGGATTTTCTGTGTCAGCTGTTTGTC-3′ (forward) and 5′-GACAAACAGCTGACACAGAAAATCCCAGAT-3′ (reverse) for p.Arg530Cys and 5′-TGACCCCTACTGTAGCCTCTGCCCTTTCAGCCCCACTGAC-3′ (forward) and 5′-AGAGGCTACAGTAGGGGTCAGTCCGGCGGGTGCCC-3′ (reverse) for the Ser495 deletion.
The PCR products were purified, digested with EcoRI and XhoI, and ligated into the pH6EX3 expression vector as previously described.9 The resulting recombinant plasmid carried the extra N-terminal sequence MSPIHHHHHHLVPRGSEASNS.
Production and Purification of Recombinant 6His-Ser495del and 6His-p.Arg530Cys FADS2
Protein production in Rosetta (DE3) cells and purification of WT or mutant FADS2 were carried out as described previously.9 Where indicated, the WT and variant proteins were incubated with a 2.5-fold molar excess of FAD at 4°C overnight, and the free cofactor was eventually removed on a PD10-column run in 40 mmol/L HEPES/Na and 5 mmol/L 2-ME (pH 7.4).
Protein Concentration and Measurements of FAD Saturation
Protein concentration was measured by the Bradford method with BSA as a standard. In an alternative procedure, the concentration of the purified FADS2 was estimated by absorbance spectra as previously described9 with the use of ε280 (56,435 mM−1 · cm−1 and 0.998 mg/mL−1 for the WT protein; 55,810 mM−1 · cm−1 and 0.997 mg/mL−1 for 6His-p.Ser495del FADS2; and 56,435 mM−1 · cm−1 and 0.999 mg/mL−1 for 6His-p.Arg530Cys FADS2) as calculated from the protein sequence with the ExpasyProtParam tool. The degree of FAD saturation (i.e. the ratio of FAD to protein monomer) was calculated from the spectra as previously described.9
Measurements of FAD Synthesis Rate
The different fluorescence properties of FAD and FMN were exploited for measuring the rate of FAD synthesis. Fluorescence time courses (excitation at 450 nm and emission at 520 nm) were followed in a LS50 Perkin Elmer spectrofluorometer. FAD and FMN fluorescence were calibrated with standard solutions, whose concentrations were calculated with ε450 = 12.2 mM−1 · cm−1 for FMN and 11.3 mM−1 · cm−1 for FAD. The rate of FAD synthesis (nmol FAD · min−1 · mg protein−1) was calculated from the rate of decrease in fluorescence, measured as the tangent to the initial part of the experimental curve. For activity measurements, purified protein fractions (10 μg protein as a monomer, unless otherwise indicated) were incubated at 37°C in 2 ml of 50 mol/L Tris-HCl (pH 7.5) containing 5 mmol/L MgCl2, 2 μmol/L FMN, 100 μmol/L ATP, and additional reagents as appropriate.
Limited Proteolysis with Trypsin
WT and mutant FADS2 proteins with or without incubation with a molar excess of FAD were used for testing the susceptibility to trypsin digestion. Samples (10 μg) were incubated with or without 0.015 mg/mL trypsin (bovine pancreas trypsin, T8003, Sigma-Aldrich) at 0°C in 40 mmol/L HEPES/Na and 5 mmol/L 2-ME (400 μL). Aliquots of 2.5 μg protein were sampled at different time points, and the reaction was stopped by the addition of 0.3 mg/mL of soybean trypsin inhibitor (T9003, Sigma-Aldrich). As an internal standard for the quantity of loaded protein in the gel, BSA (1 μg) was also added to the loading buffer solution. The products of the proteolysis reaction were separated by 12% SDS-PAGE and electro-transferred onto a nitrocellulose membrane with a Trans-Blot SD Semi-Dry Electrophoretic Transfer Cell (Bio-Rad). The immobilized proteins were incubated for 3 hr with a 3,000-fold dilution of a polyclonal antiserum against FADS.13 The bound antibodies were visualized with the aid of secondary anti-rabbit IgG antibodies conjugated with alkaline phosphatase (1:3,500 dilution). Quantitative evaluation of immuno-reactive protein bands was carried out by densitometric analysis with ImageLab software 5.1 (Bio-Rad).
Statistical Analysis
Statistical analysis was performed with the Poisson test for determining the number of loss-of-function (LOF) variants in FLAD1 and with the Student’s t test for pairwise comparisons of means in Tables 2, 3, and 4. A p value of <0.05 was considered statistically significant (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
Table 2.
Rate of FAD Synthesis in Fibroblasts from Individuals with FLAD1 Variants
| ID | Rate of FAD Synthesis (pmol/min/mg) |
|---|---|
| C1 | 4.37 ± 0.61 |
| C2 | 4.36 ± 0.4 |
| C3 | 3.66 ± 0.38 |
| S1a | 2.05 ± 0.59∗∗ |
| S2 | 1.57 ± 0.7∗∗∗ |
| S4a | 1.04 ± 0.47∗∗∗ |
Data represent the mean ± SD of three to four independent cell lysates. ∗∗p < 0.01, ∗∗∗p < 0.001.
Table 3.
FAD, FMN, and Riboflavin Quantification in Cellular and Mitochondrial Fibroblast Samples from Control and Affected Individuals
| ID | FAD | FMN | Riboflavin | FAD/FMN Ratio | |
|---|---|---|---|---|---|
| Cellular flavin content (pmol/mg) | C1 | 111.7 ± 2.8 | 13.3 ± 1.4 | 2.6 ± 0.5 | 8.5 ± 0.7 |
| C2 | 171.2 ± 18.8 | 12.4 ± 0.3 | 3.1 ± 0.1 | 13.8 ± 1.9 | |
| C3 | 105.8 ± 6.9 | 13.0 ± 0.0 | 2.1 ± 0.6 | 8.2 ± 0.5 | |
| S1a | 82.3 ± 8.6 | 11.7 ± 0.8 | 2.1 ± 0.8 | 7.0 ± 0.2 | |
| S2 | 84.3 ± 3.1 | 11.5 ± 0.6 | 2.4 ± 0.9 | 7.3 ± 0.1 | |
| S4a | 111.4 ± 6.6 | 15.8 ± 0.4 | 2.5 ± 0.5 | 7.0 ± 0.6 | |
| Mitochondrial flavin content (pmol/mg) | C1 | 2,093 | 3.0 ± 0.1 | NT | 697.7 ± 24.0 |
| C2 | 1,940 | 3.2 | NT | 606.3 | |
| C3 | 1,895 | 3.2 | NT | 592.2 | |
| S4a | 1,310 ± 70∗∗ | 2.0 ± 0.7 | NT | 655.0 ± 195.7 |
For cellular flavin content, data represent the mean ± SD of two independent cell lysates. NT, not tested. ∗∗p < 0.01.
Table 4.
Kinetic Properties of 6His-p.Ser495del, 6His-p.Arg530Cys, and WT FADS2
| Forward Reaction | 6His-p.Arg530Cys | 6His-p.Ser495del | WT |
|---|---|---|---|
| Km FMN (μM) | 0.39 ± 0.07 | 0.98 ± 0.23∗∗∗ | 0.32 ± 0.10 |
| Km ATP (μM) | 4.63 ± 1.00∗ | 8.02 ± 2.23 | 8.10 ± 1.37 |
| Vmax (nmol·min−1·mg−1) | 7.9 ± 3.7∗∗∗ | 10.2 ± 1.6∗∗ | 43.3 ± 12.2 |
| Mg2+50 (mM) | 0.04 ± 0.02∗ | 0.09 ± 0.01 | 0.07 ± 0.01 |
Data represent the mean ± SD of three experiments. ∗p < 0.05; ∗∗p < 0.01, ∗∗∗p < 0.001.
Results
Metabolic research centers from Australia, Austria, Denmark, England, Finland, France, Germany, Italy, and Turkey independently achieved and subsequently shared results from candidate-gene or WES analysis on families and individuals affected by riboflavin-responsive MADD and/or a suspected mitochondrial respiratory-chain disorder (Table 1). As a result of this collaborative effort, from seven unrelated families (F1–F7 in Figure 1A) we identified nine affected individuals harboring recessive mutations in one gene, FLAD1 (GenBank: NM_025207.4), coding for FADS. Detailed case reports are available in the Supplemental Note.
