Abstract
Cation transport regulator homolog 1 (Chac1) is an endoplasmic reticulum (ER) stress inducible gene that has a function as a γ-glutamyl cyclotransferase involved in the degradation of glutathione. To characterize the translation and stability of Chac1, we found that the Kozak-like sequence present in the 5′ untranslated region (5′UTR) of the Chac1 mRNA was responsible for Chac1 translation. In addition, the short form (ΔChac1), which translated from the second ATG codon, was generated in the absence of the 5′UTR. The proteasome pathway predominantly participated in the stability of the Chac1 protein; however, its expression was remarkably up-regulated by co-transfection with ubiquitin genes. Using an immunoprecipitation assay, we revealed that ubiquitin molecule was directly conjugated to Chac1, and that mutated Chac1 with all lysine residues replaced by arginine was also ubiquitinated. Finally, we showed that WT Chac1 but not ΔChac1 reduced the intracellular level of glutathione. Taken together, our results suggest that the Chac1 protein expression is regulated in translational and post-translational fashion due to the Kozak-like sequence in the 5′UTR and the ubiquitin-mediated pathways. The bidirectional roles of ubiquitination in regulating Chac1 stabilization might give us a new insight into understanding the homeostasis of glutathione under pathophysiological conditions.
The ubiquitin-proteasome pathway is a proteolytic mechanism that specifically ubiquitinates protein substrates targeted for degradation by the proteasome1. This pathway is known to associate with endocytosis of membrane proteins, specific protein-protein interactions for signal transduction, DNA repair and gene transcription2,3,4. Protein ubiquitination forms an isopeptide bond between the C-terminal carboxyl group of ubiquitin and the ε-amino group of lysine residues on target proteins. The successive linkage of ubiquitin to a protein substrate with free ubiquitins through its lysine residues forms a polyubiquitin chain. In the course of ubiquitin conjugation, a ubiquitin activating enzyme (E1) phosphorylates the carboxylic group of ubiquitin and binds the phospho-ubiquitin to the active cysteine site. This is followed by the transfer of the activated ubiquitin to the cysteine residue of ubiquitin conjugating enzyme (E2) through a trans(thio)esterification reaction. Through the assistance of ubiquitin ligase (E3), the ubiquitin molecule is transferred to a lysine residue on the protein substrate. It is presumed that more than 600 types of E3 ubiquitin ligases exist in the human genome and that each one of them recognizes specific substrates, suggesting that different types of ubiquitin-conjugation reactions play key roles in regulating a variety of cellular events, including signal transduction, gene transcription and the cell cycle5.
A ubiquitin molecule contains seven lysine residues (K6, K11, K27, K29, K33, K48 and K63); thus, it is considered that there are seven different types of isopeptide linkages. Polyubiquitin chains are usually composed of only one type of isopeptide linkage; however, more than two types of isopeptide linkages (i.e., heterogeneous chains) have been identified6,7,8. It is reported that the K48-linked polyubiquitin chain is recognized by the proteasome, leading to the degradation of the substrate protein6,9. On the other hand, the K63-linked polyubiquitin chain participates in endosomal trafficking, signal transduction, and DNA repair but not in protein degradation2,3,10. In addition, the heterogeneous chains formed by certain pairs of E2 and E3 are reported to synthesize non-degradable proteins8. Moreover, certain proteasome degradation processes are independent of lysine residues on the substrate protein (i.e., non-canonical ubiquitination)11. For instance, the degradation of p21, ERK3 and Cyclin G1 by the proteasome is mediated by the ubiquitination at the N-terminal methionine instead of lysine residues12,13,14,15. Furthermore, the hydroxyl groups of serine and threonine residues and the thiol group of cysteine residues are found to be potential sites of ubiquitination16,17,18,19.
Chac1 was first identified as a novel ER stress inducible gene in human aortic endothelial cells treated with oxidized phospholipids20,21. Until now, it has been reported that various stimuli triggering ER stress up-regulated Chac1 mRNA21,22,23,24,25,26,27,28. Activating transcription factor 4 (ATF4) plays a key role in regulating the expression of Chac1 by directly binding to the ATF4-consensus sequence in the 5′ flanking region of the Chac1 gene28,29,30. ATF4 is among the three canonical ER resident stress sensors, PERK-ATF4, IRE1-sXBP1 and ATF6. Moreover, the expression level of Chac1 mRNA is increased in breast and ovarian cancers associated with poor prognosis, and knockdown of Chac1 results in decreased cell migration31. Chac1 mRNA is also up-regulated in Shigella-infected HeLa cells and duck hepatitis A virus genotype C-infected duck liver32,33. In regard to functional capability, it has been reported that Chac1 functions as a γ-glutamyl cyclotransferase that specifically degrades glutathione by its catalytically active residue at E116 in the mouse and E115 in the human protein30,34,35. Furthermore, Chac1 plays an important role in regulating neurogenesis by Notch deglycination at the E1669 residue36,37. Until now, studies regarding the transcriptional regulation and functional characterization of Chac1 have been actively performed; however, its translational and post-translational regulatory mechanisms are still poorly elucidated. We previously reported that Chac1 protein was stabilized by the treatment of a proteasome inhibitor28. We therefore investigated expression mechanisms of the mouse Chac1 protein by focusing on its 5′UTR region as well as ubiquitination and proteasome-mediated degradation.
Results
The expression level of the Chac1 protein is regulated by the proteasome pathway
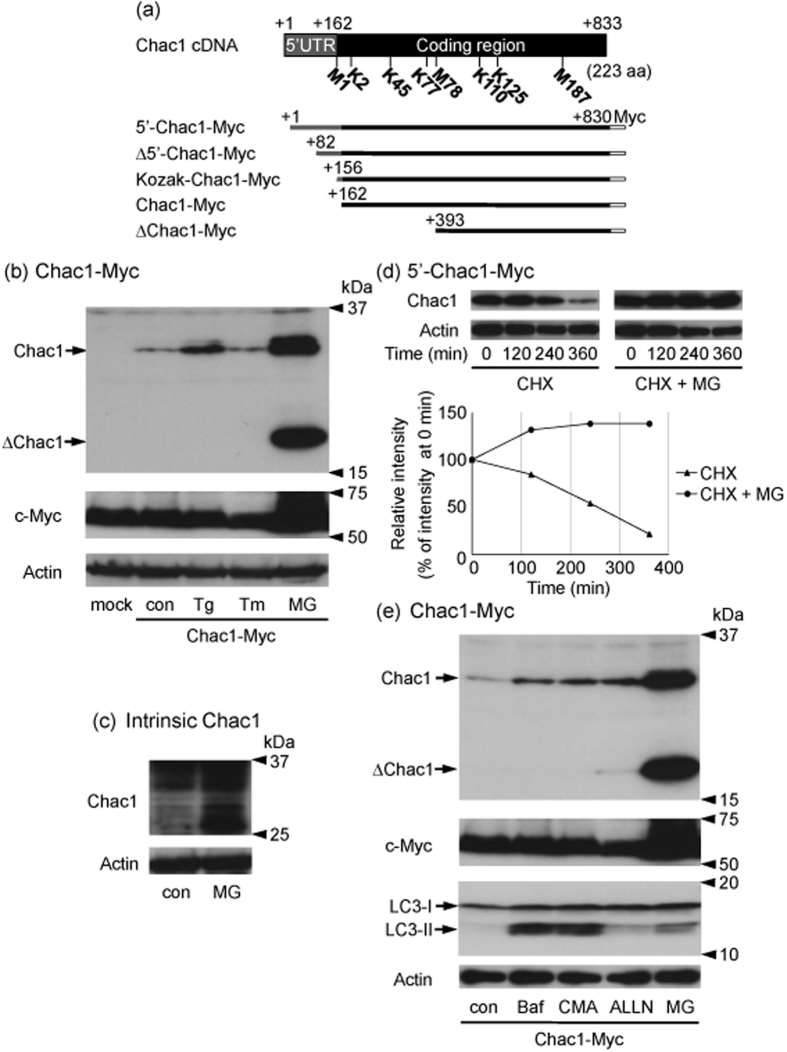
