Abstract
Evaluating the pathogenicity of a variant is challenging given the plethora of types of genetic evidence that laboratories consider. Deciding how to weigh each type of evidence is difficult, and standards have been needed. In 2015, the American College of Medical Genetics and Genomics (ACMG) and the Association for Molecular Pathology (AMP) published guidelines for the assessment of variants in genes associated with Mendelian diseases. Nine molecular diagnostic laboratories involved in the Clinical Sequencing Exploratory Research (CSER) consortium piloted these guidelines on 99 variants spanning all categories (pathogenic, likely pathogenic, uncertain significance, likely benign, and benign). Nine variants were distributed to all laboratories, and the remaining 90 were evaluated by three laboratories. The laboratories classified each variant by using both the laboratory’s own method and the ACMG-AMP criteria. The agreement between the two methods used within laboratories was high (K-alpha = 0.91) with 79% concordance. However, there was only 34% concordance for either classification system across laboratories. After consensus discussions and detailed review of the ACMG-AMP criteria, concordance increased to 71%. Causes of initial discordance in ACMG-AMP classifications were identified, and recommendations on clarification and increased specification of the ACMG-AMP criteria were made. In summary, although an initial pilot of the ACMG-AMP guidelines did not lead to increased concordance in variant interpretation, comparing variant interpretations to identify differences and having a common framework to facilitate resolution of those differences were beneficial for improving agreement, allowing iterative movement toward increased reporting consistency for variants in genes associated with monogenic disease.
Introduction
The assessment of pathogenicity of genetic variation is one of the more complex and challenging tasks in the field of clinical genetics. It is now clear that enormous genetic variation exists in the human population. Most of this variation, including very rare variants, is unlikely to contribute substantively to human disease. For example, a typical genome sequence and reference genome have about 3.5 million differences, of which 0.6 million are rare or novel.1 As such, the challenge of interpreting the clinical significance of this variation is well recognized as a barrier to furthering genomic medicine.2, 3
We have previously reported both inconsistencies across laboratories in the classification of Mendelian-disease variants and high discordance in the use of a single simple classification system, whereby reviewers showed a bias toward overestimating pathogenicity.4 Furthermore, recent analyses of variant classifications in ClinVar showed that for the 11% (12,895/118,169) of variants with two or more submitters, interpretations differed in 17% (2,229/12,895).3 Inconsistency of the classification of variants across professional genetics laboratories has been reported elsewhere.5 These data highlight the need for a more systematic and transparent approach to variant classification.
Laboratories performing and reporting the results of clinical genetic testing are now tasked with considering a plethora of types of genetic evidence, some applicable to all genes and others specific to individual genes and diseases. To date, laboratories have developed their own methods of variant assessment because the prior American College of Medical Genetics and Genomics (ACMG) variant-reporting guidelines did not address the weighting of evidence for variant classification.6 Some laboratories assign points to types of evidence and generate a score,7 and others define specific combinations of evidence that allow them to arrive at each classification category8 or use a Bayesian framework to combine data types into a likelihood ratio.9 Still others have simply relied on expert judgment of the individual body of evidence on each variant to make a decision.
Deciding how to categorize and weigh each type of evidence is challenging, and guidance has been needed. Making the task even more challenging is that the true pathogenicity is not known for most variants, and it is therefore difficult to validate approaches to variant assessment, particularly for addressing variants that have limited evidence. However, combining the collective experience of experts in the community to begin to build a more systematic and transparent approach to variant classification is essential, and this has led the ACMG and Association for Molecular Pathology (AMP) to develop a framework for evidence evaluation. The initial framework was published in early 201510 and focused on variants in genes associated with Mendelian disease.
The ACMG-AMP guidelines defined 28 criteria (each with an assigned code) that address evidence such as population data, case-control analyses, functional data, computational predictions, allelic data, segregation studies, and de novo observations. Each code is assigned a weight (stand-alone, very strong, strong, moderate, or supporting) and direction (benign or pathogenic), and then rules guide users to combine these evidence codes to arrive at one of five classifications: pathogenic (P), likely pathogenic (LP), variant of uncertain significance (VUS), likely benign (LB), or benign (B). In some cases, the strength of individual criteria can be modified at the discretion of the curator, and the overall classification can be modified with expert judgment. As an example, a minor allele frequency (MAF) greater than the disease prevalence but less than 5% is coded benign strong (BS1); this is considered strong evidence against pathogenicity for a highly penetrant monogenic disorder and supports a LB classification when it is combined with at least one supporting line of evidence against pathogenicity (BP1–BP6). If BS1 is combined with another strong line of evidence against pathogenicity (BS2–BS4), this supports a B classification. Conversely, a variant predicted to be null (PVS1) would be classified as LP if it is absent from population databases (PM2) or P if it is observed to be de novo with confirmed paternity and maternity (PS2). If not enough lines of evidence are invoked to classify a variant as P, LP, LB, or B, or there are valid but contradictory lines of evidence, a variant is interpreted as a VUS.
We set out to evaluate how the ACMG-AMP guidelines compare to individual laboratory approaches to variant classification and explore the variance in the use and interpretation of the pathogenicity criteria. Nine laboratories participating in the Clinical Sequencing Exploratory Research (CSER) consortium evaluated the use of the new ACMG-AMP guidelines and in-house interpretations to assess inter-laboratory concordance by either method of variant classification. Our goals were to evaluate consistency of the use of the ACMG-AMP codes and subsequent pathogenicity classification. Further, we used these criteria to analyze the basis for discordance and sought to reconcile differing implementations with an eye to guidance clarification.
