Abstract
Motivation: Medical Subject Headings (MeSH) indexing, which is to assign a set of MeSH main headings to citations, is crucial for many important tasks in biomedical text mining and information retrieval. Large-scale MeSH indexing has two challenging aspects: the citation side and MeSH side. For the citation side, all existing methods, including Medical Text Indexer (MTI) by National Library of Medicine and the state-of-the-art method, MeSHLabeler, deal with text by bag-of-words, which cannot capture semantic and context-dependent information well.
Methods: We propose DeepMeSH that incorporates deep semantic information for large-scale MeSH indexing. It addresses the two challenges in both citation and MeSH sides. The citation side challenge is solved by a new deep semantic representation, D2V-TFIDF, which concatenates both sparse and dense semantic representations. The MeSH side challenge is solved by using the ‘learning to rank’ framework of MeSHLabeler, which integrates various types of evidence generated from the new semantic representation.
Results: DeepMeSH achieved a Micro F-measure of 0.6323, 2% higher than 0.6218 of MeSHLabeler and 12% higher than 0.5637 of MTI, for BioASQ3 challenge data with 6000 citations.
Availability and Implementation: The software is available upon request.
Contact: zhusf@fudan.edu.cn
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
Medical Subject Headings (MeSH) is a comprehensive controlled vocabulary, which has been developed and maintained by National Library of Medicine (NLM), resulting in already 27 455 MeSH main headings (MHs) (http://www.nlm.nih.gov/pubs/factsheets/mesh.html) by 2015. One important usage of MeSH is to index citations in MEDLINE (NCBI Resource Coordinators, 2015; Nelson et al., 2004), to catalog documents, books as well as audiovisuals recorded in NLM. Currently one citation in MEDLINE is indexed by approximately 13 MHs on average. MeSH has also been used in many other applications in biomedical text mining and information retrieval, such as query expansion (Lu et al., 2010; Stokes et al., 2010), document clustering (Gu et al., 2013; Huang et al., 2011b; Zhu et al., 2009a,b) and document searching (Peng et al., 2015). Thus accurate MeSH indexing of biomedical documents is crucial for the biomedical researchers in formulating novel scientific hypothesis and discovering new knowledge.
Currently, human curators in NLM assign most relevant MeSH headings to documents, resulting in that 806 326 MEDLINE citations were indexed in 2015 (http://www.nlm.nih.gov/bsd/bsd_key.html). This work is very precious but clearly laborious, since to index an article, curators need to review the full text of the corresponding MEDLINE article, which is time consuming and prohibitively expensive For example, the average cost of annotating one MEDLINE article is estimated to be around $9.4 (Mork et al., 2013), meaning a huge total cost for indexing around one million documents per year. Also MEDLINE is rapidly growing, it would be more challenging for manual annotation to index all coming documents on time.
To address this problem, NLM has developed an automatic MeSH indexing software, Medical Text Indexer (MTI), to assist MeSH curators. MTI recommends suitable MHs to each MEDLINE citation using the title and abstract as input (Aronson et al., 2004; Mork et al., 2014). MTI consists of two main components: MetaMap Indexing (MMI) and PubMed Related Citations (PRC). MMI extracts biomedical concepts from title and abstract, and then map them to corresponding MHs, while PRC attempts to find similar MEDLINE citations using a modified k-nearest neighbor (KNN) algorithm, PubMed Related Articles (PRA) (Lin and Wilbur, 2007). The MHs of these similar citations are then extracted and combined with the MHs by MMI. After some post-processing steps, such as applying indexing rules, a ranked list of MHs is recommended to the MeSH indexers.
From a machine learning viewpoint, automatic MeSH indexing can be considered as a large-scale multi-label classification problem (Liu et al., 2015), where each MH is a class label and each citation (instance) has multiple MHs. To address this multi-label classification problem, there are two main challenging aspects on the MeSH (label) and citation (instance) sides. First, on the MeSH (label) side, a large number of MHs have a highly biased distribution. For example, out of all 27 455 MHs, the most frequent MH, ‘Humans’, appears more than eight million times in the whole MEDLINE citations with abstracts, while the 20 000th frequent MH, ‘Hypnosis, Anesthetic’, appears only around 200 times. In addition, the number of annotated MHs for each citation varies greatly, ranging from more than 30 to less than 5. These aspects make the problem very challenging to estimate an effective and efficient prediction model for multi-label classification. Second, on the citation (instance) side, complicated semantics of biomedical documents cannot be effectively captured by a simple bag of words (BOW) approach, because a huge number of domain phrases, concepts and abbreviations exist in the biomedical literature. For example, similar concepts can be represented by different words, while the same word may have very different meanings depending upon the contexts. More concretely, ‘malignancy’, ‘tumor’ and ‘cancer’ are all very close concepts to each other, while ‘CAT’ can represent different genes, depending on organisms (Chen et al., 2004). Similarly, the same abbreviation is used as totally different concepts occasionally. For example, ‘CCC’ stands for Continuous Curvilinear Capsulorhexis in one citation (PMID:25291748), but Continuous Circular Course in another citation (PMID:23618326). However, simple BOW representation ignores the order of words and can hardly capture word semantics. In fact, using BOW, it would be very hard to distinguish different concepts represented by the same word, and also difficult to build connections between two different words representing similar concepts. Thus similar citations based on BOW representation may have totally different MHs. Table 1 gives a typical example, where a citation of interest is PMID:25236620, an article about cytopathology fellowship. Surprisingly, if one uses BOW, the most similar citation to PMID:25236620 among three articles in Table 1 becomes PMID:23416813, which is about the diagnosis of adult orbital masse by different techniques, although these two citations share only one MH, ‘Humans’. The reason that this inaccurate similarity between two citations exists is: the term ‘cytopathology’ appears frequently in PMID:25236620 and also ‘cytopathologically’ and ‘cytopathological’ appear many times in PMID:23416813. These three terms have the same stemmed form, causing them to be regarded as the same term, and therefore leading to a very high similarity to these two citations in terms of BOW.
Table 1.
