Abstract
Because microbial communities play a key role in carbon (C) and nitrogen (N) cycling, changes in the soil microbial community may directly affect ecosystem functioning. However, the effects of N deposition and management practices on soil microbes are still poorly understood. We studied the effects of these two factors on soil microbial biomass carbon (MBC) and community composition in Moso bamboo plantations using high-throughput sequencing of the 16S rRNA gene. Plantations under conventional (CM) or intensive management (IM) were subjected to one of four N treatments for 30 months. IM and N addition, both separately and in combination, significantly increased soil MBC while decreasing bacterial diversity. However, increases in soil MBC were inhibited when N addition exceeded 60 kg N∙ha−1∙yr−1. IM increased the relative abundances of Actinobacteria and Crenarchaeota but decreased that of Acidobacteria. N addition increased the relative abundances of Acidobacteria, Crenarchaeota, and Actinobacteria but decreased that of Proteobacteria. Soil bacterial diversity was significantly related to soil pH, C/N ratio, and nitrogen and available phosphorus content. Management practices exerted a greater influence over regulation of the soil MBC and microbial diversity compared to that of N deposition in Moso bamboo plantations.
As the most abundant organisms on earth, microbes play a key role in natural ecosystems, including the biogeochemical cycles of carbon (C) and nitrogen (N) and the biodegradation or stabilization of environmental contaminants1,2,3. The soil microbial biomass has an important role in nutrient cycling and is therefore essential for plant growth4. Changes in the soil microbial community may directly affect soil ecosystem functioning, particularly C and N cycling5. In recent years, high-throughput sequencing technologies have been widely used to analyze the species composition and functional diversity of microbial populations under different fertilization regimes6,7,8. These technologies provide a more detailed description of microbial communities than traditional methods, such as cloning libraries, denaturing gradient gel electrophoresis (DGGE), and phospholipid fatty acid (PLFA) analysis and has proven to be a very powerful technique in microbial ecology research9. Because soil microbes are sensitive to environmental change10,11, a comprehensive analysis of the microbial community structure and microbial responses to environmental change using high-throughput sequencing technologies could improve our understanding of biogeochemical cycles in natural or managed ecosystems12.
Moso bamboo (Phyllostachys pubescens Mazel ex H. de Lehaie), distributed widely in subtropical China13, grows rapidly and is the most important source of non-wood forest products in China14,15. Under typical conventional management (CM), trunks and shoots of Moso bamboo are harvested regularly without any other management interventions15. In recent decades, intensive management (IM) practices, such as removing understory vegetation, plowing, and fertilization, have been widely adopted in order to maximize economic returns13,15. It has been reported that these IM practices alter the soil microbial biomass and community diversity16,17,18. For example, Liu et al.19 determined via polymerase chain reaction-denaturing gradient gel electrophoresis (PCR-DGGE) that fertilizer application significantly increased rhizosphere microbial diversity in a Moso bamboo forest. In contrast, Sun et al.20 observed that long-term conventional management of bamboo plantations did not significantly alter soil microbial biomass and diversity. However, few studies have used high-throughput sequencing technologies to assess the effects of management practices on soil microbial biomass and community diversity in Moso bamboo plantations.
Over the last three decades, the average bulk deposition of N has rapidly increased across China21, particularly in subtropical China22,23. Moso bamboo plantations are primarily distributed across the region with the greatest N deposition in China, according to both current estimates and future predictions24. Research has shown that N addition may reduce soil microbial biomass25,26 and community diversity27. Compton et al.28 and Frey et al.29 found that chronic N supplementation decreased soil microbial biomass at the Harvard Forest. However, Paul et al.30 found that the microbial biomass was greater under high N input (300 kg N∙ha−1∙yr−1) than under low N input (100 kg N∙ha−1∙yr−1). Johnson et al.31 also observed that N addition (80 kg N∙ha−1∙yr−1) increased the soil microbial biomass over seven years in a simulated N deposition study. Boxman et al.32 found that N deposition did not affect soil microbial biomass. The reasons for these differences in results remain unclear and require further study. The lack of understanding of the effects of N deposition on soil microbial quantity, community composition, and microbial diversity in Moso bamboo plantations with different management practices limits our ability to predict the complex response of plantation ecosystems to global environmental changes.
In the present study, we employed high-throughput sequencing to analyze the effects of nitrogen deposition and management practices on soil microbial community and diversity in Moso bamboo plantations. The objectives of this study were to test the following hypotheses: (1) IM practices increase soil microbial biomass carbon (MBC) and microbial diversity; (2) N deposition increases soil MBC and diversity; and (3) the combination of IM and N deposition exerts a greater effect on soil MBC and community composition than each practice independently.
Results
Soil microbial biomass carbon
Through assessment of soil samples from CM and IM plots in the Moso bamboo plantation, we found that soil MBC content was significantly higher under IM than under CM when no N was added (Fig. 1). Soil MBC content increased significantly with the addition of N under both CM and IM, but decreased significantly when the N addition rate exceeded 30 kg N∙ha−1∙yr−1 (N30 treatment) under IM and 60 kg N∙ha−1∙yr−1 (N60 treatment) under CM (Fig. 1).
Figure 1. Soil microbial biomass carbon (MBC) contents under conventional management (CM) or intensive management (IM) and three N addition treatments.N30: 30 kg N∙ha−1∙yr−1; N60: 60 kg N∙ha−1∙yr−1; N90: 90 kg N∙ha−1∙yr−1.
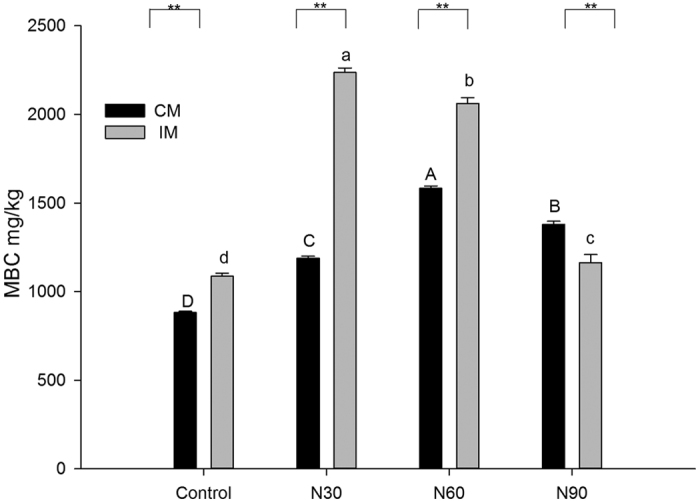
Lowercase letters indicate significant differences between N addition treatments under IM(P < 0.05). Capital letters indicate significant differences between N addition treatments under CM (P < 0.05).Asterisks indicate significant differences between CM and IM at identical N addition rates (*P < 0.05, **P < 0.01). Values represent means of three replicates, and error bars indicate standard errors.