The mouse Chac1 protein consists of 223 amino acids (aa). The Chac1 mRNA is coded from position 162 to 833 (+162/+833) following the 5′UTR (+1/+161) as found in the NCBI database under accession NM_026929 (Fig. 1a). We previously reported that the Chac1 protein is stabilized by the proteasome inhibitor, MG132, in Neuro2a cells transiently transfected with a Chac1 construct (+162/+833)28. We next focused on the 5′UTR region of the Chac1 mRNA, especially the Kozak-like sequence (GGCACC) just before the translation start site (Fig. 1a). First, we constructed Chac1 expression vectors having the entire 5′UTR (+1/+830; 5′-Chac1-Myc), a truncated 5′UTR (+82/+830; Δ5′-Chac1-Myc), the Kozak-like sequence just before the translation start site (+156/+830; Kozak-Chac1-Myc), and a vector lacking the 5′UTR (+162/+830; Chac1-Myc) (Fig. 1a and Supplementary Fig. S1). To investigate the expression of the Chac1 protein, a Chac1 vector having a Myc-epitope at the C-terminus (Chac1-Myc) was transiently transfected into HEK293 cells. The cells were subsequently treated with the ER stress inducers (thapsigargin (Tg) and tunicamycin (Tm)) or MG132 (Fig. 1b). Consistent with our previous report, the Chac1 protein was stabilized by MG132, but not by the other agents. To confirm whether intrinsic Chac1 protein is stabilized by MG132, we investigated its expression in HEK293 cells in the presence or absence of MG132 using a Chac1 antibody (Fig. 1c). As a result, the expression of intrinsic Chac1 was actually increased by treatment with MG132. We further investigated Chac1 expression in the presence or absence of cycloheximide (CHX), an inhibitor of protein translation, to determine whether the increased Chac1 expression by MG132 was due to the inhibition of proteasomal degradation. Chac1 protein was degraded in the presence of CHX in a time dependent manner, whereas it was markedly stabilized by the treatment of a combination of CHX and MG132 (Fig. 1d). As MG132 was reported to activate autophagy38, we examined whether autophagy takes part in regulating the expression level of Chac1 protein. HEK293 cells expressing Chac1-Myc were treated with MG132, lysosomal acidification inhibitors (Bafilomycin A1 (Baf) or Concanamycin A (CMA)), or the calpain I inhibitor N-Acetyl-L-leucyl-L-leucyl-L-norleucinal (ALLN). As shown in Fig. 1e, Chac1 was stabilized by MG132 treatment, whereas treatments with Baf, CMA or ALLN showed small effects. Under the same conditions, the autophagic marker LC3 conjugated to PE (PE-conjugated LC3-II)39, was dramatically up-regulated by treatment with Baf and CMA, whereas the effect of MG132 was to a lesser extent (Fig. 1e). In parallel, the expression of the intrinsic c-Myc protein, which is a well-known substrate of proteasomal degradation40,41, was also stabilized only by treatment with MG132. As a result, the c-Myc protein was stabilized by MG132 but not by the other agents (Fig. 1e). These data indicate that the Chac1 protein is predominantly regulated by the proteasome pathway in a post-translational manner.
Figure 1. The Chac1 protein is regulated by the proteasome pathway.
(a) Schematic representation of the Chac1 constructs showing the positions of methionine and lysine used in this study (NCBI accession No. NM_026929). (b) HEK293 cells were transfected with pcDNA3.1 (mock) or Chac1-Myc. After 24 h, cells were treated with vehicle (con), 0.1 μM thapsigargin (Tg), 2 μg/mL tunicamycin (Tm) or 20 μM MG132 (MG) for 12 h, and the resultant cell lysates were analyzed by western blot using c-Myc or Actin antibodies according to Materials and Methods. (c) HEK293 cells were treated with vehicle (con) or 20 μM MG for 12 h, and the resultant cell lysates were analyzed by western blot using Chac1 or Actin antibodies. (d) HEK293 cells were transfected with 5′-Chac1-Myc. After 24 h, cells were treated with 10 μg/mL cycloheximide (CHX) in the presence or absence of MG for the indicated times, and the resultant cell lysates were analyzed by western blot using c-Myc or Actin antibodies. Band intensities were analyzed by ImageJ software (National Institutes of Health, U.S.A.). (e) HEK293 cells were transfected with Chac1-Myc. After 24 h, the cells were treated with vehicle (con), 50 nM Bafilomycin A1 (Baf), 50 nM Concanamycin A (CMA), 10 μM N-Acetyl-L-leucyl-L-leucyl-L-norleucinal (ALLN) or 20 μM MG for 12 h, and the resultant cell lysates were analyzed by western blot using c-Myc, LC3 or Actin antibodies. All experimental data were obtained from three independent cultures.
Chac1 translation is enhanced by the Kozak-like sequence in the 5′ untranslated region and the short form of Chac1 is translated from the second methionine codon
To investigate whether the 5′UTR of Chac1 is responsible for the translation and stabilization of the protein, we transfected HEK293 cells with each of the Chac1 constructs shown in Fig. 1a. As a result of transfection, the expression level of Chac1 was markedly up-regulated in the presence of the Kozak-like sequence before the translation start site (Fig. 2a). The Chac1 protein was usually detected at approximately 30 kDa by western blot; however, another short form of Chac1 (ΔChac1) was detected at approximately 17 kDa when HEK293 cells were treated with MG132 (Figs 1b,e and 2a). As Chac1 mRNA has two additional methionine codons in its coding region (Fig. 1a), we speculated that ΔChac1 could be translated from the downstream methionine codons of a prospective translation start site. Thus, we made four mutant Chac1 constructs by replacing each methionine with isoleucine (Chac1 (M1I, M78I and M187I)-Myc) and deleting the upstream region from the second ATG codon (+393/+830; ΔChac1-Myc) (Figs 1a and 2b). We then evaluated the expression level of each construct in the presence of MG132. As shown in Fig. 2bΔChac1 was detected in HEK293 cells overexpressing Chac1 (WT)-Myc, Chac1 (M1I)-Myc, Chac1 (M187I)-Myc and ΔChac1 (+393/+830)-Myc but not in cells transfected with Chac1 (M78I)-Myc (Fig. 2b). This suggested that ΔChac1 might be translated from the second ATG codon of the Chac1 mRNA.
Figure 2. Chac1 translation is enhanced by the Kozak-like sequence in the 5′ untranslated region and the short form of Chac1 is translated from the second methionine codon.
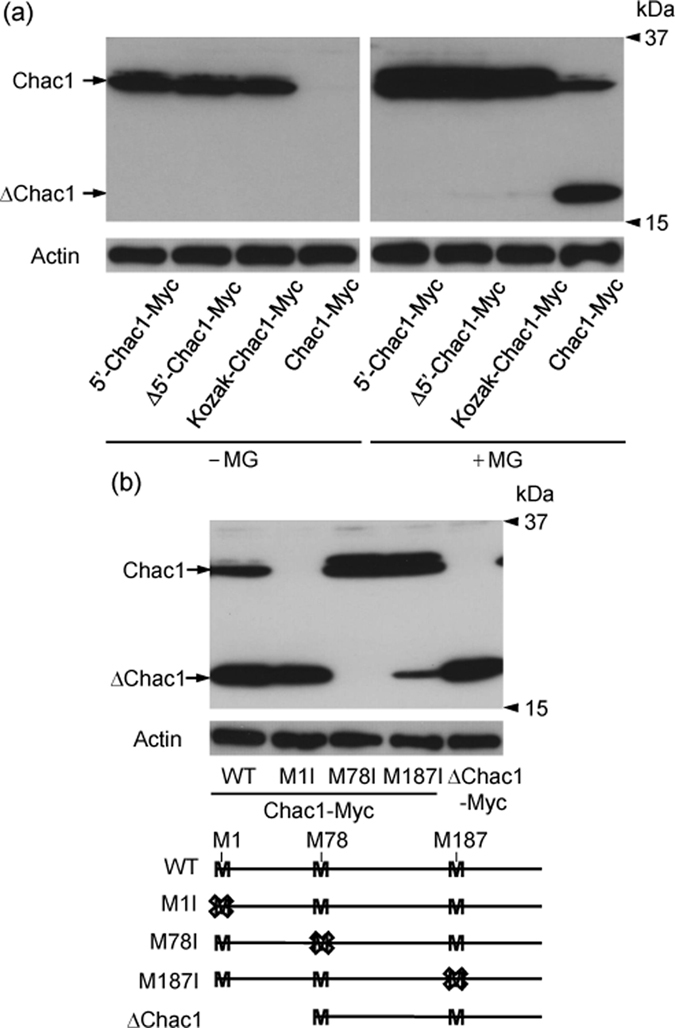
(a) HEK293 cells were transfected with 5′-Chac1-Myc, Δ5′-Chac1-Myc, Kozak-Chac1-Myc or Chac1-Myc. After 24 h, the cells were treated with vehicle (−MG) or 20 μM MG for 12 h, and the resultant cell lysates were analyzed by western blot using c-Myc or Actin antibodies. (b) HEK293 cells were transfected with Chac1-Myc (WT, M1I, M78I or M187I) or ΔChac1-Myc. After 24 h, the cells were treated with 20 μM MG for 12 h, and the resultant cell lysates were analyzed by western blot using c-Myc or Actin antibodies. All experimental data were obtained from three independent cultures.