Typical example to show how well D2V-TFIDF works
| PMID | Title | MH | BOW | DSR-BOW | DSR | MH |
|---|---|---|---|---|---|---|
| 25236620 | Cytopathology fellowship milestones. | Accreditation; Clinical Competence; Cytodiagnosis; Education, Medical, Graduate; Fellowships and Scholarships; Humans; Pathology; United States. | – | – | – | – |
| 23416813 | Comparison of computed tomographic and cytopathological findings in the evaluation of adult orbital mass. | Adult; Aged; Aged, 80 and over; Biopsy, Fine-Needle; Eye Neoplasms; Humans; Middle Aged; Orbital Diseases; Orbital Neoplasms; Orbital Pseudotumor; Prospective Studies; Sensitivity and Specificity; Tomography, X-Ray Computed; Young Adult. | 0.5032 | 0.3620 | 0.2208 | 0.0476 |
| 23597252 | Fellowship training in pediatric pathology: a guide for program directors. | Education, Medical, Graduate; Fellowships and Scholarships; Humans; Pathology; Pediatrics. | 0.2315 | 0.3930 | 0.5545 | 0.4444 |
| 24576024 | The pathology milestones and the next accreditation system. | Accreditation; Clinical Competence; Education, Medical, Graduate; Humans; Pathology; United States. | 0.43813 | 0.4935 | 0.5489 | 0.7500 |
BOW is ‘bag of words’, DSR is ‘deep semantic representation’ (equivalent to ‘document to vector’ (D2V)) and MH is ‘MeSH main heading’. The last four columns show the similarity scores against PMID:25236620.
Many studies have been carried out to tackle the challenging problem of automatic MeSH indexing based on different principles, such as k-nearest neighbor (KNN) (Trieschnigg et al., 2009), Naive Bayes (Jimeno-Yepes et al., 2012b), support vector machine (SVM) (Jimeno-Yepes et al., 2012a), Learning to Rank (LTR) (Huang et al., 2011a; Liu et al., 2015; Mao and Lu, 2013), deep learning (Jimeno-Yepes et al., 2014; Rios and Kavuluru, 2015) and multi-label learning (Liu et al., 2015; Tsoumakas et al., 2013). MeSHLabeler is a state-of-the-art automatic MeSH indexing algorithm, which won the first place in the large-scale MeSH indexing task of both BioASQ2 and BioASQ3 competition (http://bioasq.org) (Liu et al., 2015; Tsatsaronis et al., 2015). To address the distribution bias problem on the MH side, MeSHLabeler improves the performance of indexing MeSH by using a large number of different types of evidence regarding MH. These evidences are nicely integrated by using the framework of LTR. However, MeSHLabeler as well as other cutting-edge methods have not considered the problem on the citation (instance) side, and even MeSHLabeler still uses classic BOW representations, such as unigram and bigram. Recently, from the context of machine learning, the concept of dense semantic representation, such as Word2Vec (W2V), Word2Phrase (W2P) and Document2Vec (D2V), has been proposed to capture semantic and context information of text (Bengio et al., 2003; Le and Mikolov, 2014; Mikolov et al., 2013; Mitchell and Lapata, 2010; Socher et al., 2012, 2013). This new concept brings an opportunity to improve the performance of automatic MeSH indexing from the citation side.
Specifically, we have developed DeepMeSH to address the large-scale MeSH indexing problem. Instead of using rather shallow BOW representation, DeepMeSH incorporates deep semantic representation into MeSHLabeler to improve the performance of automatic indexing MeSH over large-scale document data. In particular, DeepMeSH uses a new dense semantic representation, D2V-TFIDF, which has both features of ‘document to vector’ (D2V) and ‘term frequency with inverse document frequency’ (TFIDF), meaning that D2V-TFIDF is more effective than individual D2V and TFIDF to find similar MEDLINE documents. Again Table 1 shows a typical result of using D2V-TFIDF. Regarding the citation in question, PMID:25236620, if we use dense semantic representation (DSR) only, PMID:23597252 can be selected as a highly similar citation, while if we consider both DSR and BOW (which is equivalent to D2V-TFIDF), another citation PMID:24576024 is more similar to PMID:25236620 than the other two citations. Importantly, this result is consistent with the similarity computed by using MHs only, as shown in the last column of Table 1. PMID:24576024 has the largest number of common MHs with PMID:25236620 among three articles in Table 1. Another point is that we use not only simple but rather diverse evidence in terms of dense semantic representation, following the framework of MeSHLabeler. That is, DeepMeSH takes advantage of new dense semantic representation to address the problem of the instance side and the MeSHLabeler framework to address the challenge on the label side. We validated the performance advantage of DeepMeSH by using BioASQ3 benchmark data with 6000 citations. DeepMeSH achieved the Micro F-measure of 0.6323, which is around 12% higher than that of 0.5637 by MTI and 2% higher than that of 0.6218 by MeSHLabeler.
2 Related work
Many studies for the problem of automatic MeSH indexing have used a relatively small- or middle-sized training data, or focus on only a small number of MHs. For example, NLM researchers explored the performance of several different machine learning algorithms, such as SVM, naive Bayes and AdaBoost, over a dataset of only around 300 000 citations (Jimeno-Yepes et al., 2012b, 2013). Rios and Kavuluru (2015) build Convolutional Neural Network (CNN) models for 29 MHs using around a further smaller dataset of 9000 citations. A clear limitation of these studies is that their approaches cannot be generalized to large-scale MeSH indexing in practice.
The BioASQ challenge provides a more realistic and practical benchmark to advance the design of effective algorithms for large-scale MeSH indexing (Tsatsaronis et al., 2015). Many effective algorithms have emerged through the BioASQ challenge, such as MetaLabeler (Tsoumakas et al., 2013), L2R (Huang et al., 2011a; Mao and Lu, 2013) and MeSHLabeler (Liu et al., 2015). However, all of them use the traditional shallow BOW representation. This is inadequate for capturing the semantic and context information of MEDLINE citations precisely, and therefore limits the performance of these models. Recently to address the problem of BOW representation, dense semantic representation for texts has been proposed in the machine learning domain (Bengio et al., 2003; Le and Mikolov, 2014; Mikolov et al., 2013). The performance of dense representation has, however, not yet been examined well in large-scale MeSH indexing, with one exception, in which weighted ‘word to vector’ (W2V) for MeSH indexing was explored (Kosmopoulos et al., 2015). The approach by (Kosmopoulos et al., 2015) is however rather primitive and not thorough enough to build a totally new approach for large-scale MeSH indexing. That is, they first use KNN to find similar citations using a new semantic representation and then the citations with high precision are just added to the results of MTI, meaning a kind of addition to MTI. In fact, the performance of such method was Micro F-measure of 0.575 on BioASQ2 data, which is only around 1% improvement of 0.57 by MTI. However this slight improvement sheds light on the possibility of exploring more effective representation for citations and developing efficient methods for integrating such representation to improve the performance of large-scale MeSH indexing.
