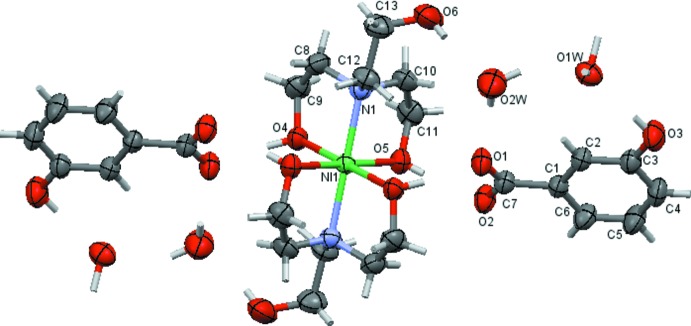
In the molecular cation of the title compound, the NiII ion is located on an inversion centre and is coordinated by two tridentate triethanolamine ligands. Two 3-hydroxybenzoate counter-anions and four lattice water molecules give rise to the formation on an intricate system of hydrogen bonds.
Keywords: crystal structure, 3-hydroxybenzoic acid, triethanolamine, hydrogen bonding
Abstract
The reaction of 3-hydroxybenzoic (m-hydroxybenzoic) acid (MHBA), triethanolamine (TEA) and Ni(NO3)2 in aqueous solution led to formation of the hydrated title salt, [Ni(C6H15NO3)2](C7H5O3)·4H2O. In the complex cation, the Ni2+ ion is located on an inversion centre. Two symmetry-related TEA ligands occupy all coordination sites in an N,O,O′-tridentate coordination, leading to a slightly distorted NiN2O4 octahedron. Two ethanol groups of each TEA ligand form two five-membered chelate rings around Ni2+, while the third ethanol group does not coordinate to the metal atom. Two MHBA− anions in the benzoate form are situated in the outer coordination sphere for charge compensation. An intricate network of hydrogen bonds between the free and coordinating hydroxy groups of the TEA ligands, the O atoms of the MHBA− anions and the water molecules leads to the formation of a two-dimensional structure extending parallel to (010).
Chemical context
Triethanolamine (TEA) is a substance with relatively low antimicrobial (Zardini et al., 2014 ▸) and plant-growth-stimulating (Loginov et al., 2012 ▸) activities. However, it is a well-known compound owing to technical applications as a curing agent for epoxy and rubber polymers, adhesives and antistatic agents, and as a corrosion inhibitor in metal-cutting (Ashton Acton, 2013 ▸). The interaction of metal ions with TEA can result in the formation of complexes in which it demonstrates monodentate (Kumar et al., 2014 ▸), bidentate (Long et al., 2004 ▸), tridentate (Mirskova et al., 2013 ▸; Haukka et al., 2005 ▸) or tetradentate (Zaitsev et al., 2014 ▸; Langley et al., 2011 ▸) binding modes. TEA ligands are also able to interact as bridging ligands between two metal cations (Sharma et al., 2014 ▸) or as bridging ligands to form one-dimensional polymeric structures (Custelcean & Jackson, 1998 ▸). Moreover, there are metal complexes in which TEA molecules are non-coordinating and are consequently situated outside the actual coordination spheres (Ilyukhin et al., 2013 ▸; Manos et al., 2012 ▸).
In contrast to the other two biologically active isomers of hydroxybenzoic acid, namely o-hydroxybenzoic (salicylic) and p-hydroxybenzoic (paraben) acid, m-hydroxybenzoic acid (MHBA) has no specific biological action. Nevertheless, MHBA is a component of castoreum, the exudate from the castor sacs of the mature North American beaver, used in perfumery and folk medicine (Müller-Schwarze & Houlihan, 1991 ▸). Most metal complexes of MHBA are in their mixed-ligand form in which mono- (Ma et al., 2013 ▸; Köse et al., 2012 ▸) or bidentate (Thompson et al., 2015 ▸; Zaman et al., 2012 ▸) coordination through the carboxylic oxygen atoms take place. The latter coordination mode may give rise to the generation of polymeric metal complexes (Koizumi et al., 1984 ▸; Koziol et al., 1990 ▸). There are also structures in which MHBA molecules are non-coordinating (Zaman et al., 2013 ▸) or simultaneously coordinating and non-coordinating (Li et al., 2008 ▸).