In the IM plots, the maximum soil MBC content (2237.3 mg∙kg−1) was observed in the N30 treatment and was 105% greater than that observed in the control. In the CM plots, the maximum soil MBC content (1583.0 mg∙kg−1) was observed in the N60 treatment and was 79.6% greater than that in the control. While the soil MBC contents of the N addition treatments differed significantly from each other, all were significantly greater than that of the control treatment.
Soil microbial community composition
More than 35,000 valid reads were obtained for each replicate via a sequence optimization process and quality filtering. The median sequence length of each read was 253 bp. A total of 68,531 operational taxonomic units (OTUs) representing 41 phyla and 266 genera were detected using 97% identity as the cutoff. The dominant phyla were Proteobacteria (34.9%), Acidobacteria (27.7%), Verrucomicrobia (8.5%), and Actinobacteria (7%) (Fig. 2). In addition, 22 phyla were detected at relatively low abundances and accounted for less than 1% of the observed phyla. The dominant genera were DA101 (4.3%), Rhodoplanes (1.8%), Candidatus Koribacter (1.7%), Candidatus Solibacter (1.4%), and Rhodanobacter (1.4%).
Figure 2. Taxonomic distribution of soil microbial communities in the Moso bamboo plantation under all treatments.
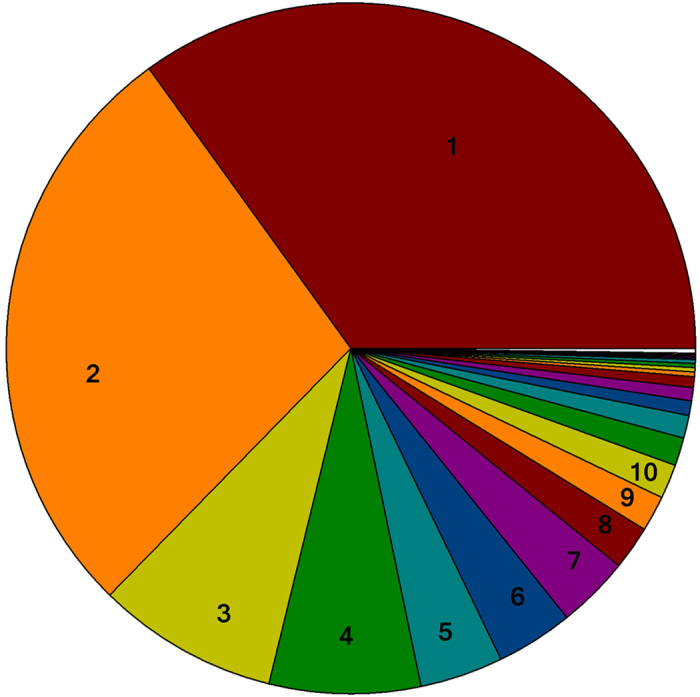
Numbers indicate the ten most prevalent phyla: (1) Proteobacteria, (2) Acidobacteria, (3) Verrucomicrobia, (4) Actinobacteria, (5) Chloroflexi, (6) Planctomycetes, (7) Gemmatimonadetes, (8) AD3, (9) Crenarchaeota, and (10) Others, including sequences that could not be classified into any known group.
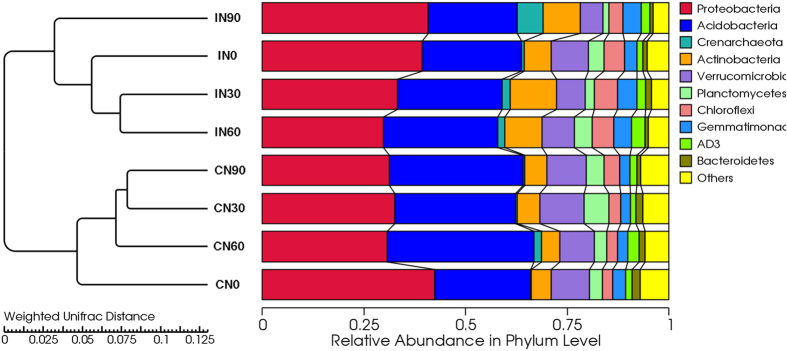
Although fewer OTUs were detected in the CM plots (54,439) than in the IM plots (58,187), a greater number of phyla were observed under the CM treatments (39) than under the IM treatments (36). Under both CM and IM treatments, Proteobacteria, Acidobacteria, Verrucomicrobia, Actinobacteria, Chloroflexi, Planctomycetes, Gemmatimonadetes, AD3, Crenarchaeota, and Bacteroidetes contributed to a large proportion of phyla (Fig. 3). The relative abundances of Actinobacteria and Proteobacteria were higher under IM than under CM, whereas Acidobacteria, Crenarchaeota, and Verrucomicrobia exhibited the opposite trend.
Figure 3. Comparison of the soil bacterial communities at the phylum level in the Moso bamboo plantation under all treatments.
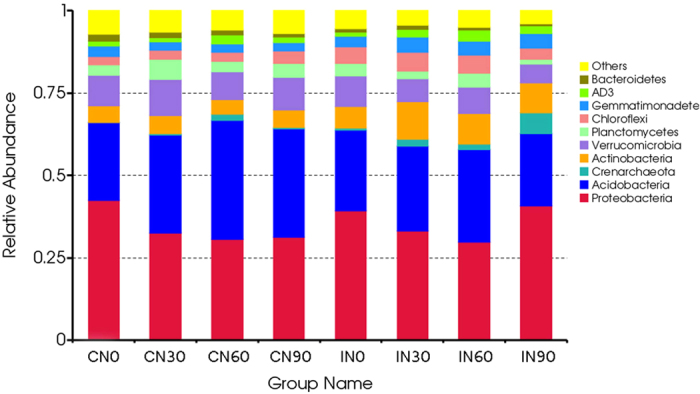
The relative abundances of the dominant bacterial groups in the soil differed depending on the nitrogen deposition and management practices. Relative abundances are based on the proportional frequencies of DNA sequences that could be classified. CN0: conventional management with no added N; CN30: conventional management with 30 kg N∙ha−1∙yr−1; CN60: conventional management with 60 kg N∙ha−1∙yr−1; CN90: conventional management with 90 kg N∙ha−1∙yr−1; IN0: intensive management with no added N; IN30: intensive management with 30 kg N∙ha−1∙yr−1; IN60: intensive management with 60 kg N∙ha−1∙yr−1; IN90: intensive management with 90 kg N∙ha−1∙yr−1.
