Abstract
Curry-Jones syndrome (CJS) is a multisystem disorder characterized by patchy skin lesions, polysyndactyly, diverse cerebral malformations, unicoronal craniosynostosis, iris colobomas, microphthalmia, and intestinal malrotation with myofibromas or hamartomas. Cerebellar medulloblastoma has been described in a single affected individual; in another, biopsy of skin lesions showed features of trichoblastoma. The combination of asymmetric clinical features, patchy skin manifestations, and neoplastic association previously led to the suggestion that this could be a mosaic condition, possibly involving hedgehog (Hh) signaling. Here, we show that CJS is caused by recurrent somatic mosaicism for a nonsynonymous variant in SMO (c.1234C>T [p.Leu412Phe]), encoding smoothened (SMO), a G-protein-coupled receptor that transduces Hh signaling. We identified eight mutation-positive individuals (two of whom had not been reported previously) with highly similar phenotypes and demonstrated varying amounts of the mutant allele in different tissues. We present detailed findings from brain MRI in three mutation-positive individuals. Somatic SMO mutations that result in constitutive activation have been described in several tumors, including medulloblastoma, ameloblastoma, and basal cell carcinoma. Strikingly, the most common of these mutations is the identical nonsynonymous variant encoding p.Leu412Phe. Furthermore, this substitution has been shown to activate SMO in the absence of Hh signaling, providing an explanation for tumor development in CJS. This raises therapeutic possibilities for using recently generated Hh-pathway inhibitors. In summary, our work uncovers the major genetic cause of CJS and illustrates strategies for gene discovery in the context of low-level tissue-specific somatic mosaicism.
Main Text
The multiple-congenital-anomalies disorder Curry-Jones syndrome (MIM: 601707) was first presented (in abstract form) by Cynthia Curry and Marilyn Jones at the David W. Smith Workshop on Malformations and Morphogenesis in 1987. These authors described two unrelated individuals with the shared features of unilateral coronal craniosynostosis, cutaneous syndactyly, bilateral preaxial polydactyly of the feet, and unusual streaky skin lesions. Subsequently, the term Curry-Jones syndrome (CJS) was applied to this condition.1, 2
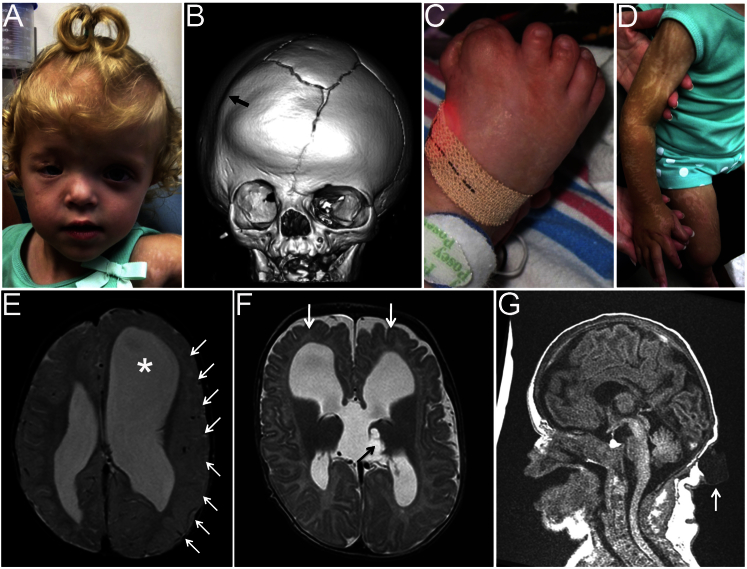
The first formal publication on CJS was by Temple et al.3 and included detailed clinical descriptions of the two original subjects and three further unrelated individuals. Four more simplex cases have since been added to the literature; each affected individual had abnormal skin patches and preaxial polydactyly of the feet.4, 5, 6 Additional features found in most individuals have included ectopic hair growth, abnormalities of brain development, coloboma and/or microphthalmia, coronal suture synostosis, cutaneous syndactyly, and intestinal malrotation and/or obstruction (reviewed by Grange et al.6). Figures 1A–1D show the major craniofacial, limb, and dermatological features of CJS in a previously unreported individual (subject 8 in our series); MRI scans of three individuals (Figures 1E–1G) illustrate the diversity of cerebral malformations (described in more detail below). Mild intellectual disability has been present in some individuals. In a single individual, a desmoplastic medulloblastoma (World Health Organization grade IV) of the right cerebellum, found incidentally on a follow-up brain MRI scan at 17 months, was treated successfully by surgical resection, chemotherapy, and radiotherapy.6 In two individuals, biopsies of active skin lesions were reported to show features of either trichoblastoma6 or nevus sebaceus.3 In addition, odontogenic keratocysts have been found in one individual, and lesions in the bowel, identified histologically as hamartomas or myofibromas, were reported in three instances.3, 6
Figure 1.
Clinical and Radiological Features and Brain Imaging of Individuals with CJS Caused by SMO Mutations
(A–D) Subject 8 at 2 years of age. (A) Facial features show asymmetry, frontal bossing, and scarring of the right eyelid. (B) Computed-tomographic head scan at 7 months of age shows right coronal synostosis (arrow), a bifurcated sagittal suture, and an anterior fontanellar bone. (C) The right foot is medially deviated with a duplicated hallux and partial cutaneous syndactyly of digits 1–3. (D) The skin has linear streaks of hypopigmentation with atrophy; this was not visible at birth but had developed by the age of 6 months.
(E–G) Brain MRI. (E) At 1 year, 5 months old, subject 5 shows left-sided hemimegalencephaly with extensive cortical malformation (arrows) and ventriculomegaly (asterisk). (F) At 3 months old, subject 6 shows subtle abnormalities of the gyri and cortex (white arrows); a cyst in the left thalamus (black arrow) connects with a third-ventricle midline cyst. (G) At 1 day old, subject 8 shows a thin corpus callosum, a mildly hypoplastic cerebellum, and an occipital cystic lesion pathologically confirmed to represent a lymphangiomatous malformation (white arrow).
