Abstract
Nonsyndromic patent ductus arteriosus (PDA) is a common congenital heart defect (CHD) with both inherited and acquired causes, but the disease mechanisms have remained elusive. Using combined genome-wide linkage analysis and whole-exome sequencing (WES), we identified independent mutations in PRDM6, which encodes a nuclear protein that is specific to vascular smooth muscle cells (VSMC), has histone methyl transferase activities, and acts as a transcriptional suppressor of contractile proteins. In vitro assays showed that the mutations cause loss of function either by intracellular redistribution of the protein and/or by alteration of its methyltransferase activities. Wild-type embryonic ductus arteriosus (DA) exhibited high levels of PRDM6, which rapidly declined postnatally as the number of VSMCs necessary for ductus contraction increased. This dynamic change suggests that PRDM6 plays a key role in maintaining VSMCs in an undifferentiated stage in order to promote their proliferation and that its loss of activity results in premature differentiation and impaired remodeling of the DA. Our findings identify PRDM6 mutations as underlying genetic causes of nonsyndromic isolated PDA in humans and implicates the wild-type protein in epigenetic regulation of ductus remodeling.
Introduction
Patent ductus arteriosus (PDA [MIM: 607411]) is the second most common congenital heart defect (CHD) and affects up to 5,000 live births in the United States.1, 2 During fetal life, the ductus arteriosus (DA) shunts blood from the pulmonary artery to the aorta, bypassing the lungs. Its abrupt closure at birth, which establishes the mature circulatory pattern, is the result of a protracted and dramatic process of vascular remodeling. According to results from animal studies, the remodeling of the DA is a complex process involving the migration of neural-crest-derived cells into the subendothelial space and their transformation to vascular smooth muscle cells (VSMCs), as well as extracellular-matrix accumulation and formation of subintimal cushions. Subsequently, an increase in vasoactive peptides such as endothelin A1, a decrease in prostaglandin E2 levels,3, 4 and ultimately VSMC contraction result in closure of the vessel.5, 6 Experimental in vivo and in vitro studies have shown that failure in any step of this process can result in persistent patency of the DA and thus lead to pulmonary arterial hypertension, pulmonary edema, and congestive heart failure in humans.7
The molecular mechanisms underlying DA remodeling in humans are not understood. The disease can be acquired, and congenital rubella syndrome is the best-known cause.8 The increased familial recurrence, the autosomal-dominant inheritance of rare syndromic and nonsyndromic PDA, and the development of the trait in transgenic mice are strong pieces of evidence supporting the sizable role of genetics in the development of this condition. These studies have implicated the central role of neural-crest-derived VSMCs and genes regulating their function in remodeling of the DA.9, 10 Genetic determinants of nonsyndromic PDA in humans are not known. We ascertained 35 individuals affected by nonsyndromic familial PDA and extended their kindreds. Except for a large African-American kindred with sufficient power for linkage, all other affected individuals were white subjects from nuclear kindreds (Figure S1). All were products of healthy full-term pregnancies; maternal rubella had been excluded. They all had undergone cardiac catheterization at the time of recruitment and had no other vascular anomalies. All but one were living subjects, and all had undergone surgical or transvenous device closure for isolated PDA. Combined genome-wide linkage analysis, whole-exome sequencing (WES), and gene-set enrichment analysis (GSEA) were performed for the identification of disease-associated mutations.
Material and Methods
Ascertainment and Recruitment of PDA Subjects
The study protocol was approved by the Human Investigation Committee of the Yale University School of Medicine, the institutional review boards at the University of Nebraska and University of Kansas, and the Committee for Ethics and Research Conduction at the Isfahan Cardiovascular Research Center. Consent was obtained from all subjects. Records were reviewed for documentation of diagnosis, age of diagnosis, clinical findings, treatment, and family history. Subjects underwent physical examinations, and venous blood samples were collected. Blood samples were processed locally by either traditional phenol-chloroform extraction or salt extraction.
Analysis of Linkage
Genome-wide analysis of linkage was performed with the Affymetrix GeneChip 10K Array. Additional markers were typed in selected intervals for maximizing informativeness for the linked interval. These markers were genotyped by PCR with specific fluorescently labeled oligonucleotide primers and genomic DNA as a template. The resulting amplified products were fractionated by electrophoresis on denaturing polyacrylamide gels with an ABI3700 instrument and analyzed with GeneScan 2.1 and Genotyper 1.1.1 software. Two independent investigators scored all genotypes. Analysis of linkage was performed with GeneHunter 2.1. For allele frequencies, mean frequencies of 300 ethnically matched control individuals were used.
Targeted Sequence Capture
Genomic DNA was captured on exomes at the W.M. Keck Facility at Yale University as described earlier.2 The Roche NimbleGen 2.1M Human Exome Array covers 34.0 Mb of genomic sequence and approximately 180,000 exons of 18,673 protein-coding genes. In brief, DNA was fragmented and ligated to linkers and then fractionated by agarose gel electrophoresis. Extracted DNA was amplified by PCR and hybridized to the capture arrays. Bound genomic DNA was eluted, purified, and amplified by ligation-mediated PCR. The PCR products were purified and subjected to DNA sequencing on the Illumina platform.
Sequencing
Captured libraries were sequenced on the Illumina Genome Analyzer, and then image analysis and base calling were performed. Sequence reads were mapped to the human reference genome (UCSC Genome Browser hg19) with the MAQ program SAMtools. Resulting sequence data were processed with MAQ software. SAMtools was used to detect single-nucleotide variants, which were subsequently filtered against the reference genome as described earlier.11 Filters were applied against published databases. A Perl-based computer script was used for annotating variants according to their effect on the protein, novelty, conservation, and tissue expression. Variants were considered non-conservative if the substituted amino acid was conserved in all species or had a maximum of one conserved substitution. Sanger sequencing was carried out for most variants of interest, including all variants in PRDM6 (PR domain 6). Gene Ontology (GO) was used for GSEA. Variants were filtered if they had allele frequencies greater than 0.01% in the Exome Aggregation Consortium (ExAC) Browser and if they were considered not damaging by PolyPhen and SIFT software. Mutations in genes with expression levels 2 SDs below average in the heart, cardiomyocytes, smooth muscle cells, and aorta were excluded from analysis. The expression level from each tissue was based on Affymetrix GNF_U133A (as were expression data from the UCSC Genome Browser). The 1000 Genomes Project and an exome database of 2,000 healthy white subjects were used as controls. Population structure was excluded by principal-component analysis (Figure S3).