Ubiquitin overexpression stabilizes Chac1 protein in HEK293 cells
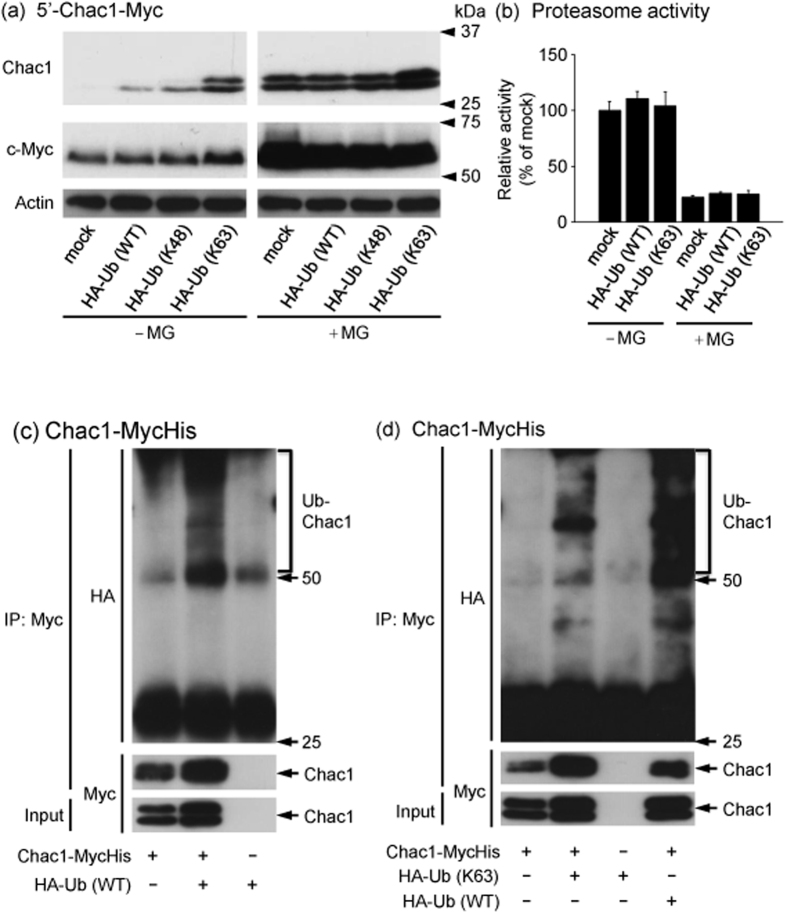
Because Chac1 was degraded by the proteasome pathway, we investigated whether the degradation of the Chac1 protein is facilitated by ectopically expressed ubiquitin. 5′-Chac1-Myc was transfected together with N-terminal HA-tagged wild type ubiquitin (HA-Ub (WT)) or single-lysine mutant ubiquitin with K48 (HA-Ub (K48)) or K63 (HA-Ub (K63)) as the only lysine residue in the ubiquitin molecule42. Contrary to our expectations, Chac1 was stabilized by the co-transfection of each HA-Ub construct, and the effect of HA-Ub (K63)-overexpression was prominent (Fig. 3a). The increase in Chac1-Myc expression by HA-Ub (WT) co-expression was almost at the same level as observed by HA-Ub (K48). On the other hand, the expression of intrinsic c-Myc was not elevated by the transfection of each type of HA-Ub construct (Fig. 3a). We then investigated the proteasome activity when HA-Ub (WT or K63) was transfected into HEK293 cells (Fig. 3b). As a result, the activity was not altered by the HA-Ub overexpression, whereas it was decreased by the MG132 treatment. Next, we examined whether Chac1 was directly ubiquitinated in the HA-Ub (WT) or HA-Ub (K63)-overexpressing cells using an immunoprecipitation assay. As shown in Fig. 3c,d, immunoprecipitates from cells expressing both Chac1-MycHis and HA-Ub (WT or K63) in the presence of MG132 showed a ladder of higher molecular size bands detected by an anti-HA antibody. Interestingly, the band derived from the cells expressing HA-Ub (K63) was weaker compared to HA-Ub (WT) (Fig. 3d).
Figure 3. Chac1 is stabilized by ubiquitin co-expression and actually ubiquitinated in HEK293 cells.
(a) HEK293 cells were transiently co-transfected with 5′-Chac1-Myc and HA-Ub (WT, K48 or K63). After 24 h, the cells were treated with vehicle (−MG) or 20 μM MG for 12 h, and the resultant cell lysates were analyzed by western blot using c-Myc or Actin antibodies. (b) HEK293 cells transfected with pcDNA3.1 (mock), HA-Ub (WT or K63). After 36 h, the cells were collected and the proteasome activity was measured according to the Materials and Methods. (c) HEK293 cells were transiently co-transfected with Chac1-MycHis and HA-Ub (WT). After 24 h, the cells were treated with 20 μM MG for 12 h. Myc-tagged proteins were immunoprecipitated from the resultant cell lysates using a c-Myc antibody and Protein G Sepharose and were analyzed by western blot using HA or c-Myc antibodies. Total cell lysate (Input) was analyzed by western blot using a c-Myc antibody. (d) HEK293 cells were transiently co-transfected with Chac1-MycHis and HA-Ub (WT or K63). After 24 h, the cells were treated with 20 μM MG for 12 h. Myc-tagged proteins were immunoprecipitated from the resultant cell lysates using a c-Myc antibody and Protein G Sepharose and were analyzed by western blot using HA or c-Myc antibodies. The total cell lysate (Input) was analyzed by western blot using a c-Myc antibody. All experimental data were obtained from three independent cultures. Ub-Chac1 indicates the ubiquitinated Chac1.
Mutant Chac1 is still ubiquitinated in HEK293 cells and stabilized in a similar fashion as wild type Chac1 after substitution of all lysine residues for arginine
As Chac1 was directly ubiquitinated in HA-Ub overexpressing cells, we next investigated whether the ubiquitin molecule conjugated to the lysine residues in Chac1 as observed in many other proteins. To address this issue, we made a construct of mutated Chac1 (+1/+830; 5′-Chac1 (K0)-Myc) where all lysine residues were replaced with arginines because the mouse Chac1 protein has five lysine residues in the coding region (Fig. 1a). HEK293 cells were then transfected with 5′-Chac1 (WT)-Myc or 5′-Chac1 (K0)-Myc together with HA-Ub (WT) and incubated in the presence or absence of MG132 (Fig. 4a). The expression level of 5′-Chac1 (K0)-Myc as well as 5′-Chac1 (WT)-Myc was increased by co-transfection of HA-Ub (WT) and by MG132 treatment. Therefore, we examined whether the mutant Chac1 (K0) was ubiquitinated or not. As shown in Fig. 4b, the HA-positive molecules were detected in the Myc-immunoprecipitate derived from the cells co-expressing Chac1 (K0)-MycHis and HA-Ub (WT) in the presence of MG132.
Figure 4. Chac1 (K0) devoid of lysines is still ubiquitinated in HEK293 cells.