The etiology of CJS was previously unknown, but three clinical observations are relevant to hypotheses for causation. First, the nine previously reported individuals comprised seven males and two females with similar disease severity, making an X-linked mutation unlikely. Second, the sporadic origin of all individuals in association with patchy skin lesions and asymmetric cranial findings led Temple et al.3 to propose an underlying mosaic mutation; however, Grange et al.6 favored a germline constitutional mutation in view of the consistent presentation and bilaterality of some of the other features. Third, the occurrence of medulloblastoma in basal cell nevus syndrome (BCNS [MIM: 109400])—caused by mutations in PTCH1 (MIM: 601309), which encodes the hedgehog (Hh) receptor—as well as the overlapping cranial and limb abnormalities found in disorders caused by mutations in GLI3 (MIM: 165240), a downstream effector of the Hh pathway, led Grange et al.6 to propose that perturbation of Hh signaling could underlie CJS. However, sequencing of PTCH1 and GLI3 in DNA obtained from the blood of two individuals with CJS was normal.6 The objective of the present work was to identify the causative mutation(s) underlying CJS through whole-exome sequencing (WES) by initially using an overlap strategy in four individuals to pinpoint disease-causing variants in the same gene.
The study was approved by Oxfordshire Research Ethics Committee B (reference C02.143) and the Riverside Research Ethics Committee (reference 09/H0706/20). Participants or their parents provided informed, written consent for genetic studies. Four of five samples initially chosen for analysis were from CJS-affected individuals reported by Temple et al.;3 these comprised both eyelid tissue (sample 1-2, collected during an operation to repair a malformed upper eyelid with ectopic hairs) and fibroblasts from an affected skin biopsy (sample 1-4) from subject 1 (the individual originally described by Jones) and fibroblasts from affected skin of subjects 2 (sample 2-3) and 3 (sample 3-2), the latter of whom was the individual originally described by Curry. All fibroblast lines were analyzed between three and five passages. An additional fifth sample (4-1) was from the blood of subject 4, an unpublished individual with features overlapping CJS. The clinical features of these four individuals are summarized in Table 1.
Table 1.
Clinical Features of Subjects Diagnosed with CJS
| Subject | Gender | Craniosynostosis | Brain | Developmental Attainment | Eyes | Skin | Hair | GI Tract | Cutaneous Syndactyly | Polydactyly | Tumors | Other | Previously Reported? |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 (SMO+) | F | RC | possible cyst in trigone of right ventricle | normal | R amblyopia; skin overgrowth | waxy, streaky lesions; nevus sebaceus | ectopic patch of hair above R eye | GI bleeding; chronic constipation | L 1/2/3 (H); R 2/3 (H) | R trifid hallux | – | asymmetric face | case 4 in Temple et al.3 |
| 2 (SMO+) | M | LC | ACC; R HMEG; R VMEG | mild delay | normal | raised, linear white streaks; normal biopsy | ectopic patch of hair near eyes | intestinal obstruction | L 2/3 (H), 3/4 (F); R 2/3/4 (H), 2/3 (F) | L and R broad thumbs, preaxial polydactyly (F) | myofibromas of large bowel | lip pits; seizures | case 1 in Temple et al.3 |
| 3 (SMO+) | M | LC | partial ACC; asymmetric dilated ventricles | mild ID; IQ < 70 | L and R microphthalmia; R iris coloboma | raised, scar-like pale lesions; non-specific biopsy | ectopic patch of hair above R eye | esophageal dysmotility | L and R 1/2/3/4 (H) | L and R bifid halluces | – | asymmetric face; fused central incisor; oligodontia; freckled areas on soles of feet; esophageal dysmotility | case 3 in Temple et al.3 |
| 4 | M | bicoronal | dilated ventricles; choroid plexus cyst | mild delay | congenital glaucoma; secondary cataract | pigmentary anomalies in Blashko’s lines, most notable on limbs | chaotic hair patterning of lateral portions of eyebrows | diarrheal episodes | L and R 2/3/4 (F) | broad halluces | – | cleft palate | no |
| 5 (SMO+) | M | none | mild ACC; L HMEG; R VMEG and PMG; occipital meningocele; Chiari I malformation | mild to moderate developmental delay | L mild colobomatous microphthalmia with unusually shaped pupil | raised, linear streaks (L arm and leg, chin, and a few other areas) | ectopic hair on cheek | malrotation; intestinal pseudo-obstruction; chronic constipation; subtotal colectomy for volvulus and obstipation | L 1/2/3/4 (H), 3/4 (F); R 2/3 (H), 3/4 (F) | L and R preaxial polydactyly (F); R thumb nubbin | desmoplastic medulloblastoma of cerebellum; odontogenic keratocysts; benign colonic polyps and smooth muscle hamartoma | large anterior fontanelle; lip pits | patient 2 in Grange et al.6 |
| 6 (SMO+) | M | none | partial ACC; MEG; VMEG; Chiari I malformation | mild delay (1–2 years behind academically) | L and R iris colobomas | hypopigmented streaky lesions | ectopic patch of hair near eyes | malrotation | L 1/2/3 (H), 1/2 (F); R 2/3 (H) | L and R preaxial polydactyly (H), preaxial polydactyly (F) | trichoblastoma; smooth muscle hamartomas of GI tract | mildly asymmetric face; lip pit | patient 1 in Grange et al.6 |