Table 1.
Pre-treatment Genetic, Clinical, Biochemical, and Histopathological Findings in Individuals with FLAD1 Variants
| ID | Sex | Consanguineous | FLAD1 Variants (GenBank:NM_025207.4) | FADS1 Variants (GenBank:NP_079483.3) | Affected Domain | Age of Onset | Status | Responsive to Riboflavin | Increased Blood Acylcarnitinesa | Elevated Organic Acidsa |
Muscle Investigations |
||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Respiratory-Chain Activitiesb | Other Mitochondrial Enzyme Activities | Histology | |||||||||||
| S1a | F | yes | c.[1484_1486delCCT]; [1484_1486delCCT] | p.[Ser495del];[Ser495del] | FADS | 32 hr | died at 3 days | NT | C5, C8, C14, C5-DC | ethylmalonic, adipic, suberic, and dehydrosebacic acids, hexanoylglycine | within control range | NR | vacuoles in muscle fibers (EM not done) |
| S1b | M | yes | c.[1484_1486delCCT]; [1484_1486delCCT] | p.[Ser495del];[Ser495del] | FADS | 3 months | alive at 22 years | yes | C4 | ethylmalonic and methylsuccinic acids | NR | NR | NR |
| S2 | F | no | c.[568_569dupGC]; [1588C>T] | p.[Val191Glnfs∗10]; [Arg530Cys] | MPTb (fs), FADS | 20 years | alive at 44 years | yes | C8, C10, C10:1 | ethylmalonic and glutaric acids, lactate | CII↓, CIII↓, CIV↓ | CS↑ | lipid-storage myopathy, faint COX staining |
| S3 | F | no | c.[836delT]; [1588C>T] | p.[Phe279Serfs∗45]; [Arg530Cys] | MPTb (fs), FADS | 44 years | alive at 56 years | yes | C5, C8, C10, C10:1, C14 | ethylmalonic acid, tiglylglycine | CI+III↓, CII+III↓ | CS↑ | lipid-storage myopathy, muscle beta oxidation (C4: 132%; C8: 15%; C16: 56%) |
| S4a | F | yes | c.[526_537delinsCA]; [526_537delinsCA] | p.[Ala176Glnfs∗8]; [Ala176Glnfs∗8] | MPTb (fs) | 4 months | alive at 8 years | yes | C4, C6, C8, C10, C14, C14:1, C14:2, C16:1, C18, C18:1, C18:2 | adipic, suberic, ethylmalonic, and methylsuccinic acids, ketosis | CI↓, CII↓ | PDH↓ | lipid-storage myopathy |
| S4b | F | yes | c.[526_537delinsCA]; [526_537delinsCA] | p.[Ala176Glnfs∗8]; [Ala176Glnfs∗8] | MPTb (fs) | 8 months | died at 16 years, 2 months | discontinued (side effects) | normal or C4-OH, C5, C6, C8, C10, C14:1, C16:1 | NR | NR | NR | NR |
| S5 | M | yes | c.[401_404delTTCT]; [401_404delTTCT] | p.[Phe134Cysfs∗8]; [Phe134Cysfs∗8] | MPTb (fs) | 6 months | died at 6 months | NT | C3, C5, C6, C8:1 | adipic acid | CI↓, CII+III↓ | CS↑ | lipid-storage myopathy, faint COX staining |
| S6 | F | no | c.[324delG]; [498delC;508T>C] | p.[Arg109Alafs∗3]; [Ser167Profs∗20; Phe170Leu] | MPTb (fs) | birth | died at 9 months | no | NR | NR | CI↓, CII+III↓ | CS↑ | lipid-storage myopathy, absent SDH staining, several NADH- and COX-negative fibers |
| S7 | F | yes | c.[401_404delTTCT]; [401_404delTTCT] | p.[Phe134Cysfs∗8]; [Phe134Cysfs∗8] | MPTb (fs) | 2 months | died at 4 months | NT | C4, C6, C8, C10, C10:1, C12, C14:1, C16, C16:1, C18:1 | ethylmalonic and glutaric acids | NR | NR | lipid-storage myopathy, faint SDH staining, normal COX |
Abbreviations are as follows: ↓, decrease; ↑, increase; C4, butyrylcarnitine; C4-OH, OH-butyrylcarnitine; C5, isovalerylcarnitine; C5-DC, glutarylcarnitine; C6, hexanoylcarnitine; C8, octanoylcarnitine; C10, decanoylcarnitine; C10:1, decenoylcarnitine; C12, dodecanoylcarnitine; C14, tetradecanoylcarnitine; C14:1, tetradecenoylcarnitine; C14:2, tetradecadienoylcarnitine; C16, hexadecanoylcarnitine; C16:1, hexadecenoylcarnitine; C18, octadecanoylcarnitine; C18:1, octadecenoylcarnitine; C18:2, octadecdienoylcarnitine; CI–CIV, complexes I–IV; COX, cytochrome c oxidase; CS, citrate synthase; EM, electron microscopy; F, female; FADS, FAD synthase domain; fs, frameshift; M, male; MPTb, molybdopterin binding domain; NR, not reported; NT, not tested; PDH, pyruvate dehydrogenase; and SDH, succinate dehydrogenase.
Actual values are reported in Table S3.
Actual values are reported in Table S5.
Genetic Analyses
In family F1, both siblings (S1a and S1b) were found to carry a homozygous mutation resulting in an in-frame deletion of serine 495 (c.1484_1486delCCT [p.Ser495del]) in the FADS domain. Individuals S2 and S3 were found to carry the same missense mutation, c.1588C>T (p.Arg530Cys), affecting the FADS domain and a frameshift variant in exon 2 on the other allele. The four additional individuals were all found to carry biallelic frameshift variants in exon 1 or 2. Carrier testing showed that each parent analyzed was heterozygous for one allele (Figure 1A). None of the predicted LOF variants was present in an in-house database with WES data from approximately 6,000 individuals or in >120,000 alleles from the Exome Aggregation Consortium (ExAC) Browser (accessed January 2016). Only variants c.1588C>T (p.Arg530Cys) and c.508T>C (p.Phe170Leu) were recorded in the ExAC Browser; c.1588C>T was recorded as heterozygous three times in 121,410 alleles, and c.508T>C was recorded as heterozygous two times in 121,408 alleles. Of note, the affected individuals can be distinguished by their symptoms and the identified FLAD1 variants (Table 1 and the Supplemental Note). Whereas subjects S1b, S2, and S3 (who showed a milder clinical course and a remarkable response to riboflavin) carried at least one allelic variant that changes or deletes a single amino acid in the FADS domain, subject S4a (who showed only a partial response to riboflavin) and subjects S4b, S5, S6, and S7 (who had an early onset and lethal disease) all carried two predicted LOF variants affecting the MPTb domain.
FADS activity is expected to be essential for human metabolism, and no other protein with analogous function has been reported. Therefore, FLAD1 variants predicted to result in complete LOF were unexpected.
Residual FADS Activity and Flavin Content in FLAD1 Mutant Cells
The mild increase in metabolites derived from deficient multiple acyl-CoA dehydrogenases argues for only a partial defect in FAD synthesis and levels (Table S3). In accordance with this, mild but significant decreases in the rate of FAD synthesis were measured in cultured fibroblasts derived from the affected individuals, whose activity was 50% (S1a), 38% (S2), and 25% (S4a) of that of control individuals (Table 2). Analysis of flavin content in the same cell extracts and in blood from S4a and S4b supported remaining FADS activity in all investigated subjects and revealed FAD, FMN, and riboflavin amounts within the control range but increased amounts of FMN in erythrocytes (Table 3 and Figure S3). Only when enriched mitochondrial fractions from subject S4a’s fibroblasts were investigated were amounts of FAD and FMN found to be slightly but significantly reduced (Table 3). In accordance with decreased availability of mitochondrial FAD, subject S4a’s fibroblast samples had reduced amounts of ETFDH and complex II, two mitochondrial dehydrogenases that use FAD as a co-factor (Figure S4). The amount of FAD in a skeletal-muscle biopsy from S5 (who, like S4a, carried biallelic frameshift variants in exon 2) was 145 nmol/L, slightly below the control range (174–471 nmol/L). Whereas reduced FADS activity in S1a and S2 can be explained by missense mutations, the residual FADS activity and the relatively high content of FAD in S4a and S5 suggest alternative mechanisms that allow expression of functional FADS from FLAD1 variants predicted to result in complete LOF.