Material and Methods
CSER is a National Human Genome Research Institute (NHGRI)- and National Cancer Institute (NCI)-funded consortium exploring the clinical use of genomic sequencing, developing best practices, and identifying obstacles to implementation. It is composed of nine clinical U-award sites focusing on all aspects of clinical sequencing, the ClinSeq project,11 and nine R-award sites focusing on ethical, legal, and social implications. Eight of the nine clinical U-award sites and ClinSeq participated in this exercise. These included laboratories performing exome and/or genome sequencing for the following projects: BASIC3 (Baylor College of Medicine, Houston), PediSeq (Children’s Hospital of Philadelphia), CanSeq (Dana Farber Cancer Institute, Boston), HudsonAlpha Institute for Biotechnology, MedSeq (Brigham and Women’s Hospital and Partners Healthcare, Boston), NextGen (Oregon Health Sciences University, Portland), NCGENES (University of North Carolina, Chapel Hill), and NEXT Medicine (University of Washington, Seattle). Eight of the nine sites were accredited by the Clinical Laboratory Improvement Amendments (CLIA).
Selection and Classification of Variants
Each site nominated 11 variants identified in their sequencing projects for this exercise. Submitted variants were single-nucleotide substitutions or small indels (<22 bp) in genes thought to be associated with Mendelian disease. Each site was instructed to provide a range of variants in each classification category with varying degrees of difficulty. Accepted classifications were B, LB, VUS, LP, and P. Each variant submission also included whether it was identified as a diagnostic result or an incidental finding. Any internal evidence that the submitting laboratory used to classify the variant—for example, the phenotype and family history of the proband or whether parental testing identified the variant as de novo—was also provided to all laboratories. Nine variants (two P, two LP, two VUS, two LB, and one B) were selected for distribution to all laboratories without the submitting laboratory’s classification; half were identified as incidental findings, half were identified as diagnostic findings, and one was from a carrier screen. The remaining 90 variants were randomly distributed to at least two other laboratories, enabling classifications from at least three laboratories for each variant. Each laboratory was asked to classify the pathogenicity by applying both their internal process and then the ACMG-AMP system. They were asked to document which ACMG-AMP criteria were invoked for the ACMG-AMP classification and note whether they found the classification of each variant difficult, moderate, or easy. Time taken for categorizing the variant was requested but not consistently recorded.
Application of Automated Tool for Calculation of Overall Classification from Evidence Codes
In order to assess whether ACMG-AMP evidence codes were combined appropriately by the variant curator, we developed a pathogenicity calculator that combines the provided codes to generate a final classification. We used this calculator to compare the calculated ACMG-AMP classification based on tabulating the evidence codes provided by the laboratory with the final ACMG-AMP classification submitted by the laboratory. We shared these data with sites for consideration during consensus discussions and manually verified the results to identify which discrepancies were due to errors by the submitting laboratory and which were due to the use of judgment in overruling the ACMG-AMP classification.
Analysis of Intra- and Inter-laboratory Concordance
Descriptive statistics summarized the intra-laboratory classification concordance between the ACMG-AMP system and the laboratory’s own process and the inter-laboratory concordance both for each laboratory’s own process and for the ACMG-AMP system across laboratories. Additionally, we quantified the level of agreement. To do this, we considered the five-tier classification system in the following order—B, LB, VUS, LP, and P—and defined a one-step level of disagreement to be a range of classifications from one category to the next ordered category (e.g., from VUS to LP or LP to P); the maximum level included four steps (i.e., B to P). In addition, we tracked disagreements that were more likely to lead to medical-management differences (P or LP versus any of VUS, LB, and B) and disagreements less likely to affect clinical decision making (e.g., VUS versus LB or B, or confidence differences, such as B versus LB or P versus LP). To quantify the overall level of absolute agreement on ACMG-AMP and laboratory criteria within sites and agreement between sites using ACMG-AMP or laboratory criteria, we calculated Krippendorff’s alpha (K-alpha); ranging from 0 to 1, this generalized measure of absolute agreement corrects for chance responding and can handle any number of raters, scale of measurement, and missing data. Because it focuses on disagreement, it overcomes many of the weaknesses associated with other agreement measures, such as Cohen’s kappa.12, 13, 14, 15, 16 In general, values of 0.80 and above are considered evidence of good agreement.14 We also calculated 95% confidence intervals (CIs) for K-alpha by using bootstrapping with 20,000 replications.17
Two variants were excluded from the quantitative analyses and are not represented in Figures 1A and 1B; however, they are represented in the overall concordance shown in Figures 1C and 2. One variant was a low-penetrance allele (c.3920T>A [p.Ile1307Lys] [GenBank: NM_001127510.2] in APC [MIM: 611731]) for which several laboratories did not assign an ACMG-AMP classification, and the other variant (c.1101+1G>T [GenBank: NM_001005463.2] in EBF3 [MIM: 607407]) was a predicted loss-of-function variant in a gene for which there is no known association with disease. Neither of these two variants was relevant to this analysis of classifying high-penetrance variants for Mendelian conditions, for which the ACMG-AMP guidelines are intended. In addition, the two laboratories that had key personnel involved in the development of the ACMG-AMP recommendations were excluded from one study-wide sensitivity analysis to evaluate whether familiarity with the system affects concordance. Lastly, we performed a second sensitivity analysis by excluding the classifications of the submitting laboratory to determine the dependence of these results on a single laboratory and whether classification in a real case setting rather than only for the comparison study affects results.
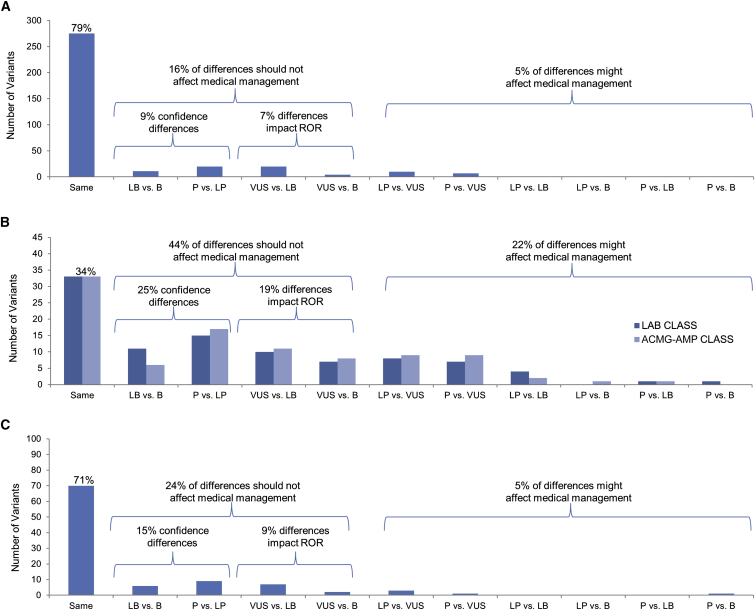
Figure 1.