No significant differences were observed in the number of OTUs among the N addition treatments. Under CM, N addition decreased the relative abundances of Proteobacteria and Bacteroidetes but increased the relative abundance of Acidobacteria. Under IM, N addition increased the relative abundances of Crenarchaeota and Actinobacteria but decreased the relative abundance of Proteobacteria, with the exception of the N90 (90 kg N∙ha−1∙yr−1) treatment.
Soil microbial community and diversity
The Chao1 index, which reflects the species richness of a community, was significantly greater under CM than under IM treatment, regardless of N treatment. Index values were significantly lower in treatments with added N compared to that of the control under both CM and IM (Fig. 4).
Figure 4. Chao 1 index of soil microbial diversity under conventional management (CM) or intensive management (IM) and three N addition treatments.
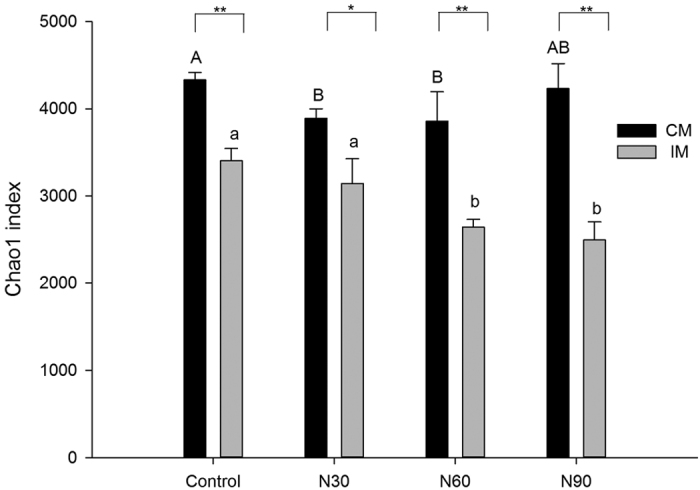
N30: 30 kg N∙ha−1∙yr−1; N60: 60 kg N∙ha−1∙yr−1; N90: 90 kg N∙ha−1∙yr−1. Lowercase letters indicate significant differences between N addition treatments under IM(P < 0.05). Capital letters indicate significant differences between N addition treatments under CM(P < 0.05).Asterisks indicate significant differences between CM and IM at the same N addition rate (*P < 0.05, **P < 0.01). Values represent means of three replicates, and error bars indicate standard errors.
Cluster analysis of the soil microbial communities divided the bacterial communities into two groups (Fig. 5). All samples from the CM treatments clustered in group 1, and all samples from the IM treatments clustered in group II, regardless of N addition. This grouping was further confirmed by principal coordinate analysis (PCoA) (Fig. 6). Differences in microbial community structure were primarily due to a combination of nitrogen deposition and management practices (57.73%), with management practices alone accounting for 36.26% of the variation and nitrogen addition accounting for 21.47%.
Figure 5. UniFrac UPGMA (unweighted pair group method with arithmetic mean) cluster analysis of microbial communities in soil samples from Moso bamboo plantation under different nitrogen deposition and management practice treatments.
Figure was constructed based on Illumina sequencing data. CN0: conventional management with no added N; CN30: conventional management with 30 kg N∙ha−1∙yr−1; CN60: conventional management with 60 kg N∙ha−1∙yr−1; CN90: conventional management with 90 kg N∙ha−1∙yr−1; IN0: intensive management with no added N; IN30: intensive management with 30 kg N∙ha−1∙yr−1; IN60: intensive management with 60 kg N∙ha−1∙yr−1; IN90: intensive management with 90 kg N∙ha−1∙yr−1.
Figure 6. Principal coordinate analysis (PCoA) plot based on 16S rRNA gene sequencing of 24 samples.
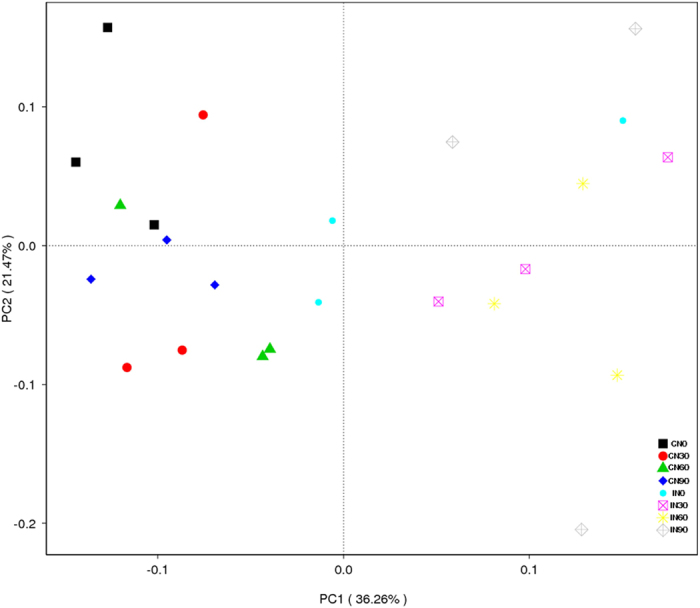
Scatter plot shows principal coordinate 1 (PC1) versus principal coordinate 2 (PC2). Percentages shown are percentages of variation explained by the components. CN0: conventional management with no added N; CN30: conventional management with 30 kg N∙ha−1∙yr−1; CN60: conventional management with 60 kg N∙ha−1∙yr−1; CN90: conventional management with 90 kg N∙ha−1∙yr−1; IN0: intensive management with no added N; IN30: intensive management with 30 kg N∙ha−1∙yr−1; IN60: intensive management with 60 kg N∙ha−1∙yr−1; IN90: intensive management with 90 kg N∙ha−1∙yr−1.
Relationship between microbial diversity and soil properties
The relationships between the α-diversity of soil microbes and soil properties were analyzed (Table 1). Chao1 and Shannon index values were significantly positively correlated with soil pH, and the Chao1 index was significantly negatively correlated with soil NO3− and NH4+ concentrations. OTU number was also significantly positively correlated with pH but was negatively correlated with total N (TN), NO3−, and NH4+ concentrations. MBC was significantly negatively correlated with pH. Canonical correspondence analysis (CCA) demonstrated a correlation between primary soil properties and the microbial community; this showed that soil pH, C/N ratio, and available phosphorus (AP) and TN concentrations had the biggest impacts on the microbial community (Fig. 7).
Table 1. Pearson’s correlation coefficients between soil properties and soil microbial community indicators.
| OTUs | Chao1 | Shannon | MBC | |
|---|---|---|---|---|
| pH | 0.844** | 0.785** | 0.588** | −0.553** |
| SOC | −0.222 | −0.219 | −0.172 | 0.169 |
| TN | −0.475* | −0.403 | −0.299 | −0.079 |
| C/N | 0.348 | 0.262 | 0.14 | 0.095 |
| AP | −0.163 | −0.192 | −0.164 | −0.018 |
| NO3− | −0.644** | −0.564** | −0.404 | −0.018 |
| NH4+ | −0.661** | −0.502* | −0.392 | −0.1 |
OTUs: operational taxonomic units (97% identity); MBC: microbial biomass carbon; SOC: soil organic carbon; TN: total nitrogen; AP: available phosphorus.