Immunoblot Analysis of FLAD1 Mutant Cells
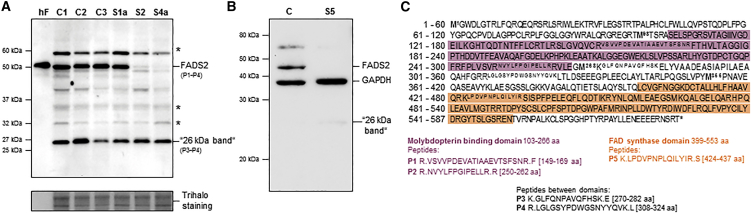
In order to investigate the molecular consequences of the identified FLAD1 variants, we performed immunoblot analyses for FADS in cell extracts from cultured fibroblasts (from S1a, S2, and S4a) or from liver tissue (from S5) (Figure 2). Using antibodies recognizing the FADS C terminus, containing the FADS domain, we detected a number of bands, including three high-intensity bands that were expressed in both fibroblast and liver tissue: ∼60 and ∼50 kDa bands corresponding to the expected mitochondrial (isoform 1 [FADS1]) and cytosolic (isoform 2 [FADS2]) proteins, respectively (Figure 1B), and a ∼26 kDa band. Mass spectrometry confirmed only the ∼50 and 26 kDa bands to be FADS, and only peptides located downstream of the MPTb domain encoded by exon 2 were detected in the 26 kDa band (Figure 2A and 2C). As predicted by the identified variants, individuals S4a and S5, both homozygous for frameshift variants in exon 2, showed no detectable FADS2. In contrast, subject S1a, homozygous for a single amino acid deletion (p.Ser495del) in the FADS domain, showed FADS2 amounts within the control range. Notwithstanding, in subject S2, in whom one allele is predicted to cause an amino acid change (p.Arg530Cys) in the FADS domain and a late-onset riboflavin-responsive MADD phenotype, immunoblot analysis also revealed severely decreased amounts of FADS2. The observation that all mutations affect only the FADS2 band argues for unspecific cross-reactive signals at 60 and 26 kDa. Nonetheless, the findings of unique FADS peptides in the 26 kDa band suggest that at least a minor amount of isoform(s) might add to the observed residual FADS activity. A faint 26 kDa band was repeatedly observed in liver tissue from S5, but not in control liver (Figure 2B), suggesting that compensatory increases in FADS isoforms are masked by unspecific signals in fibroblast cells (Figure 2A). However, because of the limited amount of liver tissue, we were not able to verify the identity of the 26 kDa band. Two shorter isoforms (isoform 3 [GenBank: NM_00114891] and isoform 4 [GenBank: NM_001184892]) have been reported in RefSeq (accessed January 2016). It is not likely that the 26 kDa protein corresponds to any of these, because both short isoforms contain sequences specific to the MPTb domain. This argues for alternative mechanisms by which residual FADS activity is produced in FLAD1 mutant cells.
Figure 2.
Analysis of FADS Proteins
(A and B) Protein extract from cultured fibroblasts (A) or a liver biopsy (B) were separated by SDS-PAGE and immunoblotted with polyclonal antibodies against the C-terminal end of the human FADS. Subjects (S1a, S2, S4a, and S5) and healthy control individuals (C1, C2, C3, and C) are indicated on top of the gels. Purified human FADS2 is indicated by “hF.” Trihalo staining or GAPDH were used as loading controls. The numbers (P1–P4) on the right side of the immunoblot in (A) correspond to FADS peptides detected by mass spectrometry (illustrated in C). Bands marked with an asterisk represent non-specific anti-FADS antibody bindings as defined by mass spectrometry.
(C) The amino acid sequence of human FADS1 (GenBank: NP_079483.3) is shown in one-letter symbols. The MPTb domain and the FADS domain are shaded in violet and orange, respectively, and the four Met residues upstream of the FADS domain are written in bold, and their protein positions are indicated. Peptides selected as identifiers of FADS and for use in the mass-spectrometry analysis are written in superscript and further listed below with their analysis number (P1–P5) and protein domain. The signal from peptide 5 was treated as a nonsignificant finding because it was detected at the boarder of the detection limit of the SRM analysis in all analyzed gel bands. The analysis was performed on gel bands processed from two independent cell lysates, and each peptide mixture was injected and analyzed by msass spectrometry twice.
Alternative FLAD1 Transcripts and Activation of Downstream ATG Start Sites Might Rescue LOF Variants
Because most of the investigated individuals carried frameshift variants in exon 2, we were interested in the mutation load of FLAD1. We therefore divided FLAD1 into two parts: one 1,117 bp fragment covering exons 1 and 2 and coding for the MPTb domain and a second 647 bp fragment covering exons 3–7 and coding for the FADS domain. Although the ExAC Browser records an equal distribution of missense and synonymous variants among the first (0.178 variants/bp) and second (0.156 variants/bp) parts of the gene, it lists 47 alleles with predicted LOF variants affecting exon 1 or 2 and only two LOF variants affecting exons 3–7. This significant accumulation of LOF variants in exons 1 and 2 (47 LOF variants within 1,117 bp) rather than in exons 3–7 (two LOF variants within 647 bp, p = 10−7, Poisson test) suggests alternative mechanisms resulting nonetheless in functional FADS.
It has been shown that the FADS domain alone, without the MPTb domain, is able to mediate FADS activity.11 Because the frameshift variants affect the MPTb domain, we speculated whether additional splice variants or a re-initiation of translation using a downstream ATG codon could encode an active FADS. A prerequisite for re-initiation of translation would be a stable FLAD1 transcript. qRT-PCR analysis from subject S4a’s fibroblast cells with frameshift variants on both alleles revealed stable FLAD1 transcript levels, which were reduced only by about 50% in comparison to control levels (Figure S1), indicating that the transcripts were only partially degraded by nonsense-mediated decay. Likewise, sequence analysis of FLAD1 RT-PCR products amplified from individual S2’s fibroblasts showed heterozygosity for the c.1588C>T variant, indicating that the C allele containing c.568_569dupGC in exon 2 was stably expressed (data not shown).
The human FLAD1 transcript contains two in-frame downstream ATG codons located between the frameshift variants and the region encoding the FADS domain (Figures 1B and 3A). Theoretically, these truncated FADS proteins, starting at Met268 or Met355, could be translated from the variant FLAD1 transcripts and thereby ensure cytosolic FAD synthesis. On the basis of the idea of truncated products with an active FADS domain, three different constructs were expressed in primary fibroblast cells from a control individual: the full-length construct starting at Met98, a second construct starting at Met268, and a third construct at Met355. All of the FLAD1 constructs were expressed and resulted in stable proteins easily detected by immunoblotting (Figure 3C). When analyzing FADS activity in cell extracts, we found that expression of the Met268 construct resulted in a FAD synthesis rate higher than that of control cells (data not shown). These data support the possibility that re-initiation of translation using an in-frame ATG codon would result in a stable protein containing an active FADS domain. However, the Met268 and Met355 proteins did not migrate with a mass similar to that of any of the FADS bands detected by immunoblot analysis of case and control fibroblasts, including the 26 kDa band (Figure 3C).
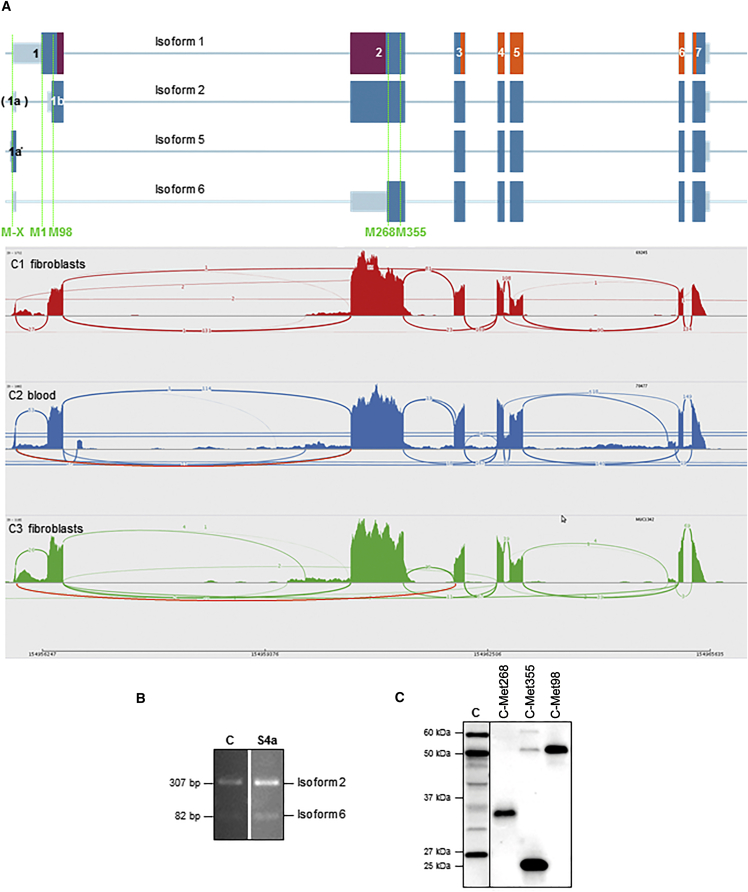
Figure 3.