Distribution of Variant-Classification Comparisons according to the Extent of Differences across a Five-Tiered Classification Scheme
(A) Intra-laboratory concordance between laboratory and ACMG-AMP classification systems. This graph compares each site’s use of the ACMG-AMP rules to their own laboratory classification methods.
(B) Inter-laboratory concordance of 97 variants. This graph compares the same calls, based on either the ACMG-AMP rules or the site’s rules, between laboratories.
(C) Inter-laboratory concordance after consensus efforts. This graph shows a final comparison of calls between sites after consensus-building efforts.
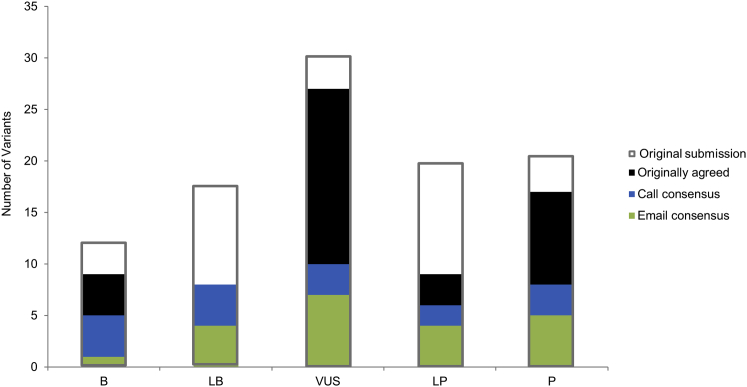
Figure 2.
Distribution of 99 Variants Submitted for Assessment
Gray outlines illustrate the distribution of variant classifications submitted for assessment. Green bars indicate those calls that were agreed upon after initial review, blue bars indicate those calls agreed upon after email exchange, and black bars indicate those calls agreed upon after discussion on conference calls.
Analysis of ACMG-AMP Lines of Evidence
We analyzed the lines of evidence used for each variant classification to identify how commonly specific evidence codes and classification rules were used across all of the variants, the overall agreement in the pattern of ACMG-AMP codes used across sites for each variant, and the consistency with which each ACMG-AMP code was used within each variant. These were determined with a frequency table, the mean of coefficient of variation (CV) values across variants with each ACMG-AMP code, and K-alpha values of ACMG-AMP codes within each variant. Descriptive statistics of how often the strength of each line of evidence was modified during variant interpretation were also calculated.
Consensus Discussions
The variants with discrepant classifications based on the ACMG-AMP guidelines were discussed via phone conferences (n = 23) or via email (n = 43). Variants were chosen for discussion by phone conference if they were interpreted by all nine laboratories or if they were discrepant by more than one category of disagreement. The laboratory that submitted each of these 23 variants presented the lines of evidence used by all laboratories and the rationale for using, not using, or altering the strength of a particular evidence code. Once all evidence was discussed, each laboratory was asked to provide a final classification. For variants for which only one laboratory was discordant for only a one-level difference, the discordant laboratory was asked to re-review their classification in light of the evidence used and classifications made by the other laboratories. The discordant laboratory then provided either a change or a decision to retain the original classification, including the rationale in both scenarios by email. During phone conferences and via email, laboratories had the opportunity to share any internal data that could have contributed to discordance.
Results
Intra-laboratory Classification Concordance between Unique Laboratory Criteria and the ACMG-AMP System
The intra-laboratory comparison of the laboratory process and the ACMG-AMP system for the 347 paired variant assessments is summarized in Figure 1A. The classifications matched for 275 of 347 (79%) variant assessments. Eleven of the 347 paired variant assessments (3.2%) differed by greater than one level. Overall, in 48 of the 72 (67%) discordant calls, the ACMG-AMP system calls were closer to VUS. Specifically, a classification of B or LB was more likely to result from using the laboratories’ own criteria than from using the ACMG-AMP criteria. The K-alpha value for agreement within laboratories ranged from 0.77 to 1.00 (average = 0.91; seven of nine laboratories had K-alpha > 0.90).
Inter-laboratory Concordance in Classification
Considering the inter-laboratory classification for 97 variants, there was no statistically significant difference in concordance across laboratories between classifications based on laboratory criteria and those based on ACMG-AMP criteria (lab K-alpha = 0.76, 95% CI = [0.73, 0.80]; ACMG-AMP K-alpha = 0.72, 95% CI = [0.68, 0.76]). In other words, implementation of the ACMG-AMP criteria did not yield more consistent variant classification among these laboratories. All laboratories reviewing the variant (three to nine) agreed for 33 (34%) when they used either the ACMG-AMP system or their own criteria. No significant difference was found in inter-laboratory concordance when the two laboratories that contributed to the ACMG-AMP classification recommendations were removed from the analysis (K-alpha lab = 0.77, 95% CI = [0.73, 0.80]; K-alpha ACMG-AMP = 0.70, 95% CI = [0.66, 0.74]) or when the site that submitted the variant classifications was removed from the analysis (K-alpha lab = 0.76, 95% CI = [0.71, 0.80]; K-alpha ACMG-AMP = 0.75, 95% CI = [0.71, 0.78]). The distribution of types of disagreement among laboratories using each method is shown in Figure 1B. A total of 43/194 (22%) classifications had category differences that are more likely to influence medical decision making (P or LP versus VUS, LB, or B), the majority of which (33) were P or LP versus VUS. An additional 36 classifications (19%) involved differences between VUS and LB or B, which could have an impact on results reported by the laboratory given that many laboratories do not report LB or B results and that reporting VUS results could result in a more lengthy disclosure process and uncertainty of follow-up. The remaining 25% of variant classifications were differences in the confidence of calls (P versus LP or B versus LB), which are unlikely to have an impact on clinical care.