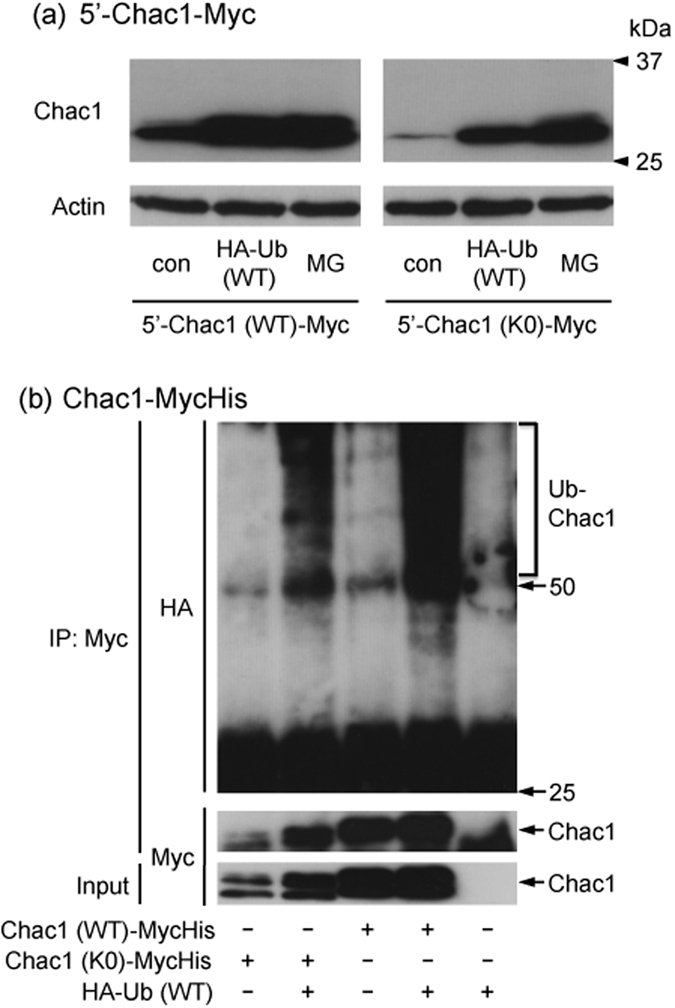
(a) HEK293 cells were transiently co-transfected with 5′-Chac1 (WT or K0)-Myc and HA-Ub (WT). After 24 h, the cells were treated with vehicle (con) or 20 μM MG for 12 h, and the resultant cell lysates were analyzed by western blot using c-Myc or Actin antibodies. (b) HEK293 cells were transiently co-transfected with Chac1 (WT or K0)-MycHis and HA-Ub (WT). After 24 h, the cells were treated with 20 μM MG for 12 h. Myc-tagged proteins were immunoprecipitated from the resultant cell lysates using a c-Myc antibody and Protein G Sepharose and were analyzed by western blot using HA or c-Myc antibodies. The total cell lysate (Input) was analyzed by western blot using c-Myc antibody. Figure 4a,b data were obtained from three and two independent cultures, respectively. Ub-Chac1 indicates the ubiquitinated Chac1.
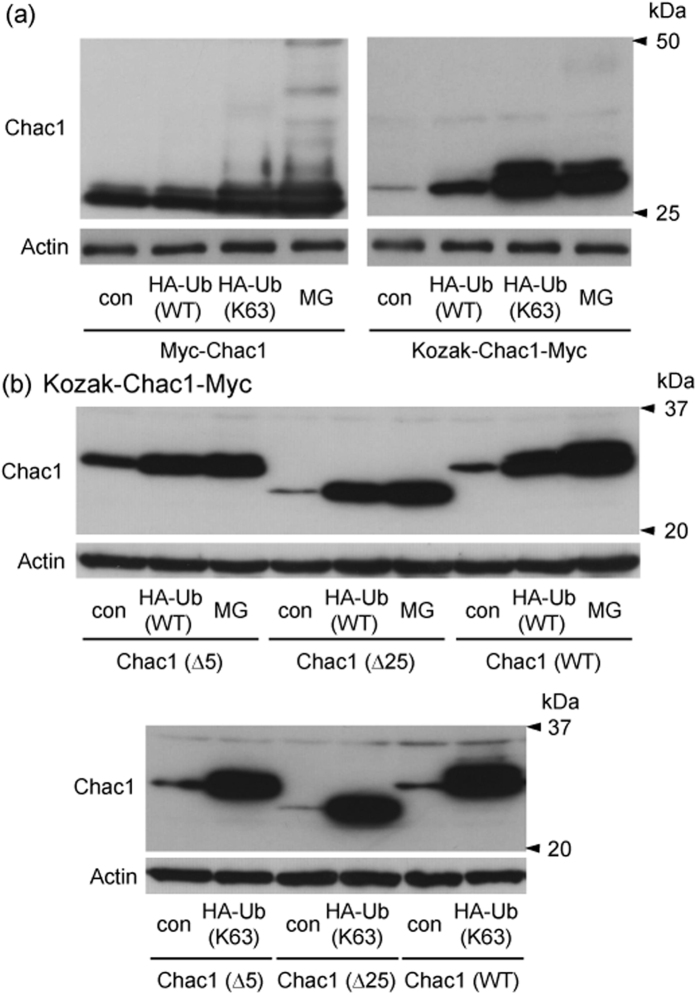
The N-terminal region of Chac1 is not associated with either stabilization by ubiquitin co-expression or degradation by the proteasome pathway
It has been reported that the degradation of p21, ERK3 and Cyclin G1 through the proteasome pathway is triggered by ubiquitination at the N-terminal methionine instead of lysine residues12,13,14,15. As these proteins were stabilized by the addition of the Myc-epitope to the N-terminal methionine, we constructed N-terminal Myc-tagged Chac1 (+162/+833; Myc-Chac1) with the Kozak sequence (CCCACC) just before the Myc-epitope sequence. We then examined whether the expression levels of Myc-Chac1 and Kozak-Chac1-Myc were affected by co-transfection of HA-Ub (WT or K63) or by MG132 treatment. As shown in Fig. 5a, the expression level of Myc-Chac1 was much higher than that of Kozak-Chac1-Myc in the absence of MG132, and the co-transfection of HA-Ub (K63) and the MG132 treatment also increased each of the expression levels, respectively. A ladder of higher molecular weight bands were observed in both cases of HA-Ub (K63) co-transfection and MG132 treatment in the Myc-Chac1-expressing cells. The Chac1 protein has a lysine residue at the position next to the first methionine (2 aa) and its N-terminal region contains a proline-rich region (10–24 aa), which is well conserved between the mouse and human genes. Therefore, we investigated whether the N-terminal region of Chac1 contributes to its stabilization. We constructed two types of N-terminal deletion constructs: Kozak-Chac1 (Δ5)-Myc and Kozak-Chac1 (Δ25)-Myc, lacking 2–6 aa and 2–26 aa, respectively. As shown in Fig. 5b, the expression of each truncated protein was similarly influenced by Ub-overexpression and MG132 treatment compared with WT Chac1.
Figure 5. The N-terminal region of Chac1 is not associated with either stabilization by ubiquitin co-expression or degradation by the proteasome pathway.
(a) HEK293 cells were transiently co-transfected with Myc-Chac1 or Kozak-Chac1 (WT)-Myc in addition to HA-Ub (WT or K63). After 24 h, the cells were treated with vehicle (con) or 20 μM MG for 12 h, and the resultant cell lysates were analyzed by western blot using c-Myc or Actin antibodies. (b) HEK293 cells were transiently co-transfected with Kozak-Chac1 (Δ5, Δ25 or WT)-Myc in addition to HA-Ub (WT or K63). After 24 h, the cells were treated with vehicle (con) or 20 μM MG for 12 h, and the resultant cell lysates were analyzed by western blot using c-Myc or Actin antibodies. All experimental data were obtained from three independent cultures.
Overexpression of Chac1 but not ΔChac1 significantly decreases the intracellular level of glutathione
It has been reported that Chac1 specifically degrades intracellular glutathione through its catalytic active site at E116 in mice or E115 in humans30,34,35. Finally, to determine whether ΔChac1 translated from the second methionine codon has catalytic activity, we transfected ΔChac1-Myc, Chac1-Myc and 5′-Chac1-Myc into HEK293 cells. As shown in Fig. 6a, the expression level of each type of Chac1 protein was found to be quite different. Chac1-Myc and 5′-Chac1-Myc significantly decreased the intracellular level of glutathione to a similar extent (**p < 0.01), whereas ΔChac1-Myc did not degrade glutathione even though it contains a catalytic active site (E116) (Fig. 6b).
Figure 6. Chac1 overexpression significantly decreases the level of intracellular glutathione whereas ΔChac1 fails to change it.