3 Methods
3.1 Overview
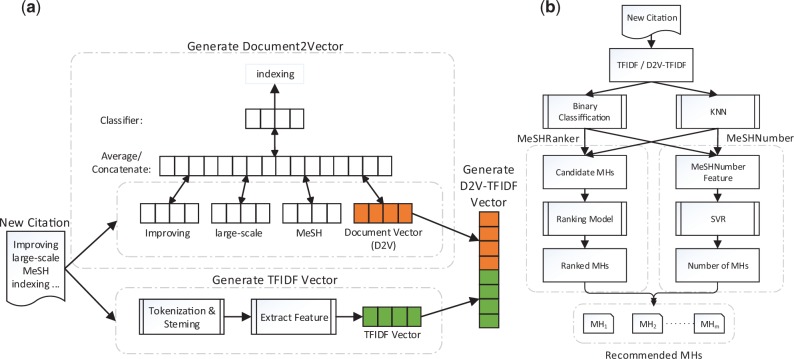
The MeSH indexing problem is to assign a certain number of MHs from the whole MHs list, which contains more than 27 000 terms, to a new MEDLINE citation. MeSHLabeler solves this problem by integrating multiple types of evidence generated from BOW representation in the framework of LTR. In contrast, by keeping the same, efficient framework of LTR, DeepMeSH integrates another type of strong evidence generated from dense semantic representation. Specifically, for each citation, it first generates a dense semantic vector, D2V and the classical TFIDF vector, and then concatenates both to have the final vector representation, D2V-TFIDF. We train binary classifiers of each MHs with D2V-TFIDF and also KNN models. These trained models are finally used to recommend suitable MHs in the framework of LTR. Figure 1 shows the entire framework of DeepMeSH.
Fig. 1.
The work flow of (a) generating D2V-TFIDF and (b) DeepMeSH
3.2 Preliminary background
3.2.1. Citation representation: TFIDF and D2V
In BOW representation, each citation can be represented with a vector consisting of all terms in a controlled vocabulary. Term frequency-inverse document frequency (TFIDF) is the most widely used scheme to weight each term. TF is the term frequency in a document and IDF is the inverse document frequency in the corpus. The idea behind TFIDF is that terms that occur more frequently in a particular document and also occur more in a subset of documents only should be emphasized more. The weight of each term can then be computed by the product of TF and IDF.
Document to vector (D2V) is a recently developed methodology to realize dense semantic representation for documents (Le and Mikolov, 2014). Given a document, both the document and words in the document are represented by a dense continuous vector, which are called ‘document embedding’ (DE) and ‘word embedding’ (WE). They are concatenated together to predict the next word in the given context. In this representation, the ordering of words when appearing in the document is kept, which makes D2V different from TFIDF. In addition, no label information is required in D2V representation learning.
Word embedding (WE) can be further weighted by IDF as follows:
| (1) |
where Wi is the word embedding of the ith word and IDFi is the IDF score of the ith word. We refer to the weight obtained by Eq. (1) ‘weighted word embedding’ (WWE). In addition, phrase embedding (PE) can be obtained by treating phrase as single token in the training. Specifically, Word2Phrase (W2P) method identifies phrases using a mutual information based approach and then learns its vector representation (Mikolov et al., 2013).
Also document embedding can be generated directly from word or phrase embedding results. For example, we can compute the average of the word embedding of each word in a document, as the document embedding. In our experiments, we use two different ways to obtain document embeddings: W2V, which is document embedding obtained in the above manner from word embedding (WE), and weighted word to vector (WW2V), which is also obtained similarly from WWE. Additionally, we use W2P and weighted W2P (WW2P) obtained from phrase embedding (PE) and weighted phrase embedding (WPE) for document embeddings.
3.2.2. Regular methods for large-scale MeSH indexing
Using global evidence: binary relevance
The main idea of binary relevance is to convert the problem of predicting MHs for a given citation to multi-label classification and further to a series of binary classification problems (Zhang and Zhou, 2014), meaning a binary classifier for one label (MH). Given a test instance (citation), we use all binary classifiers to predict labels, i.e. MHs. MetaLabeler adopts the idea of binary relevance, and further train a regression model to predict the number of MHs. Given a test citation, assuming K be the number of predicted MHs, all candidate MHs are ranked with respect to relevance scores predicted by MH classifiers. Finally, top K MHs are recommended.
Originally the relevance scores predicted by different MH classifiers were not comparable theoretically, while this problem was solved by using ‘normalized relevance’ (Liu et al., 2015).
2. Using local evidence: k-nearest neighbor (KNN)
For each test citation, we find k-nearest indexed neighbors based on cosine similarity of their feature vectors. Then, these neighbor citations and their similarities are used to score the candidate MHs. The score of each MH can be calculated as follow:
| (2) |
where is the number of most similar citations, Si is the similarity score of the ith citation and Bi is a binary variable to indicate if the candidate MH is annotated in the ith citation or not.
3.2.3. MeSHLabeler
MeSHLabeler consists of two major components, MeSHRanker and MeSHNumber.
MeSHRanker
Given a test citation, MeSHRanker generates a candidate MH list firstly, and then ranks all candidate MHs by considering multiple types of evidence. The evidences can be mainly classified into five groups: (i) Global evidence comes from all MH binary classifiers and their improved algorithm with normalized relevance score, which we call ‘global evidence’, since the whole MEDLINE citations are used to train a binary classifier for each MH. (ii) Local evidence refers to the scores from the most similar citations obtained by KNN. (iii) MeSH dependency is specific to each candidate MH, by which the MH-MH pair correlation by considering their co-occurrence in MEDLINE is used. (iv) Pattern matching extracts the MHs and their synonyms from the abstract and title of citation directly. (v) Indexing rule comes from the result of MTI.
2. MeSHNumber
For each test citation, MeSHNumber predicts the number of MHs by considering multiple features. These features are as follows: (i) The number of annotated MHs of citations from the same journal recently. (ii) The number of annotated MHs of k-nearest neighbor citations. (iii) The highest scores of the MH binary classifiers. (iv) The highest scores of MeSHRanker. (v) The MH number predicted by MetaLabeler. (vi) The number of MHs recommended by MTI.
Given a new citation, MeSHRanker returns a ranked MH list and MeSHNumber predicts the number of MHs, m. Then the top m MHs in the ranked list is returned as the final recommendation.
3.3 Proposed method: DeepMeSH
3.3.1. Generating TFIDF, D2V and D2V-TFIDF
Figure 1(a) illustrates the manner of generating TFIDF, D2V and D2V-TFIDF, given a citation. TFIDF includes both unigrams and bigrams, which are generated from the title and abstract of a citation. Both W2V and D2V are trained by using neural network with one hidden layer based on stochastic gradient descent and back propagation.