Consensus Discussions
The interpretation of 33/99 (34%) variants was identical across all sites that used the ACMG-AMP guidelines. After either emails or conference calls among the reporting laboratories, consensus on variant classifications based on the ACMG-AMP guidelines was achieved for 70/99 (71%) variants. Twenty-one of the discrepant variants were resolved via email, and the remaining 16 were resolved during phone conferences. The distribution and sources of variant-interpretation consensus can be found in Figure 2; gray outlines show the original distribution of submitted variant interpretations. Figure 1C shows the distribution of types of disagreement among laboratories after the consensus effort. Of the 29 variants that remained discordant, 25 involved only one level of difference (15 differed in confidence differences, three differed between LP and VUS, and seven differed between VUS and LB). Of the four variants with greater than one level of difference, two involved a difference between P and VUS, LB, or B. The final classifications for the 70 variants for which consensus was achieved, and the range of classifications for the remaining 29 discordant variants, are presented in Table S1.
Consensus discussions led to the clarification of the correct use of several ACMG-AMP lines of evidence, some of which included errors in the appropriate use of the rules already described in the guidelines (Table 1). Although the ACMG-AMP guidelines suggest a VUS classification when conflicting pathogenic and benign lines of evidence are identified, some laboratories allowed one line of conflicting benign evidence of only a supporting level (e.g., computational predictions) to override otherwise strong evidence of pathogenicity. In these cases, consensus discussion led to the use of expert judgment, as described in the ACMG-AMP guidelines, for appropriately disregarding the limited conflicting evidence, such as computational predictions. For two variants, achieving concordant interpretations required one laboratory’s internal data. It was difficult to resolve the two variants that were excluded from the intra- and inter-laboratory analyses because the ACMG-AMP rules were not designed for low-penetrance variants (risk alleles) or variants in genes not clearly associated with the disorder. Some discrepancies in classification occurred because laboratories were interpreting the same variant for two different associated conditions, which have different disease frequencies. This led to a discordant use of the rules related to allele frequency.
Table 1.
ACMG-AMP Rule Clarifications and Suggestions for Modification
| Rule | Description | Clarifications and/or Suggestions |
|---|---|---|
| PVS1 | variant predicted null where LOF is a mechanism of disease | do not apply to variants that are near the 3′ end of the gene and escape nonsense-mediated decay |
| PS1 | variant with the same amino acid change as a previously established pathogenic variant, regardless of nucleotide change | does not include the same variant being assessed because it is not yet pathogenic, and the rule is intended for variants with a different nucleotide change |
| PS2 | de novo variant with confirmed maternity and paternity | apply this rule as moderate or supporting if the variant is mosaic and its frequency in tissue is consistent with the phenotype |
| PS3 | variant shown to have a deleterious effect by a well-established functional study | reduce the strength for assays that are not as well validated or linked to the phenotype |
| PM1 | variant located in a mutational hotspot and/or critical and well-established functional domain | not meant for truncations; more clarification is needed for applying this rule |
| PM2, BS1 | variant absent in population databases or with an allele frequency too high for the disease | cannot assume longer indels would be detected by next-generation sequencing |
| use a published control dataset if its size is at least 1,000 individuals | ||
| cannot be applied for low-quality calls or non-covered regions | ||
| must define the condition and inheritance pattern | ||
| PM3 | for recessive disorders, variant in trans with a pathogenic variant | invoke this rule as supporting if the phase is not established |
| can upgrade if more than one proband is reported | ||
| PM4 | protein-length-changing variant | applicable for in-frame deletions, insertions, or stop-loss variants, but not frameshifts, nonsense, and splice variants |
| PM5 | novel missense variant at amino acid with different pathogenic missense change | ensure pathogenicity of previously reported variant |
| suggest changing “novel” to “different” because some variants that are not novel might require assessment with this rule | ||
| PP3, BP4 | variant with multiple lines of computational evidence | all lines must agree |
| PP4 | the patient’s phenotype or family history is highly specific to the genotype | not meant to be used for genetically heterogeneous conditions or conditions with unsolved etiology |
| not typically applied for an analysis of incidental findings, but it could be applied for prior observations | ||
| PP5, BP6 | variant called pathogenic or benign by a reputable source | only applicable when evidence is not available (e.g., Sharing Clinical Reports Project) |
| BS2 | variant observed in a healthy adult for a disorder with full penetrance at an early age | populations might not have been screened or excluded for the phenotype |
| BP1 | variant in a gene in which truncations primarily cause disease | clarify the meaning of “primary”; suggest >90% |
| BP2, BP5 | variant in trans with a dominant pathogenic variant (BP2) or in an individual with an alternate molecular basis for disease (BP5) | clarify that one should apply BP2 when the pathogenic variant is seen in the same gene as the variant being evaluated and apply BP5 when the pathogenic variant is in a different gene |
Errors in Using the ACMG-AMP System
Our implementation of a computational tool to assess accuracy of combining the ACMG evidence codes used by the laboratories showed that for 16 out of 353 (5%) variant assessments, the ACMG-AMP codes listed by the laboratories did not support the classification chosen. When the laboratories were queried on these discrepancies, 9 of the 16 were due to tabulation errors, whereas judgment was used to override the ACMG-AMP rules for 7 of the 16 variants. The tabulation errors suggest that using computational tools to calculate the classification will lead to a modest increase in the accuracy of applying the rules.
ACMG-AMP Lines of Evidence Invoked and Modified in Strength
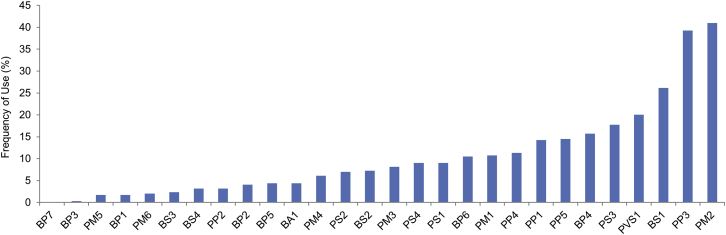
The frequency with which each ACMG-AMP code was invoked is listed in Figure 3. All lines of evidence were used at least once, except for BP7 (a silent, or synonymous, variant) given that no silent variants were submitted. Sixteen lines were used in fewer than 10% of variants, seven were used in 11%–18% of variants, and four were used in over 20% of all variant classifications: PVS1 (20%, predicted to be truncating), BS1 (26%, allele frequency is too high), PP3 (39%, computational evidence), and PM2 (41%, absent in population databases).