To the best of our knowledge, metal complexes on the basis of MHBA and ethanolamines have not yet been obtained and structurally characterized. Here, the synthesis and structure of [Ni(C6H15NO3)2](C7H5O3)2·4H2O is reported.
Structural commentary
The asymmetric unit of the title compound contains one half of the complex nickel(II) cation (the other part being completed by inversion symmetry), one MHBA− counter-anion and two water molecules (Fig. 1 ▸). Two symmetry-related TEA ligand molecules coordinate in a N,O,O′-tridentate binding mode to the metal cation, giving rise to a slightly distorted octahedral NiN2O4 coordination environment. One hydroxyl group of each ethanol substituent is not involved in the coordination and is directed away from the coordination centre. As a result of symmetry requirements, the nitrogen atoms are in trans-positions of the coordination polyhedron, giving rise to a linear N—Ni—N angle. The Ni—N bond length is 2.1158 (13) Å, and the Ni—O4 and Ni—O5 bond lengths are 2.0734 (11) and 2.0636 (12) Å, respectively. The N—Ni—O angles range from 82.22 (5) to 97.78 (5)° and the O—Ni—O angles from 89.94 (5) to 90.06 (5)°. Since the TEA ligands coordinate in their neutral form, charge compensation is required by two MHBA− anions. They are in their benzoate form and are located in the outer coordination sphere, with the carboxylate group tilted by 14.1 (2)° relative to the aromatic ring. The water molecules are also non-coordinating.
Figure 1.
The molecular entities in the title structure, with displacement ellipsoids drawn at the 50% probability level. The parts of the asymmetric unit are identified by labelled atoms; all other atoms are generated by the symmetry operation (−x + 1, −y, −z + 1).
Supramolecular features
The supramolecular structure features an intricate network of intermolecular O—H⋯O hydrogen bonds (Table 1 ▸), including four cyclic motifs of different sizes. The MHBA− anion is connected to the complex cation by a pair of rather strong hydrogen bonds [D⋯A = 2.579 (2) and 2.638 (2) Å, respectively] within a  (8) motif (Etter, 1990 ▸) (Fig. 2 ▸). This ‘cation–anion’ hydrogen-bonded unit is further associated to the other moieties through formation of an 11-membered ring between the non-coordinating hydroxyl group O6 and water molecule O2W. Three additional hydrogen bonds, O2W⋯O1, O3⋯O1W and O2W⋯O1W, lead to the same
(8) motif (Etter, 1990 ▸) (Fig. 2 ▸). This ‘cation–anion’ hydrogen-bonded unit is further associated to the other moieties through formation of an 11-membered ring between the non-coordinating hydroxyl group O6 and water molecule O2W. Three additional hydrogen bonds, O2W⋯O1, O3⋯O1W and O2W⋯O1W, lead to the same  (11) graph-set motif, in each case with hydrogen bonds of medium strength (Table 1 ▸). The fourth cyclic motif has graph-set notation
(11) graph-set motif, in each case with hydrogen bonds of medium strength (Table 1 ▸). The fourth cyclic motif has graph-set notation  (12) and consists of a centrosymmetric 12-membered cycle between two unique water molecules and the non-coordinating hydroxyl group O6 (Fig. 3 ▸). Together, the above-mentioned hydrogen-bonding interactions give rise to a two-dimensional supramolecular structure extending parallel to (010).
(12) and consists of a centrosymmetric 12-membered cycle between two unique water molecules and the non-coordinating hydroxyl group O6 (Fig. 3 ▸). Together, the above-mentioned hydrogen-bonding interactions give rise to a two-dimensional supramolecular structure extending parallel to (010).