Alternative FLAD1 Transcripts and Their Predicted Products
(A) Analysis of transcriptome data for FLAD1 from two unrelated fibroblast control samples (C1 and C3) and one human blood sample (C2) revealed several possible transcripts, summarized as Sashimi plots (bottom). Schematic diagram shows isoform transcript structures with MPTb and FADS domains marked in violet and orange, respectively (top). The most abundant transcript (isoform 2) corresponds to the sequence reported in RefSeq (GenBank: NM_201398.2) but has an alternative intron in exon 1 and lacks the predicted MTS. Isoforms 5 and 6 represent transcripts without exons 1b and/or 2, where the identified FLAD1 frameshift variants are located, but both are able to express an active FADS domain. Isoform 5 can use the Met marked as M-X in the very beginning of the transcript, whereas isoform 6 utilizes Met268. In RNA from fibroblasts or blood, we did not identify a single transcript resulting in isoforms 1 (GenBank: NM_025207.4), 3 (GenBank: NM_00114891), or 4 (GenBank: NM_001184892) as reported in RefSeq. Isoforms 3 and 4 are not shown.
(B) PCR products of cDNA from control subjects and subject S4a confirmed the presence of non-degraded FLAD1 mRNA (all details are provided in Figure S1). The primer pair is able to amplify either isoform 2 or isoform 6 of the transcript, resulting in 307 or 84 bp products, respectively.
(C) Immunoblot analysis of the different FADS isoforms (Met98, Met268, and Met355) overproduced in fibroblasts derived from a healthy control individual shows their stability and detectability by anti-FADS antibody. A control fibroblast sample (C) is shown for comparison.
We next investigated the possibility of alternative transcripts by analyzing RNA sequencing data generated from two fibroblast samples (C1 and C3) or a whole-blood RNA sample (C2). Figure 3A summarizes in Sashimi plots all detected RNA splice variants. In both tissues, FADS2 (comprising exons 1a and 1b) represents the most abundant transcript; the next most-abundant transcript is missing exon 1a but still encodes the same cytosolic protein isoform of 54 kDa. In none of the investigated samples did we find evidence of FADS1, which covers all of exon 1 and retains intron 1a, or isoforms 3 (GenBank: NM_00114891) and 4 (GenBank: NM_001184892) reported in RefSeq. These observations are in agreement with the immunoblotting experiments (Figure 2). In control fibroblast C3, 27 out of 28 reads starting with exon 1a continued in exon 1b; however, one paired read detected a transcript variant covering exon 1a, including an ATG codon directly connected in frame to the open reading frame in exon 3 (isoform 5 in Figure 3A). Isoform 5, missing exons 1b and 2, has the potential to encode a 26 kDa functional FADS missing the MPTb domain. In addition, we found a substantial number (11) of mRNA reads directly connecting exons 1a and 2 (whereas 114 reads connected exons 1b and 2). None of them included the ATG codon in exon 1a. This isoform, now isoform 6, is predicted to code for the FADS starting with Met268. In order to validate the presence of isoform 6, we performed an RT-PCR experiment by using primers specific to exons 1a and 2. Unexpectedly, we found substantial amounts of both isoforms 2 and 6 (Figure 3B). None of the identified frameshift variants in the ExAC Browser or the frameshift variants presented here, which are predicted to be pathogenic considering isoforms 1 and 2, would affect the FADS encoded by isoform 6. Immunoblot analysis using case and control fibroblasts failed to detect the predicted FADS proteins starting with Met268 or Met355, but it did identify a 26 kDa band containing two FADS peptides located downstream of the MPTb domain (Figures 2 and 3C). The FADS uniqueness of these peptides to the human proteome, as confirmed by bioinformatics analyses, strongly suggests that the 26 kDa band indeed contains FADS (see Subjects and Methods). We speculate that the 26 kDa band is based on isoform 5 and/or a proteolytically processed version of the Met268 isoform 6 and has an active FADS domain that allows residual FADS activity, even in cells with frameshift variants.
FLAD1 Missense Variants and Responsiveness to Riboflavin
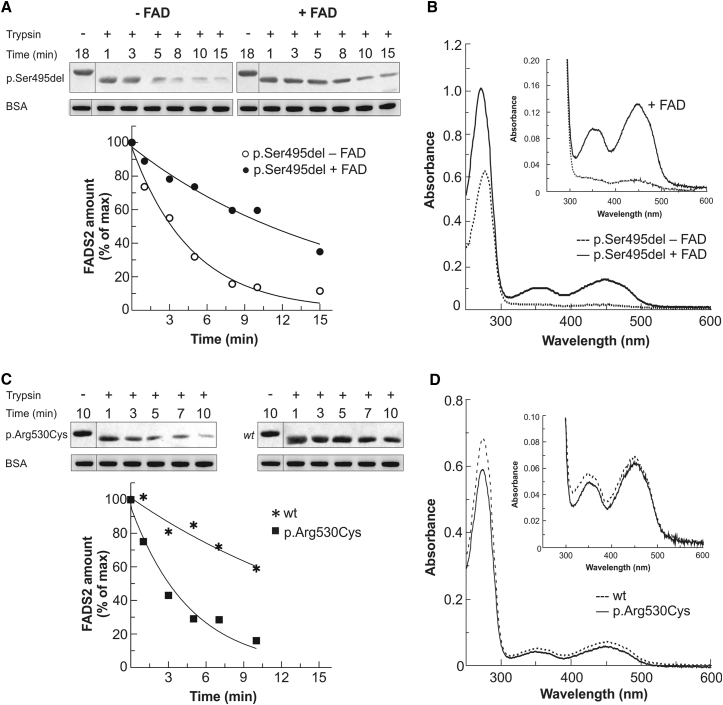
Natural human FADS, a putative component of FAD delivery machinery, behaves as a FAD-binding protein.9 Subjects S1b, S2, and S3 were all found to carry FADS proteins (p.Ser495del and p.Arg530Cys) with a potentially altered ability to bind to and deliver flavin cofactor to nascent client apo-flavoenzymes, as well as altered stabilization requirements. To investigate this further, we overproduced His-tagged p.Ser495del and p.Arg530Cys mutant FADS2 proteins in E. coli and characterized protein stability and activity with and without in vitro reconstitution with FAD. As illustrated in Figure 4, purified variant p.Ser495del and p.Arg530Cys FADS2 proteins showed higher sensitivity to proteolytic degradation than did WT FADS2 (Figures 4A and 4C), indicating that these polypeptides are less tightly folded and consequently are more susceptible to degradation by cellular proteases. Spectrophotometric analysis of the purified WT, p.Ser495del, and p.Arg530Cys FADS2 proteins revealed that the p.Ser495del protein, but not the p.Arg530Cys protein, contained less bound flavin (Figures 4B and 4D). In accordance with previous studies showing that WT FADS2 is able to bind free FAD,9 in vitro reconstitution with a molar excess of FAD resulted in a fully cofactor-bound p.Ser495del protein and rescued proteolytic degradation (Figures 4A and 4B). Supplementation of flavin in the p.Arg530Cys protein did not change its spectrophotometric properties and did not increase resistance to proteolytic degradation (data not shown).
Figure 4.