Figure 3.
Frequency of Use for Each ACMG Line of Evidence
We further analyzed sources of discordance in the use of the ACMG-AMP codes to identify those criteria commonly used in an inconsistent manner. For variants where at least one site invoked BA1 (the allele frequency is >5% and too high to cause the disorder), we only evaluated concordance of the use of BA1. This was due to the fact that if a site selected BA1, they did not need to evaluate any other codes. For rules invoked more than ten times overall, PP4 (the phenotype is highly specific to the gene) was used the most inconsistently among the laboratories for a given variant (mean CV = 1.74). This is not surprising given the subjective nature of deciding how specific a phenotype is to a given gene. The most consistently applied rule was PVS1 (null variant where loss of function [LOF] is a known mechanism of disease; mean CV = 0.55). The mean and SD of the CV for all lines of evidence used are available in Table S2.
We also evaluated which criteria laboratories had increased or decreased in evidence strength and found that a total of nine lines of evidence were modified at least once. Three criteria were increased in strength (PM3, PP1, and BP2), and seven were decreased in strength (PVS1, PS1, PS2, PS3, PS4, PM3, and BS1). Co-segregation data supporting pathogenicity (PP1) was the most commonly modified line of evidence, whereby laboratories increased the strength from supporting to moderate or strong for nine interpretations on the basis of the strength of the segregation evidence available in the literature or from the laboratory’s internal data. The other most common examples of modified strength included the following: PVS1 (a predicted null variant in a gene where LOF is a known mechanism of disease) was downgraded from very strong four times, PS2 (well-established functional studies show a deleterious effect) was downgraded three times, and BS1 (MAF is too high for the disorder) was downgraded three times.
Specific Variant Examples
The GLA (MIM: 300644) c.639+919G>A variant (GenBank: NM_000169.2), which has been reported in individuals with late-onset Fabry disease (MIM: 301500) and reduced alpha-galactosidase A enzyme activity,18, 19 was classified by three laboratories. Ranging from VUS to P, the interpretations based on ACMG-AMP rules were discordant; however, all sites agreed on the classification of P when they used internal rules. This variant was absent from 528 race-matched control individuals across two studies.18, 19, 20 A functional study also supported an effect of this variant on splicing.19 All three laboratories invoked PS3 (a well-established functional study [clinical alpha-galactosidase enzyme testing] showed a deleterious effect of the variant) and PP1 (evidence of segregation), and one site increased the strength of this line of evidence from supporting to moderate on the basis of three families cited in the literature. PS4 (the prevalence of the variant in affected individuals is statistically greater than that in control individuals) was invoked by the one laboratory that called the variant P by using both their own rules and the ACMG-AMP criteria. Upon further discussion, the group agreed that this rule was applicable on the basis of a single publication citing a significant p value and other studies showing a statistical increase but requiring manual calculation. PVS1 (predicted-null variant in a gene where LOF is a known mechanism of disease) was also applied by all three laboratories after the group decided to downgrade the strength from very strong to strong because of the minor retention of wild-type transcript and the fact that the variant was a deep intronic variant for which a functional study was needed to demonstrate its impact on splicing. Three lines of evidence originally invoked by only one laboratory each were discarded: (1) PM4 (protein-length-changing variant) because this rule is only applicable for in-frame deletions, insertions, and stop-loss variants; (2) PP5 (a reputable source calls the variant pathogenic) because the reputable source’s evidence was available for review by the curators and; (3) PP3 and BP4 (multiple lines of computational evidence agree) because this rule applies only when all predictions agree and not simply when some agree and others do not. All three sites came to a consensus that this variant is P on the basis of both their internal laboratory criteria and the ACMG-AMP criteria.
The group reviewed the c.1529C>T (p.Ala510Val) variant (GenBank: NM_003119.2) in SPG7 (MIM: 602783), which is associated with autosomal-recessive spastic paraplegia (MIM: 607259), a disease that is known to have a variable, but generally adult, age of onset.21 It was interpreted by all nine laboratories and had a range of pathogenicity classifications from LB to P. This variant was observed in 0.4% (267/66,688) of chromosomes of European ancestry (EU) in the ExAC Browser and has been found to have a 3%–4% heterozygote frequency in the UK and an estimated homozygote frequency of 20–40 per 100,000 individuals.22 Shared data included that the variant was observed in the heterozygous state in 3 of the first 50 participants sequenced by the submitting laboratory’s CSER study. The frequency of SPG7-associated spastic paraplegia is conservatively estimated to be 2–6 per 100,000 individuals according to the higher estimate; this yields an estimated frequency of 0.8% for all associated alleles. Multiple publications have cited the identification of homozygous or compound-heterozygous (including this variant) affected individuals.22, 23, 24 It is notable that the laboratories that concluded this was a VUS or LB variant considered the BS1 criteria (the variant is more common than the disease, adjusted for the autosomal-recessive inheritance pattern). Two of the laboratories that concluded the variant was P according to the ACMG-AMP rules used the PM2 criteria (the variant is absent from control individuals or has an extremely low frequency if recessive). The remainder of the laboratories did not use any rules on population-frequency data. An additional line of evidence with conflicting use was PS1 (the variant results in the same amino acid change as a previously established pathogenic variant, regardless of nucleotide change). This rule was invoked by four of nine laboratories; however, after clarification that the intent of this rule, as described in the ACMG-AMP guidelines, is to be applied only for a “different” nucleotide change (i.e., something other than SPG7 c.1529C>T that still leads to p.Ala510Val), all laboratories agreed that this rule was not applicable. Consensus was not reached for this variant largely because of discordance in applying the population-frequency lines of evidence. Some groups continued to weigh the published literature evidence of pathogenicity, whereas other groups concluded that it could not be a high-penetrance variant given its allele frequency. Given a perceived deficit of affected homozygotes relative to affected compound heterozygotes, some felt it might have low penetrance unless it is paired with a more deleterious variant. Resolving the role of this variant in disease might ultimately require a better understanding of the penetrance and possible role of modifiers, and classifying the pathogenicity of lower-penetrance variants was outside the scope of the ACMG-AMP guidelines.