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1W—H1WA⋯O2i | 0.83 (3) | 1.89 (3) | 2.711 (2) | 168 (3) |
| O1W—H1WB⋯O6ii | 1.06 (5) | 1.63 (3) | 2.674 (2) | 168 (4) |
| O2W—H2WA⋯O1iii | 0.85 | 1.95 | 2.775 (2) | 165 |
| O2W—H2WB⋯O1W iii | 0.85 | 2.07 | 2.830 (2) | 149 |
| O3—H3⋯O1W | 0.82 | 1.96 | 2.775 (2) | 177 |
| O4—H4⋯O1iii | 0.87 (2) | 1.72 (2) | 2.579 (2) | 169 (2) |
| O5—H5⋯O2iii | 0.74 (3) | 1.90 (3) | 2.638 (2) | 175 (3) |
| O6—H6⋯O2W iii | 0.82 | 1.93 | 2.728 (3) | 165 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  .
.
Figure 2.
Different ring motifs generated by hydrogen bonds (shown as dashed lines). Symmetry codes refer to Table 1 ▸.
Figure 3.
The packing of the molecular entities in the crystal structure (shown as dashed lines). For clarity, H atoms have been omitted.
Database survey
A survey of the Cambridge Structural Database (CSD) (Groom & Allen, 2014 ▸) showed that coordination complexes of TEA or MHBA with many metals including those of the s-, d-, p-, and f-block elements have been documented. 50 entries correspond to structures in which TEA molecules are ligating, including 21 examples in a tetradentate mode (e.g. AKEXET, GEGTIV, IBOCOR, JOMDAS, LAKYAX, RUQSUR, SUTZIQ) and two polymeric structures (GOCVEZ, CUMSAE, CUMSAE01). The combination of tri- and tetradentate coordination modes is observed in five cases (MEVQIN, MEVQOT, EYIPAD, LAKYAX, MUCBIV). There is only one structure with TEA in a monodentate mode (KISMUW) and one with a bidentate mode (QAJDIP). The most frequently encountered tridentate coordination mode is also observed in the title compound and reported for 22 entries (e.g. ASUGEA, CABTEF, DAYPOJ, FOVKIL, ETOLNI, GUQXEV, IGALOR).
There are 40 entries for MHBA coordination complexes in the CSD. For 14 entries, the MHBA molecules occupy a coordination sphere in the form of mixed-ligand complexes in monodentate coordination (e.g. GIMLEU, MEZFIG, NESFOH, SEZJOX), while bidentate coordination (e.g. MIQYIV, SISTAQ, WINFIJ, YIQQIZ) is found in twelve cases and a combination of the two modes only for entries CIVGOF and KIDBEE. Polymeric metal complex formation is reported for seven structures (CIWPIH, COSLAX, COSLIF, KIDBOO, COSKUQ, COSLEB, KIDBII). It should be noted that the hydroxyl group of the MHBA molecule is involved in coordination neither in discrete nor in polymeric complexes. For five entries, MHBA molecules are situated in the outer spheres (GANZAY, LAMMOD, MEWBOH, NIWJAF and WEJNIJ), as is the case in the title compound.
Synthesis and crystallization
To an aqueous solution (2.5 ml) of Ni(NO3)2 (0.091 g, 0.5 mmol) was slowly added an ethanol solution (5 ml) containing TEA (132 µl) and MHBA (0.138 g, 1 mmol) under constant stirring. A light-green crystalline product was obtained at room temperature by solvent evaporation after 25 days.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. C-bound hydrogen atoms were placed in calculated positions and refined in the riding-model approximation, with C—H = 0.93 and 0.97 Å for aromatic and methylene hydrogen atoms, respectively, and with U iso(H) = 1.2U eq(C). O-bound hydrogen atoms were found from difference maps. Those attached to water molecule O1W and to hydroxy O atoms O4 and O5 were refined freely whereas those attached to water molecule O2W and hydroxy atoms O3 and O6 were refined with constrained O—H distances of 0.85 and 0.82 Å, respectively. For all O-bound hydrogen atoms, U iso(H) = 1.5U eq(O).