Mutagenesis and Adenoviral Constructs
pcDNA3.1 plasmid containing mouse wild-type Prdm6 with FLAG tag was kindly provided by Dr. Eric Olson. PRDM6 amino acid substitutions p.Arg549Gln (c.1646G>A) and p.Cys263Ser (c.788G>C) were generated with the QuikChange II Site-Directed Mutagenesis Kit (Agilent Technologies) according to the manufacturer’s instructions. Adenovirus expression plasmid pAd/CMV/V5-DEST (Invitrogen, Life Technologies) separately encoding FLAG-PRDM6-WT, FLAG-PRDM6-p.Arg549Gln, and FLAG-PRDM6-p.Cys263Ser was prepared with Gateway Cloning technology (Life Technologies) according to the manufacturer’s instructions.
Antibodies
Antibodies to α-smooth muscle actin (α-SMA), myosin heavy chain 11 (MYH11), CD31, GM130, importin-β, and H3 pan were purchased from Abcam. Antibodies to phalloidin and GAPDH were purchased from Cell Signaling Technology. Antibodies to H4K20me2, H3K9me2, and H4 pan were purchased from EMD Millipore. Antibodies to PRDM6, FLAG, and Calnexin were purchased from Novus Biologicals, Sigma-Aldrich, and Enzo Life Sciences, respectively.
Immunofluorescence Staining
Wild-type mouse DAs were micro-dissected and embedded in optimal cutting temperature (OCT) compound at three different developmental stages: embryonic days 14.5 (E14.5) and 17.5 (E17.5) and postnatal day 0.5 (P0.5).12 The differences in fluorescence intensities were compared with MATLAB software.
Cells were seeded in a glass-bottom petri dish. HEK293 cells were transfected with Lipofectamine 2000, and human aortic VSMCs were infected with adenovirus. 48 hr later, cells were fixed in 4% paraformaldehyde and were permeabilized in 0.1% Triton X-100 and blocked with 3% BSA in PBS after appropriate washes. Specimens were subsequently incubated with primary antibodies followed by fluorescence-conjugated secondary antibodies at room temperature and were mounted with ProLong Gold Antifade Mountant with DAPI (Invitrogen). Images were acquired with a Zeiss LSM510 confocal microscope.
Immunoblotting
Human aortic VSMCs (passage 3) transfected with adenovirus were lysed with cell-lysis buffer (Cell Signaling Technology) for extraction of total protein or with the EpiQuik Total Histone Extraction Kit (EpiGentek) for obtaining histone protein according to the manufacturers’ instructions. Lysates were processed and applied to SDS-PAGE and further immunoblotted with target primary antibodies followed by appropriate horseradish-peroxidase-conjugated secondary antibodies. Enhanced chemiluminescence reagents were applied for blot development. ImageJ software was used for quantification of the band intensities.
GSEA
GSEA is a systematic analysis frequently used for identifying disease-causing mutations for complex traits.13, 14, 15 For this analysis, we assigned to GO pathways all non-conservative deleterious variants with allele frequencies less than 0.01% (<1:10,000) in the ExAC Browser. After exclusion of population stratification, 2,000 exomes of European descent were used as controls. Subsequently, we used Fisher’s exact test to compare the ratio of identified variants and total number of bases sampled in a given pathway to the same ratio obtained from the control population. p values for these comparisons were corrected for multiple comparisons as previously described, and p < 2.7 × 10−4 was considered significant. The estimated odds ratio of these variants for the trait was calculated. We rejected the null hypothesis that these pathways could be over-represented in a small sample by chance by applying a randomization test of exome variants of 32 randomly selected individuals from the control exome database with a p value of 4 × 10−51.
Statistical Analysis
All in vitro experiments were carried out in triplicate. Data are expressed as means ± SEM. Comparisons between two groups were performed with Fisher’s exact test. Statistical significance is defined as p < 0.05. The expected number of genes with more than one previously unreported non-conservative and deleterious mutation was determined by Monte Carlo simulation (104 iterations) specifying the total number of nonsynonymous and deleterious mutations and 21,000 genes of observed coding length.
Results
Linkage Analysis Maps the Disease-Associated Mutation to 5q23 in a Large PDA-Affected Kindred
Kindred PDA-101 (Figure 1A) is a large African-American family, first reported in 1965.16 To our knowledge, this is the largest reported nonsyndromic-PDA-affected kindred with sufficient power for linkage (see Supplemental Note). In sum, a total of nine individuals with PDA were identified in the family; all had a definitive diagnosis of PDA, were born at term as the product of normal gestation, and could trace their ancestry to a single pair of founders. We found no subjects with PDA or other CHD anomalies in other branches of the kindred, arguing against multifactorial determination (complex trait). These findings strongly supported autosomal-dominant transmission of PDA with incomplete penetrance, thereby prompting further genetic study.