*P < 0.05, **P < 0.01.
Figure 7. Canonical correspondence analysis (CCA) of the relative abundances of dominant microbial and soil environmental factors.
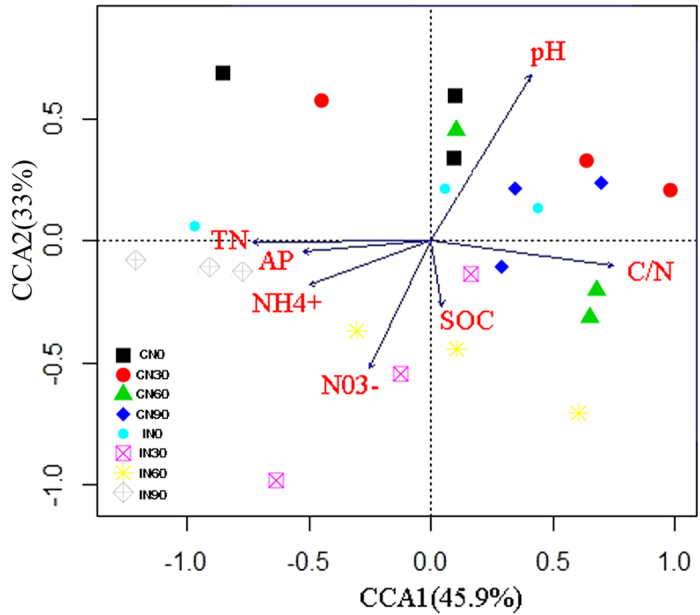
Soil factors indicated in red text include pH, SOC (soil organic carbon), TN (total nitrogen), AP (available phosphorus), NH4 + , NO3−, and C/N ratio. Arrow lengths indicate the strength of the relationship between the soil property and the overall microbial community. CN0: conventional management with no added N; CN30: conventional management with 30 kg N∙ha−1∙yr−1; CN60: conventional management with 60 kg N∙ha−1∙yr−1; CN90: conventional management with 90 kg N∙ha−1∙yr−1; IN0: intensive management with no added N; IN30: intensive management with 30 kg N∙ha−1∙yr−1; IN60: intensive management with 60 kg N∙ha−1∙yr−1; IN90: intensive management with 90 kg N∙ha−1∙yr−1.
Discussion
Effect of management practices on soil microbial biomass and community diversity
In this study, we found significantly higher levels of MBC in the IM treatments compared to the CM treatments without N addition (Figs 1 and 4), indicating that IM practices led to a significant increase in soil microbial biomass. This partially supports our first hypothesis, that IM practices increase soil MBC and diversity. IM practices such as fertilization may provide abundant nutrients for the growth of microorganisms and thus increase soil MBC33,34. Fertilization can also increase the productivity of Moso bamboo35, resulting in increased litter and nutrient return, which may contribute to microbial growth. Similar results were observed by Yu et al.17.
The second part of this hypothesis, that IM increases microbial diversity, was not borne out by the results. In fact, we found that IM significantly reduced soil microbial diversity. Similarly, He et al.18 observed that after 15 years of IM, the abundance of N2-fixing bacteria decreased in Moso bamboo forests. A comparable result was also observed in a Castanea mollissima forest16. Some management practices, such as plowing and weeding, may reduce the diversity of the aboveground plant community32, alter soil conditions, and destroy original habitat, potentially creating an unfavorable environment for some microbes and reducing microbial diversity. In addition, the osmotic potential in the soil solution of the IM plots may have become toxic due to the introduction of additional ions via fertilizer36, further reducing microbial abundance.
Previous studies found that fertilization changed the microbial community’s structure and composition37,38,39. Our results found that Actinobacteria were more prevalent with IM treatment, particularly with N addition. Actinobacteria can depolymerize the polyphenols in litter into small, soluble molecules and thus play an important role in the decomposition of lignin. The abundance of Actinobacteria therefore affects soil enzyme activity40. This result partially explains our previous finding that decomposition of leaf litter and lignin in Moso bamboo forests occurs more rapidly under IM than CM24. Acidobacteria, in contrast, were more prevalent in CM plots; these microbes are generally oligotrophic, consistent with their significantly lower abundances in the nutrient-rich rhizosphere and agricultural soils compared to that of bulk soil5,41. Crenarchaeota are a dominant group of microorganisms that govern NH4+ oxidation42. The decrease in the abundance of Crenarchaeota under IM that we observed may further affect the nitrifying process and N2O emissions in the soil of Moso bamboo forests.
Effect of nitrogen deposition on soil microbial biomass and community diversity
Our results demonstrated that a low amount of added N increased the soil MBC, but when N addition exceeded a certain threshold (30 kg N∙ha−1∙yr−1 for IM and 60 kg N∙ha−1∙yr−1 for CM), a sharp decrease in soil MBC occurred (Fig. 1). This partially supports our second hypothesis, that N addition increases soil MBC and microbial diversity, but it is clear that the relationship is dependent on the amount of N added. We previously observed a limited amount of N in the soil of Moso bamboo plantations under IM, even though fertilization provided some N43. It is also known that N deposition can increase soil microbial biomass in an N-limited region31,44. Additional N input may directly increase available N and indirectly increase organic matter by enhancing plant productivity, thus promoting the growth of microorganisms26. However, excessive N input has negative effects on microbial activity. Many studies have reported that long-term nitrogen deposition reduced soil MBC25,26. Nitrogen saturation induced by excess N input can decrease soil pH, leading to leaching of magnesium and calcium and mobilization of aluminum45. When this happens, microbes may become magnesium- or calcium-limited or suffer aluminum toxicity26, resulting in reduced microbial biomass. Our results indicate that the N input threshold for Moso bamboo plantations may be 60 kg N∙ha−1∙yr−1 (Fig. 1).
As for the effect of N addition on microbial diversity, we found that added N significantly decreased soil microbial community diversity, particularly under IM (Fig. 4). Similarly, Frey et al.29 observed that the diversity of the ectomycorrhizal fungal community was reduced under N addition (50 kg N∙ha−1∙yr−1) compared to that of control areas in the Harvard Forest. Compton et al.28 also observed that N addition strongly influenced the DNA profiles of the microbial community. A meta-analysis revealed that the abundances of microbes decreased under N addition and that these reductions were more evident when higher total amounts of N were added and for longer durations26. In our study, the main reason for this reduced microbial diversity appears to be a reduction in soil pH induced by N fertilization, particularly with IM treatment. Previous studies have also found that fertilization treatment reduces soil pH39,46.