| 7 (SMO+) | M | none | normal | normal | normal | white, patchy skin behind knees | abnormal hair growth on shoulders and limbs | normal | R 1/2/3/4 (H) | L and R preaxial polydactyly (F); R: broad thumb | – | plagiocephaly | Thomas et al.5 |
| 8 (SMO+) | F | RC | partial ACC; moderate cerebral asymmetry consistent with L HMEG; PMG | mild delay | normal; dysmorphic R eyelid | linear areas of hypopigmented skin atrophy (extremities and trunk) | – | malrotation; intermittent pseudo-obstruction; serosal nodules in the appendix, mesentery, and duodenum | L and R variable 1/2/3 (H and F) | L and R duplicated thumbs and halluces | scalp lymphangiomas: benign hamartomatous lesion with features of lymphangioma and nevus sebaceus; serosal hamartomas in duodenum, appendix, and mesentery | accessory bone at anterior fontanelle; wormian bones of L posterior skull; L leg longer than R; lumbar scoliosis | no |
| 9 | F | none | ACC; dilated ventricles; macrocephaly | psychomotor delay | bilateral colobomas of iris, retina, and choroid; strabismus; nystagmus | hypopigmented patch on trunk | high frontal hairline | chronic constipation | L and R 3/4 (H) | L and R duplicated thumbs and halluces | lipomyelomeningocele | rudimentary sacral vertebrae | no |
| 10 (SMO+) | F | LC | ACC; abnormal cortical dysplasia of R perirolandic region | psychomotor delay | corneal clouding; eyelid irregularity and ectopic hairs | mostly R-sided hypo- and hyperpigmented swirling skin lesions; increased hair in hyperpigmented areas | ectopic hairs; hirsutism | dysmotility; malrotation; multiple hamartomas of small intestine | R 1/2 (H) | L and R duplicated thumbs and halluces | multiple mesenteric hamartomas with smooth muscle bundles with inter-myenteric ganglia | grade 2 hydronephrosis | no |
Abbreviations are as follows: F, female; M, male; LC, left coronal; RC, right coronal; R, right; L, left; ACC, agenesis of the corpus callosum; MEG, megalencephaly; HMEG, hemimegalencephaly; VMEG, ventriculomegaly; PMG, polymicrogyria; ID, intellectual disability; GI, gastrointestinal; H, hands; and F, feet.
After DNA extraction, we performed WES by using the SeqCap EZ Human Exome Library v.2.0 (NimbleGen) on a HiSeq 2000 (Illumina). Paired-end reads (100 bp) were mapped to hs37d5 with Stampy v.1.0.22,7 and after removal of artifacts, the average coverage was >30× over 82% of the exome. We used Platypus8 to call variants, and assuming that CJS is caused by very rare autosomally located variant(s), we prioritized the data by excluding common variants present in either our in-house database of solved cases or the NHLBI Exome Sequencing Project (ESP) Exome Variant Server (later revised to the Exome Aggregation Consortium [ExAC] Browser for reanalysis). We compared across CJS individuals for hits in the same gene. This analysis did not yield any genes with rare coding variants shared by three or four individuals, and only three genes had rare coding variants shared by two individuals, but none were strong candidates (Table S1).
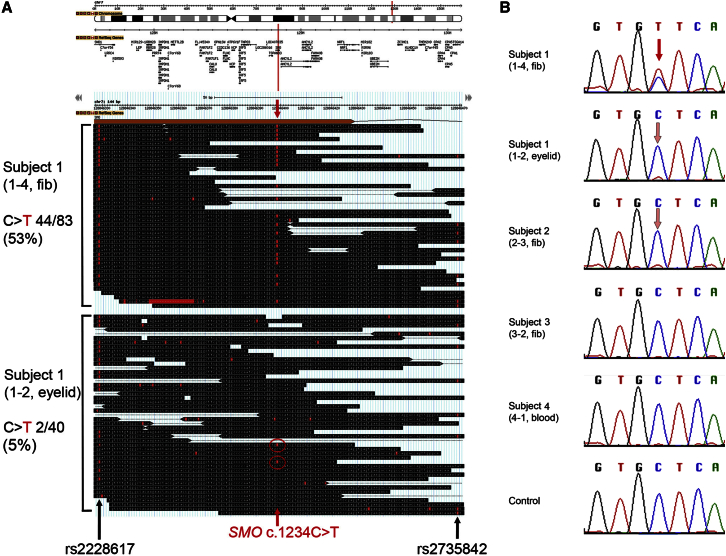
As an alternative approach, we compared the data from the two tissues separately sequenced from subject 1, given that mosaicism for a pathogenic variant could be detectable through differences in allele frequencies between datasets. Variants were ranked according to a somatic p value score generated by the software tool MiG,9 which compares variant and reference read counts between two datasets (Figure S1). The top hit was a nonsynonymous substitution in SMO (MIM: 601500; GenBank: NM_005631.4): c.1234C>T (p.Leu412Phe), present in 44 of 83 sequence reads from fibroblast sample 1-4. Notably, this variant was not called in the exome data from eyelid sample 1-2, although a low level of the same variant (2 of 40 sequence reads) was apparent on manual examination of the reads (Figure 2A). The fortuitous presence of two heterozygous flanking SNPs (rs2228617 and rs2735842; dbSNP137) in this individual enabled the two SMO alleles to be easily distinguished. We noted that when sequence reads included both the c.1234 position and one of the flanking SNPs, the mutant c.1234T reads were always in cis with the variant allele of the SNP in both samples. However, for the eyelid sample 1-2, the converse was not true, supporting the conclusion that the c.1234C>T mutation was mosaic in this sample (see Figure 2). SMO encodes smoothened (SMO), a frizzled G-protein-coupled receptor that plays a key role in transducing Hh signaling. Hh binding relieves patched-mediated suppression of SMO to allow transduction of the signal, probably mediated by conformational changes within the 7-transmembrane bundle.11 Hence, the variant matched all three criteria (autosomal, mosaic, and affecting Hh signaling) for the characteristics of a candidate CJS-associated gene on the basis of clinical deduction.3, 6 Adding further weight to the conclusion that this was the pathogenic variant, the Catalogue of Somatic Mutations in Cancer (COSMIC) revealed that SMO c.1234C>T is a known mutation hotspot in multiple tumor types (discussed further below). Dideoxy-sequencing of SMO exon 6 in sample 1-4 confirmed the presence of the C>T variant in an apparently heterozygous state, but the mutant peak was barely visible in sample 1-2 (Figure 2B).