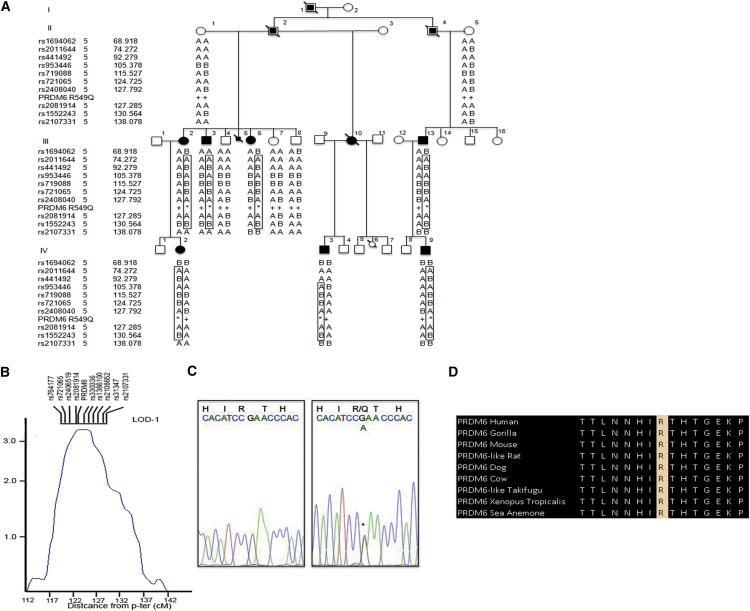
Figure 1.
PRDM6 p.Arg549Gln in a Large PDA-Affected Kindred
(A) Pedigree PDA-101 is shown. Individuals with PDA are indicated by black symbols, individuals without PDA are shown as unfilled symbols, and individuals with unknown PDA status are shown with partially filled squares. Circles represent females, squares represent males, and symbols with a slash through them indicate deceased subjects. Haplotypes on 5q23 are shown under each individual.
(B) Multipoint LOD scores for linkage of PDA to 5q23. SNPs tightly linked to the location of PRDM6 in the LOD-1 region are shown. The maximum LOD score is indicated, and the location of PRDM6 is shown. The LOD score peak occurs at θ = 0 with marker rs2081914, and the LOD-1 interval spans 10 cM.
(C) The DNA sequence of a segment flanking c.1646G>A in PRDM6 is shown from an unaffected kindred member (left) and a heterozygous mutation carrier (right). This single-base substitution (asterisk) changes the wild-type arginine to glutamine at amino acid 549 (p.Arg549Gln).
(D) A portion of the PRDM6 amino acid sequence flanking Arg549 is shown from diverse vertebrate species.
In order to map the disease-associated mutation in PDA-101, we performed genome-wide SNP genotyping in all family members by using the Affymetrix GeneChip 10K Array according to the manufacturer’s protocol. We used two pre-specified models of the trait locus to compare the inheritance of marker alleles to the inheritance of PDA: (1) a conservative model specified the trait locus as autosomal dominant with 90% penetrance, a phenocopy rate of 1%, and a disease allele frequency of 0.001, and (2) a stringent model specified 99% penetrance, 0.1% phenocopies, and an allele frequency of 0.0001. Pairwise analysis of linkage demonstrated positive LOD scores for linkage to a cluster of markers in chromosomal region 5q23. The maximum pairwise LOD score was 2.57 (at a recombination fraction [θ] of 0 with the marker rs2081914). Multipoint analysis confirmed linkage to this chromosomal segment, yielding a maximum LOD score of 3.12 for linkage of the trait locus (Figure 1B), PDA1, to rs2081914 at θ = 0. No other interval in the genome yielded a LOD score greater than 1.0, providing strong evidence that PDA1 lies in the 5q23 interval. Changing estimates of penetrance of the trait locus from 80% to 95% and the frequency of mutant genes did not have substantial effects on the LOD score or the disease locus (data not shown). Linkage analysis of only affected individuals yielded similar results, such that no other genomic interval was shared among all studied subjects. Review of the segregating haplotypes demonstrated that all affected individuals share a haplotype in this interval (Figure 1A). These findings indicate significant linkage of PDA to 5q23 in this kindred.
A Missense Mutation in PRDM6 Underlies Autosomal-Dominant PDA
The LOD-1 support interval, which approximates the 95% confidence interval for the disease locus, spans a10 cM (10 Mb) interval flanked by loci rs721065 and rs2108652 and contains 42 annotated genes. Four remotely related members of this kindred (subjects III-2, IV-9, III-11, and IV-6; Figure 1A) were screened for mutations by WES on the Roche NimbleGen 2.1M Human Exome Array. We achieved 95% coverage of the reference genome at a minimum depth of 20 reads. After quality filtering, we found a mean of 18,000 exomic variations per individual. We used SAMtools and the Genome Analysis Toolkit to detect polymorphic sites. The remainder (an average of 400 variants) were filtered against 1000 Genomes, the National Heart, Lung, and Blood Institute (NHLBI) Exome Sequencing Project (ESP) Exome Variant Server (ESP6500, including 2,300 African Americans), the ExAC Browser, and the Yale 5,000 exome database (including 2,000 healthy control individuals). These variants were further filtered by conservation as follows. If the identified amino acid change was seen in 0 or 1 of 44 vertebrate species, the change was considered non-conservative. After this filtering, there remained only one single non-conservative PRDM6 variant (c.1646G>A [p.Arg549Gln] [GenBank: NM_001136239.1]) that tightly linked to the location of the maximum LOD score within the shared haplotype and perfectly segregated with the disease (Figure 1C). Other mutations in the recombination interval were either intronic or common polymorphisms (Table S1). Arg549 resides in the protein’s fourth zinc-finger domain, which is critical for nuclear localization of the protein and is highly conserved in orthologs from humans to sea anemones (Figure 1D). The mutation was present in all other affected individuals, was absent in all unaffected family members and all exome databases (including the ExAC Browser), and was predicted to be deleterious by PolyPhen and SIFT.