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | [Ni(C6H15NO3)2](C7H5O3)2·4H2O |
| M r | 703.37 |
| Crystal system, space group | Monoclinic, P21/n |
| Temperature (K) | 293 |
| a, b, c (Å) | 8.40515 (12), 21.4397 (3), 9.48944 (14) |
| β (°) | 106.1835 (15) |
| V (Å3) | 1642.27 (4) |
| Z | 2 |
| Radiation type | Cu Kα |
| μ (mm−1) | 1.50 |
| Crystal size (mm) | 0.32 × 0.14 × 0.12 |
| Data collection | |
| Diffractometer | Agilent Xcalibur Ruby |
| Absorption correction | Multi-scan (CrysAlis PRO; Agilent, 2014 ▸) |
| T min, T max | 0.912, 1.000 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 12555, 3399, 3066 |
| R int | 0.030 |
| (sin θ/λ)max (Å−1) | 0.629 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.038, 0.115, 1.03 |
| No. of reflections | 3399 |
| No. of parameters | 226 |
| No. of restraints | 3 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.26, −0.44 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989016005521/wm5282sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989016005521/wm5282Isup2.hkl
CCDC reference: 1471925
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
This work was supported by Grants of the Center of Science and Technology, Uzbekistan
supplementary crystallographic information
Crystal data
| [Ni(C6H15NO3)2](C7H5O3)2·4H2O | F(000) = 748 |
| Mr = 703.37 | Dx = 1.422 Mg m−3 |
| Monoclinic, P21/n | Cu Kα radiation, λ = 1.54184 Å |
| a = 8.40515 (12) Å | Cell parameters from 7605 reflections |
| b = 21.4397 (3) Å | θ = 4.1–75.8° |
| c = 9.48944 (14) Å | µ = 1.50 mm−1 |
| β = 106.1835 (15)° | T = 293 K |
| V = 1642.27 (4) Å3 | Prism, light-green |
| Z = 2 | 0.32 × 0.14 × 0.12 mm |
Data collection
| Agilent Xcalibur Ruby diffractometer | 3399 independent reflections |
| Radiation source: Enhance (Cu) X-ray Source | 3066 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.030 |
| Detector resolution: 10.2576 pixels mm-1 | θmax = 75.9°, θmin = 4.1° |
| ω scans | h = −10→10 |
| Absorption correction: multi-scan (CrysAlis PRO; Agilent, 2014) | k = −23→26 |
| Tmin = 0.912, Tmax = 1.000 | l = −11→7 |
| 12555 measured reflections |
Refinement
| Refinement on F2 | Hydrogen site location: mixed |
| Least-squares matrix: full | H atoms treated by a mixture of independent and constrained refinement |
| R[F2 > 2σ(F2)] = 0.038 | w = 1/[σ2(Fo2) + (0.0717P)2 + 0.3396P] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.115 | (Δ/σ)max < 0.001 |
| S = 1.03 | Δρmax = 0.26 e Å−3 |