The variant with the largest range of discordance (P to B) after consensus efforts based on the ACMG-AMP guidelines was BTD (MIM: 609019) c.1330G>C (p.Asp444His) (GenBank: NM_000060.2), which was interpreted by three laboratories. BTD is associated with autosomal-recessive biotinidase deficiency (MIM: 253260), and this variant was detected in one allele of an unaffected individual. The interpretations based on the ACMG-AMP guidelines originally ranged from VUS to P; however, after consensus efforts, the laboratory that classified the variant originally as VUS changed their interpretation to B, whereas the other two laboratories kept their interpretations as LP and P. This variant has been identified in multiple biotinidase-deficiency-affected individuals who have variants associated with profound deficiency on the other allele.25, 26 It was observed in 5.4% of chromosomes of Finnish ancestry and 4.15% of EU chromosomes in the ExAC Browser, and there were 83 reported instances of homozygosity. The laboratory that interpreted this variant as B invoked the MAF > 5% (BA1) as standalone criteria and noted the presence of homozygotes in population databases. Like SPG7, this might represent an allele that is more likely to be pathogenic when it is found to be compound heterozygous with a more deleterious allele than when it is found to be homozygous. This is a general problem with recessive disorders, and it might make consideration of the genotype rather than the pathogenicity of each allele more important for disease prediction. The laboratories classifying this variant as P and LP used expert judgment to overrule the use of BA1, which supports a B classification. They cited multiple reputable laboratories that have interpreted this variant as pathogenic and evidence that individuals who are homozygous and compound heterozygous for this variant might have a more mild form of the disease. Consensus efforts brought the group further from agreement on this variant; however, this highlights the importance of employing expert judgment when making interpretations, as well as the challenges that stem from using the ACMG-AMP guidelines to interpret the clinical significance of variants that might be associated with lower penetrance and mild presentations of disease.
The Supplemental Note and Table 1 describe in detail the criteria-specific clarifications that resulted from common usage errors, as well as the challenges and discussion topics related to each of the ACMG-AMP evidence categories that are utilized in implementing the ACMG-AMP guidelines. This material is designed to further clarify numerous rules found in the ACMG-AMP publication. In addition, more general recommendations and additional resources that could increase consistency of the usage of ACMG-AMP rules across laboratories are defined in Box 1.
Box 1. Recommendations and Additional Resources for Increasing Consistency in the Usage of ACMG-AMP Rules.
-
•
Develop disease-specific allele-frequency thresholds to enable lowering of the stand-alone benign criteria from a MAF of ≥5% to values specific to each disorder.
-
•
Establish a resource of all genes to define whether LOF is a known mechanism of disease.
-
•
Make recommendations for which computational algorithms are best in practice.
-
•
Better define “well-established” functional data and/or distribute a resource that lists functional assays that meet the well-established threshold. Also define when to use reduced strength of the rule.
-
•
Develop quantitative thresholds of evidence for and against segregation of different strengths.
-
•
Promote the development of software tools that automate computable aspects of the ACMG-AMP guidelines to improve accurate use.
Discussion
Interpreting the pathogenicity of a genetic variant requires evaluating a large number of heterogeneous types of evidence to arrive at a unitary descriptor of pathogenicity. Given the complexity of the data and uncertainty regarding the validity or utility of some of the data used for these interpretations, it is unsurprising that there would be variation among laboratories regarding these determinations. To that end, the ACMG-AMP system for classifying variant pathogenicity10 is an important first step in efforts to improve the consistency of variant classification among laboratories. The guidelines include standardized terminology for classifying variants associated with monogenic diseases and a defined series of evidence types that can be used in pathogenicity assessment, enabling a record of the specific evidence type and strength used for determining pathogenicity. This enhances transparency and facilitates resolution of discrepancies in variant interpretation. It also forms a basis for iteratively building on the evidence as new data become available over time.
This study systematically evaluated the implementation of the ACMG-AMP guidelines in the medical practice of variant assessment. Nearly all ACMG-AMP lines of evidence were used, and PVS1 (predicted truncating), BS1 (allele frequency too high), PP3 (computational evidence), and PM2 (absent in population databases) (Figure 3) reflect the spectrum of variants chosen for the exercise and the most available types of data. We identified differences in the application of the criteria but no difference in classification concordance between the ACMG-AMP system and the laboratory method. In part, the discordance in applying the ACMG-AMP guidelines was due to the subjective process of deciding when certain criteria are met. However, the guidelines provided a valuable framework for subsequent discussion of evidence, often leading to resolution of differences in variant interpretation; achieving this would have been more difficult if each laboratory relied on an independent method for variant assessment. The differences in both the intra- and inter-laboratory analyses identified points of confusion and inaccurate use of the ACMG-AMP criteria, as well as areas where expert judgment is required and additional guidance is needed. It should be noted that this was the first time that most sites had worked with the ACMG-AMP guidelines, and thus familiarity and systems for implementation of the criteria were still evolving. In addition, because the variants were distributed as a pilot evaluation of the ACMG-AMP guidelines and not for clinical reporting, not all sites subjected the variants to their typical CLIA process of review, which includes final review by a board-certified laboratory geneticist or an equivalently trained individual. Thus, the level of discordance reported here might have been inflated by the atypical workflows being deployed. In contrast, the resolution of the discordant variants involved multiple board-certified geneticists and others with long-standing experience in variant assessment, documenting the importance of this level of training in variant interpretation. These study results underscore the need for training in the use of genetic resources, evaluation of variant evidence, and application of the ACMG-AMP guidelines, even among experienced professionals. This study identified areas of confusion regarding the ACMG-AMP criteria, and these will be useful in developing training materials and further guidance for variant assessment. As recently described,3 the Clinical Genome Resource (ClinGen) consortium is developing tools to aid variant classification based on the ACMG-AMP guidelines and will make this information public through both community availability of the tool and documentation of applied codes with variants submitted to ClinVar.