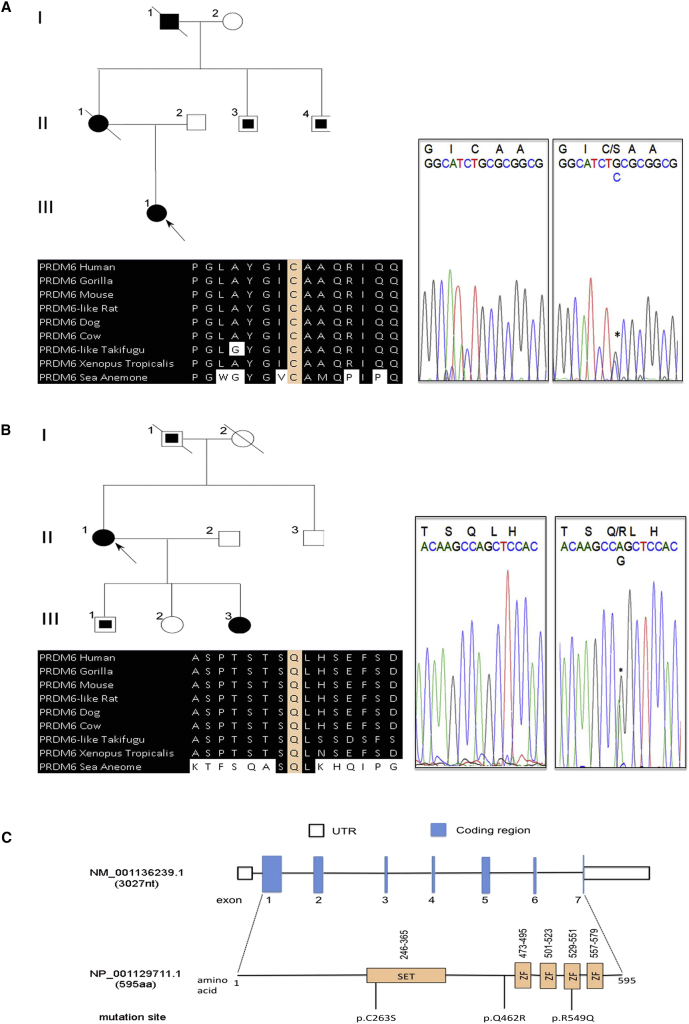
Examination of the exome data in the remaining PDA-affected individuals (n = 34) led to the identification of two additional non-conservative mutations in PRDM6 (c.788G>C [p.Cys263Ser] and c.1385A>G [p.Gln462Arg]) in two unrelated subjects (Figures 2A and 2B). Amino acid substitution p.Gln462Arg was also present in the affected daughter of the subject. No segregation analysis could be carried out in the kindred affected by p.Cys263Ser, because the affected mother and maternal grandfather of the index subject had died before sample collection. Statistical analysis using two random control subjects from the NHLBI Exome Variant Server showed that recurrent mutations in this gene in this number of samples is unlikely to have occurred by chance (p < 4.74 × 10−6, Monte Carlo simulation). It is noteworthy that no single nonsynonymous mutation in PRDM6 was identified in exomes of our 2,000 healthy white control individuals. Amino acid substitutions p.Cys263Ser and p.Gln462Arg were also absent in all other public exome databases (including the ExAC Browser) and were predicted to be deleterious by PolyPhen and SIFT. Cysteine at codon 263 and glutamine at codon 462 are highly conserved among all orthologs and paralogs (Figures 2A and 2B). Cys263 resides in the highly conserved SET domain, which is involved in chromatin regulation and determines how the enzyme catalyzes the transfer of the methyl from S-adenosyl methionine to the histone residue.17 A schematic of the PRDM6 domains and the location of the three independent amino acid substitutions are shown in Figure 2C.
Figure 2.
Independent Non-conservative Mutations of PRDM6 in Subjects with PDA
(A and B) A pedigree with a DNA segment flanking the PRDM6 c.788G>C mutation (resulting in p.Cys263Ser) and a portion of the protein flanking Cys263 in diverse vertebrate species are shown in (A). A pedigree with a DNA segment flanking the PRDM6 c.1385A>G mutation (resulting in p.Arg549Gln) and a portion of the protein flanking Gln462 in diverse vertebrate species are shown in (B). Individuals with PDA are indicated by black symbols, individuals without PDA are shown as unfilled symbols, and individuals with unknown PDA status are shown with partially filled squares. Circles represent females, squares represent males, and symbols with a slash through them indicate deceased subjects.
(C) A schematic of PRDM6 with functional domains and the location of the three independent mutations are shown. Intronic regions are not drawn to scale. ZF denotes zinc finger.