The N-induced reduction in microbial diversity may affect the structure of the microbial community47. CM is not conducive to nutrient accumulation, therefore, N addition resulted in a decrease in the relative abundances of eutrophic taxa (including members of the Proteobacteria and Bacteroidetes) and an increase in the abundances of oligotrophic taxa (mainly Acidobacteria) (Fig. 3). Under IM, the relative abundances of Proteobacteria and Crenarchaeota were higher in the N90 treatments than in the control, whereas Acidobacteria and Verrucomicrobia exhibited the opposite trend. Again, this may be because the relative abundances of eutrophic taxa (including members of the Proteobacteria and Bacteroidetes) increased, whereas those of oligotrophic taxa (mainly Acidobacteria) decreased in high-N plots48. Similar effects have been also observed in previous studies28,49. Actinobacteria, an important microbe in lignin decomposition40, exhibited a greater relative abundance in the N30 treatment than in the N90 treatment under both IM and CM, thus providing a reasonable explanation for our previous observation that a low level of N addition facilitates the decomposition of Moso bamboo leaf litter, while a high level suppresses decomposition24.
Interactive effect of nitrogen deposition and management practices on soil microbial biomass and community diversity
Our third hypothesis was that the combination of management practices and N deposition would exert greater effects on soil microbial biomass and diversity than either practice independently. Two-way ANOVA demonstrated that both management practices and N deposition had significant positive effects on soil MBC (Fig. 1 and Table 2) but significant negative effects on soil microbial community diversity (Fig. 4 and Table 3). These patterns were true for the two variables, both alone and in combination, supporting our third hypothesis. PCoA and cluster analyses demonstrated that management practices had a greater impact on soil microbial diversity than nitrogen deposition (Figs 5 and 6). IM practices such as plowing, weeding, and fertilization alter the structural and physicochemical properties of soil more than the addition of nitrogen alone. Moreover, IM has been performed at the site for approximately 15 years, whereas the N addition experiment was only conducted for 2.5 years. The cumulative effects of IM practices on soil microbes were therefore greater than those of N addition owing to the short duration of the experiment.
Table 2. Two-way ANOVA indicating the effects of N addition and management type (intensive or conventional) on soil microbial biomass carbon.
| Source of variation | SS | df | MS | F | P-value |
|---|---|---|---|---|---|
| N addition | 2742792.2 | 3 | 914264.1 | 1574.6 | <0.0001 |
| Management type | 861123.7 | 1 | 861123.7 | 1483.0 | <0.0001 |
| Interaction | 1264365.6 | 3 | 421455.2 | 725.8 | <0.0001 |
| Within | 9290.4 | 16 | 580.7 | ||
| Total | 4877572.0 | 23 |
SS: sum-of-squares; df: degrees of freedom; MS: mean square.
Table 3. Two-way ANOVA indicating the effects of N addition and management type (intensive or conventional) on the Chao 1 index of soil microbial community composition.
| Source of variation | SS | df | MS | F | P-value |
|---|---|---|---|---|---|
| N addition | 1295767.9 | 3 | 431922.6 | 9.4 | 0.0008 |
| Management type | 8005801.7 | 1 | 8005802 | 175.2 | <0.0001 |
| Interaction | 840578.7 | 3 | 280192.9 | 6.1 | 0.006 |
| Within | 730924.5 | 16 | 45682.8 | ||
| Total | 10873072.9 | 23 |
SS: sum-of-squares; df: degrees of freedom; MS: mean square.
Relationship between microbial diversity and soil properties
Previous research indicated that soil pH may strongly influence the soil microbial community composition and bacterial diversity. Soil pH is thought to be a good predictor of bacterial community composition28,37,39,50,51,52,53,54. The results of this study demonstrated that bacterial diversity and community composition were mainly correlated with soil pH (Fig. 7 and Table 1). A strong negative relationship between bacterial diversity and soil pH was also observed by Lauber et al.55 and has been supported by other studies8,50. Our results also demonstrated that N, available P, and the C/N ratio are important parameters for shaping microbial community structure (Fig. 7). In accordance with this, previous studies observed that bacterial community structure was closely correlated with the NO3− concentration39,56,57. Additionally, Zhao et al.58 observed that Betaproteobacteria and Deltaproteobacteria abundances were positively correlated with available soil P content. Chu et al.59 observed that the C/N ratio was significantly correlated with the relative abundances of various dominant phyla in Arctic tundra soils, consistent with the abundances and composition of soil bacterial communities in the Changbai Mountains60. These results indicate that there are significant correlations between the diversity and composition of microbial communities and soil properties. Therefore, management practices and N addition may affect microbial diversity through changes in soil parameters.
In the present study, measurement of the soil microbial biomass included both bacteria and fungi, whereas assessment of the V4 hypervariable region of the 16S rRNA gene only reflected changes in the bacterial community composition. A previous study revealed that N deposition also inhibits fungal community composition26. Therefore, future studies should explore the effects of N deposition and management practices on fungal diversity.
In conclusion, we found that while both intensive management and N addition increased soil microbial biomass and reduced microbial diversity on a Moso bamboo plantation, management practices exerted a greater influence over regulation of these parameters than N deposition. These results not only have practical implications for the management of Moso bamboo forests, but also serve to broaden our understanding of the effect of human interventions and N deposition on soil microbial communities.
Methods
Study site
The study site was located in Qingshan Town, Lin’an City (30°14′N, 119°42′E), Zhejiang Province, China. The area has a monsoonal subtropical climate with a mean annual precipitation of 1,420 mm and a mean annual temperature of 15.6 °C, ranging from 24 °C in July to 3 °C in January. The area receives an average of approximately 1,847 hours of sunshine per year and features an average of 230 frost-free days per year.
Experimental design and measurement
Detailed information on the experimental design can be found in Song et al.24. Briefly, CM and IM Moso bamboo plantations were established at the study site. The CM plantation was originally established in the late 1970 s, and the IM plantation was established based on the CM area in 2001. CM practices included only the selective and regular harvest of bamboo trunks and shoots, which also took place in the IM area24. The IM area included additional management practices, such as plowing, weeding with herbicides, and fertilization. Specifically, in September of each year, 450 kg∙ha−1 of compound fertilizer (15:6:20 N:P2O5:K2O) was manually and evenly scattered on the soil surface, followed by deep plowing to 0.3 m. There was a greater abundance and biomass of plant species in the CM area than in the IM area24.
