The unequal C—C bond lengths in the six-membered ring of the C6H7N2O2 + cation of the title compound can be understood in terms of two separate delocalized systems.
Keywords: conjugation, anti-aromatic, hydrogen bonds, layered structure, crystal structure
Abstract
In the cation of the title hydrated molecular salt, C6H7N2O2 +·Cl−·H2O, the six-membered ring shows unequal bond lengths consistent with delocalization of electrons over two separate 6π systems with single bonds between them. In the crystal, the components are linked by N—H⋯Cl, N—H⋯O, O—H⋯Cl and O—H⋯O hydrogen bonds, generating double layers propagating in (100).
Chemical context
In the course of our ongoing studies (Plater & Harrison, 2013 ▸, 2014a
▸,b
▸; Plater & Jackson, 2014 ▸) on new conjugated products obtained from the oxidation of aromatic amines, we attempted the oxidation of 1,2,4,5-tetraaminobenzene, 1. As long ago as 1887, it was demonstrated (Nietzki & Hagenbach, 1887 ▸) that this compound undergoes aerial oxidation to form 2,5-diamino-1,4-benzoquinonediimine, 2. More recently, Braunstein et al. (2003 ▸) have studied the oxidation of compound 1 and the related compound 2,4-diaminoresorcinol, 3, to synthesize (1E)-N-(2,2-dimethylpropyl)-5-[(2,2-dimethylpropyl)amino]-2-hydroxy-4-oxocyclohexa-2,5-dien-1-iminium chloride, 4, which generates the zwitterion 5 when treated with base.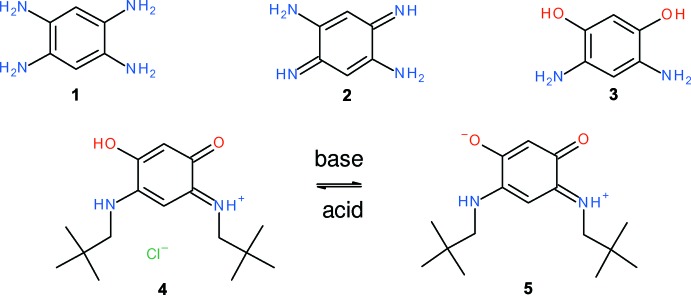
By careful oxidation of the tetrahydrochloride salt of amine 1 with potassium dichromate, we isolated and crystallized the chloride salt of the parent 3-amino-4-hydroxy-6-oxocyclohexa-2,4-dien-1-iminium cation, 8, as a monohydrate [C6H7N2O2
+·Cl−·H2O, (I)] in the form of purple needles. This reaction must proceed via the elusive intermediate 6 which spontaneously hydrolyses. The first hydrolysis product should be intermediate 7. This contains a conjugated iminium salt and a vinylogous amide, which must hydrolyse rapidly, possibly because of the stability of the acidic enol formed. It appears to be a rapid hydrolysis for an amide under mild conditions and so stabilization of a tetrahedral intermediate by the positive iminium salt might occur.
Structural commentary
The asymmetric unit of (I) consists of one essentially planar C6H7N2O2
+ cation (r.m.s. deviation for the non-hydrogen atoms = 0.028 Å), a chloride counter-ion and a water molecule of crystallization (Fig. 1 ▸). Despite being a nominal 6π aromatic system, the bond lengths of the C1–C6 ring in (I) are far from equal and are split into three groups of two: the shortest are C1—C6 [1.354 (5)] and C3—C4 [1.381 (5)], followed by C4—C5 [1.406 (5)] and C1—C2 [1.436 (5) Å]. Finally, the C2—C3 [1.532 (4)] and C5—C6 [1.500 (5) Å] lengths are those expected for a C—C σ bond.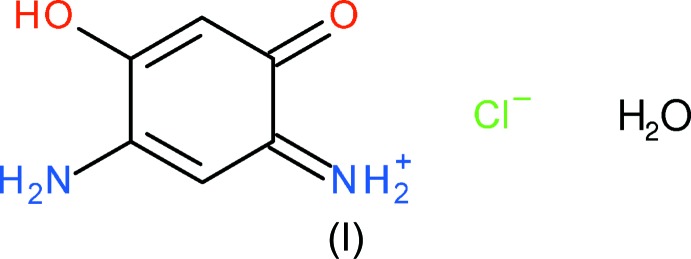
Figure 1.