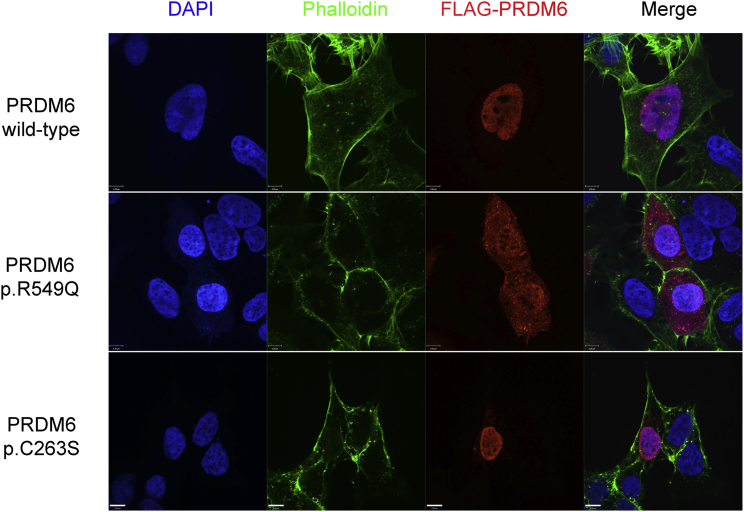
PRDM6 c.1646G>A Impairs Nuclear Localization of the Encoded Protein
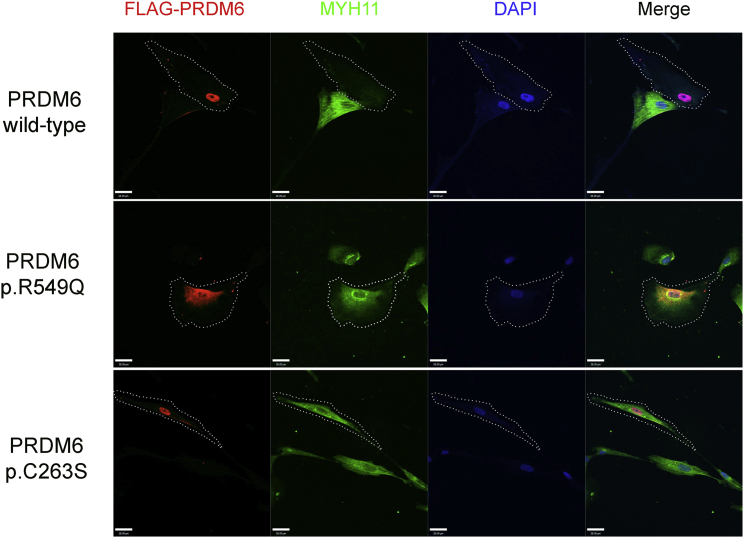
PRDM6 is a nuclear protein that functions as a histone methyltransferase and suppresses the transcription of VSMC contractile proteins.18 We overexpressed constructs encoding wild-type PRDM6 and PRDM6 variants p.Arg549Gln and p.Cys263Ser, which lie in critical domains in HEK293 cells and human aortic VSMCs. Arg549 resides in a protein segment that has previously been shown to be critical for nuclear localization.18 Furthermore, in silico analysis designated this amino acid as an RNA binding site. Functional in vitro characterization of PRDM6 p.Arg549Gln in HEK293 cells and human aortic VSMCs showed that the variant protein significantly loses its capability for nuclear localization (Figures 3 and 4). Co-staining with markers of the endoplasmic reticulum (calnexin), Golgi apparatus (GM130), and mitochondria (Cox IV) and the nuclear transporter importin-β excluded retention of the protein on any of these organelles (Figure S2). We subsequently used an adenovirus vector to overexpress DNA encoding wild-type PRDM6 and PRDM6 variants p.Arg549Gln and p.Cys263Ser in human aortic VSMCs. Immunofluorescence (IF) showed that, compared to the empty vector, wild-type PRDM6 suppressed MYH11, but PRDM6 p.Arg549Gln and p.Cys263Ser largely failed to reduce its amount (Figure 4). Given that the function of the wild-type protein is to silence the transcription of VSMCs, this finding suggests loss of function.
Figure 3.
The Effect of PRDM6 Amino Acid Substitutions on Its Nuclear Localization
Immunofluorescence staining of wild-type PRDM6, PRDM6 p.Arg549Gln (p.R549Q), and p.Cys263Ser (p.C263S) in HEK293 cells. Whereas wild-type PRDM6 and p.Cys263Ser were localized in the nucleus, p.Arg549Gln was predominantly localized in the cytoplasm. Colors are as follows: blue, DAPI; green, phalloidin; and red, PRDM6 with FLAG tag. Scale bars represent 7 μm.
Figure 4.
The Effect of PRDM6 Amino Acid Substitutions on MYH11 Cellular Levels
Immunostaining demonstrates the localization of PRDM6 (red) in relationship to the nucleus (blue) and the staining for MYH11 in human aortic smooth muscle cells. Cells infected with wild-type PRDM6, PRDM6 p.Arg549Gln (p.R549Q), and PRDM6 p.Cys263Ser (p.C263S) are shown in the top, middle, and bottom rows, respectively. Wild-type PRDM6 suppressed MYH11. Substitution p.Arg549Gln was largely retained in the cytoplasm, whereas p.Cys263Ser was mainly expressed in the nucleus. In smooth muscle cells overexpressing DNA encoding p.Arg549Gln and p.Cys263Ser, MYH11 staining was comparable to that in non-transfected cells. Dotted lines delineate transfected cells. Scale bars represent 28 μm.
PRDM6 Substitutions Exhibit Loss of Function in Histone Modification and Suppression of VSMC Contractile Proteins
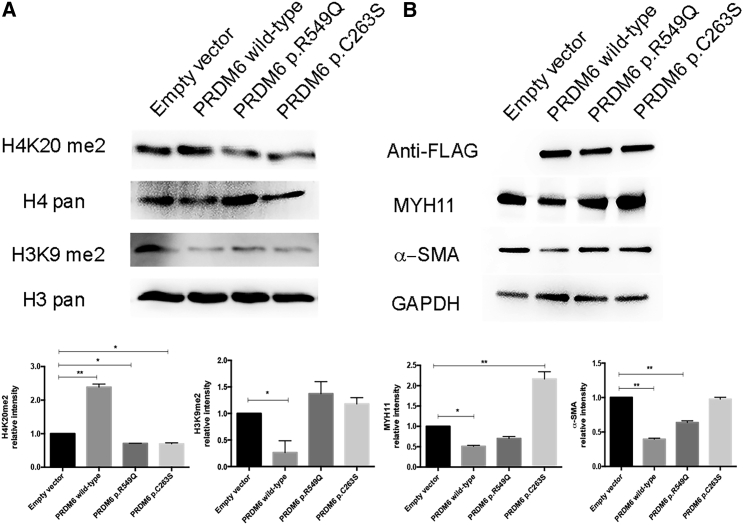
PRDM6 has been implicated in histone lysine methylation and transcriptional suppression of contractile protein in smooth muscle cells.18 We examined the effects of the amino acid substitution on H3K9 and H4K20 dimethylation in human aortic VSMCs by using an adenovirus vector to overexpress DNA encoding wild-type PRDM6 and PRDM6 variants p.Arg549Gln and p.Cys263Ser. Wild-type PRDM6 reduced dimethylation of H3K9 and increased dimethylation of H4K20, whereas both variant PRDM6 proteins had completely opposite effects (Figure 5A).
Figure 5.