As described above, our discussion and consensus building led to a decrease in variant discordance from 66% to 29% of the 99 variants analyzed. This underscores the importance of not only having a standardized approach to variant assessment but also sharing variant interpretations for identifying and potentially resolving discordance. Given the rarity of most variants causative for monogenic disease, sharing data and comparing interpretations are imperative for ensuring the greatest opportunity for informed and collaborative variant interpretation. It is important to also reflect on the goals of variant assessment. Although numerous variants have robust evidence that can unequivocally allow classification into discreet categories without debate, many other variants have limited or conflicting evidence, making it difficult to accurately classify these variants. Indeed, for 29 (29%) of the 99 variants assessed in this study, a consensus classification was not achieved, and 5 of the 29 involved a difference between the categories of P or LP and VUS, LB, or B, which could affect medical management. This finding highlights that classifying sequence variants is similar to other fields of medicine in which practitioners can legitimately differ in their assessments of pathogenicity of a laboratory finding. By defining and applying formal criteria that parse these heterogeneous data types, we can better understand and analyze these legitimate differences in expert opinion and at the same time reduce errors and discrepancies.
Conflicts of Interest
Most authors are employed by clinical-service providers. G.P.J. is a scientific advisory board member for ActX, a genetic-testing company. R.C.G. has received compensation from Invitae, Prudential, Illumina, AIA, Helix, and Roche for advisory services or speaking. S.E.P. serves on the scientific advisory board of Baylor Miraca Genetic Laboratory. Baylor College of Medicine and Miraca Holdings Inc. have formed a joint venture with shared ownership and governance of the Baylor Miraca Genetics Laboratories, which perform exome sequencing.
Acknowledgments
This work was funded by the National Human Genome Research Institute (NHGRI) and National Cancer Institute (UO1HG0006546, U01HG006485, U01HG006500, U01HG006492, UM1HG007301, UM1HG007292, UM1HG006508, U01HG006487, U01HG006507, U01HG007307, U41HG006834, R21HG006596, R01HG006600, P50HG007257, R01HG006600, R01HG004500, R01CA154517, R01HG006618, R21HG006594-01, R01HG006615-01, R21HG006612, and 5R21HG006613-02) and additional support from the participating laboratories. This research was supported in part by the Intramural Research Program of the NIH NHGRI. We thank the following individuals for contributing to variant assessment at the sites: Hana Zouk, PhD; Steven M. Harrison, PhD; Jillian G. Buchan, PhD; Jessica Booker, PhD; Jim Evans, MD, PhD, Kate Foreman, MS; Gloria Haskell, PhD; Kristy Lee, MS; Julianne O’Daniel, MS; Bradford Powell, MD, PhD; Bryce Seifert, PhD; and Karen Weck, MD.
Published: May 12, 2016
Footnotes
Supplemental Data include a Supplemental Note, Supplemental Acknowledgments, and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.03.024.
Contributor Information
Gail P. Jarvik, Email: pair@u.washington.edu.
Heidi L. Rehm, Email: hrehm@partners.org.
Web Resources
1000 Genomes, http://www.1000genomes.org/
ClinGen Pathogenicity Calculator, http://calculator.clinicalgenome.org/site/cg-calculator
CSER Consortium, https://cser-consortium.org/
ExAC Browser, http://exac.broadinstitute.org
Gene Tests, http://www.ncbi.nlm.nih.gov/books/NBK1107/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
Sharing Clinical Reports Project, https://www.clinicalgenome.org/data-sharing/sharing-clinical-reports-project-scrp/
Supplemental Data
References
- 1.Kohane I.S., Hsing M., Kong S.W. Taxonomizing, sizing, and overcoming the incidentalome. Genet. Med. 2012;14:399–404. doi: 10.1038/gim.2011.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Evans B.J., Burke W., Jarvik G.P. The FDA and genomic tests--getting regulation right. N. Engl. J. Med. 2015;372:2258–2264. doi: 10.1056/NEJMsr1501194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rehm H.L., Berg J.S., Brooks L.D., Bustamante C.D., Evans J.P., Landrum M.J., Ledbetter D.H., Maglott D.R., Martin C.L., Nussbaum R.L., ClinGen ClinGen--the Clinical Genome Resource. N. Engl. J. Med. 2015;372:2235–2242. doi: 10.1056/NEJMsr1406261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amendola L.M., Dorschner M.O., Robertson P.D., Salama J.S., Hart R., Shirts B.H., Murray M.L., Tokita M.J., Gallego C.J., Kim D.S. Actionable exomic incidental findings in 6503 participants: challenges of variant classification. Genome Res. 2015;25:305–315. doi: 10.1101/gr.183483.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yorczyk A., Robinson L.S., Ross T.S. Use of panel tests in place of single gene tests in the cancer genetics clinic. Clin. Genet. 2015;88:278–282. doi: 10.1111/cge.12488. [DOI] [PubMed] [Google Scholar]
- 6.Richards C.S., Bale S., Bellissimo D.B., Das S., Grody W.W., Hegde M.R., Lyon E., Ward B.E., Molecular Subcommittee of the ACMG Laboratory Quality Assurance Committee ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genet. Med. 2008;10:294–300. doi: 10.1097/GIM.0b013e31816b5cae. [DOI] [PubMed] [Google Scholar]