The molecular structure of (I) showing 50% displacement ellipsoids. Hydrogen bonds are shown as double-dashed lines.
The short C3—C4 and C4—C5 bonds correlate with the approximately equal C3—N1 [1.320 (4)] and C5—N2 [1.306 (4) Å] bond lengths, which imply equal delocalization of the positive charge of the cation over atoms N1 and N2, mediated via the C—N and C—C bonds between them. In terms of the ‘oxygen side’ of the cation, the C6—O2 bond [1.320 (4) Å] is short for a C—O single bond whereas C2—O1 [1.227 (4) Å] is slightly lengthened for a nominal C=O double bond. This in combination with the C1—C2 and C1—C6 bond lengths again implies a degree of delocalization over these five atoms. However, the long C2—C3 and C5—C6 bonds imply little, if any, conjugation between the two delocalized components (O2/C6/C1/C2/O1 and N2/C5/C4/C3/N1) of the cation.
The cation features two intramolecular N—H⋯O hydrogen bonds, viz. N1—H2n⋯O1 and N2—H4n⋯O2 (Table 1 ▸), which both close S(5) rings.
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H2n⋯O1 | 0.81 (5) | 2.33 (5) | 2.653 (4) | 105 (4) |
| N2—H4n⋯O2 | 0.84 (4) | 2.23 (5) | 2.595 (4) | 107 (4) |
| N1—H1n⋯Cl1 | 0.80 (5) | 2.45 (5) | 3.238 (3) | 169 (4) |
| N1—H2n⋯O3i | 0.81 (5) | 2.25 (5) | 3.011 (4) | 156 (4) |
| N2—H3n⋯Cl1ii | 0.93 (4) | 2.22 (4) | 3.149 (3) | 177 (4) |
| N2—H4n⋯Cl1iii | 0.83 (5) | 2.44 (5) | 3.231 (3) | 158 (4) |
| O2—H1o⋯O3 | 0.92 (5) | 1.65 (5) | 2.548 (4) | 165 (4) |
| O3—H1w⋯O1iv | 0.88 (5) | 1.98 (5) | 2.801 (4) | 154 (4) |
| O3—H2w⋯Cl1v | 1.00 (5) | 2.11 (5) | 3.116 (3) | 176 (4) |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  ; (iv)
; (iv)  ; (v)
; (v)  .
.
Supramolecular features
In the crystal, the components are linked by N—H⋯Cl, N—H⋯O, O—H⋯Cl and O—H⋯O hydrogen bonds (Table 1 ▸). If the cation and chloride anion are considered together, then [001] chains arise (Fig. 2 ▸) in which adjacent cations are related to each other by c-glide symmetry. Each link in the chain comprises two cations and two anions and  (12) loops are apparent.
(12) loops are apparent.
Figure 2.
Detail of the crystal structure of (I) showing the formation of [001] chains of cations and chloride ions linked by N—H⋯Cl hydrogen bonds. Symmetry codes as in Table 1 ▸.
When the cation and water molecule are considered together, an [001] chain also arises (Fig. 3 ▸). The water molecule plays a key role in terms of both accepting hydrogen bonds from O2 and N1 and donating a hydrogen bond to O1 (it also acts as a donor to the chloride ion). The end result is a chain featuring  (12) loops (counted via the intramolecular N1—H2n⋯O1 hydrogen bond).
(12) loops (counted via the intramolecular N1—H2n⋯O1 hydrogen bond).
Figure 3.
Detail of the crystal structure of (I) showing the formation of [001] chains of cations and water molecules linked by O—H⋯O and N—H⋯O hydrogen bonds. Symmetry codes as in Table 1 ▸.