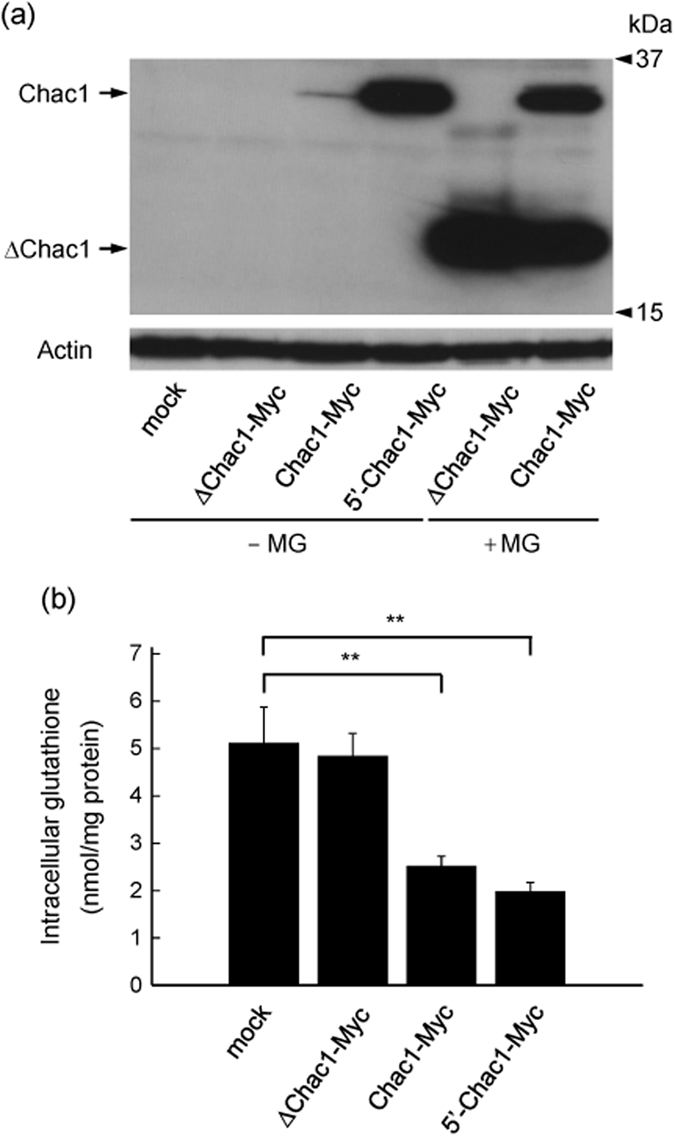
(a) HEK293 cells transfected with pcDNA3.1 (mock), ΔChac1-Myc, Chac1-Myc or 5′-Chac1-Myc. After 24 h, the cells were treated with vehicle (-MG) or 20 μM MG for 12 h, and the resultant cell lysates were analyzed by western blot using c-Myc or Actin antibodies. Experimental data were obtained from three independent cultures. (b) HEK293 cells were transfected with pcDNA3.1 (mock), ΔChac1-Myc, Chac1-Myc or 5′-Chac1-Myc. After 36 h, cells were collected, and the level of intracellular glutathione was measured. Values represent the mean ± SD obtained from three independent cultures. **p < 0.01 using one way-ANOVA followed by Tukey’s test.
Discussion
In this study, we show that the Kozak-like sequence in the 5′UTR region of the Chac1 mRNA increases the expression level of the Chac1 protein and is responsible for the translation of the protein from the first methionine codon. In the absence of this sequence, the second ATG codon of the Chac1 mRNA can become a translation start site to produce the short form (ΔChac1). Our results also indicate that the expression level of the Chac1 protein is predominantly regulated by the proteasome pathway. Though it is well known that protein degradation by the proteasome pathway is followed by the sequential conjugation of ubiquitin molecules catalyzed by specific ubiquitin ligases, the co-transfection of the ubiquitin constructs WT, K48 or K63 increased the expression level of the Chac1 protein (Fig. 3). Surprisingly, the expression levels of the mutant Chac1 protein of K0 as well as those of Δ5 and Δ25 are up-regulated by ubiquitin co-expression (Figs 4 and 5). We also reveal that the intracellular level of glutathione is significantly decreased by the overexpression of full-length Chac1 but not ΔChac1 containing the active catalytic site E116.
We previously reported that the Chac1 protein is stabilized by MG132 treatment in Neuro2a cells transiently transfected with the Chac1 construct (+162/+833)28. Consistent with our previous report, the expression level of the Chac1-Myc protein was elevated by MG132 in overexpressing HEK293 cells and the intrinsic Chac1 protein was also increased in the presence of MG132 (Fig. 1b,c). Thus, endogenous Chac1 as well as exogenous one was similarly regulated by the proteasome pathway. In contrast to the MG132-mediated stabilization of Chac1 in a post-translational manner (Fig. 1d), only a small portion of Chac1 was found to be degraded by autophagy because treatment with Baf and CMA, which are known to attenuate autophagic degradation through the inhibition of lysosomal acidification, slightly increased the expression level of Chac1 (Fig. 1e). Treatment with ALLN under the present experimental conditions may reflect its influence on proteasomal activity as ALLN is commonly used as a calpain I inhibitor and is reported to suppress trypsin-like, chymotrypsin-like and caspase-like peptidases in the proteasomes43,44,45.
In this study, we detected two proteins around 30 kDa as Chac1. We therefore investigated an effect of a N-glycosylation inhibitor, Tm, on the expression of Chac1 protein, but the band shift was not observed (Fig. 1b). Additionally, each type of Chac1 having a Myc-epitope at the N- or C-terminus showed two bands by western blot (Fig. 5a), suggesting that the Chac1 protein could not be cleaved around the either terminal. Considering the molecular size of ubiquitin, neither of bands seems to be ubiquitin-adducts. It is reported that various kinds of proteins are subject to a variety of post-translational modifications (e.g., phosphorylation, acetylation and so on) to modulate their functions. Therefore, we intend to characterize the two bands in more detail to understand the enzymatic regulation of Chac1 in our future study.
It has been reported that the Kozak sequence adjacent to the ATG codon facilitates translation46. Comparing the nucleotide sequences of Chac1 5′UTR among mouse, rat and human, the Kozak-like sequence and N-terminal region of these Chac1 genes are well conserved (Supplementary Fig. S1), and we revealed that the Kozak-like sequence plays an important role in facilitating the expression of Chac1 protein. With a view to revealing whether Chac1 is translationally regulated by the Kozak-like sequence, we compared the amount of Chac1 mRNA derived from each of the transfected genes (Supplementary Fig. S2). As a result, the Chac1 mRNAs from Kozak-Chac1-Myc or Chac1-Myc were expressed to the same extent, however, the expression levels of 5′-Chac1-Myc or Δ5′-Chac1-Myc were lower than those of Kozak-Chac1-Myc or Chac1-Myc, respectively. These results indicated that other regions of Chac1 5′UTR in addition to its Kozak-like sequence might contribute to the Chac1 expression. We here demonstrated that the Kozak-like sequence has a crucial role in enhancing the Chac1 translation, but the regulation of Chac1 expression by its 5′UTR seems complicated.