Characterization of Recombinant 6His-p.Arg530Cys and 6His-p.Ser495del FADS2 Proteins
(A and B) The trypsin sensitivity (A) and degree of FAD saturation (B) of 6His-p.Ser495del FADS2 are reported in comparison with those of the same protein after reconstitution with a 2.5-fold molar excess of FAD. In (B), spectra of both 6His-p.Ser495del FADS2 purified in its apoform (0.58 mg/mL protein concentration, dashed line) and the reconstituted holoform (0.64 mg/mL protein concentration, black line) are reported.
(C and D) The trypsin sensitivity (C) and degree of FAD saturation (D) of 6His-p.Arg530Cys FADS2 are reported in comparison with those of WT FADS2. In (D), spectra of both 6His-p.Arg530Cys FADS2 (0.44 mg/mL protein concentration, straight line) and WT protein (0.5 mg/mL protein concentration, dashed line) are reported.
Trypsin sensitivity was analyzed by immunoblotting. The control represents protein treated under the same condition but in the absence of trypsin (−). The slower migration of the control band might reflect that the 6His tag is rapidly removed upon addition of trypsin. BSA was added to the loading buffer as an internal standard. In the graph, the time course of the limited proteolysis is reported as a percentage of the control amount (arbitrarily set to 100%). The degree of FAD saturation was estimated from the UV/Vis absorbance spectra.
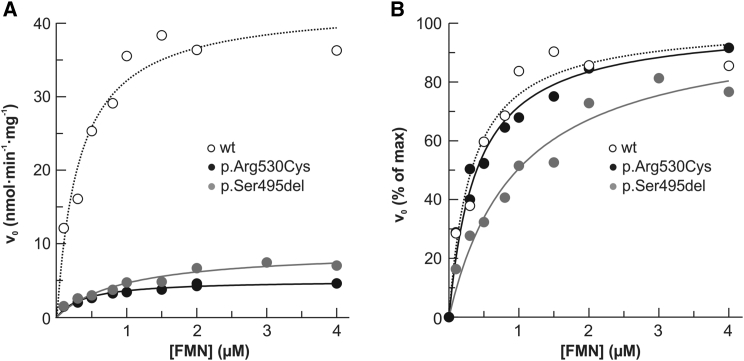
When fully saturated with a molar excess of FAD, both mutant FADS2 proteins still showed severely less catalytic activity than did the WT protein (Figure 5). These kinetic studies suggest that even though in vitro reconstitution with FAD can rescue the stability of the p.Ser495del protein, it cannot correct its maximum in vitro catalytic activity. Nevertheless, because of its increased Km value for FMN (inset in Figure 4B), the p.Ser495del protein is expected to be more sensitive to physiological changes in cellular amounts of riboflavin than the WT counterpart.
Figure 5.
Kinetic Characterization of Recombinant 6His-p.Arg530Cys and 6His-p.Ser495del FADS2 Proteins
The dependence of FMN concentration on the rate of FAD synthesis catalyzed by WT (0.17 nmol), 6His-Arg530Cys (0.17 nmol), or 6His-Ser495del (0.18 nmol) FADS2. The FAD synthesis catalyzed by WT FADS2 is presented as open symbols. Mutant FADS is presented as closed symbols. The rate of FAD synthesis was measured in the presence of 100 μmol/L ATP and 5 mmol/L MgCl2. v0 was measured by the initial rate of fluorescence decrease (excitation at 450 nm and emission at 520 nm) and expressed in nmol FMN · min−1 · mg−1 mutant FADS2 (A) and as a percentage of the Vmax value (set arbitrarily to 100%) (B). Data points are fitted according to the Michaelis-Menten equation.
After identification of MADD or FLAD1 variants, riboflavin treatment was initiated in subjects S1b, S2, S3, S4a, and S6. In agreement with the expected molecular actions from the in vitro studies, individuals S1b, S2, and S3 showed responsiveness to riboflavin in vivo, including improvement of cardiac and muscular functions, some decreases in acylcarnitine species, and normalization of urine organic acids (Table S3). A mild improvement of hypotonia was observed in subject S4a, whereas no evident beneficial effect was reported in subject S6.
Discussion
We describe nine affected individuals from seven independent families affected by MADD and/or multiple-respiratory-chain deficiency and mutations in FLAD1, which encodes the enzyme FADS. The individuals show a metabolic myopathy with lipid storage and respiratory-chain deficiency, resembling what is seen in individuals with ETFDH defects and a late-onset riboflavin-responsive MADD,28 yet they do not exhibit the progressive neurological signs, involving motor, sensory, or cranial neuronopathies that are seen in the more recently identified individuals diagnosed with Brown-Vialetto-Van Laere syndrome (MIM: 211530), involving defects in RFVT-encoding genes.32, 35, 51, 52, 53 When investigated, individuals with FLAD1 variants showed no hearing or visual impairment, and their peripheral-nerve conduction velocity was normal. Typical clinical phenotypes were hypotonia and swallowing and speech difficulties. Scoliosis was reported in three individuals, and many developed respiratory insufficiencies or arrest. Cardiomyopathy was reported only in a single individual, and two required pacemaker implantation because of tachycardia or sudden cardiac arrest. Most subjects presented with disease in early infancy, and four died within the first year of life or in late childhood. Four subjects are still alive, and three (S1b, S2, and S3, aged 22, 44, and 56 years, respectively) differ from the others because they have gene variants affecting a single amino acid in the FADS domain, share a milder clinical course, and are responsive to treatment with riboflavin.
The clinical heterogeneity is mirrored by an allelic series of variants, including transcript-specific frameshift variants and variants affecting only a single amino acid. FAD is an essential cofactor, and FADS is the only known human enzyme catalyzing the synthesis of FAD from FMN. The unexpected FLAD1 frameshift variants led us to discover previously undescribed transcript isoforms. FLAD1 codes for a protein consisting of MPTb and FADS domains. All of the identified frameshift variants affect the MPTb domain. Early in evolution, these two domains appeared as separate proteins, encoded by two different genes, whereas in mammals they appear as one protein containing two domains.1 However, the FADS domain was shown to function independently from the other domain.11 Indeed, we have discovered FLAD1 isoforms that code for only the FADS domain (isoforms 5 and 6 in Figure 3). The existence of these isoforms might explain why affected individuals with biallelic FLAD1 frameshift variants still harbor substantial FADS activity (Table 2). However, despite residual FADS activity, homozygous carriers of frameshift variants are affected, and their disease seems more severe than that observed in individuals harboring single amino acid changes in the FADS domain (Table 1). This could be explained by at least four different mechanisms. (1) the mRNA expression levels of FADS isoforms 5 and 6 were found to be low, and although we did not find conclusive evidence of their protein products in fibroblasts from case and control individuals, protein mass spectrometry of the 26 kDa band supports their existence (Figures 2 and 3). Determining the exact identity of the 26 kDa band will require further studies. (2) We only studied blood and fibroblast transcriptomes, and we have no information about the isoform expression pattern in the affected (muscle) tissue. (3) Although, there might be sufficient FADS activity, the isoforms lack the uncertain but potentially important activity of the MPTb domain. Indeed, it has been recently suggested that the MPTb domain possess FAD hydrolase activity10 and might be needed for optimal FAD release to apoproteins.9, 12, 54 Addressing this question will require more experiments. (4) Finally, even though our protein and transcriptome data do not support substantial expression of the mitochondrial FLAD1 isoform 1, the mitochondrial isoform might be expressed in other tissues or in other experimental conditions where direct interaction between FADS and apoproteins is needed to ensure sufficient or catalytically controlled flavinylation of mitochondrial proteins.9, 54 Loss of isoforms 1 and 2 could result in disturbed subcellular distribution of FADS. Isoforms 5 and 6 do not harbor a MTS. Although some polypeptides can reach mitochondria without a cleavable MTS,55 the frameshift variants in the MPTb domain most likely result in deficiency of mitochondrial FADS. Mitochondrial biogenesis and function in these cells would then depend on supply from cytosolic FAD synthesis. In accordance with this idea, cellular FAD amounts within or slightly below the control range were observed in all investigated subjects. Significant decreases in FAD content were found only in enriched mitochondrial fractions from subject S4. Moreover, the lipid storage and decreased activity of respiratory-chain enzymes in muscle homogenates from several individuals (Tables 1, S4, and S5) points specifically to a mitochondrial defect. In subject S4a, this was accompanied by decreased amounts of ETFDH and the SDHA flavocomponent of complex II (Figure S4). Similarly, we would expect amounts of other mitochondrial flavoproteins, such as acyl-CoA dehydrogenases, to be decreased as well.20, 21, 22, 23 Even though this was not investigated, all reported individuals with variant FLAD1 genotypes showed MADD or ethylmalonic and/or adipic aciduria (Tables 1 and S3), which argues for a more general impairment in flavinylation of mitochondrial proteins under these conditions. Decreased activity of complex II combined with MADD biochemistry was also observed in a recently reported subject with riboflavin-responsive exercise intolerance due to a defect in mitochondrial FAD transport.33
Variants affecting only a single amino acid were detected in four families. The c.508T>C (p.Phe170Leu) missense variant identified in F6 in cis with a frameshift variant most likely does not contribute to the FADS deficiency. In families F2 and F3, we discovered a c.1588C>T (p.Arg530Cys) missense variant, and in a family F1, we found a 3 bp deletion (c.1484_1486del [p.Ser495del]); both of these affect the FADS domain. In vitro studies demonstrated that these variants result in impaired but detectable FADS activity (Figure 5). At least for the deletion variants, we could show that incubation with a molar excess of FAD significantly improved protein stability (Figure 4), arguing for a chaperone-like action of FAD, similar to what has been reported in a number of other metabolic disorders caused by gene variations (mainly of the missense type) in proteins using FAD as a cofactor.15, 16, 17, 18, 19, 56, 57 Concluding on the effectiveness of riboflavin treatment in individuals with different types of FLAD1 variants will require further in vitro studies of FLAD1 genotypes and a larger group of individuals with longer follow up. However, because there are no reported side effects of riboflavin, all FLAD1-defective individuals should initially be treated with riboflavin no matter their type of mutation. The fatal outcome of the untreated brother of an older sister (S1a), who responded to riboflavin, stresses the importance of early diagnosis and treatment in these disorders.