For this study, we established 12 CM plots and 12 IM plots in November 2012. Each plot had an area of 20 × 20 m and was surrounded by a 20-mbuffer zone. In accordance with the local background atmospheric N deposition rate of 30–37 kg N∙ha−1∙yr−1 61,62,63, N addition treatments included a low-N treatment (30 kg N∙ha−1∙yr−1, N30), a medium-N treatment (60 kg N∙ha−1∙yr−1, N60), and a high-N treatment (90 kg N∙ha−1∙yr−1, N90). Three replicate plots per treatment were randomly placed in each management area. Beginning in January 2013, following a widely used method for simulating N deposition64,65, NH4NO3 was dissolved in 10 L water to the proper final N concentration and sprayed evenly on the forest floor with an electric sprayer at the beginning of every month. An equivalent amount of N-free water was sprayed on each control plot to control for the effect of the added water24.
Soil sampling
In July 2015, ten soil cores at a depth of 0–20 cm were randomly collected from each plot and mixed together. Samples were sieved through 2-mm mesh to remove roots, plant residues, and stones. A portion of each soil sample was collected in a 50-mL centrifuge tube, placed in a cooler, and transferred to the laboratory. The tubes were archived at −80 °C until being used for molecular analysis. The remaining samples were used to measure the microbial biomass (from field-moist soil) and were then air-dried to determine the physicochemical properties of the soil.
Analysis of soil microbial biomass and physiochemical properties
MBC generally accounts for 40–50% of the microbial biomass and 1–5% of soil organic carbon, and is thus regarded as an important indicator of soil microbial biomass66. We therefore measured MBC to reflect the dynamics of soil microbial biomass. MBC was estimated using the chloroform fumigation-extraction method67. The pH of a soil-water (1:2.5 w/v) suspension was measured using a pH meter (FE20, Mettler Toledo, Switzerland) after shaking for 30 min. Soil organic carbon (SOC) and TN were determined using an elemental analyzer (Elementar Vario EL III, Germany). AP in the soil was extracted with sodium bicarbonate and determined using the molybdenum blue method68. The soil KCl-extractable NO3− and NH4+ were determined by extraction with 2 M KCl, followed by steam distillation and titration69.
DNA extraction and library construction
Microbial DNA was extracted from the soil samples (0.25 g of wet weight) using an Ezup Column Soil DNA Purification Kit (Sangon Biotech, Shanghai, China) according to the manufacturer’s instructions. Total DNA was evaluated on a 1.0% agarose gel, and the DNA concentration and quality (A260/A280) of the extracts were estimated visually using a NanoDrop ND-1000 UV-V spectrophotometer (Thermo Scientific, Rockwood, TN, USA).
16S rRNA gene amplification and sequencing
The V4 hypervariable region of the 16S rRNA gene was amplified from each soil sample using the PCR primers 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) and a sample tagging approach70,71. PCRswere performed in 30-μL reactions with 15 μL of Phusion® High-Fidelity PCR Master Mix (New England BioLabs, Ipswich, MA, USA), 0.2 μM forward and reverse primers, and approximately 10 ng of template DNA. Thermal cycling conditions consisted of initial denaturation at 98 °C for 1 min; 30 cycles of denaturation at 98 °C for 10 s, annealing at 50 °C for 30 s, and elongation at 72 °C for 30 s; and a final extension at 72 °C for 5 min. PCR products were purified with a Gene JET Gel Extraction Kit (Thermo Scientific) and combined in equimolar ratios with the quantitative DNA binding method to create a DNA pool that was subsequently used for sequencing from the adaptor. High-throughput sequencing of the 16S rRNA tag-encoded gene was performed on the Illumina MiSeq platform at Novogene (Beijing, China).
Bioinformatics analysis
Sequencing reads were assigned to each sample according to the unique barcode tags. Sequences were analyzed with the QIIME (Quantitative Insights Into Microbial Ecology) software package and UPARSE pipeline70. The reads were first filtered by QIIME quality filters. The default settings for Illumina processing in QIIME were used. Then, the UPARSE pipeline was used to discern OTUs with 97% identity. For each OTU, a representative sequence was selected and a taxonomic group was assigned using the Ribosomal Database Project (RDP) classifier. The species richness of each sample was estimated by rarefaction analysis; the Chao1 index of each library was determined as described previously72.
Statistical analysis
One-way analysis of variance (ANOVA) including post-hoc correction for multiple comparisons using the Bonferroni method was used to determine the statistical significance of differences in MBC and the Chao1 index for each sampling event among the four experimental treatments in the two plantation management schemes. Two-way ANOVA was performed to assess the combined effect of N deposition and management practices on microbial biomass and diversity. The Pearson correlation coefficient between soil properties and α-diversity was also calculated. Analyses were conducted using SPSS (Statistical Package for the Social Sciences) 18.0 for Windows (SPSS Inc., Chicago, Illinois, USA).
QIIME was used to calculate the weighted UniFrac. PCoA and UPGMA (unweighted pair group method with arithmetic mean) clustering were conducted on the weighted UniFrac based on a protocol published previously73. In addition, a CCA was performed to identify the abiotic factors with the most impact on bacterial community composition, and these results were used to construct a soil property matrix for variation partitioning analysis in R v.2.8.1 with the vegan package.
Additional Information
How to cite this article: Li, Q. et al. Nitrogen deposition and management practices increase soil microbial biomass carbon but decrease diversity in Moso bamboo plantations. Sci. Rep. 6, 28235; doi: 10.1038/srep28235 (2016).
Acknowledgments
This study was funded by the National Natural Science Foundation of China (Grant No. 31270517, 31470529), “948” Project of the State Forestry Bureau of China (Grant No. 2013-4-55), and Pandeng Project for Young & Middle-aged Discipline Leaders of Zhejiang Province (Grant No. pd2013234).
Footnotes
Author Contributions X.S. designed the study. Q.L. and H.G. performed the data collection and analysis. Q.L., X.S., H.G. and F.G. interpreted the results and wrote the paper.