Despite the increased expression of Chac1 by MG132, Chac1 was also up-regulated by ubiquitin co-expression, and it actually became ubiquitinated in HEK293 cells (Fig. 3). On the other hand, we observed that the ubiquitin-overexpression did not influence the proteasome activity. It is therefore unclear how Chac1 ubiquitination determines the fate of degradation or stabilization. In contrast to the transfected Chac1, the amount of intrinsic c-Myc remained almost unchanged by ubiquitin co-expression even though it was up-regulated by MG132 (Fig. 3a). Considering the higher transfection efficiency of HEK293 cells, we exclude the possibility that the overexpressed ubiquitin is not enough to increase the intrinsic c-Myc expression. These results imply that the stabilizations of Chac1 and c-Myc are regulated in a different manner even though both are proteasomal substrates. In addition, the overexpression of HA-Ub (K63) showed a prominent effect on Chac1 expression; however, the K63-ubiquitination of Chac1 detected by immunoprecipitation was weaker than the WT-ubiquitination. Chac1 stabilization by the HA-Ub (K63) co-expression might involve an unknown mechanism that remains to be determined. In some cases, such as the NF-κB and Nrf2 pathways, it has been demonstrated that proteasomal degradation of regulatory factors followed by their ubiquitination play a key role in the stabilization of the associated functional molecules47,48. On the other hand, it has been reported that the degradation and stabilization of the same protein [proteasome substrates such as Myc by SCF (Fbw7) and SCF (β-TrCP) and Cryptochromes by FBXL3 and FBXL21, respectively] are regulated by different ubiquitin ligase complexes through different linkage types of polyubiquitination49,50. These phenomena may explain why Chac1 was stabilized by ubiquitin co-expression despite its predominant degradation in the proteasome. Therefore, identification of the Chac1-specific ubiquitin ligases is necessary to uncover its post-translational regulation. Moreover, it has been reported that E3-E2 pairs, such as the U-box E3 ubiquitin ligase CHIP with the E2 ubiquitin conjugating enzyme UbcH5, form different types of polyubiquitin chains that contain all seven possible ubiquitin-isopeptide linkages and that the heterogeneous linkage inhibits the substrate protein degradation by the proteasome8. Therefore, further studies using different ubiquitin constructs may give an insight into understanding which type of polyubiquitin chain conjugates to stabilize or degrade the Chac1 protein.
Here, we demonstrated that the Chac1 mutant, where all lysine residues were replaced with arginine, met a similar fate as WT Chac1 under the same conditions of proteasome inhibition and Ub-overexpression. We furthermore constructed a Chac1 (K0) gene having a Myc-epitope in which a lysine residue was substituted with an arginine one (Chac1 (K0)-Myc(KR)His) though a Myc-epitope is often used to characterize protein ubiquitination and degradation as well as this study. This additional experiment showed that the expression of this lysineless Chac1 (K0)-Myc(KR)His is up-regulated by ubiquitin-overexpression and actually ubiquitinated in the presence of MG132 (Supplementary Fig. S3). These results support this finding that Chac1 ubiquitination might be actually lysine-independent. It was previously reported that the lysine substituted mutants of p21, ERK3 and Cyclin G1 were still ubiquitinated and degraded through the proteasome pathway in a similar fashion12,13,14,15. Collectively, Chac1 may be regulated by the non-canonical ubiquitination pathway. It has been reported that the N-terminal methionine of some substrate proteins is specifically ubiquitinated and that the addition of a Myc-epitope to the N-terminal methionine stabilizes the modified ubiquitin substrates11,12,13,14,15. In this study, the N-terminal Myc-tagged Chac1 (Myc-Chac1) and Kozak-Chac1-Myc were stabilized by the HA-Ub (K63) co-transfection and MG132 treatment, whereas the HA-Ub (WT) co-transfection hardly affected the expression level of Myc-Chac1 (Fig. 5a). On the other hand, Chac1 lacking the N-terminal region (Δ5 and Δ25) still responded to the overexpression of both HA-Ub (WT) and HA-Ub (K63), as well as MG132 treatment (Fig. 5b). These data suggest that the N-terminal region of Chac1 is not strongly associated with both stabilization through ubiquitination and degradation through the proteasome pathway even though Myc-Chac1 showed a slightly different result in this study. We also examined the property of Kozak-Chac1 without any tags, which was quite similar to that of C-terminal tagged Chac1 (Kozak-Chac1-Myc) (data not shown).
In the current study, the specific signals corresponding to ubiquitinated Chac1 were broadly detected around larger molecular weight by western blot, however its ubiquitinated forms remain to be determined. It has been reported that the hydroxyl groups of serine and threonine residues and even the thiol group of cysteine residues are potential sites for ubiquitination16,17,18,19. Therefore, these types of ubiquitination may participate in the stabilization and the proteasomal degradation of the Chac1 protein. We also analyzed the expression levels of serially deleted Chac1 (1–77 aa, 1–130 aa, 1–186 aa, 78–223 aa) and found that all deleted Chac1 proteins were stabilized under these experimental conditions (data not shown). Collectively, it is considered that there seems to be multiple ubiquitinated sites in the Chac1 protein molecule. Therefore, further characterization of the polyubiquitin chains and identification of the modified residues in Chac1 might give new insights into understanding the post-translational regulation of Chac1.
Under our present experimental conditions, the translation efficiency of 5′-Chac1-Myc was higher than that of Chac1-Myc; however, the difference in expression patterns as detected by western blot did not reflect the level of intracellular glutathione (Fig. 6). It has been reported that glutathione inhibits the enzymatic activity of γ-glutamylcysteine synthetase, the rate-limiting enzyme of glutathione biosynthesis; however, the reduction of glutathione releases γ-glutamylcysteine synthetase from this feedback inhibition51,52. Likewise, there might be some unknown mechanisms regulating Chac1 activity. Therefore, the expression level of Chac1 was not simply correlated with the level of intracellular glutathione as observed in this study. In contrast, ΔChac1 failed to degrade intracellular glutathione despite containing a catalytically active residue (E116). This result suggests that the N-terminal region of Chac1 (1–77 aa) is indispensable for the enzymatic activity of γ-glutamyl cyclotransferase to sustain its proper conformation and/or interaction with glutathione. Therefore, further analysis of the 3D protein structure of Chac1 is necessary to understand the function of this enzyme.
In conclusion, we have shown that the 5′UTR of Chac1 plays an important role in expressing the full-length and short form of Chac1, and the expression level of Chac1 protein is predominantly regulated by its ubiquitination. As Chac1 is known to attenuate the intracellular level of glutathione responsible for healing several pathological conditions induced by oxidative stress53,54,55, uncovering characteristic features of Chac1 will contribute to a novel therapeutic agent against oxidative stress-related diseases.
Materials and Methods
Materials
Thapsigargin (Tg), tunicamycin (Tm), cycloheximide (CHX), bafilomycin A1 (Baf), and N-Acetyl-L-leucyl-L-leucyl-L-norleucinal (ALLN) were obtained from Sigma-Aldrich (U.S.A.). MG132 (MG) and Suc-LLVY-MCA were purchased from Peptide Institute (Japan). Concanamycin A (CMA) was obtained from Wako (Japan). Antibodies against Chac1, c-Myc (9E10), HA, LC3 and Actin were purchased from Abcam (UK), Santa Cruz Biotechnology (U.S.A.), Clontech Laboratories (U.S.A.), Medical & Biological Laboratories (Japan) and Calbiochem (U.S.A.), respectively.
Construction of plasmids
Mouse Chac1 cDNA (NCBI accession No. NM_026929) was amplified from a C57BL/6 mouse brain-derived cDNA library by polymerase chain reaction (PCR) and the Chac1 coding region (+162/+833) was cloned into pcDNA3.1 (pcDNA3.1-Chac1). To detect Chac1 protein by the c-Myc antibody, a Myc-epitope was directly fused to the C-terminus of Chac1 in pcDNA3.1-Chac1 to generate pcDNA3.1-Chac1-Myc by PCR (+162/+830; Chac1-Myc) using anti-sense primer 5′-TCACAGATCCTCTTCTGAGATGAGTTTTTGTTCGGTCAGTGCCAGAGGC-3′ (Fig. 1a). Four Chac1 expression constructs comprising the entire 5′ untranslated region (5′UTR) (+1/+830; 5′-Chac1-Myc), a truncated 5′UTR (+82/+830; Δ5′-Chac1-Myc), the Kozak-like sequence just before the translation start site (+156/+830; Kozak-Chac1-Myc) and a short form of mouse Chac1 translated from the second methionine (+393/+830; ΔChac1-Myc) were generated by PCR using pcDNA3.1-Chac1-Myc as the template (Fig. 1a). We further constructed three mutants (M1I, M78I and M187I; Fig. 2b) using pcDNA3.1-Chac1-Myc as the PCR template to replace methionine with isoleucine residues and mutant 5′-Chac1 (K0)-Myc (Fig. 4a) using pcDNA3.1-5′-Chac1-Myc as the PCR template to replace all lysine residues in the coding region with arginine. In addition, we generated Chac1 (WT, K0)-MycHis constructs by subtracting the stop codon and the 5′UTR from pcDNA3.1-5′-Chac1 (WT, K0)-Myc by PCR and cloning the resultant cDNAs into pcDNA3.1-MycHis. Two N-terminal deletion constructs lacking the 2–6 amino acid (aa) (Kozak-Chac1 (Δ5)-Myc) and 2–26 aa (Kozak-Chac1 (Δ25)-Myc) sequences of Chac1 were generated by PCR using pcDNA3.1-Kozak-Chac1-Myc as the template (Fig. 5b). To construct the N-terminal Myc-tagged Chac1 (Myc-Chac1; Fig. 5a), the pcDNA3.1-Chac1 coding region was subcloned into a pCMV-Myc vector using EcoR I to fuse the Myc-epitope to the Chac1 N-terminus, followed by subcloning into pcDNA3.1 using Xba I. The N-terminal HA-tagged wild type and single-lysine mutant ubiquitin constructs were generously provided by Dr. Kah-Leong LIM42.