When all components are considered together, (100) double sheets result (Fig. 4 ▸), with the water-O3—H2w⋯Cl1 hydrogen bond providing the key link between the sheets. Overall, the chloride ion accepts four hydrogen bonds (three N—H⋯Cl and one O—H⋯Cl interactions) in an irregular geometry.
Figure 4.
The packing in (I) viewed along [001] showing the formation of (100) double layers.
Database survey
The compound (1E)-N-(2,2-dimethylpropyl)-5-[(2,2-dimethylpropyl)amino]-2-hydroxy-4-οxocyclohexa-2,5-dien-1-iminium chloride chloroform monosolvate (CCDC refcode: VASVER; Braunstein et al., 2003 ▸) was noted in the chemical context section above: these authors discuss its electronic structure in detail including its potentially anti-aromatic character. The crystal structure of the parent unprotonated zwitterion 3-oxo-4-amino-6-iminiophenolate monohydrate (HAZQUV; Yang et al., 2005 ▸) is known as are those of a number of its alkylated/functionalized derivatives (Braunstein et al., 2009 ▸; Tamboura et al., 2009 ▸; Kauf & Braunstein, 2011 ▸) and metal complexes (Paretzki et al., 2010 ▸). The carbon–carbon bond lengths in the six-membered ring in all these compounds are similar to those seen in (I).
Synthesis and crystallization
1,2,4,5-Benzenetetraamine tetrahydrochloride (200 mg, 0.7 mmol) in distilled water (75 ml) was treated with an excess of K2Cr2O7 (140 mg, 0.48 mmol, 0.6 eq) and stirred at room temperature for 24 h. The brown mixture was neutralized with NaHCO3 giving a brown or red precipitate, which was then extracted with CH2Cl2 (10 × 50 ml). The yellow extracts were combined, decanted, then stirred with methanol (50 ml) containing five drops of conc. HCl(aq). The yellow solution turned purple. This was evaporated to dryness, then the product was dissolved in methanol (50 ml) to yield a red solution and recrystallized by slow evaporation to leave the title compound (15 mg, 8%) as purple needles: m.p. > 473 K; λmax (ethanol)/nm 503 (log ∊ 2.90) and 325(3.99); ν (diamond anvil)/cm−1 2953br, 1688s, 1547vs, 1401vs, 1310vs, 1251vs, 1141vs, 871vs, 853s, 711vs, 654vs, 579vs, 454s and 420s; m/z (orbitrap ASAP) 139.0498 (M +, 100%), C6H7N2O2 requires 139.0502. The UV/visible spectrum of (I) is shown in Fig. 5 ▸.
Figure 5.
UV/visible spectrum of (I) (2.3 × 10−4 M solution in ethanol).
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. The C-bound H atoms were geometrically placed (C—H = 0.95 Å) and refined as riding atoms. The N- and O-bound H atoms were located in difference maps and their positions were freely refined. The constraint U iso(H) = 1.2U eq(carrier) was applied in all cases. The crystal studied was found to be a twin with the components related by a 180° rotation about [001].
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C6H7N2O2·Cl·H2O |
| M r | 192.60 |
| Crystal system, space group | Monoclinic, P21/c |
| Temperature (K) | 100 |
| a, b, c (Å) | 6.3070 (7), 14.9614 (18), 8.9198 (11) |
| β (°) | 93.457 (1) |
| V (Å3) | 840.15 (17) |
| Z | 4 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.42 |
| Crystal size (mm) | 0.11 × 0.04 × 0.03 |
| Data collection | |
| Diffractometer | Rigaku Mercury CCD |
| Absorption correction | – |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 3102, 3102, 2789 |
| R int | ? |
| (sin θ/λ)max (Å−1) | 0.651 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.072, 0.159, 1.22 |
| No. of reflections | 3102 |
| No. of parameters | 131 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.57, −0.40 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989016005107/sj5497sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989016005107/sj5497Isup2.hkl
CCDC reference: 1470620
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
We thank the EPSRC National Crystallography Service (University of Southampton) for the data collection and the EPSRC National Mass Spectrometry Service (University of Swansea) for the HRMS data.