The procedure of generating these representations for a given citation is: (i) we first generate vectors of D2V and TFIDF, (ii) normalize the two feature vectors independently by using the unit length and (iii) finally concatenate the two normalized vectors into a longer vector, which we call D2V-TFIDF.
3.3.2. Using regular MeSH indexing methods with D2V and D2V-TFIDF
D2V is a dense semantic representation generated by considering semantic and context information in text, meaning that D2V can find semantic similar citations even without shared words. This would be helpful for identifying general MHs that are semantically related to many citations. On the other hand, TFIDF is a sparse representation that is useful for mapping some very specific MHs strictly. These two representations are complement to each other. D2V-TFIDF concatenates the sparse and dense features together, and thus includes both raw text information and semantic information, providing diverse evidence for MeSH indexing.
BCD2V-TFIDF: Binary Classification using D2V-TFIDF
Using the binary relevance approach, we can train binary classifiers for any MH using D2V-TFIDF features. In fact in our experiment, we used the latest one million MEDLINE citations to train linear kernel SVM for each MH.
2. KNND2V-TFIDF: KNN using D2V-TFIDF
We can build KNN for any MH using D2V-TFIDF. In our experiments, we first used the D2V-TFIDF vector for a given citation to compute the cosine similarity score against the latest one million MEDLINE citations. We then selected the k nearest citation for MHs. Score can be computed using Eq. (2).
3. KNND2V: KNN using D2V
Similar to KNND2V-TFIDF, D2V can be used to retrieve k-nearest citations and again Score can be computed using Eq. (2).
3.3.3. DeepMeSH: incorporate deep semantic information into MeSHLabeler
Step 1: generate candidate MeSHs with different representations
The number of all MHs reaches 27 000, and most of them are irrelevant MHs to a particular citation. So we generate candidate MHs only, which are considered to be more suitable to a given citation, by using the following three sources:
D2V-TFIDF: The top in the ranked list by BCD2V-TFIDF or top in the ranked list by .
TFIDF: The top in the ranked list by .
PRA: PRA is a modified KNN method by NLM (Lin and Wilbur, 2007), and the top MHs by PRA.
We should note that this process of focusing on a small number of MHs reduces both computation time and false positives, resulting in performance improvement. These three sources contain individually unique information and complement to each other, so that merging them would be reasonable to have a set of candidates covering diverse evidence.
Step 2: use MeSHRanker to rank MeSHs with D2V and D2V-TFIDF related features
The candidate MHs are ranked by the LTR framework of MeSHRanker. The difference from MeSHRanker is using not only the raw text information (considered in MeSHRanker) but also the semantic information, by using D2V. The input features used here are classified into four types:
BCD2V-TFIDF score
KNND2V-TFIDF and KNND2V scores
MeSH frequency rankWe count the times of MH appearing in the MEDLINE, and then rank MHs in the descending order. We add this information as a feature into MeSHRanker that learns better to each candidate MH.
Similarities of nearest neighbor by KNND2V-TFIDF and KNND2V
These two features indicate if the neighbor citations found by KNND2V-TFIDF and KNND2V are credible.
Step 3: select the top ranked MHs to recommend as output
MeSHNumber provides the number to be recommended, m, and the top m MHs in the ranked list by MeSHRanker are returned as the final MHs to be recommended.
4 Experiments
4.1 Data
We downloaded 23 343 329 citations of MEDLINE/PubMed from NLM in Nov 2014, before the BioASQ3 challenge. 13 156 128 indexed citations with both abstracts and titles were locally stored as training data. For generating classical TFIDF features, we used BioTokenizer to preprocess MEDLINE raw text (Jiang and Zhai, 2007). Both unigram and bigram features were used to represent each citation. Similar to other work (Liu et al., 2015; Tsoumakas et al., 2013), the features that appear less than 6 times were removed. Finally, we obtained 112 674 unigram and 1 873 030 bigram features. Each citation was then represented by a very sparse vector of 1 985 704 dimension with the TFIDF weighting scheme.
We sorted all downloaded citations by time. The latest 10 000 citations were used as validation set for tuning parameters of binary classifiers. In addition, next latest 1 000 000 citations were used for learning D2V and D2V-TFIDF representation and their corresponding classifiers, since generating deep semantic representation is time consuming, particularly if we use the whole MEDLINE. Also in preliminary experiments, we found that the performance gain by using the whole MEDLINE is very marginal. On the other hand, TFIDF based binary classifiers, BCTFIDF, were trained with the whole MEDLINE, because improved classification performance on the low frequency MHs can be achieved with more training instances.
From BioASQ3, we obtained another dataset of 49 774 distinct citations, which were randomly divided into three parts: MeSHRanker training set (with 23 774 citations), MeSHNumber training set (with 20 000 citations) and test set (with 6000 distinct citations). The same test set was used for all methods, including binary relevance approaches, KNN, MeSHLabeler and DeepMeSH.
4.2 Implementation and parameter setting
Several open source tools were used in the implementation of DeepMeSH: RankLib (http://sourceforge.net/p/lemur/wiki/RankLib/) to implement our LTR model, LambdaMart (Burges, 2010) and LibSVM to implement support vector regression (SVR) (Chang and Lin, 2011). Logistic regression and Linear SVM were implemented by using LIBLINEAR (Fan et al., 2008). We used gensim (http://radimrehurek.com/gensim) for the implementation of W2V, W2P and D2V. We first transformed all text into lowercase. The continuous bag of Words (CBOW) mode was then used to generate dense semantic representation. The dimensions of all dense vectors were set to 200.
For all KNN, MH scores were computed from top 20 nearest neighbors. and were set to 45 and 20, respectively. and were set to 50 and 40, respectively, following (Liu et al., 2015).
A server with four Intel XEON E5-4650 2.7 GHz CPU and 128 GB RAM was used. Learning all binary classifiers (including D2V-TFIDF and TFIDF) for MHs and DeepMeSH required around one week and two hours, respectively, and annotating new citations needed around two or three hours over ten thousands citations.
4.3 Performance evaluation measure
Let K denote the size of all labels (MeSH headings), and N be the number of instances (citations). Let yi and be the true and predicted label for instance i, respectively. We mainly use three different metrics based on F-measure to evaluate the performance of different models.
F-measure: EBF
EBF is the standard F-measure which can be computed as the harmonic mean of standard precision (EBP) and recall (EBR), as follows:
| (3) |
where
where
We note that we can compute EBP and EBR by summing EBPi and EBRi, respectively, over all instances.