| 3399 reflections | Δρmin = −0.44 e Å−3 |
| 226 parameters | Extinction correction: SHELXL2014 (Sheldrick, 2015), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| 3 restraints | Extinction coefficient: 0.0013 (3) |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Ni1 | 0.5000 | 0.0000 | 0.5000 | 0.03393 (15) | |
| O4 | 0.64514 (15) | 0.07781 (5) | 0.49590 (13) | 0.0419 (3) | |
| H4 | 0.658 (3) | 0.0931 (8) | 0.4149 (15) | 0.063* | |
| O5 | 0.34872 (15) | 0.02307 (7) | 0.29528 (13) | 0.0436 (3) | |
| O2 | 0.61445 (15) | −0.13123 (6) | 0.82440 (16) | 0.0516 (3) | |
| N1 | 0.38809 (17) | 0.06100 (6) | 0.61955 (15) | 0.0393 (3) | |
| O1 | 0.34416 (15) | −0.11360 (6) | 0.76086 (14) | 0.0499 (3) | |
| C7 | 0.4724 (2) | −0.14099 (8) | 0.83791 (18) | 0.0404 (3) | |
| O3 | 0.1335 (2) | −0.23264 (8) | 1.1285 (2) | 0.0679 (4) | |
| H3 | 0.0720 | −0.2045 | 1.0868 | 0.102* | |
| C3 | 0.2777 (2) | −0.23037 (8) | 1.0902 (2) | 0.0470 (4) | |
| C2 | 0.3022 (2) | −0.18851 (8) | 0.98698 (19) | 0.0417 (4) | |
| H2 | 0.2181 | −0.1609 | 0.9419 | 0.050* | |
| C10 | 0.4602 (3) | 0.04530 (10) | 0.77728 (19) | 0.0513 (4) | |
| H10A | 0.3929 | 0.0133 | 0.8049 | 0.062* | |
| H10B | 0.4571 | 0.0820 | 0.8362 | 0.062* | |
| C1 | 0.4506 (2) | −0.18721 (7) | 0.94995 (18) | 0.0391 (3) | |
| C9 | 0.6038 (3) | 0.13094 (9) | 0.5707 (3) | 0.0576 (5) | |
| H9A | 0.6147 | 0.1687 | 0.5179 | 0.069* | |
| H9B | 0.6797 | 0.1337 | 0.6685 | 0.069* | |
| C6 | 0.5756 (3) | −0.22894 (9) | 1.0159 (2) | 0.0529 (5) | |
| H6A | 0.6756 | −0.2287 | 0.9919 | 0.063* | |
| C8 | 0.4288 (3) | 0.12535 (9) | 0.5813 (3) | 0.0559 (5) | |
| H8A | 0.4134 | 0.1541 | 0.6552 | 0.067* | |
| H8B | 0.3526 | 0.1372 | 0.4880 | 0.067* | |
| C4 | 0.4027 (3) | −0.27183 (9) | 1.1560 (2) | 0.0576 (5) | |
| H4A | 0.3879 | −0.3001 | 1.2257 | 0.069* | |
| C11 | 0.6364 (3) | 0.02244 (11) | 0.8101 (2) | 0.0569 (5) | |
| H11A | 0.7093 | 0.0573 | 0.8081 | 0.068* | |
| H11B | 0.6700 | 0.0043 | 0.9075 | 0.068* | |
| O6 | 0.1021 (3) | 0.06378 (8) | 0.7862 (2) | 0.0809 (6) | |
| H6 | 0.0736 | 0.0271 | 0.7755 | 0.121* | |
| C5 | 0.5489 (3) | −0.27099 (10) | 1.1178 (3) | 0.0623 (5) | |
| H5A | 0.6319 | −0.2993 | 1.1614 | 0.075* | |
| C12 | 0.2050 (2) | 0.05169 (10) | 0.5721 (2) | 0.0502 (4) | |
| H12A | 0.1833 | 0.0079 | 0.5847 | 0.060* | |
| H12B | 0.1663 | 0.0606 | 0.4679 | 0.060* | |
| C13 | 0.1006 (3) | 0.08968 (11) | 0.6485 (3) | 0.0642 (5) | |
| H13A | 0.1428 | 0.1320 | 0.6627 | 0.077* | |