In conclusion, our studies have identified FLAD1 variants as a cause of potentially treatable myopathies associated with MADD. This further establishes the heterogeneous etiology of MADD; some have ETF and/or ETFDH variants, and others have defects in genes responsible for cellular uptake of riboflavin, synthesis of flavin cofactors, or mitochondrial FAD transport. Because there are still unsolved cases of riboflavin-responsive MADD, it can be expected that the list of involved genes will still grow in the future. Besides being important for the pathomechanism of FLAD1 defects, our findings also stress that care should be taken in the interpretation of predicted LOF variants in large-scale sequencing projects. Sometimes LOF variants affect only specific isoforms, and such findings do not allow conclusions that the particular gene is not essential.
Acknowledgments
We thank all affected individuals and their families for their participation and for providing important samples for the present research study. We thank Dr. Francesco De Cillis for his participation in the early stages of this work, Dr. James Pitt for biochemical evaluation of individuals S1a and S1b, Dr. Norma Romero for muscle histology analysis of individual S7, and Dr. Maria Teresa Balsimelli, Dr. Carla Cozzolino, Prof. Vincenzo Nigro, and Dr. Filippo Maria Santorelli for their clinical care and biochemical evaluation of individual S6. The study was supported by the Aarhus County Research Initiative, the Danish Council of Independent Medical Research (4004-00548 to R.K.J.O.), the John and Birthe Meyer Foundation (N.G.), the European Commission FP7-PEOPLE-ITN Mitochondrial European Educational Training Project (GA 317433 to R.H., J.A.M., and H.P.), the Neuromuscular Research Association Angela Maria Nieddu (L.V.), the Telethon Genetic BioBank (GTB12001D to E.P.), the EuroBioBank network (E.P.), the Telethon Foundation (grant GGP15041 to D.G.), the Italian Ministry of Health (GR2010–2316392 to D.G.), the Programma Operativo Nazionale “Ricerca e Competitività” 2007–2013 (01_00937 to M.B.), the European Research Council (309548 to R.H.), the UK Medical Research Council (G1000848 to R.H.), the German Bundesministerium für Bildung und Forschung (BMBF) through the German Network for Mitochondrial Disorders (mitoNET; 01GM1113C to T.M. and H.P.), the E-Rare project GENOMIT (01GM1207 to T.M. and H.P.), and the Juniorverbund in der Systemmedizin “mitOmics” (FKZ 01ZX1405C to T.B.H.).
Published: June 2, 2016
Footnotes
Supplemental Data include a Supplemental Note, four figures, and five tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.04.006.
Contributor Information
Rikke K.J. Olsen, Email: rikke.olsen@clin.au.dk.
Maria Barile, Email: maria.barile@uniba.it.
Accession Numbers
The accession numbers for the nine FLAD1 variants reported in this paper are ClinVar: SCV000266355, SCV000266356, SCV000266357, SCV000266358, SCV000266359, SCV000266360, SCV000266361, SCV000266362, and SCV000266363.
Web Resources
ClinVar, http://www.ncbi.nlm.nih.gov/clinvar/
ExAC Browser, http://exac.broadinstitute.org/
ExPasy ProtParam, http://web.expasy.org/protparam/
Integrative Genomics Viewer, https://www.broadinstitute.org/igv/
MutationTaster, http://www.mutationtaster.org/
OMIM, http://www.omim.org/
Skyline, https://skyline.gs.washington.edu/labkey/wiki/home/software/Skyline/
UCSC Genome Browser, http://www.genome.ucsc.edu/
UniProt, http://www.uniprot.org/
Supplemental Data
References
- 1.Barile M., Giancaspero T.A., Brizio C., Panebianco C., Indiveri C., Galluccio M., Vergani L., Eberini I., Gianazza E. Biosynthesis of flavin cofactors in man: implications in health and disease. Curr. Pharm. Des. 2013;19:2649–2675. doi: 10.2174/1381612811319140014. [DOI] [PubMed] [Google Scholar]
- 2.Joosten V., van Berkel W.J. Flavoenzymes. Curr. Opin. Chem. Biol. 2007;11:195–202. doi: 10.1016/j.cbpa.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 3.Lienhart W.D., Gudipati V., Macheroux P. The human flavoproteome. Arch. Biochem. Biophys. 2013;535:150–162. doi: 10.1016/j.abb.2013.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yonezawa A., Inui K. Novel riboflavin transporter family RFVT/SLC52: identification, nomenclature, functional characterization and genetic diseases of RFVT/SLC52. Mol. Aspects Med. 2013;34:693–701. doi: 10.1016/j.mam.2012.07.014. [DOI] [PubMed] [Google Scholar]
- 5.Depeint F., Bruce W.R., Shangari N., Mehta R., O’Brien P.J. Mitochondrial function and toxicity: role of B vitamins on the one-carbon transfer pathways. Chem. Biol. Interact. 2006;163:113–132. doi: 10.1016/j.cbi.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 6.McCormick D.B. A trail of research on cofactors: an odyssey with friends. J. Nutr. 2000;130(2S, Suppl):323S–330S. doi: 10.1093/jn/130.2.323S. [DOI] [PubMed] [Google Scholar]
- 7.Brizio C., Galluccio M., Wait R., Torchetti E.M., Bafunno V., Accardi R., Gianazza E., Indiveri C., Barile M. Over-expression in Escherichia coli and characterization of two recombinant isoforms of human FAD synthetase. Biochem. Biophys. Res. Commun. 2006;344:1008–1016. doi: 10.1016/j.bbrc.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 8.Galluccio M., Brizio C., Torchetti E.M., Ferranti P., Gianazza E., Indiveri C., Barile M. Over-expression in Escherichia coli, purification and characterization of isoform 2 of human FAD synthetase. Protein Expr. Purif. 2007;52:175–181. doi: 10.1016/j.pep.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 9.Torchetti E.M., Bonomi F., Galluccio M., Gianazza E., Giancaspero T.A., Iametti S., Indiveri C., Barile M. Human FAD synthase (isoform 2): a component of the machinery that delivers FAD to apo-flavoproteins. FEBS J. 2011;278:4434–4449. doi: 10.1111/j.1742-4658.2011.08368.x. [DOI] [PubMed] [Google Scholar]
- 10.Giancaspero T.A., Galluccio M., Miccolis A., Leone P., Eberini I., Iametti S., Indiveri C., Barile M. Human FAD synthase is a bi-functional enzyme with a FAD hydrolase activity in the molybdopterin binding domain. Biochem. Biophys. Res. Commun. 2015;465:443–449. doi: 10.1016/j.bbrc.2015.08.035. [DOI] [PubMed] [Google Scholar]
- 11.Miccolis A., Galluccio M., Giancaspero T.A., Indiveri C., Barile M. Bacterial over-expression and purification of the 3‘phosphoadenosine 5’phosphosulfate (PAPS) reductase domain of human FAD synthase: functional characterization and homology modeling. Int. J. Mol. Sci. 2012;13:16880–16898. doi: 10.3390/ijms131216880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miccolis A., Galluccio M., Nitride C., Giancaspero T.A., Ferranti P., Iametti S., Indiveri C., Bonomi F., Barile M. Significance of redox-active cysteines in human FAD synthase isoform 2. Biochim. Biophys. Acta. 2014;1844:2086–2095. doi: 10.1016/j.bbapap.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 13.Torchetti E.M., Brizio C., Colella M., Galluccio M., Giancaspero T.A., Indiveri C., Roberti M., Barile M. Mitochondrial localization of human FAD synthetase isoform 1. Mitochondrion. 2010;10:263–273. doi: 10.1016/j.mito.2009.12.149. [DOI] [PubMed] [Google Scholar]