Figure 2.
Identification of a Mosaic SMO c.1234C>T Mutation
(A) GBrowse10 visualization of exome sequence data from two tissues of subject 1. The upper panel shows the location of SMO in chromosomal region 7q32.1, and the middle and lower panels display a 144 bp alignment of sequencing reads aligned to exon 6 and the following intron; bases that match the reference sequence are boxed in black, and variants are in red. The red arrows indicate the position of the c.1234C>T mutation (indicated by a “t” within a red box), which was present in 53% of reads in tissue 1-4 (middle panel; not all reads are shown) but only in 5% of reads in tissue 1-2 (lower panel). Flanking heterozygous SNPs are indicated by black arrows.
(B) Dideoxy-sequence traces for the c.1234C>T mutation. (Top) In sample 1-4, the mutation appears to be present in the heterozygous state, and there is no evidence of dilution by non-mutant cells (red arrow). (Middle) In samples 1-2 and 2-3, a small proportion of the mutant c.1234T allele is suspected to be present on the basis of both the presence of a small T peak and the reduced relative height of the normal C peak (pink arrows; compare with the control at bottom). By contrast, the mutation does not appear to be present in samples 3-2 and 4-1.
In light of this finding, we scrutinized the exome sequence data for SMO from the other three CJS individuals in greater detail. The identical c.1234C>T variant was present in 2.7% (6 of 219) of reads from sample 2-3 (Figure S2) but was absent in the other two samples (0 of 57 reads from sample 3-2 and 0 of 179 reads from sample 4-1); no other suspicious SMO variant was detected in the exome sequences of subjects 3 or 4.
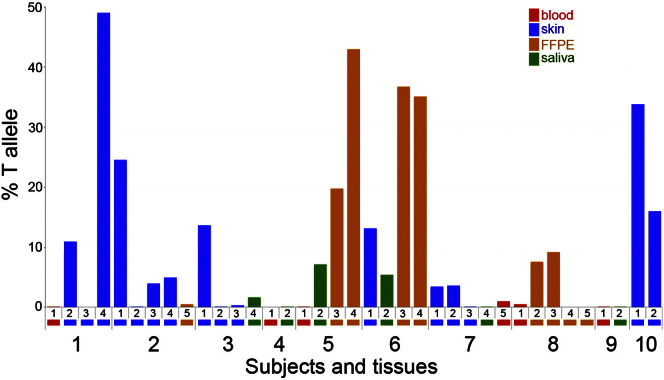
To gather further evidence that mosaic SMO mutations cause CJS, we collected and analyzed additional tissue samples from subjects 1–4 together with a further six individuals with CJS (subjects 5 and 6 reported by Grange et al.,6 subject 7 reported by Thomas et al.,5 and three unpublished individuals; clinical features are summarized in Table 1). Material from affected regions was prioritized and included archival formalin-fixed paraffin-embedded (FFPE) sections, such as those illustrated for affected skin and gut hamartoma in Grange et al.6 (Table S2). We used the Ion Torrent PGM to perform deep sequencing of SMO exon 6 in a total of 37 different tissue samples from the ten CJS subjects (including the previously sequenced samples) and three control samples. Table S3 lists the primer sequences and conditions used for SMO (GenBank: NG_023340.1) amplification and sequencing. Deep sequencing of DNA from subject 1 confirmed a ∼50% frequency for c.1234C>T in fibroblast sample 1-4 and showed that the variant was present at an 11% frequency in eyelid sample 1-2 (Figure 3 and Table S2). Surprisingly, we did not identify any mutant c.1234T alleles in the affected skin sample (1-3) from which the fibroblasts constituting sample 1-4 were derived, possibly indicating positive selection of mutant cells during growth in culture. In subject 2, the exome finding suggesting that the tested sample (2-3) was positive for an SMO mutation was validated, and c.1234C>T variant frequencies ranging from 4%–25% were detected in three of five samples, confirming mosaicism for the identical SMO mutation in this individual.
Figure 3.
Quantification of Tissue Mosaicism for the SMO c.1234C>T Mutation
Plot of deep sequencing data shows the proportion of the mutant c.1234T allele in tissue samples collected from subjects 1–9. Similar tissue sources are grouped by color, indicated by a colored rectangle below the corresponding sample number. Individual tissue identifications along the horizontal axis correspond to the numbering listed in Table S2.
Deep sequencing of SMO in the samples used for exome sequencing (3-2 and 4-1) confirmed that the c.1234C>T mutation was essentially undetectable, given that the data were indistinguishable (by Fisher’s exact test) from the control samples (Table S2). However, a new skin-biopsy sample from subject 3 was positive (14%), and a low level (1.6%) was detected in saliva. Furthermore, five of the six additional CJS individuals (subjects 5–8 and 10) were also mosaic for c.1234C>T (Figure 3 and Table S2). FFPE material from an occipital meningocele and cerebellar medulloblastoma were available from subject 5 (corresponding to patient 2 in Grange et al.6), and analysis showed relatively high levels of mosaicism for the c.1234T allele at 20% and 43%, respectively. Similarly, the abdominal smooth muscle tumor sections from subject 6 (patient 1 in Grange et al.;6 samples 6-3 and 6-4) contained the c.1234T allele at 35%–37%. In subject 7, a mildly affected individual,5 we detected c.1234T at 3%–4% in skin (sample 7-1, taken from a hairy area of the inner leg) and associated fibroblasts (7-2), as well as a femoral bone marrow sample (7-5, 1%). Finally, we identified the c.1234T variant in samples from two unpublished individuals, subjects 8 (Figures 1A–1D and 1G and Supplemental Note) and 10 (Table 1). FFPE samples from the colon (8-2) and cecum (8-3) collected from subject 8 during investigation of intestinal malrotation and mesenteric masses (which contained hamartomatous nodules consisting of disorganized bundles of mature smooth muscle intermixed with nerve fibers and ganglion cells) contained the c.1234T allele at levels of 8%–9%. In subject 10, an affected skin sample and derived fibroblasts contained c.1234T at 33% and 16%, respectively (Figure 3).