Macro F-measure: MaF
MaF is the harmonic mean of macro-average precision (MaP) and macro-average recall (MaR) as follows:
| (4) |
The macro-average precision and recall are obtained by first computing the precision for each label (MH) separately and then averaging over all labels, as follows:
where
Micro F-measure: MiF
MiF is the harmonic mean of micro-average precision (MiP) and micro-average recall (MiR), as follows:
| (5) |
where
In addition, average label similarity is defined to measure the semantic similarity between one citation and a set of citations. Given a citation x, let be a set of citations (for example, by KNN), the average label similarity between X and x can be computed as follows:
| (6) |
where , i.e. the number of nearest neighbors, Y is the set of true MHs of citation x and Yi is the set of true MHs of ith citation xi of X.
4.4 Experimental results
To compare the performance of different methods reliably, we generated 50 test datasets from all 6000 original test citations using bootstrap with replacement. For examining the model performance over large-scale data, all these 50 datasets consisted of 6000 citations. Paired t-test was then used to evaluate the statistical significance of performance improvement between the best performed method and all other methods. P-values of smaller than 0.05 are considered as statistically significant. We conducted four experiments: (i) the performance of different representations, TFIDF, W2V, WW2V, W2P, WW2P, D2V, W2V-TFIDF, WW2V-TFIDF, W2P-TFIDF, WW2P-TFIDF, D2V-TFIDF were compared using KNN. W2V is obtained by the average of ‘word embedding (WE)’ (described in Section 3.2.1) of each word over all words in the document, and WW2V is computed similarly by using ‘weighted word embedding’ (WWE), shown in Eq. (1). Similarly, we obtain W2P and WW2P form phrase embedding and weighted phrase embedding. To make a fair comparison, the best deep semantic representation was selected to perform the following experiments. (ii) The performance of binary relevance with selected representations was compared. (iii) The performance of DeepMeSH by incorporating the deep semantic information into the LTR framework of MeSHRanker. For a fair comparison, the number of MHs for all representations was predicted by MetaLabeler based on TFIDF representation. (iv) The performance of DeepMeSH with the number of MHs predicted by MeSHNumber was examined. Please note that in BioASQ challenge, the systems are evaluated by micro F-measure, MiF, which is also the focus of our system.
4.4.1. Performance comparison between different representations by KNN
The average performance of each representation over 50 test datasets by KNN is presented in Table 2. For each performance metric, the best representation that statistically significantly outperformed all other representations is highlighted in boldface (see the detailed P-values in the supplementary materials). In the top part, the performance of TFIDF and five dense semantic representations, W2V, WW2V, W2P, WW2P and D2V were directly examined, while in the bottom part, the performance of five joint representations W2V-TFIDF, WW2V-TFIDF, W2P-TFIDF, WW2P-TFIDF and D2V-TFIDF were compared. First, D2V-TFIDF achieved the best performance out of all 11 representations in all 9 measures, particularly for MiF, being 0.4831, followed by WW2P-TFIDF of 0.4814, W2P-TFIDF of 0.4797, WW2V-TFIDF of 0.4647 and W2V-TFIDF of 0.4570. Second, WW2V achieved the best MiF in six individual representations, being 0.4521, followed by WW2P with 0.4435 and TFIDF with 0.4412. Interestingly, although WW2V (WW2P) is just a weighted W2V (W2P) by using IDF, the performance difference between W2V (W2P) and WW2V (WW2P) was quite large. In fact, MiF of W2P was 0.4066, while MiF of WW2P was 0.4435. Third, based on the macro F-measure for individual representations, TFIDF achieved the best MaF of 0.3109, meaning that traditional BOW representation is suitable for finding infrequent MHs (since macro F-measure weights the performance for infrequent MHs more). Finally, the performance of all joint representations was significantly better than their component representation under all measures. For example, D2V-TFIDF achieved the highest MaF of 0.3313, which is higher than 0.3109 by TFIDF and 0.1576 by D2V. This indicates that the traditional BOW representation and dense semantic representation have a good complementary relationship, and their combination improves the performance greatly.
Table 2.
Performance comparison of KNNs with different feature representation
| Method | MiP | MiR | MiF | EBP | EBR | EBF | MaP | MaR | MaF |
|---|---|---|---|---|---|---|---|---|---|
| KNNTFIDF | 0.4369 | 0.4455 | 0.4412 | 0.4362 | 0.4547 | 0.4317 | 0.3274 | 0.2960 | 0.3109 |
| KNNW2V | 0.4133 | 0.4215 | 0.4174 | 0.4083 | 0.4216 | 0.4027 | 0.1438 | 0.1230 | 0.1326 |
| KNNWW2V | 0.4477 | 0.4565 | 0.4521 | 0.4444 | 0.4616 | 0.4394 | 0.2332 | 0.2126 | 0.2225 |
| KNNW2P | 0.4027 | 0.4106 | 0.4066 | 0.3968 | 0.4098 | 0.3914 | 0.1201 | 0.1018 | 0.1102 |
| KNNWW2P | 0.4392 | 0.4478 | 0.4435 | 0.4351 | 0.4515 | 0.4300 | 0.1970 | 0.1786 | 0.1873 |
| KNND2V | 0.4271 | 0.4355 | 0.4313 | 0.4207 | 0.4361 | 0.4156 | 0.1726 | 0.1450 | 0.1576 |
| KNNW2V-TFIDF | 0.4526 | 0.4615 | 0.4570 | 0.4516 | 0.4710 | 0.4472 | 0.3359 | 0.3027 | 0.3185 |
| KNNWW2V-TFIDF | 0.4602 | 0.4693 | 0.4647 | 0.4593 | 0.4793 | 0.4549 | 0.3412 | 0.3091 | 0.3244 |
| KNNW2P-TFIDF | 0.4750 | 0.4844 | 0.4797 | 0.4752 | 0.4951 | 0.4702 | 0.3371 | 0.3054 | 0.3205 |
| KNNWW2P-TFIDF | 0.4768 | 0.4862 | 0.4814 | 0.4764 | 0.4963 | 0.4714 | 0.3398 | 0.3095 | 0.3239 |
| KNND2V-TFIDF | 0.4784 | 0.4878 | 0.4831 | 0.4783 | 0.4983 | 0.4733 | 0.3468 | 0.3171 | 0.3313 |
Although D2V performed worse than WW2V, D2V-TFIDF outperformed WW2V-TFIDF as well as all the other representations significantly in all 9 measures. This result demonstrates high complementary advantages between D2V and TFIDF. It also indicates that, by incorporating important context information in biomedical text, documents embedding can capture semantic information most out of all these embeddings. Moreover, the better performance of WW2P-TFIDF over WW2V-TFIDF suggests that phrase embedding is more effective than word embedding in capturing semantic information. Considering the performance advantage of D2V-TFIDF and the complementarity between D2V and TFIDF, we use D2V-TFIDF, D2V and TFIDF only in the following experiments.