supplementary crystallographic information
Crystal data
| C6H7N2O2·Cl·H2O | F(000) = 400 |
| Mr = 192.60 | Dx = 1.523 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| a = 6.3070 (7) Å | Cell parameters from 1747 reflections |
| b = 14.9614 (18) Å | θ = 3.2–27.5° |
| c = 8.9198 (11) Å | µ = 0.42 mm−1 |
| β = 93.457 (1)° | T = 100 K |
| V = 840.15 (17) Å3 | Rod, purple |
| Z = 4 | 0.11 × 0.04 × 0.03 mm |
Data collection
| Rigaku Mercury CCD diffractometer | θmax = 27.6°, θmin = 2.7° |
| ω scans | h = −8→8 |
| 3102 measured reflections | k = −19→19 |
| 3102 independent reflections | l = −7→11 |
| 2789 reflections with I > 2σ(I) |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Hydrogen site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.072 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.159 | w = 1/[σ2(Fo2) + (0.0472P)2 + 1.7994P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.22 | (Δ/σ)max < 0.001 |
| 3102 reflections | Δρmax = 0.56 e Å−3 |
| 131 parameters | Δρmin = −0.39 e Å−3 |
| 0 restraints |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refined as a 2-component twin (180° rotation about [001]) |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.2403 (6) | 0.3972 (2) | 0.3386 (4) | 0.0152 (7) | |
| H1 | 0.2413 | 0.3419 | 0.2865 | 0.018* | |
| C2 | 0.2473 (6) | 0.3979 (2) | 0.4998 (4) | 0.0132 (7) | |
| C3 | 0.2536 (6) | 0.4888 (2) | 0.5792 (4) | 0.0120 (7) | |
| C4 | 0.2459 (6) | 0.5671 (2) | 0.4970 (4) | 0.0155 (7) | |
| H4 | 0.2493 | 0.6233 | 0.5468 | 0.019* | |
| C5 | 0.2332 (6) | 0.5632 (2) | 0.3392 (4) | 0.0133 (7) | |
| C6 | 0.2324 (6) | 0.4747 (2) | 0.2601 (4) | 0.0133 (7) | |
| N1 | 0.2648 (6) | 0.4838 (2) | 0.7271 (3) | 0.0165 (7) | |
| H1n | 0.265 (7) | 0.527 (3) | 0.779 (5) | 0.020* | |
| H2n | 0.270 (7) | 0.436 (3) | 0.770 (5) | 0.020* | |
| N2 | 0.2212 (6) | 0.6344 (2) | 0.2543 (4) | 0.0162 (7) | |
| H3n | 0.227 (7) | 0.690 (3) | 0.300 (5) | 0.019* | |
| H4n | 0.216 (7) | 0.629 (3) | 0.161 (5) | 0.019* | |
| O1 | 0.2459 (4) | 0.33037 (17) | 0.5778 (3) | 0.0184 (6) | |
| O2 | 0.2219 (5) | 0.48297 (18) | 0.1125 (3) | 0.0201 (6) | |
| H1o | 0.230 (7) | 0.430 (3) | 0.063 (5) | 0.024* | |
| Cl1 | 0.22638 (16) | 0.67344 (6) | 0.89828 (10) | 0.0208 (3) | |
| O3 | 0.3077 (5) | 0.34226 (17) | −0.0329 (3) | 0.0223 (6) | |
| H1w | 0.247 (8) | 0.294 (3) | −0.001 (5) | 0.027* | |