- 7.Karbassi I., Maston G.A., Love A., DiVincenzo C., Braastad C.D., Elzinga C.D., Bright A.R., Previte D., Zhang K., Rowland C.M. A Standardized DNA Variant Scoring System for Pathogenicity Assessments in Mendelian Disorders. Hum. Mutat. 2016;37:127–134. doi: 10.1002/humu.22918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thompson B.A., Spurdle A.B., Plazzer J.P., Greenblatt M.S., Akagi K., Al-Mulla F., Bapat B., Bernstein I., Capellá G., den Dunnen J.T., InSiGHT Application of a 5-tiered scheme for standardized classification of 2,360 unique mismatch repair gene variants in the InSiGHT locus-specific database. Nat. Genet. 2014;46:107–115. doi: 10.1038/ng.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldgar D.E., Easton D.F., Byrnes G.B., Spurdle A.B., Iversen E.S., Greenblatt M.S., IARC Unclassified Genetic Variants Working Group Genetic evidence and integration of various data sources for classifying uncertain variants into a single model. Hum. Mutat. 2008;29:1265–1272. doi: 10.1002/humu.20897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biesecker L.G., Mullikin J.C., Facio F.M., Turner C., Cherukuri P.F., Blakesley R.W., Bouffard G.G., Chines P.S., Cruz P., Hansen N.F., NISC Comparative Sequencing Program The ClinSeq Project: piloting large-scale genome sequencing for research in genomic medicine. Genome Res. 2009;19:1665–1674. doi: 10.1101/gr.092841.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayes A.F., Krippendorff K. Answering the call for a standard reliability measure for coding data. Commun. Methods Meas. 2007;1:77–89. [Google Scholar]
- 13.Krippendorff K. Reliability in content analysis: Some common misconceptions and recommendations. Hum. Commun. Res. 2004;30:411–433. [Google Scholar]
- 14.Krippendorff K. Second Edition. Sage; 2004. Content analysis: An introduction to its methodology. [Google Scholar]
- 15.Brennan R.L., Prediger D.J. Coefficient kappa: Some uses, misuses, and alternatives. Educ. Psychol. Meas. 1981;41:687–699. [Google Scholar]
- 16.Zwick R. Another look at interrater agreement. Psychol. Bull. 1988;103:374–378. doi: 10.1037/0033-2909.103.3.374. [DOI] [PubMed] [Google Scholar]
- 17.Krippendorff, K. (2013). Bootstrapping Distributions for Krippendorff’s Alpha for Coding of Predefined Units: Single-Valued cα and multi-valued mvα. http://web.asc.upenn.edu/usr/krippendorff/boot.c-Alpha.pdf.
- 18.Lin H.Y., Huang C.H., Yu H.C., Chong K.W., Hsu J.H., Lee P.C., Cheng K.H., Chiang C.C., Ho H.J., Lin S.P. Enzyme assay and clinical assessment in subjects with a Chinese hotspot late-onset Fabry mutation (IVS4c+c919G→A) J. Inherit. Metab. Dis. 2010;33:619–624. doi: 10.1007/s10545-010-9166-7. [DOI] [PubMed] [Google Scholar]
- 19.Ishii S., Nakao S., Minamikawa-Tachino R., Desnick R.J., Fan J.Q. Alternative splicing in the alpha-galactosidase A gene: increased exon inclusion results in the Fabry cardiac phenotype. Am. J. Hum. Genet. 2002;70:994–1002. doi: 10.1086/339431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hwu W.L., Chien Y.H., Lee N.C., Chiang S.C., Dobrovolny R., Huang A.C., Yeh H.Y., Chao M.C., Lin S.J., Kitagawa T. Newborn screening for Fabry disease in Taiwan reveals a high incidence of the later-onset GLA mutation c.936+919G>A (IVS4+919G>A) Hum. Mutat. 2009;30:1397–1405. doi: 10.1002/humu.21074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Casari G., De Fusco M., Ciarmatori S., Zeviani M., Mora M., Fernandez P., De Michele G., Filla A., Cocozza S., Marconi R. Spastic paraplegia and OXPHOS impairment caused by mutations in paraplegin, a nuclear-encoded mitochondrial metalloprotease. Cell. 1998;93:973–983. doi: 10.1016/s0092-8674(00)81203-9. [DOI] [PubMed] [Google Scholar]
- 22.Roxburgh R.H., Marquis-Nicholson R., Ashton F., George A.M., Lea R.A., Eccles D., Mossman S., Bird T., van Gassen K.L., Kamsteeg E.J., Love D.R. The p.Ala510Val mutation in the SPG7 (paraplegin) gene is the most common mutation causing adult onset neurogenetic disease in patients of British ancestry. J. Neurol. 2013;260:1286–1294. doi: 10.1007/s00415-012-6792-z. [DOI] [PubMed] [Google Scholar]
- 23.Elleuch N., Depienne C., Benomar A., Hernandez A.M., Ferrer X., Fontaine B., Grid D., Tallaksen C.M., Zemmouri R., Stevanin G. Mutation analysis of the paraplegin gene (SPG7) in patients with hereditary spastic paraplegia. Neurology. 2006;66:654–659. doi: 10.1212/01.wnl.0000201185.91110.15. [DOI] [PubMed] [Google Scholar]
- 24.van Gassen K.L., van der Heijden C.D., de Bot S.T., den Dunnen W.F., van den Berg L.H., Verschuuren-Bemelmans C.C., Kremer H.P., Veldink J.H., Kamsteeg E.J., Scheffer H., van de Warrenburg B.P. Genotype-phenotype correlations in spastic paraplegia type 7: a study in a large Dutch cohort. Brain. 2012;135:2994–3004. doi: 10.1093/brain/aws224. [DOI] [PubMed] [Google Scholar]
- 25.Norrgard K.J., Pomponio R.J., Swango K.L., Hymes J., Reynolds T.R., Buck G.A., Wolf B. Mutation (Q456H) is the most common cause of profound biotinidase deficiency in children ascertained by newborn screening in the United States. Biochem. Mol. Med. 1997;61:22–27. doi: 10.1006/bmme.1997.2597. [DOI] [PubMed] [Google Scholar]
- 26.Hymes J., Stanley C.M., Wolf B. Mutations in BTD causing biotinidase deficiency. Hum. Mutat. 2001;18:375–381. doi: 10.1002/humu.1208. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.