References
- Fierer N. & Jackson R. B. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. USA 103, 626–631 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J. L., Bohannan B. J. M. & Whitaker R. J. Microbial biogeography: from taxonomy to traits. Science 320, 1039–1043 (2008). [DOI] [PubMed] [Google Scholar]
- He Z., Nostrnd J., Deng Y. & Zhou J. Development and applications of functional gene microarrays in the analysis of the functional diversity, composition, and structure of microbial communities. Front. Environ. Sci. Engin. China 5, 1–20 (2011). [Google Scholar]
- Brookes P. C., Landman A., Pruden G. & Jenkinson D. S. Chloroform fumigation and the release of soil nitrogen: a rapid direct extraction method to measure microbial biomass nitrogen in soil. Soil Biol. Biochem. 17, 837–842 (1985). [Google Scholar]
- López-Lozano N. E., Heidelberg K. B., Nelson W., Garcia-Oliva F. & Eguiarte L. E. Microbial secondary succession in soil microcosms of a desert oasis in the Cuatro Cienegas Basin, Mexico. Peer J. 1, e47 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallin S., Jones C. M., Schloter M. & Philippot L. Relationship between N cycling communities and ecosystem functioning in a 50-year-old fertilization experiment. ISME J. 3, 597–605 (2009). [DOI] [PubMed] [Google Scholar]
- Ye L. & Zhang T. Bacterial communities in different sections of a municipal wastewater treatment plant revealed by 16S rDNA 454 pyrosequencing. Appl. Microbiol. Biotechnol. 97, 2681–2690 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J. et al. Responses of bacterial communities in arable soils in a rice-wheat cropping system to different fertilizer regimes and sampling times. PLoS One 9, e85301 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch L. F. W. et al. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 1, 283–290 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank D. A. & Groffman P. M. Ungulate vs. landscape control of soil C and N processes in grasslands of Yellowstone National Park. Ecology 79, 2229–2241 (1998). [Google Scholar]
- Ramette A. & Tiedje J. M. Biogeography: an emerging cornerstone for understanding prokaryotic diversity, ecology, and evolution. Microb. Ecol. 53, 197–207 (2007). [DOI] [PubMed] [Google Scholar]
- Fierer N., Nemergut D., Knight R. & Craine J. M. Changes through time: integrating microorganisms into the study of succession. Res. Microbiol. 161, 635–642 (2010). [DOI] [PubMed] [Google Scholar]
- Song X. et al. Carbon sequestration by Chinese bamboo forests, and their ecological benefits: assessment of potential, problems, and future challenges. Environ. Rev. 19, 418–428 (2011). [Google Scholar]
- Li R., Werger M. J. A., During H. J. & Zhong Z. Biennial variation in production of new shoots in groves of the giant bamboo Phyllostachys pubescens in Sichuan, China. Plant Ecol. 135, 103–112 (1998). [Google Scholar]
- Zhou G., Jiang P. & Xu Q. Carbon fixing and transition in the ecosystem of bamboo stands. (Science Press, 2010). [Google Scholar]
- Xu Q., Jiang P., Wu Q., Wang J. & Wu J. Effects of intensive management on soil microbial biomass and functional diversity in Castanea mollissima stands. Scientia Silave Sinicae 43, 15–19 (2007). [Google Scholar]
- Yu W., Jiang Z., Liu M. & Zhao X. Effect of different land use patterns on soil microbial biomass carbon. Chin. J. Soil. Sci. 39, 282–286 (2008). [Google Scholar]
- He D. et al. Effect of intensive management on abundance and composition of soil N2-fixing bacteria in the Phyllostachys heterocycla stand. Chin. J. Appl. Ecol. 26, 2961–2968 (2015). [PubMed] [Google Scholar]
- Liu W., Deng G. H., Zhang J. C. & Yang Q. P. Influence of fertilizer application on rhizosphere microbial diversity of phyllostachys edulis based on PCRDGGE analysis. J. Northeast For. Univ. 39, 50–53 (2011). [Google Scholar]
- Sun D. D., Xu Q. F., Tian T. & Liu B. R. Investigation on soil microbial biomass and structure in Phyllostachys edulis plantations with increasing cultivation time. Scientia Silave Sinicae 47, 181–186 (2011). [Google Scholar]
- Liu. X. et al. Enhanced nitrogen deposition over China. Nature 494, 459–462 (2013). [DOI] [PubMed] [Google Scholar]
- Reay D. S., Dentener F., Smith P., Grace J. & Feely R. A. Global nitrogen deposition and carbon sinks. Nat. Geosci. 1, 430–437 (2008). [Google Scholar]
- Lü C. & Tian H. Spatial and temporal patterns of nitrogen deposition in China: synthesis of observational data. J. Geophys. Res. 112, 10–15 (2007). [Google Scholar]
- Song X., Zhou G., Gu H. & Qi L. Management practices amplify the effects of N deposition on leaf litter decomposition of the Moso bamboo forest. Plant Soil 395, 391–400 (2015). [Google Scholar]
- Li L., Zeng D., Yu Z., Fan Z. & Mao R. Soil microbial properties under N and P additions in a semi-arid, sandy grassland. Biol. Fertil. Soils 46, 653–658 (2010). [Google Scholar]
- Treseder K. K. Nitrogen additions and microbial biomass: A meta analysis of ecosystem studies. Ecol. Lett. 11, 1111–1120 (2008). [DOI] [PubMed] [Google Scholar]
- Ramirez K. S., Lauber C. L., Knight R., Bradford M. A. & Fierer N. Consistent effects of N fertilization on soil bacterial communities in contrasting systems. Ecology 91, 3463–3470 (2010). [DOI] [PubMed] [Google Scholar]
- Compton J. E., Watruda L. S., Porteousa L. A. & DeGroud S. Response of soil microbial biomass and community composition to chronic nitrogen additions at Harvard forest. For. Ecol. Manage. 196, 143–158 (2004). [Google Scholar]
- Frey S. D., Knorr M., Parrent J. L. & Simpson R. T. Chronic nitrogen enrichment affects the structure and function of the soil microbial community in temperate hardwood and pine forests. For. Ecol. Manage. 196, 159–171 (2004). [Google Scholar]
- Paul J. W. & Beauchamp E. G. Soil microbial biomass C, N mineralization and N uptake by corn in dairy cattle slurry and urea amended soils. Can. J. soil sci. 76, 469–472 (1996). [Google Scholar]
- Johnson D., Leake J. R., Lee J. A. & Campbell C. D. Changes in soil microbial biomass and microbial activities in response to 7 years simulated pollutant nitrogen deposition on a heathland and two grasslands. Environ. Pollut. 103, 239–250 (1998). [Google Scholar]
- Boxman A. W. et al. Vegetation and soil biota response to experimentally changed nitrogen inputs in coniferous forest ecosystems of the NITREX project. For. Ecol. Manage. 101, 65–79 (1998). [Google Scholar]
- Liu E. K. et al. Biological properties and enzymatic activity of arable soils affected by long-term different fertilization systems. J. Plant Ecol. (Chinese Version) 32, 176–182 (2008). [Google Scholar]
- Wang W. et al. Impact fertilization on soil organic carbon and soil microbial biomass carbon in Dendrocalamus farinosus forest. J. Soil Water Conserv. 27, 269–288 (2013). [Google Scholar]