4.4.2. Performance comparison between different representations by binary relevance
Table 3 shows the comparison results on the average performance of different representations on 50 test datasets using binary classification (see the detailed P-values in the supplementary materials). We used linear SVM for BCD2V and BCD2V-TFIDF, because of the performance advantage over logistic regression (logistic regression was used for BCTFIDF only). BCnormTFIDF is an alternative baseline, which normalizes the prediction score of BCTFIDFbefore ranking the candidate MHs. Experimental results show that BCD2V-TFIDF achieved the best MiF of 0.6033, followed by BCnormTFIDFwith 0.5772 and BCTFIDFwith 0.5638. The large performance difference between BCnormTFIDFand BCD2V-TFIDF highlights the advantage of D2V-TFIDF over D2V and TFIDF.
Table 3.
Comparison of binary relevance approaches with different features
| Method | MiP | MiR | MiF | EBP | EBR | EBF | MaP | MaR | MaF |
|---|---|---|---|---|---|---|---|---|---|
| BC D2V | 0.4395 | 0.4482 | 0.4438 | 0.4339 | 0.4519 | 0.4294 | 0.1627 | 0.1706 | 0.1666 |
| BC | 0.5584 | 0.5694 | 0.5638 | 0.5575 | 0.5892 | 0.5556 | 0.4662 | 0.4970 | 0.4811 |
| BCnorm | 0.5716 | 0.5829 | 0.5772 | 0.5704 | 0.5991 | 0.5667 | 0.4463 | 0.4402 | 0.4432 |
| BCD2V-TFIDF | 0.5974 | 0.6092 | 0.6033 | 0.5983 | 0.6280 | 0.5943 | 0.4741 | 0.4633 | 0.4686 |
4.4.3. Performance improvement of MeSHRanker by incorporating deep semantic representation
Table 4 shows the average performance of MeSHRanker on the 50 test datasets by incorporating evidence generated by deep semantic representation (see the detailed P-values in the supplementary materials). The results of MTIDEF (the default version of MTI) and BCD2V-TFIDF are presented as baselines. We first added the D2V related features into the input features of LTR in MeSHRanker, which corresponds to incorporate Step 2 of DeepMeSH into MeSHRanker (See Section 3.3.3). This feature infusion improved MiF of MeSHRanker from 0.6126 to 0.6216. We then focused on selecting D2V related candidates using Step 1 of DeepMeSH, by which the performance was further improved to 0.6224. Note that then DeepMeSH can be generated by adding Step 3 (MeSHNumber, See Section 3.3.3) to MeSHRanker further.
Table 4.
Performance improvement of MeSHRanker by incorporating deep semantic representation
| Method | MiP | MiR | MiF | EBP | EBR | EBF | MaP | MaR | MaF |
|---|---|---|---|---|---|---|---|---|---|
| MTIDEF | 0.5753 | 0.5526 | 0.5637 | 0.5838 | 0.5737 | 0.5566 | 0.4939 | 0.5140 | 0.5037 |
| BCD2V-TFIDF | 0.5974 | 0.6092 | 0.6033 | 0.5983 | 0.6280 | 0.5943 | 0.4741 | 0.4633 | 0.4686 |
| MeSHRanker | 0.6067 | 0.6187 | 0.6126 | 0.6091 | 0.6400 | 0.6053 | 0.5249 | 0.5400 | 0.5323 |
| + Step 2 of DeepMeSH | 0.6156 | 0.6278 | 0.6216 | 0.6180 | 0.6492 | 0.6141 | 0.5361 | 0.5476 | 0.5418 |
| + Steps 1 and 2 of DeepMeSH | 0.6164 | 0.6286 | 0.6224 | 0.6188 | 0.6501 | 0.6149 | 0.5380 | 0.5505 | 0.5442 |
4.4.4. Performance comparison of DeepMeSH with MeSHLabeler
Table 5 shows the average performance of DeepMeSH on the 50 test datasets, comparing with MeSHLabeler and MTIDEF. DeepMeSH achieved better performances than the two competing methods in all nine measures. For example, DeepMeSH achieved the best MiF of 0.6323, around 2% improvement over that of 0.6218 by MeSHLabeler and around 12% higher than that of 0.5637 by MTIDEF. Note that all these performance improvements are statistically significant. These results demonstrate the advantage of deep semantic representation in large scale MeSH indexing, which is the crucial difference between DeepMeSH and MeSHLabeler.
Table 5.
Performance comparison of DeepMeSH with MTIDEF and MeSHLabeler (P-values are shown in the parentheses)
| Method | MiP | MiR | MiF | EBP | EBR | EBF | MaP | MaR | MaF |
|---|---|---|---|---|---|---|---|---|---|
| MTIDEF | 0.5753 | 0.5526 | 0.5637 | 0.5838 | 0.5737 | 0.5566 | 0.4939 | 0.5140 | 0.5037 |
| (2.67E-85) | (1.28E-75) | (1.12E-86) | (1.16E-81) | (1.76E-73) | (8.17E-85) | (3.89E-69) | (2.19E-41) | (3.39E-63) | |
| MeSHLabeler | 0.6457 | 0.5995 | 0.6218 | 0.6480 | 0.6200 | 0.6145 | 0.5304 | 0.5216 | 0.5259 |
| (9.87E-54) | (1.43E-52) | (4.67E-60) | (3.24E-54) | (1.97E-50) | (1.49E-59) | (3.92E-48) | (5.25E-31) | (4.18E-45) | |
| DeepMeSH | 0.6589 | 0.6077 | 0.6323 | 0.6623 | 0.6280 | 0.6251 | 0.5432 | 0.5262 | 0.5346 |
4.5 Result analysis
We explore the situation and the reason why deep semantic representation, D2V-TFIDF, works well, from diverse viewpoints.