The Effect of PRDM6 Amino Acid Substitutions on the Amounts of H4K20me2, H3K9me2, MYH11, and α-SMA
Immunoblot shows the amounts of histones H4K20me2 and H3K9me2 (A) and MYH11 and α-SMA (B) in wild-type human aortic VSMCs transfected with wild-type PRDM6, PRDM6 p.Arg549Gln (p.R549Q), and PRDM6 p.Cys263Ser (p.C263S). Cells transfected with empty vector were used as controls. The lower panel shows the relative intensities by densitometry. Error bars denote the SEM.
Immunoblot analysis revealed that PRDM6 p.Arg549Gln and p.Cys263Ser were associated with higher amounts of MYH11 and α-SMA than was the wild-type (Figure 5B). Because PRDM6 amino acid substitutions might cause a dominant-negative effect by altering dimerization, we assessed this property of wild-type and variant PRDM6 in vitro by using native PAGE gel (Figure S4). No change in dimerization was identified between wild-type and variant PRDM6. These findings provide strong evidence supporting the functional effects of the amino acid substitutions and provide strong links to disease pathogenesis.
Interestingly, we also observed an increased burden of mutations in other histone-modifying genes in the remaining PDA subjects. A GSEA (see Material and Methods) revealed that previously unreported deleterious mutations in histone-modification pathways (Tables S2 and S3) were significantly associated with PDA in 15 of 32 affected individuals (estimated odds ratio = 14.95, CI = 8.63–25.50). Although GSEA does not establish causality, the finding is intriguing and relevant given a priori association between the histone modifier PRDM6 and PDA.
Evaluation of PRDM6 Amounts in DA Smooth Muscle Cells
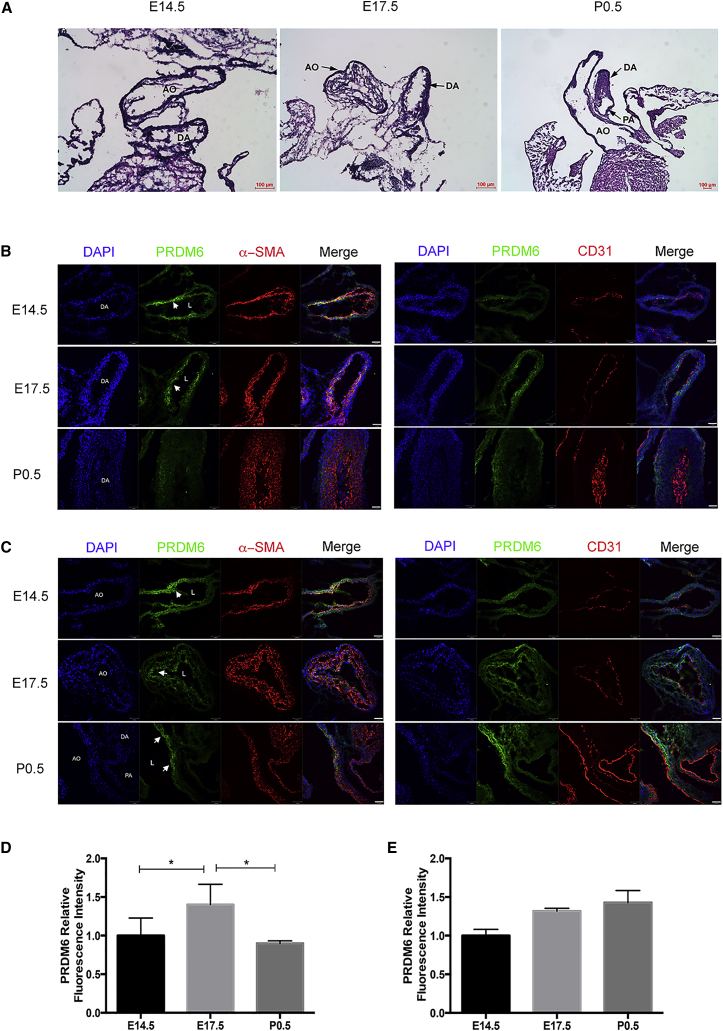
The pressing question is how reduced function of PRDM6 causes disease even though it results in higher amounts of contractile proteins. We examined the amounts of PRDM6 in wild-type DA and aorta in different stages of development by IF. Examination of mouse DA in E14.5, E17.5, and P0.5 embryos (Figure 6A) showed dynamic changes in staining of PRDM6 in DA smooth muscle cells—staining was strong prenatally but declined rapidly postnatally (Figure 6B)—whereas the aorta showed no significant change in PRDM6 staining before or after birth (Figure 6C). This reduction in the amount of ductus PRDM6 was not due to increased apoptosis (Figure S5). Despite a previous report, there was no detectable staining of PRDM6 in CD31+ (endothelial) cells. Taken together, these findings suggest that PRDM6 maintains VSMCs in an undifferentiated state during embryonic life, a function most likely required for their proliferation. The rapid postnatal decline of PRDM6 amounts can also be explained for the necessary VSMC differentiation enabling these cells to contract and occlude DA lumen. PRDM6 loss of function in humans and mice causes premature differentiation and reduced proliferation of VSMCs, resulting in impaired cushion formation and patency of the vessel. In conclusion, our study has identified PRDM6 mutations as the underlying causes of inherited nonsyndromic PDA and implies impaired epigenetic regulation in the pathogenesis of PDA.
Figure 6.
Dynamic Changes in PRDM6 Abundance in Mouse DA and Great Vessels
(A) H&E staining of the aorta (AO), ductus arteriosus (DA), and pulmonary artery (PA) at E14.5, E17.5, and P0.5.
(B and C) IF staining of PRDM6, α-SMA, and CD31 at E14.5, E17.5, and P0.5. (B) PRDM6 was detectable in smooth muscle cells, but not in endothelia cells, of the DA at E14.15 and E17.5. Its amounts were diminished in the DA at P0.5. (C) PRDM6 was persistently detected in smooth muscle cells of the aorta at all three embryonic stages without significant variation. Scale bars represent 37 μm.