- 14.Higgins C.L., Muralidhara B.K., Wittung-Stafshede P. How do cofactors modulate protein folding? Protein Pept. Lett. 2005;12:165–170. doi: 10.2174/0929866053005782. [DOI] [PubMed] [Google Scholar]
- 15.Saijo T., Tanaka K. Isoalloxazine ring of FAD is required for the formation of the core in the Hsp60-assisted folding of medium chain acyl-CoA dehydrogenase subunit into the assembly competent conformation in mitochondria. J. Biol. Chem. 1995;270:1899–1907. doi: 10.1074/jbc.270.4.1899. [DOI] [PubMed] [Google Scholar]
- 16.Cornelius N., Frerman F.E., Corydon T.J., Palmfeldt J., Bross P., Gregersen N., Olsen R.K. Molecular mechanisms of riboflavin responsiveness in patients with ETF-QO variations and multiple acyl-CoA dehydrogenation deficiency. Hum. Mol. Genet. 2012;21:3435–3448. doi: 10.1093/hmg/dds175. [DOI] [PubMed] [Google Scholar]
- 17.Henriques B.J., Rodrigues J.V., Olsen R.K., Bross P., Gomes C.M. Role of flavinylation in a mild variant of multiple acyl-CoA dehydrogenation deficiency: a molecular rationale for the effects of riboflavin supplementation. J. Biol. Chem. 2009;284:4222–4229. doi: 10.1074/jbc.M805719200. [DOI] [PubMed] [Google Scholar]
- 18.Haack T.B., Danhauser K., Haberberger B., Hoser J., Strecker V., Boehm D., Uziel G., Lamantea E., Invernizzi F., Poulton J. Exome sequencing identifies ACAD9 mutations as a cause of complex I deficiency. Nat. Genet. 2010;42:1131–1134. doi: 10.1038/ng.706. [DOI] [PubMed] [Google Scholar]
- 19.Schiff M., Haberberger B., Xia C., Mohsen A.W., Goetzman E.S., Wang Y., Uppala R., Zhang Y., Karunanidhi A., Prabhu D. Complex I assembly function and fatty acid oxidation enzyme activity of ACAD9 both contribute to disease severity in ACAD9 deficiency. Hum. Mol. Genet. 2015;24:3238–3247. doi: 10.1093/hmg/ddv074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Veitch K., Draye J.P., Vamecq J., Causey A.G., Bartlett K., Sherratt H.S., Van Hoof F. Altered acyl-CoA metabolism in riboflavin deficiency. Biochim. Biophys. Acta. 1989;1006:335–343. doi: 10.1016/0005-2760(89)90021-0. [DOI] [PubMed] [Google Scholar]
- 21.Ross N.S., Hansen T.P. Riboflavin deficiency is associated with selective preservation of critical flavoenzyme-dependent metabolic pathways. Biofactors. 1992;3:185–190. [PubMed] [Google Scholar]
- 22.Nagao M., Tanaka K. FAD-dependent regulation of transcription, translation, post-translational processing, and post-processing stability of various mitochondrial acyl-CoA dehydrogenases and of electron transfer flavoprotein and the site of holoenzyme formation. J. Biol. Chem. 1992;267:17925–17932. [PubMed] [Google Scholar]
- 23.Gianazza E., Vergani L., Wait R., Brizio C., Brambilla D., Begum S., Giancaspero T.A., Conserva F., Eberini I., Bufano D. Coordinated and reversible reduction of enzymes involved in terminal oxidative metabolism in skeletal muscle mitochondria from a riboflavin-responsive, multiple acyl-CoA dehydrogenase deficiency patient. Electrophoresis. 2006;27:1182–1198. doi: 10.1002/elps.200500687. [DOI] [PubMed] [Google Scholar]
- 24.Watmough N.J., Frerman F.E. The electron transfer flavoprotein: ubiquinone oxidoreductases. Biochim. Biophys. Acta. 2010;1797:1910–1916. doi: 10.1016/j.bbabio.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 25.Gempel K., Topaloglu H., Talim B., Schneiderat P., Schoser B.G., Hans V.H., Pálmafy B., Kale G., Tokatli A., Quinzii C. The myopathic form of coenzyme Q10 deficiency is caused by mutations in the electron-transferring-flavoprotein dehydrogenase (ETFDH) gene. Brain. 2007;130:2037–2044. doi: 10.1093/brain/awm054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olsen R.K., Olpin S.E., Andresen B.S., Miedzybrodzka Z.H., Pourfarzam M., Merinero B., Frerman F.E., Beresford M.W., Dean J.C., Cornelius N. ETFDH mutations as a major cause of riboflavin-responsive multiple acyl-CoA dehydrogenation deficiency. Brain. 2007;130:2045–2054. doi: 10.1093/brain/awm135. [DOI] [PubMed] [Google Scholar]
- 27.Horvath R. Update on clinical aspects and treatment of selected vitamin-responsive disorders II (riboflavin and CoQ 10) J. Inherit. Metab. Dis. 2012;35:679–687. doi: 10.1007/s10545-011-9434-1. [DOI] [PubMed] [Google Scholar]
- 28.Grünert S.C. Clinical and genetical heterogeneity of late-onset multiple acyl-coenzyme A dehydrogenase deficiency. Orphanet J. Rare Dis. 2014;9:117. doi: 10.1186/s13023-014-0117-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wen B., Dai T., Li W., Zhao Y., Liu S., Zhang C., Li H., Wu J., Li D., Yan C. Riboflavin-responsive lipid-storage myopathy caused by ETFDH gene mutations. J. Neurol. Neurosurg. Psychiatry. 2010;81:231–236. doi: 10.1136/jnnp.2009.176404. [DOI] [PubMed] [Google Scholar]
- 30.Bosch A.M., Abeling N.G., Ijlst L., Knoester H., van der Pol W.L., Stroomer A.E., Wanders R.J., Visser G., Wijburg F.A., Duran M., Waterham H.R. Brown-Vialetto-Van Laere and Fazio Londe syndrome is associated with a riboflavin transporter defect mimicking mild MADD: a new inborn error of metabolism with potential treatment. J. Inherit. Metab. Dis. 2011;34:159–164. doi: 10.1007/s10545-010-9242-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ho G., Yonezawa A., Masuda S., Inui K., Sim K.G., Carpenter K., Olsen R.K., Mitchell J.J., Rhead W.J., Peters G., Christodoulou J. Maternal riboflavin deficiency, resulting in transient neonatal-onset glutaric aciduria Type 2, is caused by a microdeletion in the riboflavin transporter gene GPR172B. Hum. Mutat. 2011;32:E1976–E1984. doi: 10.1002/humu.21399. [DOI] [PubMed] [Google Scholar]