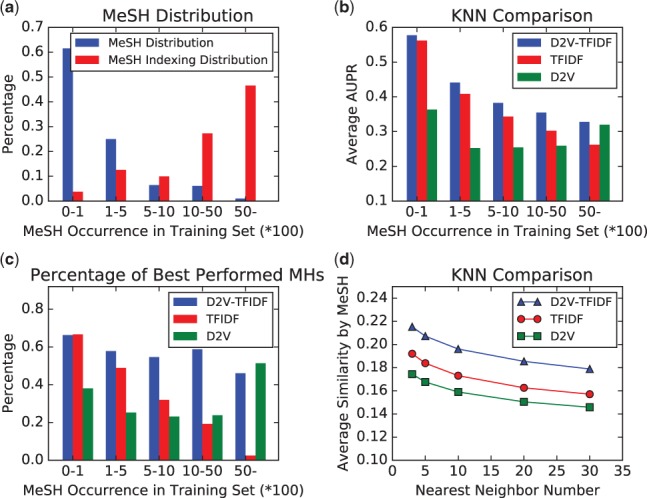
4.5.1. Distribution of MHs
We counted the number of occurrence of MHs in the training set (1 000 000 citations) of D2V. By using the number of occurrences, we split the MHs into five groups: [0, 100), [100, 500), [500, 1000), [1000, 5000) and [5000, 1 000 000), where [0, 100) means the group of MHs, each having the number of occurrences between more than zero to 100. Figure 2(a) shows the MeSH distribution and MeSH indexing distribution of the five groups. MeSH distribution is the ratio of MHs that appear in each group to all MHs, and the MeSH indexing distribution is the ratio of occurrence of MH in each group to all occurrence of all MHs in the whole training set. We can easily see that most of MHs occur very rarely: more than 60% MHs occur less than 100 times, and the sum of annotation with these MHs is just 4% of all annotation. On the other hand, only 1% MHs occur more than 5000 times, and their total number of occurrence is 47%.
Fig. 2.
Comparison with different representation
4.5.2. Performance comparison among different MH groups
We compared the performance (by Average AUPR:Area under the Precision-Recall curve, averaged over MHs in each group) of the three representations: D2V, TFIDF and D2V-TFIDF, using KNN on the five MH groups, generated in the last section. Figure 2(b) shows that KNND2V-TFIDF achieved the best on all groups. KNND2V outperformed KNNTFIDFin the most frequent MH group, and KNNTFIDFoutperformed KNND2V in other groups. The difference between KNND2V and KNNTFIDFwas the largest in the rare MH group. In summary, D2V performed better than TFIDF for frequent MHs, while TFIDF performed better than D2V for rare MHs, implying the complementary advantages between D2V and TFIDF, which made D2V-TFIDF perform the best in all MH groups.
4.5.3. Ratio of best performed MHs
We further compared the three representations, by the number (ratio) of MHs which achieved the best performance by some representation (with KNN) to all MHs in each group. Figure 2(c) shows that between KNND2V and KNNTFIDF, KNND2V outperformed KNNTFIDFin the most frequent group of MHs, while KNNTFIDF outperformed KNND2V in infrequent groups. This result further emphasizes that would be the most balanced representation among the three.
4.5.4. Finding similar citations
Figure 2(d) shows the performance comparison among the three representations for finding similar citations, where AvgSimilarity of the corresponding MH is plotted against the number of nearest neighbors. The curve of KNND2V-TFIDF was always above the other two representations clearly, indicating that similar citations found by KNND2V-TFIDF have more common MHs and are more trustable than other two representations.
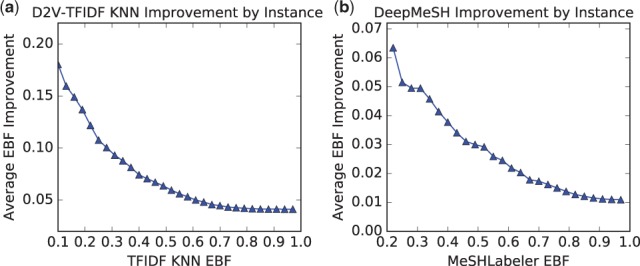
4.5.5. Performance improvement examples
Figure 3(a) shows the improved values of average EBF by KNND2V-TFIDF over KNNTFIDFfor citations with EBF of KNNTFIDFlower than a certain cut-off (which is shown in x-axis). This figure clearly shows the performance improvement is larger for citations with lower EBF. Also Figure 3(b) shows the improvement of average EBF by DeepMeSH over MeSHLabeler for again citations with lower MeSHLabeler scores than a certain cut-off (corresponding to the x-axis value). This also shows the improvement is larger for citations with smaller EBF values.
Fig. 3.
Performance improvement examples
4.5.6. Performance improvement in different languages
We further analyzed the performance improvement by DeepMeSH from MeSHLabeler for each language. Since the datasets on non-English language are too small to draw any conclusion, we only show the result on English in Table 6. We can see that the performance improvement in English looks marginal but was clearly significant, and also overall performance improvement was significant.
Table 6.
Improvement of DeepMeSH on EBF over MeSHLabeler for languages
| Language | Occurrences | MeSHLabeler | DeepMeSH | p-value |
|---|---|---|---|---|
| English | 5021 | 0.6143 | 0.6251 | 2.58E–31 |
| All | 6000 | 0.6139 | 0.6248 | 2.35E–37 |
The P-values in the last column are by paired t-test with Bonferroni multiple test correction.
5 Conclusion and discussion
We have proposed an effective solution for large-scale MeSH indexing, for which the official solution, MTI, as well as all state-of-the-art methods, use BOW representation, while BOW cannot capture rich semantic information in large-scale biomedical documents. We developed DeepMeSH, which effectively utilizes dense semantic representation, and achieves around 12 and 2% improvement over MTI and MeSHLabeler in both MiF and EBF. This improvement is especially valuable, because MeSHLabeler already integrates a variety of diverse evidence. The high performance of DeepMeSH can be attributed to two factors: (i) the deep semantic representation, D2V-TFIDF, that integrates the power of both dense representation, D2V, and sparse representation, TFIDF. (ii) ‘learning to rank’ which integrates diverse evidence smoothly and effectively.
An interesting discovery from our experiments on exploring the reason of achieving the improved performance of D2V-TFIDF is the complementarity between sparse and dense semantic representation. This is especially true of TFIDF and D2V, where TFIDF performed well on rare MHs, and D2V achieved high performance on frequent MHs. It is not surprising that D2V-TFIDF, which can enjoy the complementary advantage between TFIDF and D2V, outperformed TFIDF and D2V under all performance measures. On the other hand, as also shown by the result of (Kosmopoulos et al., 2015), which improved the performance of MTI only slightly, applying dense semantic representation only is not necessarily a good strategy. Our experiments further clarified what kind of situations can have the most benefit from that new representation, and citations annotated worse by sparse representation gained the performance improvement more. This result is reasonable and consistent with other experimental results we obtained.
Also our experiments indicate that D2V-TFIDF is a very useful representation for finding semantically similar citations. Finding similar citations is a core task in biomedical text mining for knowledge discovery, such as document searching, document clustering and query expansion. Currently, the most widely used method for finding similar citations practically in life science is PRA by NLM, which is based on sparse representation. Thus our new representation will be useful for many applications including searching similar citations and may find more promising applications as well.