In total, we identified the identical SMO c.1234C>T mutation in tissues from eight unrelated CJS-affected individuals, including the two originally described by Curry and Jones. In all but one sample (1-4, in which the mutation was originally identified by exome sequencing), the c.1234T allele was present at a level substantially below 50%, indicating tissue mosaicism. Importantly for diagnosis, we had the greatest success in detecting the mutation in affected tissues obtained by invasive procedures. The mutation was not reliably detected in blood samples from three mutation-positive individuals (levels of c.1234T ranged from 0.03% to 0.5%, which we believe is below the threshold for clinical diagnosis [1%]; Figure 3 and Table S2). We had greater success with saliva samples (mean = 3.6% and range = 0.11%–7.1% in four mutation-positive individuals), but in every individual, a different tissue sample showed a higher mutation level (Figure 3 and Table S2). This suggests that the optimum strategy for detecting mosaic mutations in CJS is to sample affected skin and tissue from internal organs if available. In subjects 4 and 9, from whom only blood and saliva samples were available, levels of the c.1234C>T mutation were 0.11% or lower. Although the results for some samples differed from those of control samples on formal statistical testing, they were deemed indeterminate for clinical purposes. Deep sequencing of the entire SMO coding region did not identify any alternative variants (data not shown). Analysis of tissue samples from affected regions would be required for determining whether these two individuals in fact carry the canonical mutation or have a distinct genetic basis for their phenotype.
The finding of widespread mosaicism in CJS suggests that it arises post-zygotically early during embryonic development. A somewhat later acquisition of the mutation is predicted to cause isolated pathology of individual organs, so we explored this possibility in the context of the skin. We performed deep sequencing of SMO in 14 isolated trichoblastoma samples and a single nevus sebaceus sample, given that analysis of skin biopsies from subjects 1 and 6 had previously demonstrated nevus sebaceus3 and trichoblastoma,6 respectively. Although we detected rare SNPs, we did not find potentially pathological variants (Table S4).
We sought to further define the neuroanatomical features of CJS by reviewing recent MRI scans of three mutation-positive individuals (subjects 5, 6, and 8). A diverse range of phenotypes were present in this small sample; prominent features are illustrated in Figures 1E–1G, and the complete phenotype is summarized in Table S5 and presented in Figure S3. Abnormal findings included hemimegalencephaly (HMEG) with cortical dysplasia, white-matter abnormalities and polymicrogyria, abnormalities of the corpus callosum, ventriculomegaly, occipital meningocele, and a Chiari type I malformation. Given the phenotypic overlap with isolated megalencephaly and/or HMEG and the previous observation of associated mutations in components of the PI3K-AKT pathway,12, 13, 14 it would be of interest to test such mutation-negative brain samples for variants in SMO.
Although no constitutional mutations in SMO have previously been described, several hotspots of somatic mutation are evident in COSMIC, and it is striking that the most frequent of these is the identical c.1234C>T transition (note, this does not occur in the context of a CpG dinucleotide, so intrinsic hypermutability15 is not predicted). Notably, the SMO c.1234C>T mutation has been identified in ameloblastoma,16, 17 medulloblastoma,18, 19 meningioma,20, 21 and basal cell carcinoma (BCC);22, 23 moreover, it has been reported as the oncogenic driver in some of these tumors.16, 23 The altered amino acid Leu412 locates to transmembrane helix 5 of SMO within one of three pivot regions that, by analogy with the β2 adrenergic receptor,24 are likely to have a key role in the conformational changes required for receptor activation. Supporting this, the presence of mutant p.Leu412Phe leads to constitutive activation in the absence of Hh ligand in Smo−/− mouse embryonic fibroblasts16, 22 and increased cell proliferation in ameloblast-lineage cells.16
Vertebrates have three Hh ligands, sonic hedgehog (SHH), desert hedgehog (DHH), and Indian hedgehog (IHH), which remove inhibition of SMO by binding to the receptor patched (encoded by PTCH1 or PTCH2 [MIM: 603673]). Downstream effectors of Hh signal transduction, notably the transcription factors GLI2 and GLI3, are normally tethered by SUFU (suppressor of fused homolog) at the base of the primary cilium, where they are proteolytically processed to repressor forms (GLI2-R and GLI3-R, respectively). SMO activation leads to KIF7-dependent translocation toward the tip of the primary cilium and to transport of full-length activated GLI proteins (GLI2-A and GLI3-A) into the nucleus, enabling transcriptional activation (reviewed by Briscoe and Therond,25 McCabe and Leahy,26 and Arensdorf et al.27). Activation of the PI3K-AKT-mTOR and/or PKA pathways can independently lead to GLI activation, indicating the potential for significant cross-talk with the Hh pathway.28 Constitutional mutations that mimic the consequences of Hh signal activation include loss-of-function mutations in negative regulators acting at multiple stages of the Hh pathway, such as RAB23 (associated with Carpenter syndrome [MIM: 606144]),29 PTCH130, 31, 32 and SUFU33, 34 (associated with BCNS), KIF7 (associated with acrocallosal syndrome),35 and GLI3 (associated with Greig cephalopolysyndactyly).36, 37 In addition, regulatory mutations in IHH (MIM: 600726) cause Philadelphia craniosynostosis.38 Reflecting