| H13B | −0.0126 | 0.0914 | 0.5865 | 0.077* | |
| O2W | 0.9448 (2) | 0.06186 (9) | 0.2035 (2) | 0.0775 (5) | |
| H2WA | 0.8479 | 0.0746 | 0.1994 | 0.116* | |
| H2WB | 0.9679 | 0.0715 | 0.1246 | 0.116* | |
| O1W | −0.06671 (19) | −0.13462 (7) | 0.99258 (17) | 0.0543 (3) | |
| H1WA | −0.164 (4) | −0.1390 (14) | 0.942 (3) | 0.079 (9)* | |
| H1WB | −0.092 (6) | −0.103 (2) | 1.070 (6) | 0.164 (18)* | |
| H5 | 0.359 (3) | 0.0545 (13) | 0.266 (3) | 0.061 (7)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Ni1 | 0.0372 (2) | 0.0332 (2) | 0.0320 (2) | −0.00273 (13) | 0.01073 (15) | −0.00022 (13) |
| O4 | 0.0463 (6) | 0.0390 (6) | 0.0429 (6) | −0.0054 (5) | 0.0164 (5) | 0.0025 (5) |
| O5 | 0.0452 (6) | 0.0453 (7) | 0.0386 (6) | −0.0013 (5) | 0.0085 (5) | 0.0060 (5) |
| O2 | 0.0403 (6) | 0.0553 (7) | 0.0605 (8) | 0.0006 (5) | 0.0163 (6) | 0.0172 (6) |
| N1 | 0.0426 (7) | 0.0375 (7) | 0.0391 (7) | −0.0020 (5) | 0.0137 (6) | −0.0040 (5) |
| O1 | 0.0419 (6) | 0.0581 (7) | 0.0514 (7) | 0.0046 (5) | 0.0161 (5) | 0.0212 (6) |
| C7 | 0.0417 (8) | 0.0388 (8) | 0.0408 (8) | −0.0003 (6) | 0.0115 (6) | 0.0049 (6) |
| O3 | 0.0679 (10) | 0.0702 (10) | 0.0772 (10) | 0.0026 (7) | 0.0396 (8) | 0.0252 (8) |
| C3 | 0.0547 (10) | 0.0437 (9) | 0.0450 (9) | −0.0046 (7) | 0.0179 (8) | 0.0048 (7) |
| C2 | 0.0446 (9) | 0.0387 (8) | 0.0408 (8) | 0.0010 (6) | 0.0100 (7) | 0.0068 (6) |
| C10 | 0.0624 (11) | 0.0543 (10) | 0.0376 (9) | −0.0011 (8) | 0.0148 (8) | −0.0101 (7) |
| C1 | 0.0427 (8) | 0.0358 (8) | 0.0375 (8) | −0.0021 (6) | 0.0088 (6) | 0.0033 (6) |
| C9 | 0.0719 (13) | 0.0399 (9) | 0.0674 (12) | −0.0155 (8) | 0.0303 (10) | −0.0105 (8) |
| C6 | 0.0481 (10) | 0.0477 (10) | 0.0625 (12) | 0.0070 (8) | 0.0145 (8) | 0.0125 (8) |
| C8 | 0.0698 (13) | 0.0356 (9) | 0.0706 (12) | 0.0017 (8) | 0.0331 (10) | −0.0030 (8) |
| C4 | 0.0707 (13) | 0.0491 (10) | 0.0519 (11) | −0.0012 (9) | 0.0154 (9) | 0.0189 (8) |
| C11 | 0.0608 (12) | 0.0601 (11) | 0.0409 (9) | −0.0055 (9) | −0.0005 (8) | −0.0077 (8) |
| O6 | 0.1092 (15) | 0.0688 (10) | 0.0874 (12) | −0.0059 (10) | 0.0650 (12) | −0.0136 (9) |
| C5 | 0.0629 (12) | 0.0523 (11) | 0.0677 (13) | 0.0123 (9) | 0.0115 (10) | 0.0233 (10) |
| C12 | 0.0419 (9) | 0.0595 (11) | 0.0515 (10) | 0.0006 (8) | 0.0169 (8) | −0.0084 (8) |
| C13 | 0.0583 (12) | 0.0652 (13) | 0.0775 (14) | 0.0078 (10) | 0.0331 (11) | −0.0063 (11) |
| O2W | 0.0663 (10) | 0.0802 (11) | 0.0964 (14) | 0.0154 (9) | 0.0401 (10) | 0.0171 (10) |