(D and E) Relative intensities of PRDM6 in the DA (D) and aorta (E) at E14.5, E17.5, and P0.5 in seven mice. L denotes lumen. Arrowheads point toward PRDM6 staining. Error bars denote the SEM.
Discussion
CHDs are the most common developmental defects in newborns and children. Although their heritability is well established, identification of their causal mutations has been limited by their complex pattern of inheritance. PDA is one the most common inherited CHDs and is also seen as a result of environmental insults such as maternal rubella infection. It is caused by impaired embryonic remodeling of the DA, a process that has been of great interest because of its resemblance to pathological remodeling of adult vessels. As a result, extensive studies have been carried out to elucidate the underlying mechanism of DA remodeling, both in vivo and in vitro. Despite these efforts, the pathophysiology of the nonsyndromic PDA in humans has remained obscure. By using linkage analysis and WES, we identified independent mutations in PRDM6 as the underlying causes of isolated nonsyndromic autosomal-dominant PDA.
The functional significance of the encoded amino acid substitutions was established by the high conservation of the mutated bases, in vitro functional studies, and tissue-specific and dynamic changes of PRDM6 amounts in the murine embryonic DA. Arg549 is a highly conserved amino acid that resides in a protein region that has been previously shown to play a critical role in the protein’s nuclear localization.18 It is also predicted to be a RNA binding site by in silico analysis. Accordingly, in vitro analysis showed that the p.Arg549Gln substitution impairs the nuclear localization of the protein and acts as a loss-of-function variant. Cys263, which is substituted by serine, is a highly conserved amino acid that resides in the SET domain, functionally critical for its methyl transferase activities. Glu462, which is substituted by arginine, is also a highly conserved amino acid and is predicted to be damaging by SIFT and PolyPhen.
PRDM6 is a member of the PRDM family of transcriptional repressors, which contain a PR domain and multiple zinc-finger domains. It is abundantly present in the cardiac outflow tract and the DA and is involved in epigenetic regulation of VSMC contractile proteins.18 Interestingly, several PDA-affected individuals showed evidence of enrichment of previously unreported non-conservative amino acid substitutions in other histone modifiers (Tables S2 and S3). Compared to PRDM6 mutations, these variants most likely impart much smaller effects on the trait given that they occur mainly sporadically and occasionally more than once in each individual. Overall, our findings prompt the question as to whether inherited or acquired forms of PDA are caused by epigenetic dysregulation of vascular genes.
PRDM6 functions as a transcriptional repressor of myocardin, GATA-6, smooth muscle cell actins, and myosins.18 These proteins are important for VSMC differentiation and their development of contractile phenotypes.19 Thus, with the finding of PRDM6 mutations, we hypothesized that PRDM6 loss of function results in early differentiation of VSMCs and thus their reduced proliferation. A rapid decline of PRDM6 in the wild-type DA suggests that VSMC differentiation occurs in later developmental stages to promote the contraction and sufficient PRDM6 amounts necessary for proliferation of these cells. Accordingly, altered function of Tfap2b—the murine ortholog of TFAP2B (MIM: 601601), the gene associated with human syndromic PDA, known as Char syndrome (MIM: 169100)—also results in impaired development of the VSMC layer and patency of the DA in mice.20 PDA has been reported in a few subjects with thoracic aortic aneurysm and rare genetic variants in MYH11 (MIM: 132900) and ACTA2 (MIM: 611788).21, 22 Structural analyses of the aorta in these affected individuals have revealed large areas of medial degeneration with very low VSMC content. In this study, we found no coding variants in MYH11, ACTA2, or JAG1 (MIM: 187500). To our knowledge, none of these genes have been associated with isolated PDA.
In summary, our study establishes the key role of PRDM6 in regulation of VSMC plasticity during DA remodeling. In vitro functional characterization of PRDM6 and its impaired function and the identification of its epigenetic targets are important steps toward discovering novel pathways for the development of drugs against diverse vascular disorders.
Acknowledgments
This work was supported by NIH grants (1R01HL122830 and 1R01HL122822) to A.M. and the NIH Centers for Mendelian Genomics (5U54HG006504). We would like to thank Dr. Kathleen Martin from Yale University for providing us with human aortic vascular smooth muscle cells. We also thank Dr. Sekar Kathiresan at Massachusetts General Hospital for providing us with access to his whole-exome sequencing database. The authors would like to thank the National Heart, Lung, and Blood Institute Grand Opportunity (GO) Exome Sequencing Project and its ongoing studies, which produced and provided exome variant calls for comparison: the Lung GO Sequencing Project (HL-102923), the Women’s Health Initiative Sequencing Project (HL-102924), the Broad GO Sequencing Project (HL-102925), the Seattle GO Sequencing Project (HL-102926), and the Heart GO Sequencing Project (HL-103010). The authors would like to thank the Exome Aggregation Consortium and the groups that provided exome variant data for comparison. A full list of contributing groups can be found at http://exac.broadinstitute.org/about.
Published: May 12, 2016
Footnotes
Supplemental Data include a Supplemental Note, five figures, and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.03.022.