the net consequence of excessive Hh signal transduction, several clinical features of CJS overlap those of the above disorders, as previously noted.6 These include craniosynostosis39 in both Carpenter and Philadelphia craniosynostosis syndromes, preaxial polydactyly in Greig and Carpenter syndromes,40 cerebral malformations in acrocallosal syndrome,35 and odontogenic keratocysts and skin involvement in BCNS.41 We also note overlap between the cerebral features and mosaic activation of components of the PI3K-AKT-mTOR pathway, such as in Proteus syndrome (MIM: 164730), fibroadipose hyperplasia and CLOVES syndrome (MIM: 612918), and HMEG (reviewed in Keppler-Noreuil et al.,12 Hevner,13 and Jansen et al.14). To our knowledge, abnormalities of the bowel have not previously been highlighted as a frequent feature of disorders associated with activation of Hh signaling, but they appear to be common in CJS (e.g., malrotation and myofibromas or hamartomas). These bowel abnormalities are consistent with the documented role of murine SHH and IHH, produced by the endodermal epithelium, as primary factors in the patterning and organogenesis of the gut,42 where Hh signaling from the endoderm controls growth of the adjacent mesenchyme.43 Mouse embryos in which Smo is deleted from the gut mesenchyme have severely reduced proliferation and differentiation of the intestinal mesenchyme and a reduced number of smooth muscle cells and enteric neurons.44
Given the observed association between CJS and neoplastic diseases (trichoblastoma and cerebellar medulloblastoma), a particularly important consequence of identifying the activating p.Leu412Phe substitution in SMO concerns the potential pharmacotherapeutic implications for management of affected individuals. Recently, there has been considerable interest in developing SMO inhibitors to reduce activation of the Hh pathway (reviewed by Arensdorf et al.27). For example, vismodegib is a clinically approved SMO inhibitor that contacts the extracellular domain and ligand-binding pocket. Interestingly, in a recent report of a subject with segmental BCNS associated with the identical SMO p.Leu412Phe substitution,45 treatment with vismodegib for 4 months led to cessation of the appearance of new lesions and shrinkage of some existing BCCs. However, other reports have suggested that the p.Leu412Phe substitution confers resistance to vismodegib, supporting a role for Leu412 in autoinhibition and/or structural stability.22, 23 This suggests that in CJS-associated tumors, the use of additional agents, such as arsenic trioxide or other more-specific GLI inhibitors or PI3K-AKT-mTOR inhibitors, that act at later stages of the Hh signal-transduction pathway should be explored,33 and indeed there is evidence that this approach can be effective against p.Leu412Phe mutants.16, 22
In summary, these data show that the major phenotypic features of CJS are attributable to excessive activation of Hh signaling owing to a specific c.1234C>T (p.Leu412Phe) somatic mutation. The finding that all eight mutation-positive individuals are mosaic might indicate that constitutional mutations are not compatible with life, as is true for several other sporadic autosomal disorders.46 The mutation is likely to be associated with a spectrum of negative and positive selective consequences during organismal growth and homeostasis, depending on cell type, which most likely explains the apparent evolution of skin lesions and low mutation levels in blood (negative selection) and the predisposition to tumorigenesis (positive selection). The association between segmental BCNS and SMO p.Leu412Phe in a recent report45 is compatible with a later occurrence of the somatic mutation during development than in subjects with the classical CJS phenotype.
Acknowledgments
We are grateful to all the families for their participation in this study. We thank the staff at the high-throughput genomics facility at the Wellcome Trust Centre for Human Genetics (Oxford Genomics Centre) for exome sequencing and Sue Butler, John Frankland, and Tim Rostron for help with cell culture and DNA sequencing. This work was supported by the National Institute for Health Research (NIHR) Oxford Biomedical Research Centre Programme (J.C.T. and A.O.M.W.), the Medical Research Council through the Weatherall Institute of Molecular Medicine Strategic Alliance (G0902418 and MC_UU_12025), and the Wellcome Trust (project grant 093329 to A.O.M.W. and S.R.F.T. and Senior Investigator Award 102731 to A.O.M.W.). The views expressed in this publication are those of the authors and not necessarily those of the National Health Service, NIHR, or Department of Health.
Published: May 26, 2016
Footnotes
Supplemental Data include a Supplemental Note, three figures, and five tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.04.007.
Web Resources
Exome Aggregation Consortium (ExAC) Browser, http://exac.broadinstitute.org/
Leiden Open Variation Database (LOVD), http://chromium.lovd.nl/LOVD2
MiG: Multi-Image Genome, https://mig.molbiol.ox.ac.uk/mig/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
Supplemental Data
References
- 1.Cohen M.M., Jr. Craniosynostosis update 1987. Am. J. Med. Genet. Suppl. 1988;4:99–148. doi: 10.1002/ajmg.1320310514. [DOI] [PubMed] [Google Scholar]
- 2.Gorlin R.J., Cohen M.M., Jr., Levin L.S. Syndromes of the Head and Neck. Oxford University Press; 1990. Syndromes with craniosynostosis: general aspects and well-known syndromes; pp. 519–539. [Google Scholar]
- 3.Temple I.K., Eccles D.M., Winter R.M., Baraitser M., Carr S.B., Shortland D., Jones M.C., Curry C. Craniofacial abnormalities, agenesis of the corpus callosum, polysyndactyly and abnormal skin and gut development--the Curry Jones syndrome. Clin. Dysmorphol. 1995;4:116–129. [PubMed] [Google Scholar]