- 32.Haack T.B., Makowski C., Yao Y., Graf E., Hempel M., Wieland T., Tauer U., Ahting U., Mayr J.A., Freisinger P. Impaired riboflavin transport due to missense mutations in SLC52A2 causes Brown-Vialetto-Van Laere syndrome. J. Inherit. Metab. Dis. 2012;35:943–948. doi: 10.1007/s10545-012-9513-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schiff M., Veauville-Merllié A., Su C.H., Tzagoloff A., Rak M., Ogier de Baulny H., Boutron A., Smedts-Walters H., Romero N.B., Rigal O. SLC25A32 Mutations and Riboflavin-Responsive Exercise Intolerance. N. Engl. J. Med. 2016;374:795–797. doi: 10.1056/NEJMc1513610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pyle A., Smertenko T., Bargiela D., Griffin H., Duff J., Appleton M., Douroudis K., Pfeffer G., Santibanez-Koref M., Eglon G. Exome sequencing in undiagnosed inherited and sporadic ataxias. Brain. 2015;138:276–283. doi: 10.1093/brain/awu348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haack T.B., Hogarth P., Kruer M.C., Gregory A., Wieland T., Schwarzmayr T., Graf E., Sanford L., Meyer E., Kara E. Exome sequencing reveals de novo WDR45 mutations causing a phenotypically distinct, X-linked dominant form of NBIA. Am. J. Hum. Genet. 2012;91:1144–1149. doi: 10.1016/j.ajhg.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kornblum C., Nicholls T.J., Haack T.B., Schöler S., Peeva V., Danhauser K., Hallmann K., Zsurka G., Rorbach J., Iuso A. Loss-of-function mutations in MGME1 impair mtDNA replication and cause multisystemic mitochondrial disease. Nat. Genet. 2013;45:214–219. doi: 10.1038/ng.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olsen R.K., Brøner S., Sabaratnam R., Doktor T.K., Andersen H.S., Bruun G.H., Gahrn B., Stenbroen V., Olpin S.E., Dobbie A. The ETFDH c.158A>G variation disrupts the balanced interplay of ESE- and ESS-binding proteins thereby causing missplicing and multiple Acyl-CoA dehydrogenation deficiency. Hum. Mutat. 2014;35:86–95. doi: 10.1002/humu.22455. [DOI] [PubMed] [Google Scholar]
- 38.Olsen R.K., Andresen B.S., Christensen E., Bross P., Skovby F., Gregersen N. Clear relationship between ETF/ETFDH genotype and phenotype in patients with multiple acyl-CoA dehydrogenation deficiency. Hum. Mutat. 2003;22:12–23. doi: 10.1002/humu.10226. [DOI] [PubMed] [Google Scholar]
- 39.Ardissone A., Invernizzi F., Nasca A., Moroni I., Farina L., Ghezzi D. Mitochondrial leukoencephalopathy and complex II deficiency associated with a recessive SDHB mutation with reduced penetrance. Mol. Genet. Metab. Rep. 2015;5:51–54. doi: 10.1016/j.ymgmr.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spinazzi M., Casarin A., Pertegato V., Salviati L., Angelini C. Assessment of mitochondrial respiratory chain enzymatic activities on tissues and cultured cells. Nat. Protoc. 2012;7:1235–1246. doi: 10.1038/nprot.2012.058. [DOI] [PubMed] [Google Scholar]
- 41.Hansen J., Palmfeldt J., Vang S., Corydon T.J., Gregersen N., Bross P. Quantitative proteomics reveals cellular targets of celastrol. PLoS ONE. 2011;6:e26634. doi: 10.1371/journal.pone.0026634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fernández-Guerra P., Birkler R.I., Merinero B., Ugarte M., Gregersen N., Rodríguez-Pombo P., Bross P., Palmfeldt J. Selected reaction monitoring as an effective method for reliable quantification of disease-associated proteins in maple syrup urine disease. Mol. Genet. Genomic Med. 2014;2:383–392. doi: 10.1002/mgg3.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.MacLean B., Tomazela D.M., Shulman N., Chambers M., Finney G.L., Frewen B., Kern R., Tabb D.L., Liebler D.C., MacCoss M.J. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Speek A.J., van Schaik F., Schrijver J., Schreurs W.H. Determination of the B2 vitamer flavin--adenine dinucleotide in whole blood by high-performance liquid chromatography with fluorometric detection. J. Chromatogr. A. 1982;228:311–316. doi: 10.1016/s0378-4347(00)80446-6. [DOI] [PubMed] [Google Scholar]
- 45.Bafunno V., Giancaspero T.A., Brizio C., Bufano D., Passarella S., Boles E., Barile M. Riboflavin uptake and FAD synthesis in Saccharomyces cerevisiae mitochondria: involvement of the Flx1p carrier in FAD export. J. Biol. Chem. 2004;279:95–102. doi: 10.1074/jbc.M308230200. [DOI] [PubMed] [Google Scholar]
- 46.Giancaspero T.A., Locato V., de Pinto M.C., De Gara L., Barile M. The occurrence of riboflavin kinase and FAD synthetase ensures FAD synthesis in tobacco mitochondria and maintenance of cellular redox status. FEBS J. 2009;276:219–231. doi: 10.1111/j.1742-4658.2008.06775.x. [DOI] [PubMed] [Google Scholar]
- 47.Liuzzi V.C., Giancaspero T.A., Gianazza E., Banfi C., Barile M., De Giorgi C. Silencing of FAD synthase gene in Caenorhabditis elegans upsets protein homeostasis and impacts on complex behavioral patterns. Biochim. Biophys. Acta. 2012;1820:521–531. doi: 10.1016/j.bbagen.2012.01.012. [DOI] [PubMed] [Google Scholar]
- 48.Giancaspero T.A., Busco G., Panebianco C., Carmone C., Miccolis A., Liuzzi G.M., Colella M., Barile M. FAD synthesis and degradation in the nucleus create a local flavin cofactor pool. J. Biol. Chem. 2013;288:29069–29080. doi: 10.1074/jbc.M113.500066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haack T.B., Kopajtich R., Freisinger P., Wieland T., Rorbach J., Nicholls T.J., Baruffini E., Walther A., Danhauser K., Zimmermann F.A. ELAC2 mutations cause a mitochondrial RNA processing defect associated with hypertrophic cardiomyopathy. Am. J. Hum. Genet. 2013;93:211–223. doi: 10.1016/j.ajhg.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Green P., Wiseman M., Crow Y.J., Houlden H., Riphagen S., Lin J.P., Raymond F.L., Childs A.M., Sheridan E., Edwards S., Josifova D.J. Brown-Vialetto-Van Laere syndrome, a ponto-bulbar palsy with deafness, is caused by mutations in c20orf54. Am. J. Hum. Genet. 2010;86:485–489. doi: 10.1016/j.ajhg.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Johnson J.O., Gibbs J.R., Megarbane A., Urtizberea J.A., Hernandez D.G., Foley A.R., Arepalli S., Pandraud A., Simón-Sánchez J., Clayton P. Exome sequencing reveals riboflavin transporter mutations as a cause of motor neuron disease. Brain. 2012;135:2875–2882. doi: 10.1093/brain/aws161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bosch A.M., Stroek K., Abeling N.G., Waterham H.R., Ijlst L., Wanders R.J. The Brown-Vialetto-Van Laere and Fazio Londe syndrome revisited: natural history, genetics, treatment and future perspectives. Orphanet J. Rare Dis. 2012;7:83. doi: 10.1186/1750-1172-7-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Giancaspero T.A., Colella M., Brizio C., Difonzo G., Fiorino G.M., Leone P., Brandsch R., Bonomi F., Iametti S., Barile M. Remaining challenges in cellular flavin cofactor homeostasis and flavoprotein biogenesis. Front Chem. 2015;3:30. doi: 10.3389/fchem.2015.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kisslov I., Naamati A., Shakarchy N., Pines O. Dual-targeted proteins tend to be more evolutionarily conserved. Mol. Biol. Evol. 2014;31:2770–2779. doi: 10.1093/molbev/msu221. [DOI] [PubMed] [Google Scholar]
- 56.Henriques B.J., Bross P., Gomes C.M. Mutational hotspots in electron transfer flavoprotein underlie defective folding and function in multiple acyl-CoA dehydrogenase deficiency. Biochim. Biophys. Acta. 2010;1802:1070–1077. doi: 10.1016/j.bbadis.2010.07.015. [DOI] [PubMed] [Google Scholar]
- 57.Carrozzo R., Torraco A., Fiermonte G., Martinelli D., Di Nottia M., Rizza T., Vozza A., Verrigni D., Diodato D., Parisi G. Riboflavin responsive mitochondrial myopathy is a new phenotype of dihydrolipoamide dehydrogenase deficiency. The chaperon-like effect of vitamin B2. Mitochondrion. 2014;18:49–57. doi: 10.1016/j.mito.2014.09.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.