- Gao P., Qiu Y., Zhou Z., He R. & Xu J. Productivity and photosynthetic ability of Phyllostachys edulis with nitrogen fertilization. J. Zhejiang A&F Univ. 31, 697–703 (2014). [Google Scholar]
- Broadbent F. E. Effects of fertilizer nitrogen on the release of soil nitrogen. Soil Sci. Soc. Am. J. 26, 692–696 (1965). [Google Scholar]
- Rousk J. et al. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 4, 1340–1351 (2010). [DOI] [PubMed] [Google Scholar]
- Zhao J. et al. Responses of bacterial communities in arable soils in a rice-wheat cropping system to different fertilizer regimes and sampling times. PLoS One 9, e85301 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J. et al. Influence of 34-years of fertilization on bacterial communities in an intensively cultivated black soil in northeast China. Soil Biol. Biochem. 90, 42–51 (2015). [Google Scholar]
- Zhao C., Peng S., Ruan H. & Zhang Y. Effects of nitrogen deposition on soil microbes. J. Nanjing For. Univ. 39, 149–155 (2015). [Google Scholar]
- Kielak A., Pijl A. S., Veen J. A. & Kowalchuk G. A. Phylogenetic diversity of acido bacteria in a former agricultural soil. ISME J. 3, 378–382 (2009). [DOI] [PubMed] [Google Scholar]
- Leininger S. et al. Archaea predominate among ammonia oxidizing prokaryotes in soils. Nature 442, 806–809 (2006). [DOI] [PubMed] [Google Scholar]
- Song X., Gu H., Wang M., Zhou G. & Li Q. Management practices regulate the response of Moso bamboo foliar stoichiometry to nitrogen deposition. Sci. Rep. 6, 24107 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. et al. Effects of elevated nitrogen deposition on soil microbial biomass carbon in the main subtropical forests of southern China. Acta Ecol. Sinica 28, 470–478 (2008). [Google Scholar]
- Vitousek P. M. et al. Human alteration of the global nitrogen cycle: sources and consequences. Ecol. Appl. 7, 737–750 (1997). [Google Scholar]
- Guo J. H. et al. Significant acidification in major Chinese croplands. Science 327, 1008–1010 (2010). [DOI] [PubMed] [Google Scholar]
- Aber J. D., Nadelhoffer K. J., Steudler P. & Melillo J. M. Nitrogen saturation in northern forest ecosystem: hypothesis and implications. BioScience. 39, 378–386 (1989). [Google Scholar]
- Fierer N. et al. Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J. 6, 1007–1017 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell B. J., Polson S. W., Hanson T. E., Mack M. C. & Schuur E. A. G. The effect of nutrient deposition on bacterial communities in Arctic tundra soil. Environ. Microbio. 12, 1842–1854 (2010). [DOI] [PubMed] [Google Scholar]
- Shen J., Zhang L., Guo J., Jessica L. R. & He J. Impact of long-term fertilization practices on the abundance and composition of soil bacterial communities in Northeast China. Appl. Soil Ecol. 46, 119–124 (2010). [Google Scholar]
- Geisseler D. & Scow K. M. Long-term effects of mineral fertilizers on soil microorganisms: a review. Soil Biol. Biochem. 75, 54–63 (2014). [Google Scholar]
- Nicol G. W., Leininger S., Schleper C. & Prosser J. I. The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ. Microbiol. 10, 2966–2978 (2008). [DOI] [PubMed] [Google Scholar]
- Kuang J. et al. Contemporary environmental variation determines microbial diversity patterns in acid mine drainage. ISME J. 7, 1038–1050 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Hu M., Xia Y., Wen X. & Ding K. Pyrosequencing analysis of bacterial diversity in 14 wastewater treatment systems in China. Appl. Environ. Microbiol. 78, 7042–7047 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber C. L., Hamady M., Knight R. & Fierer N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbio. 75, 5111–5120 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M., Xiao T., Ning Z., Xiao E. & Sun W. Microbial community analysis in rice paddy soils irrigated by acid mine drainage contaminated water. Appl. Microbiol. Biotechno. 99, 2911–2922 (2014). [DOI] [PubMed] [Google Scholar]
- Belén C. N. R., Roberto Á., Alejandro M. & Vázquez M. P. Structure, composition and metagenomic profile of soil microbiomes associated to agricultural land use and tillage systems in argentine pampas. PLoS One 9, e99949 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J. et al. Pyrosequencing reveals contrasting soil bacterial diversity and community structure of two main winter wheat cropping systems in China. Microb. Ecol. 67, 443–453 (2014). [DOI] [PubMed] [Google Scholar]
- Chu H. et al. Soil bacterial diversity in the Arctic is not fundamentally different from that found in other biomes. Environ. Microbiol. 12, 2998–3006 (2010). [DOI] [PubMed] [Google Scholar]
- Shen C. et al. Soil pH drives the spatial distribution of bacterial communities along elevation on Changbai Mountain. Soil Biol. Biochem. 57, 204–211 (2013). [Google Scholar]
- Xie Y., Zhang S., Zhao X., Xiong Z. & Xing G. Seasonal variation patterns of NH4+-N / NO3−-N ratio and δ15 NH4+ value in rainwater in Yangtze river delta. Chin. J. Appl. Ecol. 19, 2035–2041 (2008). [PubMed] [Google Scholar]
- Cui J. et al. Atmospheric wet deposition of nitrogen and sulfur in the agroecosystem in developing and developed areas of Southeastern China. Atmos. Environ. 89, 102–108 (2014). [Google Scholar]
- Jia Y. et al. Spatial and decadal variations in inorganic nitrogen wet deposition in China induced by human activity. Sci. Rep. 4, 3763 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo J., Brown S., Xue J., Fang Y. & Li Z. Response of litter decomposition to simulated N deposition in disturbed rehabilitated and mature forests in subtropical china. Plant Soil 282, 135–151 (2006). [Google Scholar]
- Fang H., Mo J., Peng S., Li Z. & Wang H. Cumulative effects of nitrogen additions on litter decomposition in three tropical forests in southern china. Plant Soil 297, 233–242 (2007). [Google Scholar]
- Chen C., Xu Z. & Mathers N. J. Soil carbon pools in adjacent natural and plantation forests of subtropical Australia. Soil Sci. Soc. Am. J. 68, 282–291 (2004). [Google Scholar]
- He J. Z., Lu Y. H. & Fu B. J. Frontiers of soil biology. (Science Press, 2015). [Google Scholar]
- Watanabe F. S. & Olsen S. R. Test of an ascorbic acid method f or determining phosphorus in water and NaCO extracts from soil. Soil Sci. Soc. Am. Proc. 29, 677–678 (1965). [Google Scholar]
- Mulvaney R. L. Nitrogen-inorganic forms. In: Sparks D. L. (Ed.), Method of Soil Analysis. Part 3. Chemical Methods. American Society of Agronomy and Soil Science Society of America, Madison,Wisconsin (1996).
- Caporaso J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G. et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss P. D. et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuczynski J. et al. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr. Protoc. Microbiol. 1E, 5.1–20 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]