Cell culture and treatment
The human embryonic kidney cell line, HEK293, was maintained in Dulbecco’s Modified Eagle’s Medium containing 8% fetal bovine serum and 1% penicillin-streptomycin. Transfections of constructs used in this study were performed with polyethylenimine “Max” (Polysciences, U.S.A.)56. HEK293 cells were treated with Tg (0.1 μM), Tm (2 μg/mL), MG (20 μM), CHX (10 μg/mL), Baf (50 nM), CMA (50 nM) and ALLN (10 μM) for the indicated times.
Western blot analysis
Cells were lysed in homogenate butter [20 mM Tris-HCl (pH 8.0) containing 137 mM NaCl, 2 mM EDTA, 10% glycerol, 1% Triton X-100, 1 mM PMSF, 10 μg/mL leupeptin and 10 μg/mL pepstatin A]. After the protein concentration was determined, each cell lysate was dissolved in sodium dodecyl sulfate (SDS)-Laemmli sample buffer [62.5 mM Tris–HCl (pH 6.8), 2% SDS and 10% glycerol]. Equal amounts of cell lysate were separated on 12.5 or 15% SDS–polyacrylamide electrophoresis gels, immunoblotted onto polyvinylidene difluoride membranes (Merck Millipore, Germany) and identified by an ECL Detection System (GE Healthcare Bioscience, U.S.A.) or Western Blotting Substrate Plus (Thermo Fisher Scientific, U.S.A.) using antibodies against Chac1 (1:1,000), c-Myc (1:1,000), HA (1:1,000), LC3 (1:3,000) or Actin (1:5,000). Band intensities were analyzed by Image J software (National Institutes of Health, U.S.A.).
Immunoprecipitation
Transfected HEK293 cells in 60 mm dishes were harvested and lysed in lysis buffer [20 mM Tris-HCl (pH 8.0) containing 150 mM NaCl, 1% Nonidet P-40, 1 mM EDTA, 1 mM PMSF, 10 μg/mL leupeptin, and 10 μg/mL pepstatin A]. Lysates were cleared by centrifugation for 6 min at 12,000 × g and soluble proteins were immunoprecipitated using 2 μg of c-Myc antibody and Protein G Sepharose (GE Healthcare Bioscience, U.S.A.). Proteins binding to the resin were washed three times in wash buffer [20 mM Tris-HCl (pH 8.0) containing 150 mM NaCl, 0.2% Nonidet P-40, 1 mM EDTA and 1 mM PMSF], eluted by SDS-Laemmli sample buffer and subsequently analyzed by western blot.
Measurement of glutathione
The glutathione content in cells was measured fluorometrically according to the method of Hisson and Hilf57. In brief, cells were cultured in Dulbecco’s Modified Eagle’s Medium and collected by centrifugation. Cell pellets were resuspended in 0.1 M sodium phosphate buffer (pH 8.0) containing 5 mM EDTA and 25% (w/v) metaphosphoric acid solution. After centrifugation for 10 min at 10,000 × g, each supernatant was incubated with 0.1 M phosphate buffer (pH 8.0) containing 5 mM EDTA and 0.1% o-phthalaldehyde for 15 min. Fluorescence of each sample was measured by fluorospectrometry using excitation at 350 nm and emission at 420 nm.
Proteasome activity assay
The chymotryptic proteasomal activity including 26S proteasome activity in transfected HEK 293 cells was measured fluorometrically according to the method of Mark FP et al.58. In brief, the cells were cultured in Dulbecco’s Modified Eagle’s Medium and collected by centrifugation. The cell pellets were resuspended in lysis buffer [PBS containing 1% (v/v) Triton X-100] and incubated for 30 min on ice. After centrifugation for 15 min at 16,500 × g, each supernatant was incubated with assay solution [30 mM Tris-HCl (pH 7.6) containing 2 mM MgCl2, 10 mM NaCl, 10 mM KCl, 0.5 mM dithiothreitol, 1 mM ATP, and 100 μM Suc-LLVY-MCA] for 60 min at 37 °C in the presence or absence of 20 μM MG132. Fluorescence of each sample was measured by fluorospectrometry using excitation at 360 nm and emission at 450 nm.
Statistical analysis
The results were expressed as the means ± SD of the indicated number. Statistical analysis was carried out by one way-ANOVA followed by Tukey’s test. **p < 0.01 was considered to be statistically significant.
Additional Information
How to cite this article: Nomura, Y. et al. Translational and post-translational regulation of mouse cation transport regulator homolog 1. Sci. Rep. 6, 28016; doi: 10.1038/srep28016 (2016).
Supplementary Material
Acknowledgments
We thank Dr. Kah-Leong LIM (Department of Physiology, National University of Singapore) for the HA-Ub (WT, K48 and K63) plasmids, and members of the Kiuchi Laboratory for helpful discussion. This work is partly supported by the OGAWA Science and Technology Foundation (to K.O.).
Footnotes
Author Contributions Y.N. and K.O. designed and performed the experiments. Y.N., K.K. and K.O. wrote the main manuscript text and Y.N. prepared figures. All authors conceived all experiments and reviewed the manuscript.
References
- Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem 78, 477–513 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K. L. & Lim G. G. K63-linked ubiquitination and neurodegeneration. Neurobiol Dis 43, 9–16 (2011). [DOI] [PubMed] [Google Scholar]
- Deng L. et al. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103, 351–361 (2000). [DOI] [PubMed] [Google Scholar]
- Lipford J. R., Smith G. T., Chi Y. & Deshaies R. J. A putative stimulatory role for activator turnover in gene expression. Nature 438, 113–116 (2005). [DOI] [PubMed] [Google Scholar]
- Bhoj V. G. & Chen Z. J. Ubiquitylation in innate and adaptive immunity. Nature 458, 430–437 (2009). [DOI] [PubMed] [Google Scholar]
- Pickart C. M. Ubiquitin in chains. Trends Biochem Sci 25, 544–548 (2000). [DOI] [PubMed] [Google Scholar]
- Wu-Baer F., Lagrazon K., Yuan W. & Baer R. The BRCA1/BARD1 heterodimer assembles polyubiquitin chains through an unconventional linkage involving lysine residue K6 of ubiquitin. J Biol Chem 278, 34743–34746 (2003). [DOI] [PubMed] [Google Scholar]
- Kim H. T. et al. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J Biol Chem 282, 17375–17386 (2007). [DOI] [PubMed] [Google Scholar]
- Thrower J. S., Hoffman L., Rechsteiner M. & Pickart C. M. Recognition of the polyubiquitin proteolytic signal. EMBO J 19, 94–102 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan J. A., Kim H. T., Ting L., Gygi S. P. & Goldberg A. L. Why do cellular proteins linked to K63-polyubiquitin chains not associate with proteasomes. EMBO J 32, 552–565 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell G. S. & Philpott A. Non-canonical ubiquitylation: mechanisms and consequences. Int J Biochem Cell Biol 45, 1833–1842 (2013). [DOI] [PubMed] [Google Scholar]
- Bloom J., Amador V., Bartolini F., DeMartino G. & Pagano M. Proteasome-mediated degradation of p21 via N-terminal ubiquitinylation. Cell 115, 71–82 (2003). [DOI] [PubMed] [Google Scholar]
- Coulombe P., Rodier G., Bonneil E., Thibault P. & Meloche S. N-Terminal ubiquitination of extracellular signal-regulated kinase 3 and p21 directs their degradation by the proteasome. Mol Cell Biol 24, 6140–6150 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Okamoto K., Peart M. J. & Prives C. Lysine-independent turnover of cyclin G1 can be stabilized by B’alpha subunits of protein phosphatase 2A. Mol Cell Biol 29, 919–928 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A. & Ben-Saadon R. N-terminal ubiquitination: more protein substrates join In. Trends Cell Biol 14, 103–106 (2004). [DOI] [PubMed] [Google Scholar]
- Wang X., Herr R. A., Chua W. J., Lybarger L., Wiertz E. J. & Hansen T. H. Ubiquitination of serine, threonine, or lysine residues on the cytoplasmic tail can induce ERAD of MHC-I by viral E3 ligase mK3. J Cell Biol 177, 613–624 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokarev A. A., Munguia J. & Guatelli J. C. Serine-threonine ubiquitination mediates downregulation of BST-2/tetherin and relief of restricted virion release by HIV-1 Vpu. J Virol 85, 51–63 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell K. & Coscoy L. Ubiquitination on nonlysine residues by a viral E3 ubiquitin ligase. Science 309, 127–130 (2005). [DOI] [PubMed] [Google Scholar]