| H2w | 0.458 (8) | 0.334 (3) | 0.008 (6) | 0.027* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.019 (2) | 0.0121 (15) | 0.0149 (17) | 0.0016 (14) | 0.0001 (14) | −0.0023 (13) |
| C2 | 0.0134 (18) | 0.0119 (15) | 0.0146 (17) | −0.0007 (14) | 0.0013 (14) | −0.0015 (13) |
| C3 | 0.0101 (17) | 0.0119 (15) | 0.0137 (16) | 0.0012 (13) | −0.0030 (13) | −0.0036 (12) |
| C4 | 0.0170 (19) | 0.0134 (16) | 0.0161 (17) | −0.0001 (15) | 0.0001 (14) | −0.0038 (13) |
| C5 | 0.0118 (18) | 0.0113 (15) | 0.0168 (17) | −0.0006 (14) | 0.0005 (14) | 0.0021 (13) |
| C6 | 0.0105 (18) | 0.0179 (16) | 0.0112 (16) | 0.0006 (14) | −0.0014 (13) | −0.0031 (13) |
| N1 | 0.0243 (19) | 0.0150 (14) | 0.0101 (15) | 0.0015 (14) | −0.0005 (13) | −0.0001 (12) |
| N2 | 0.0246 (19) | 0.0122 (13) | 0.0120 (15) | −0.0015 (13) | 0.0038 (13) | −0.0003 (12) |
| O1 | 0.0271 (15) | 0.0137 (11) | 0.0143 (12) | 0.0001 (12) | 0.0017 (11) | 0.0015 (10) |
| O2 | 0.0335 (18) | 0.0165 (13) | 0.0103 (12) | 0.0016 (12) | 0.0008 (11) | −0.0001 (10) |
| Cl1 | 0.0343 (5) | 0.0136 (4) | 0.0144 (4) | −0.0010 (4) | −0.0010 (4) | 0.0012 (3) |
| O3 | 0.0368 (18) | 0.0133 (13) | 0.0170 (14) | −0.0015 (12) | 0.0034 (12) | 0.0015 (10) |
Geometric parameters (Å, º)
| C1—C6 | 1.354 (5) | C5—C6 | 1.500 (5) |
| C1—C2 | 1.436 (5) | C6—O2 | 1.320 (4) |
| C1—H1 | 0.9500 | N1—H1n | 0.80 (5) |
| C2—O1 | 1.227 (4) | N1—H2n | 0.81 (5) |
| C2—C3 | 1.532 (4) | N2—H3n | 0.93 (4) |
| C3—N1 | 1.320 (4) | N2—H4n | 0.83 (5) |
| C3—C4 | 1.381 (5) | O2—H1o | 0.92 (5) |
| C4—C5 | 1.406 (5) | O3—H1w | 0.88 (5) |
| C4—H4 | 0.9500 | O3—H2w | 1.00 (5) |
| C5—N2 | 1.306 (4) | ||
| C6—C1—C2 | 120.6 (3) | N2—C5—C6 | 116.6 (3) |
| C6—C1—H1 | 119.7 | C4—C5—C6 | 120.4 (3) |
| C2—C1—H1 | 119.7 | O2—C6—C1 | 126.4 (3) |
| O1—C2—C1 | 124.1 (3) | O2—C6—C5 | 112.6 (3) |
| O1—C2—C3 | 118.0 (3) | C1—C6—C5 | 120.9 (3) |
| C1—C2—C3 | 117.9 (3) | C3—N1—H1n | 122 (3) |
| N1—C3—C4 | 125.2 (3) | C3—N1—H2n | 121 (3) |
| N1—C3—C2 | 114.2 (3) | H1n—N1—H2n | 117 (4) |
| C4—C3—C2 | 120.6 (3) | C5—N2—H3n | 119 (3) |
| C3—C4—C5 | 119.6 (3) | C5—N2—H4n | 120 (3) |
| C3—C4—H4 | 120.2 | H3n—N2—H4n | 121 (4) |
| C5—C4—H4 | 120.2 | C6—O2—H1o | 114 (3) |
| N2—C5—C4 | 123.0 (3) | H1w—O3—H2w | 102 (4) |
| C6—C1—C2—O1 | 176.9 (4) | C3—C4—C5—N2 | 178.6 (4) |
| C6—C1—C2—C3 | −2.2 (5) | C3—C4—C5—C6 | −1.3 (6) |
| O1—C2—C3—N1 | 2.4 (5) | C2—C1—C6—O2 | −178.8 (4) |
| C1—C2—C3—N1 | −178.5 (3) | C2—C1—C6—C5 | 0.7 (6) |