Web Resources
ExAC Browser, http://exac.broadinstitute.org
Gene Ontology, http://geneontology.org/page/go-enrichment-analysis
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
SIFT, http://sift.jcvi.org
UCSC Genome Browser, https://genome.ucsc.edu/
Supplemental Data
References
- 1.Hoffman J.I., Kaplan S. The incidence of congenital heart disease. J. Am. Coll. Cardiol. 2002;39:1890–1900. doi: 10.1016/s0735-1097(02)01886-7. [DOI] [PubMed] [Google Scholar]
- 2.Mani A., Meraji S.M., Houshyar R., Radhakrishnan J., Mani A., Ahangar M., Rezaie T.M., Taghavinejad M.A., Broumand B., Zhao H. Finding genetic contributions to sporadic disease: a recessive locus at 12q24 commonly contributes to patent ductus arteriosus. Proc. Natl. Acad. Sci. USA. 2002;99:15054–15059. doi: 10.1073/pnas.192582999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coceani F., Armstrong C., Kelsey L. Endothelin is a potent constrictor of the lamb ductus arteriosus. Can. J. Physiol. Pharmacol. 1989;67:902–904. doi: 10.1139/y89-141. [DOI] [PubMed] [Google Scholar]
- 4.Coceani F. Control of the ductus arteriosus--a new function for cytochrome P450, endothelin and nitric oxide. Biochem. Pharmacol. 1994;48:1315–1318. doi: 10.1016/0006-2952(94)90552-5. [DOI] [PubMed] [Google Scholar]
- 5.Muti R. [Morphology and structure of the ductus arteriosus (Botallo’s) and genesis of the ligamentum arteriosum in man] Biol. Lat. 1970;22:25–40. [PubMed] [Google Scholar]
- 6.Garcia O.S. [Functional architecture of the ligamentum arteriosum in adults] Acta Anat. (Basel) 1975;91:313–320. [PubMed] [Google Scholar]
- 7.Gersony W.M. Commentary: patent ductus arteriosus and the respiratory distress syndrome--a perspective. J. Pediatr. 1977;91:624–625. doi: 10.1016/s0022-3476(77)80520-9. [DOI] [PubMed] [Google Scholar]
- 8.Toizumi M., Motomura H., Vo H.M., Takahashi K., Pham E., Nguyen H.A., Le T.H., Hashizume M., Ariyoshi K., Dang D.A. Mortality associated with pulmonary hypertension in congenital rubella syndrome. Pediatrics. 2014;134:e519–e526. doi: 10.1542/peds.2013-4184. [DOI] [PubMed] [Google Scholar]
- 9.Satoda M., Zhao F., Diaz G.A., Burn J., Goodship J., Davidson H.R., Pierpont M.E., Gelb B.D. Mutations in TFAP2B cause Char syndrome, a familial form of patent ductus arteriosus. Nat. Genet. 2000;25:42–46. doi: 10.1038/75578. [DOI] [PubMed] [Google Scholar]
- 10.Mani A., Radhakrishnan J., Farhi A., Carew K.S., Warnes C.A., Nelson-Williams C., Day R.W., Pober B., State M.W., Lifton R.P. Syndromic patent ductus arteriosus: evidence for haploinsufficient TFAP2B mutations and identification of a linked sleep disorder. Proc. Natl. Acad. Sci. USA. 2005;102:2975–2979. doi: 10.1073/pnas.0409852102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keramati A.R., Fathzadeh M., Go G.W., Singh R., Choi M., Faramarzi S., Mane S., Kasaei M., Sarajzadeh-Fard K., Hwa J. A form of the metabolic syndrome associated with mutations in DYRK1B. N. Engl. J. Med. 2014;370:1909–1919. doi: 10.1056/NEJMoa1301824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang S., Song K., Srivastava R., Dong C., Go G.W., Li N., Iwakiri Y., Mani A. Nonalcoholic fatty liver disease induced by noncanonical Wnt and its rescue by Wnt3a. FASEB J. 2015;29:3436–3445. doi: 10.1096/fj.15-271171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mootha V.K., Lindgren C.M., Eriksson K.F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 15.Zaidi S., Choi M., Wakimoto H., Ma L., Jiang J., Overton J.D., Romano-Adesman A., Bjornson R.D., Breitbart R.E., Brown K.K. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013;498:220–223. doi: 10.1038/nature12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lynch H.T., Grissom R.L., Magnuson C.R., Krush A. Patent ductus arteriosus. Study of two families. JAMA. 1965;194:135–138. [PubMed] [Google Scholar]
- 17.Rice J.C., Briggs S.D., Ueberheide B., Barber C.M., Shabanowitz J., Hunt D.F., Shinkai Y., Allis C.D. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol. Cell. 2003;12:1591–1598. doi: 10.1016/s1097-2765(03)00479-9. [DOI] [PubMed] [Google Scholar]
- 18.Davis C.A., Haberland M., Arnold M.A., Sutherland L.B., McDonald O.G., Richardson J.A., Childs G., Harris S., Owens G.K., Olson E.N. PRISM/PRDM6, a transcriptional repressor that promotes the proliferative gene program in smooth muscle cells. Mol. Cell. Biol. 2006;26:2626–2636. doi: 10.1128/MCB.26.7.2626-2636.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang J., Cheng L., Li J., Chen M., Zhou D., Lu M.M., Proweller A., Epstein J.A., Parmacek M.S. Myocardin regulates expression of contractile genes in smooth muscle cells and is required for closure of the ductus arteriosus in mice. J. Clin. Invest. 2008;118:515–525. doi: 10.1172/JCI33304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao F., Bosserhoff A.K., Buettner R., Moser M. A heart-hand syndrome gene: Tfap2b plays a critical role in the development and remodeling of mouse ductus arteriosus and limb patterning. PLoS ONE. 2011;6:e22908. doi: 10.1371/journal.pone.0022908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo D.C., Pannu H., Tran-Fadulu V., Papke C.L., Yu R.K., Avidan N., Bourgeois S., Estrera A.L., Safi H.J., Sparks E. Mutations in smooth muscle alpha-actin (ACTA2) lead to thoracic aortic aneurysms and dissections. Nat. Genet. 2007;39:1488–1493. doi: 10.1038/ng.2007.6. [DOI] [PubMed] [Google Scholar]
- 22.Zhu L., Vranckx R., Khau Van Kien P., Lalande A., Boisset N., Mathieu F., Wegman M., Glancy L., Gasc J.M., Brunotte F. Mutations in myosin heavy chain 11 cause a syndrome associating thoracic aortic aneurysm/aortic dissection and patent ductus arteriosus. Nat. Genet. 2006;38:343–349. doi: 10.1038/ng1721. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.