- 4.Mingarelli R., Mokini V., Castriota Scanderbeg A., Dallapiccola B. Brachycephalosyndactyly with ptosis, cataract, colobomas, and linear areas of skin depigmentation. Clin. Dysmorphol. 1999;8:73–75. [PubMed] [Google Scholar]
- 5.Thomas E.R., Wakeling E.L., Goodman F.R., Dickinson J.C., Hall C.M., Brady A.F. Mild case of Curry-Jones syndrome. Clin. Dysmorphol. 2006;15:115–117. doi: 10.1097/01.mcd.0000194406.85052.de. [DOI] [PubMed] [Google Scholar]
- 6.Grange D.K., Clericuzio C.L., Bayliss S.J., Berk D.R., Heideman R.L., Higginson J.K., Julian S., Lind A. Two new patients with Curry-Jones syndrome with trichoblastoma and medulloblastoma suggest an etiologic role of the sonic hedgehog-patched-GLI pathway. Am. J. Med. Genet. A. 2008;146A:2589–2597. doi: 10.1002/ajmg.a.32503. [DOI] [PubMed] [Google Scholar]
- 7.Lunter G., Goodson M. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 2011;21:936–939. doi: 10.1101/gr.111120.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rimmer A., Phan H., Mathieson I., Iqbal Z., Twigg S.R., Wilkie A.O., McVean G., Lunter G., WGS500 Consortium Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat. Genet. 2014;46:912–918. doi: 10.1038/ng.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McGowan S.J., Hughes J.R., Han Z.P., Taylor S. MIG: Multi-Image Genome viewer. Bioinformatics. 2013;29:2477–2478. doi: 10.1093/bioinformatics/btt406. [DOI] [PubMed] [Google Scholar]
- 10.Stein L.D., Mungall C., Shu S., Caudy M., Mangone M., Day A., Nickerson E., Stajich J.E., Harris T.W., Arva A., Lewis S. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang C., Wu H., Katritch V., Han G.W., Huang X.P., Liu W., Siu F.Y., Roth B.L., Cherezov V., Stevens R.C. Structure of the human smoothened receptor bound to an antitumour agent. Nature. 2013;497:338–343. doi: 10.1038/nature12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keppler-Noreuil K.M., Rios J.J., Parker V.E., Semple R.K., Lindhurst M.J., Sapp J.C., Alomari A., Ezaki M., Dobyns W., Biesecker L.G. PIK3CA-related overgrowth spectrum (PROS): diagnostic and testing eligibility criteria, differential diagnosis, and evaluation. Am. J. Med. Genet. A. 2015;167A:287–295. doi: 10.1002/ajmg.a.36836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hevner R.F. Brain overgrowth in disorders of RTK-PI3K-AKT signaling: a mosaic of malformations. Semin. Perinatol. 2015;39:36–43. doi: 10.1053/j.semperi.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jansen L.A., Mirzaa G.M., Ishak G.E., O’Roak B.J., Hiatt J.B., Roden W.H., Gunter S.A., Christian S.L., Collins S., Adams C. PI3K/AKT pathway mutations cause a spectrum of brain malformations from megalencephaly to focal cortical dysplasia. Brain. 2015;138:1613–1628. doi: 10.1093/brain/awv045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rahbari R., Wuster A., Lindsay S.J., Hardwick R.J., Alexandrov L.B., Al Turki S., Dominiczak A., Morris A., Porteous D., Smith B., UK10K Consortium Timing, rates and spectra of human germline mutation. Nat. Genet. 2016;48:126–133. doi: 10.1038/ng.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sweeney R.T., McClary A.C., Myers B.R., Biscocho J., Neahring L., Kwei K.A., Qu K., Gong X., Ng T., Jones C.D. Identification of recurrent SMO and BRAF mutations in ameloblastomas. Nat. Genet. 2014;46:722–725. doi: 10.1038/ng.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown N.A., Rolland D., McHugh J.B., Weigelin H.C., Zhao L., Lim M.S., Elenitoba-Johnson K.S., Betz B.L. Activating FGFR2-RAS-BRAF mutations in ameloblastoma. Clin. Cancer Res. 2014;20:5517–5526. doi: 10.1158/1078-0432.CCR-14-1069. [DOI] [PubMed] [Google Scholar]
- 18.Jones D.T., Jäger N., Kool M., Zichner T., Hutter B., Sultan M., Cho Y.J., Pugh T.J., Hovestadt V., Stütz A.M. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488:100–105. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pugh T.J., Weeraratne S.D., Archer T.C., Pomeranz Krummel D.A., Auclair D., Bochicchio J., Carneiro M.O., Carter S.L., Cibulskis K., Erlich R.L. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012;488:106–110. doi: 10.1038/nature11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brastianos P.K., Horowitz P.M., Santagata S., Jones R.T., McKenna A., Getz G., Ligon K.L., Palescandolo E., Van Hummelen P., Ducar M.D. Genomic sequencing of meningiomas identifies oncogenic SMO and AKT1 mutations. Nat. Genet. 2013;45:285–289. doi: 10.1038/ng.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clark V.E., Erson-Omay E.Z., Serin A., Yin J., Cotney J., Ozduman K., Avşar T., Li J., Murray P.B., Henegariu O. Genomic analysis of non-NF2 meningiomas reveals mutations in TRAF7, KLF4, AKT1, and SMO. Science. 2013;339:1077–1080. doi: 10.1126/science.1233009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Atwood S.X., Sarin K.Y., Whitson R.J., Li J.R., Kim G., Rezaee M., Ally M.S., Kim J., Yao C., Chang A.L. Smoothened variants explain the majority of drug resistance in basal cell carcinoma. Cancer Cell. 2015;27:342–353. doi: 10.1016/j.ccell.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharpe H.J., Pau G., Dijkgraaf G.J., Basset-Seguin N., Modrusan Z., Januario T., Tsui V., Durham A.B., Dlugosz A.A., Haverty P.M. Genomic analysis of smoothened inhibitor resistance in basal cell carcinoma. Cancer Cell. 2015;27:327–341. doi: 10.1016/j.ccell.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katritch V., Cherezov V., Stevens R.C. Structure-function of the G protein-coupled receptor superfamily. Annu. Rev. Pharmacol. Toxicol. 2013;53:531–556. doi: 10.1146/annurev-pharmtox-032112-135923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Briscoe J., Thérond P.P. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol. 2013;14:416–429. doi: 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- 26.McCabe J.M., Leahy D.J. Smoothened goes molecular: new pieces in the hedgehog signaling puzzle. J. Biol. Chem. 2015;290:3500–3507. doi: 10.1074/jbc.R114.617936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arensdorf A.M., Marada S., Ogden S.K. Smoothened regulation: a tale of two signals. Trends Pharmacol. Sci. 2016;37:62–72. doi: 10.1016/j.tips.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y., Ding Q., Yen C.J., Xia W., Izzo J.G., Lang J.Y., Li C.W., Hsu J.L., Miller S.A., Wang X. The crosstalk of mTOR/S6K1 and Hedgehog pathways. Cancer Cell. 2012;21:374–387. doi: 10.1016/j.ccr.2011.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jenkins D., Seelow D., Jehee F.S., Perlyn C.A., Alonso L.G., Bueno D.F., Donnai D., Josifova D., Mathijssen I.M., Morton J.E. RAB23 mutations in Carpenter syndrome imply an unexpected role for hedgehog signaling in cranial-suture development and obesity. Am. J. Hum. Genet. 2007;80:1162–1170. doi: 10.1086/518047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hahn H., Wicking C., Zaphiropoulous P.G., Gailani M.R., Shanley S., Chidambaram A., Vorechovsky I., Holmberg E., Unden A.B., Gillies S. Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell. 1996;85:841–851. doi: 10.1016/s0092-8674(00)81268-4. [DOI] [PubMed] [Google Scholar]
- 31.Johnson R.L., Rothman A.L., Xie J., Goodrich L.V., Bare J.W., Bonifas J.M., Quinn A.G., Myers R.M., Cox D.R., Epstein E.H., Jr., Scott M.P. Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science. 1996;272:1668–1671. doi: 10.1126/science.272.5268.1668. [DOI] [PubMed] [Google Scholar]
- 32.Ming J.E., Kaupas M.E., Roessler E., Brunner H.G., Golabi M., Tekin M., Stratton R.F., Sujansky E., Bale S.J., Muenke M. Mutations in PATCHED-1, the receptor for SONIC HEDGEHOG, are associated with holoprosencephaly. Hum. Genet. 2002;110:297–301. doi: 10.1007/s00439-002-0695-5. [DOI] [PubMed] [Google Scholar]
- 33.Kool M., Jones D.T., Jäger N., Northcott P.A., Pugh T.J., Hovestadt V., Piro R.M., Esparza L.A., Markant S.L., Remke M., ICGC PedBrain Tumor Project Genome sequencing of SHH medulloblastoma predicts genotype-related response to smoothened inhibition. Cancer Cell. 2014;25:393–405. doi: 10.1016/j.ccr.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith M.J., Beetz C., Williams S.G., Bhaskar S.S., O’Sullivan J., Anderson B., Daly S.B., Urquhart J.E., Bholah Z., Oudit D. Germline mutations in SUFU cause Gorlin syndrome-associated childhood medulloblastoma and redefine the risk associated with PTCH1 mutations. J. Clin. Oncol. 2014;32:4155–4161. doi: 10.1200/JCO.2014.58.2569. [DOI] [PubMed] [Google Scholar]
- 35.Putoux A., Thomas S., Coene K.L., Davis E.E., Alanay Y., Ogur G., Uz E., Buzas D., Gomes C., Patrier S. KIF7 mutations cause fetal hydrolethalus and acrocallosal syndromes. Nat. Genet. 2011;43:601–606. doi: 10.1038/ng.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vortkamp A., Gessler M., Grzeschik K.H. GLI3 zinc-finger gene interrupted by translocations in Greig syndrome families. Nature. 1991;352:539–540. doi: 10.1038/352539a0. [DOI] [PubMed] [Google Scholar]
- 37.Johnston J.J., Sapp J.C., Turner J.T., Amor D., Aftimos S., Aleck K.A., Bocian M., Bodurtha J.N., Cox G.F., Curry C.J. Molecular analysis expands the spectrum of phenotypes associated with GLI3 mutations. Hum. Mutat. 2010;31:1142–1154. doi: 10.1002/humu.21328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klopocki E., Lohan S., Brancati F., Koll R., Brehm A., Seemann P., Dathe K., Stricker S., Hecht J., Bosse K. Copy-number variations involving the IHH locus are associated with syndactyly and craniosynostosis. Am. J. Hum. Genet. 2011;88:70–75. doi: 10.1016/j.ajhg.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Twigg S.R.F., Wilkie A.O.M. A genetic-pathophysiological framework for craniosynostosis. Am. J. Hum. Genet. 2015;97:359–377. doi: 10.1016/j.ajhg.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anderson E., Peluso S., Lettice L.A., Hill R.E. Human limb abnormalities caused by disruption of hedgehog signaling. Trends Genet. 2012;28:364–373. doi: 10.1016/j.tig.2012.03.012. [DOI] [PubMed] [Google Scholar]
- 41.Athar M., Li C., Kim A.L., Spiegelman V.S., Bickers D.R. Sonic hedgehog signaling in Basal cell nevus syndrome. Cancer Res. 2014;74:4967–4975. doi: 10.1158/0008-5472.CAN-14-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramalho-Santos M., Melton D.A., McMahon A.P. Hedgehog signals regulate multiple aspects of gastrointestinal development. Development. 2000;127:2763–2772. doi: 10.1242/dev.127.12.2763. [DOI] [PubMed] [Google Scholar]
- 43.Mao J., Kim B.M., Rajurkar M., Shivdasani R.A., McMahon A.P. Hedgehog signaling controls mesenchymal growth in the developing mammalian digestive tract. Development. 2010;137:1721–1729. doi: 10.1242/dev.044586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang H., Cotton J.L., Wang Y., Rajurkar M., Zhu L.J., Lewis B.C., Mao J. Specific requirement of Gli transcription factors in Hedgehog-mediated intestinal development. J. Biol. Chem. 2013;288:17589–17596. doi: 10.1074/jbc.M113.467498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khamaysi Z., Bochner R., Indelman M., Magal L., Avitan-Hersh E., Sarig O., Sprecher E., Bergman R. Segmental Basal cell nevus syndrome caused by an activating mutation in Smoothened. Br. J. Dermatol. 2016 doi: 10.1111/bjd.14425. [DOI] [PubMed] [Google Scholar]
- 46.Happle R. The categories of cutaneous mosaicism: A proposed classification. Am. J. Med. Genet. A. 2016;170:452–459. doi: 10.1002/ajmg.a.37439. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.