Supplementary Material
Funding
This work was supported in part by the National Natural Science Foundation of China (61572139), the National Institutes of Health Big Data to Knowledge (BD2K) initiative (1U54GM114838) and National Science Foundation under grants IIS-1553568. H.M. would like to thank MEXT for KAKENHI #16H02868 and Tekes for FiDiPro.
Conflict of Interest: none declared.
References
- Aronson A. et al. (2004) The NLM indexing initiative’s medical text indexer. Stud Health Technol. Inf., 107, 268–272. [PubMed] [Google Scholar]
- Bengio Y. et al. (2003) A neural probabilistic language models. J. Mach. Learn. Res., 3, 1137–1155. [Google Scholar]
- Burges C.J.C. (2010) From RankNet to LambdaRank to LambdaMART: An overview. Technical report, Microsoft Research.
- Chang C.C., Lin C.J. (2011) LIBSVM: a library for support vector machines. ACM Trans. Intell. Syst. Technol., 2, 1–27. 27:27. [Google Scholar]
- Chen L. et al. (2004) Gene name ambiguity of eukaryotic nomenclatures. Bioinformatics, 21, 248–256. [DOI] [PubMed] [Google Scholar]
- Fan R.E. et al. (2008) Liblinear: A library for large linear classification. J. Mach. Learn. Res., 9, 1871–1874. [Google Scholar]
- Gu J. et al. (2013) Efficient semi-supervised MEDLINE document clustering with MeSH semantic and global content constraints. IEEE Trans. Cybern., 43, 1265–1276. [DOI] [PubMed] [Google Scholar]
- Huang M. et al. (2011a) Recommending mesh terms for annotating biomedical articles. J. Am. Med. Inf. Assoc., 18, 660–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X. et al. (2011b) Enhanced clustering of biomedical documents using ensemble non-negative matrix factorization. Inf. Sci., 181, 2293–2302. [Google Scholar]
- Jiang J., Zhai C. (2007) An empirical study of tokenization strategies for biomedical information retrieval. Inf. Retrieval, 10, 341–363. [Google Scholar]
- Jimeno-Yepes A. et al. (2012a). MEDLINE MeSH indexing: lessons learned from machine learning and future directions. In: Proceedings of the 2nd ACM SIGHIT International Health Informatics Symposium ACM, pp. 737–742.
- Jimeno-Yepes A. et al. (2012b) A one-size-fits-all indexing method does not exist: Automatic selection based on meta-learning. JCSE, 6, 151–160. [Google Scholar]
- Jimeno-Yepes A. et al. (2013). Comparison and combination of several mesh indexing approaches In: AMIA Annual Symposium Proceedings, vol. 2013 American Medical Informatics Association, p. 709. [PMC free article] [PubMed] [Google Scholar]
- Jimeno-Yepes A. et al. (2014). Deep belief networks and biomedical text categorisation In: Australasian Language Technology Association Workshop, p. 123. [Google Scholar]
- Kosmopoulos A. et al. (2015) Biomedical semantic indexing using dense word vectors in bioasq. J. Biomed. Seman., http://www.aueb.gr/users/ion/docs/jbms_dense_vectors.pdf [Google Scholar]
- Le Q., Mikolov T. (2014) Distributed representations of sentences and documents In: ICML, pp. 1188–1196. [Google Scholar]
- Lin J., Wilbur W. (2007) Pubmed related articles: a probabilistic topic-based model for content similarity. BMC Bioinformatics, 8, 423.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K. et al. (2015) Meshlabeler: improving the accuracy of large-scale mesh indexing by integrating diverse evidence. Bioinformatics, 12, i339–i347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z. et al. (2010) Evaluation of query expansion using MeSH in PubMed. Inf. Retrieval, 12, 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Lu Z. (2013). NCBI at the 2013 BioASQ challenge task: Learning to rank for automatic MeSH indexing. Technical report.
- Mikolov T. et al. (2013) Distributed representations of words and phrases and their compositionality In: NIPS, pp. 3111–3119. [Google Scholar]
- Mitchell J., Lapata M. (2010) Composition in distributional models of semantics. Cognit. Sci., 34, 1388–1439. [DOI] [PubMed] [Google Scholar]
- Mork J. et al. (2013) The NLM medical text indexer system for indexing biomedical literature. In: BioASQ@ CLEF. [Google Scholar]
- Mork J. et al. (2014) Recent enhancements to the NLM medical text indexer. In: CLEF (Working Notes), pp. 1328–1336.
- NCBI Resource Coordinators (2015) Database resources of the national center for biotechnology information. Nucleic Acids Res., 43, D6–D17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson S.J. et al. (2004) The MeSH translation maintenance system: structure, interface design, and implementation. Medinfo, 11, 67–69. [PubMed] [Google Scholar]
- Peng S. et al. (2015) The fudan participation in the 2015 bioasq challenge: Large-scale biomedical semantic indexing and question answering. In: CLEF (Working Notes).
- Rios A., Kavuluru R. (2015) Convolutional neural networks for biomedical text classification: application in indexing biomedical articles. In: BCB, pp. 258–267. [DOI] [PMC free article] [PubMed]
- Socher R. et al. (2012) Semantic compositionality through recursive matrix-vector spaces. In: EMNLP-CoNLL, pp. 1021–1211.
- Socher R. et al. (2013) Recursive deep models for semantic compositionality over a sentiment treebank. In: EMNLP.
- Stokes N. et al. (2010) Exploring criteria for successful query expansion in the genomic domain. Inf. Retrieval, 12, 17–50. [Google Scholar]
- Trieschnigg D. et al. (2009) MeSH Up: effective MeSH text classification for improved document retrieval. Bioinformatics, 25, 1412–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsatsaronis G. et al. (2015) An overview of the bioasq large-scale biomedical semantic indexing and question answering competition. BMC Bioinformatics, 16, 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsoumakas G. et al. (2013). Large-scale semantic indexing of biomedical publications. In: BioASQ@ CLEF.
- Zhang M., Zhou Z. (2014) A review on multi-label learning algorithms. IEEE Trans. Knowl. Data Eng., 26, 1819–1837. [Google Scholar]
- Zhu S. et al. (2009a) Enhancing MEDLINE document clustering by incorporating mesh semantic similarity. Bioinformatics, 25, 1944–1951. [DOI] [PubMed] [Google Scholar]
- Zhu S. et al. (2009b) Field independent probabilistic model for clustering multi-field documents. Inf. Process. Manag., 45, 555–570. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.