- Léon S. & Subramani S. A conserved cysteine residue of Pichia pastoris Pex20p is essential for its recycling from the peroxisome to the cytosol. J Biol Chem 282, 7424–7430 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargalovic P. S. et al. Identification of inflammatory gene modules based on variations of human endothelial cell responses to oxidized lipids. Proc Natl Acad Sci USA 103, 12741–12746 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungrue I. N., Pagnon J., Kohannim O., Gargalovic P. S. & Lusis A. J. CHAC1/MGC4504 is a novel proapoptotic component of the unfolded protein response, downstream of the ATF4-ATF3-CHOP cascade. J Immunol 182, 466–476 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower N. I. & Johnston I. A. Discovery and characterization of nutritionally regulated genes associated with muscle growth in Atlantic salmon. Physiol Genomics 42, 114–130 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magne L. et al. ATF4 and the integrated stress response are induced by ethanol and cytochrome P450 2E1 in human hepatocytes. J Hepatol 54, 729–737 (2011). [DOI] [PubMed] [Google Scholar]
- Galluzzi L. et al. Induction of endoplasmic reticulum stress response by the indole-3-carbinol cyclic tetrameric derivative CTet in human breast cancer cell lines. PLos One 7, e43249 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvik L. K. et al. The duration of gastrin treatment affects global gene expression and molecular responses involved in ER stress and anti-apoptosis. BMC Genomics 14, 429 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo J. H. et al. Farnesol activates the intrinsic pathway of apoptosis and the ATF4-ATF3-CHOP cascade of ER stress in human T lymphoblastic leukemia Molt4 cells. Biochem Pharmacol 97, 256–268 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon S. J. et al. Pharmacological inhibition of cystine-glutamate exchange induces endoplasmic reticulum stress and ferroptosis. eLife 3, e02523 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh-Hashi K. et al. Transcriptional and post-translational regulation of mouse cation transport regulator homolog 1. Mol Cell Biochem 380, 97–106 (2013). [DOI] [PubMed] [Google Scholar]
- Romanoski C. E. et al. Network for activation of human endothelial cells by oxidized phospholipids: a critical role of heme oxygenase 1. Circ Res 109, e27–41 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford R. R. et al. Human CHAC1 Protein Degrades Glutathione, and mRNA Induction Is Regulated by the Transcription Factors ATF4 and ATF3 and a Bipartite ATF/CRE Regulatory Element. J Biol Chem 290, 15878–15891 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel G. et al. Elevated mRNA expression of CHAC1 splicing variants is associated with poor outcome for breast and ovarian cancer patients. Br J Cancer 106, 189–198 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tattoli I. et al. Amino acid starvation induced by invasive bacterial pathogens triggers an innate host defense program. Cell Host Microbe 11, 563–575 (2012). [DOI] [PubMed] [Google Scholar]
- Tang C., Lan D., Zhang H., Ma J. & Yue H. Transcriptome analysis of duck liver and identification of differentially expressed transcripts in response to duck hepatitis A virus genotype C infection. PLos One 8, e71051 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A. et al. Mammalian proapoptotic factor ChaC1 and its homologues function as γ-glutamyl cyclotransferases acting specifically on glutathione. EMBO Rep 13, 1095–1101 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunoda S. et al. Intact protein folding in the glutathione-depleted endoplasmic reticulum implicates alternative protein thiol reductants. eLife 3, e03421 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi Z. et al. Botch promotes neurogenesis by antagonizing Notch. Dev Cell 22, 707–720 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi Z. et al. Botch is a γ-glutamyl cyclotransferase that deglycinates and antagonizes Notch. Cell Rep 7, 681–688 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W. K. et al. Induction of autophagy by proteasome inhibitor is associated with proliferative arrest in colon cancer cells. Biochem Biophys Res Commun 374, 258–263 (2008). [DOI] [PubMed] [Google Scholar]
- Kabeya Y. et al. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci 117, 2805–2812 (2004). [DOI] [PubMed] [Google Scholar]
- Farrell A. S. & Sears R. C. MYC degradation. Cold Spring Harb Perspect Med 4, a014365 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hann S. R. Role of post-translational modifications in regulating c-Myc proteolysis, transcriptional activity and biological function. Semin Cancer Biol 16, 288–302 (2006). [DOI] [PubMed] [Google Scholar]
- Lim K. L. et al. Parkin mediates nonclassical, proteasomal-independent ubiquitination of synphilin-1: implications for Lewy body formation. J Neurosci 25, 2002–2009 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber A. et al. Proteasome inhibitors modulate chemokine production in lung epithelial and monocytic cells. Eur Respir J 24, 40–48 (2004). [DOI] [PubMed] [Google Scholar]
- Hong M., Li M., Mao C. & Lee A. S. Endoplasmic reticulum stress triggers an acute proteasome-dependent degradation of ATF6. J Cell Biochem 92, 723–732 (2004). [DOI] [PubMed] [Google Scholar]
- Bedford L., Paine S., Sheppard P. W., Mayer R. J. & Roelofs J. Assembly, structure, and function of the 26S proteasome. Trends Cell Biol 20, 391–401 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. J Mol Biol 196, 947–950 (1987). [DOI] [PubMed] [Google Scholar]
- Li W. & Kong A. N. Molecular mechanisms of Nrf2-mediated antioxidant response. Mol Carcinog 48, 91–104 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz I. E. & Dixit V. M. Signaling to NF-kappaB: regulation by ubiquitination. Cold Spring Harb Perspect Biol 2, a003350 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov N., Schülein C., Jaenicke L. A. & Eilers M. Ubiquitylation of the amino terminus of Myc by SCF(β-TrCP) antagonizes SCF(Fbw7)-mediated turnover. Nat Cell Biol 12, 973–981 (2010). [DOI] [PubMed] [Google Scholar]
- Hirano A. et al. FBXL21 regulates oscillation of the circadian clock through ubiquitination and stabilization of cryptochromes. Cell 152, 1106–1118 (2013). [DOI] [PubMed] [Google Scholar]
- Franklin C. C. et al. Structure, function, and post-translational regulation of the catalytic and modifier subunits of glutamate cysteine ligase. Mol Aspects Med 30, 86–98 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y. et al. Interaction between the catalytic and modifier subunits of glutamate-cysteine ligase. Biochem Pharmacol 74, 372–381 (2007). [DOI] [PubMed] [Google Scholar]
- Traverso N. et al. Role of glutathione in cancer progression and chemoresistance. Oxid Med Cell Longev 2013, 972913 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D. H. & Kang S. W. Targeting cellular antioxidant enzymes for treating atherosclerotic vascular disease. Biomol Ther (Seoul) 21, 89–96 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias V., Junn E. & Mouradian M. M. The role of oxidative stress in Parkinson’s disease. J Parkinsons Dis 3, 461–491 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikiji T. et al. A highly sensitive assay of IRE1 activity using the small luciferase NanoLuc: Evaluation of ALS-related genetic and pathological factors. Biochem Biophys Res Commun 463, 881–887 (2015). [DOI] [PubMed] [Google Scholar]
- Hissin P. J. & Hilf R. A fluorometric method for determination of oxidized and reduced glutathione in tissues. Anal Biochem 74, 214–226 (1976). [DOI] [PubMed] [Google Scholar]
- Mark F. P. et al. The proteasomal subunit S6 ATPase is a novel synphilin-1 interacting protein–implications for Parkinson’s disease. FASEB J 21, 1759–1767 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.