| O1—C2—C3—C4 | −177.1 (4) | N2—C5—C6—O2 | 0.8 (5) |
| C1—C2—C3—C4 | 2.0 (5) | C4—C5—C6—O2 | −179.3 (3) |
| N1—C3—C4—C5 | −179.7 (4) | N2—C5—C6—C1 | −178.8 (4) |
| C2—C3—C4—C5 | −0.3 (5) | C4—C5—C6—C1 | 1.1 (6) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H2n···O1 | 0.81 (5) | 2.33 (5) | 2.653 (4) | 105 (4) |
| N2—H4n···O2 | 0.84 (4) | 2.23 (5) | 2.595 (4) | 107 (4) |
| N1—H1n···Cl1 | 0.80 (5) | 2.45 (5) | 3.238 (3) | 169 (4) |
| N1—H2n···O3i | 0.81 (5) | 2.25 (5) | 3.011 (4) | 156 (4) |
| N2—H3n···Cl1ii | 0.93 (4) | 2.22 (4) | 3.149 (3) | 177 (4) |
| N2—H4n···Cl1iii | 0.83 (5) | 2.44 (5) | 3.231 (3) | 158 (4) |
| O2—H1o···O3 | 0.92 (5) | 1.65 (5) | 2.548 (4) | 165 (4) |
| O3—H1w···O1iv | 0.88 (5) | 1.98 (5) | 2.801 (4) | 154 (4) |
| O3—H2w···Cl1v | 1.00 (5) | 2.11 (5) | 3.116 (3) | 176 (4) |
Symmetry codes: (i) x, y, z+1; (ii) x, −y+3/2, z−1/2; (iii) x, y, z−1; (iv) x, −y+1/2, z−1/2; (v) −x+1, −y+1, −z+1.
References
- Braunstein, P., Bubrin, D. & Sarkar, B. (2009). Inorg. Chem. 48, 2534–2540. [DOI] [PubMed]
- Braunstein, P., Siri, O., Taquet, J.-P., Rohmer, M.-M., Bénard, M. & Welter, R. (2003). J. Am. Chem. Soc. 125, 12246–12256. [DOI] [PubMed]
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Kauf, T. & Braunstein, P. (2011). Inorg. Chem. 50, 11472–11480. [DOI] [PubMed]
- Nietzki, R. & Hagenbach, E. (1887). Chem. Ber. 20, 328–338.
- Paretzki, A., Pattacini, R., Huebner, R., Braunstein, P. & Sarkar, B. (2010). Chem. Commun. 46, 1497–1499. [DOI] [PubMed]
- Plater, M. J. & Harrison, W. T. A. (2013). J. Chem. Res. (S), 37, 427–434.
- Plater, M. J. & Harrison, W. T. A. (2014a). J. Chem. Res. (S), 38, 351–355.
- Plater, M. J. & Harrison, W. T. A. (2014b). J. Chem. Res. (S), 38, 651–654.
- Plater, M. J. & Jackson, T. (2014). J. Chem. Res. (S), 38, 437–442.
- Rigaku (2015). CrysAlis PRO. Rigaku Corporation, Tokyo, Japan.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Tamboura, F. B., Cazin, C. S. J., Pattacini, R. & Braunstein, P. (2009). Eur. J. Org. Chem. pp. 3340–3350.
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
- Yang, Q.-Z., Siri, O. & Braunstein, P. (2005). Chem. Eur. J. 11, 7237–7246. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989016005107/sj5497sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989016005107/sj5497Isup2.hkl
CCDC reference: 1470620
Additional supporting information: crystallographic information; 3D view; checkCIF report