| O1W | 0.0444 (7) | 0.0649 (9) | 0.0559 (8) | 0.0029 (6) | 0.0179 (6) | −0.0039 (6) |
Geometric parameters (Å, º)
| Ni1—O4i | 2.0735 (11) | C1—C6 | 1.389 (2) |
| Ni1—O4 | 2.0734 (11) | C9—H9A | 0.9700 |
| Ni1—O5 | 2.0636 (12) | C9—H9B | 0.9700 |
| Ni1—O5i | 2.0636 (12) | C9—C8 | 1.507 (3) |
| Ni1—N1i | 2.1158 (13) | C6—H6A | 0.9300 |
| Ni1—N1 | 2.1158 (13) | C6—C5 | 1.385 (3) |
| O4—H4 | 0.869 (9) | C8—H8A | 0.9700 |
| O4—C9 | 1.435 (2) | C8—H8B | 0.9700 |
| O5—C11i | 1.427 (2) | C4—H4A | 0.9300 |
| O5—H5 | 0.74 (3) | C4—C5 | 1.375 (3) |
| O2—C7 | 1.253 (2) | C11—O5i | 1.427 (2) |
| N1—C10 | 1.488 (2) | C11—H11A | 0.9700 |
| N1—C8 | 1.491 (2) | C11—H11B | 0.9700 |
| N1—C12 | 1.491 (2) | O6—H6 | 0.8200 |
| O1—C7 | 1.266 (2) | O6—C13 | 1.417 (3) |
| C7—C1 | 1.501 (2) | C5—H5A | 0.9300 |
| O3—H3 | 0.8200 | C12—H12A | 0.9700 |
| O3—C3 | 1.359 (2) | C12—H12B | 0.9700 |
| C3—C2 | 1.386 (2) | C12—C13 | 1.521 (3) |
| C3—C4 | 1.385 (3) | C13—H13A | 0.9700 |
| C2—H2 | 0.9300 | C13—H13B | 0.9700 |
| C2—C1 | 1.387 (2) | O2W—H2WA | 0.8498 |
| C10—H10A | 0.9700 | O2W—H2WB | 0.8500 |
| C10—H10B | 0.9700 | O1W—H1WA | 0.83 (3) |
| C10—C11 | 1.508 (3) | O1W—H1WB | 1.07 (5) |
| O4—Ni1—O4i | 180.0 | C2—C1—C6 | 119.57 (16) |
| O4i—Ni1—N1i | 82.22 (5) | C6—C1—C7 | 121.18 (16) |
| O4—Ni1—N1i | 97.78 (5) | O4—C9—H9A | 109.6 |
| O4—Ni1—N1 | 82.22 (5) | O4—C9—H9B | 109.6 |
| O4i—Ni1—N1 | 97.78 (5) | O4—C9—C8 | 110.20 (15) |
| O5—Ni1—O4i | 89.95 (5) | H9A—C9—H9B | 108.1 |
| O5i—Ni1—O4 | 89.94 (5) | C8—C9—H9A | 109.6 |
| O5—Ni1—O4 | 90.05 (5) | C8—C9—H9B | 109.6 |
| O5i—Ni1—O4i | 90.06 (5) | C1—C6—H6A | 120.5 |
| O5i—Ni1—O5 | 180.0 | C5—C6—C1 | 118.99 (19) |
| O5—Ni1—N1 | 96.16 (5) | C5—C6—H6A | 120.5 |
| O5—Ni1—N1i | 83.84 (5) | N1—C8—C9 | 112.60 (16) |
| O5i—Ni1—N1i | 96.16 (5) | N1—C8—H8A | 109.1 |
| O5i—Ni1—N1 | 83.84 (5) | N1—C8—H8B | 109.1 |
| N1i—Ni1—N1 | 180.0 | C9—C8—H8A | 109.1 |
| Ni1—O4—H4 | 122.6 (12) | C9—C8—H8B | 109.1 |
| C9—O4—Ni1 | 113.85 (10) | H8A—C8—H8B | 107.8 |
| C9—O4—H4 | 104.1 (12) | C3—C4—H4A | 120.2 |
| Ni1—O5—H5 | 118 (2) | C5—C4—C3 | 119.62 (17) |
| C11i—O5—Ni1 | 110.15 (11) | C5—C4—H4A | 120.2 |
| C11i—O5—H5 | 108 (2) | O5i—C11—C10 | 110.50 (15) |
| C10—N1—Ni1 | 106.39 (11) | O5i—C11—H11A | 109.5 |
| C10—N1—C8 | 113.44 (15) | O5i—C11—H11B | 109.5 |
| C10—N1—C12 | 111.71 (14) | C10—C11—H11A | 109.5 |
| C8—N1—Ni1 | 105.96 (11) | C10—C11—H11B | 109.5 |
| C8—N1—C12 | 109.74 (15) | H11A—C11—H11B | 108.1 |
| C12—N1—Ni1 | 109.32 (10) | C13—O6—H6 | 109.5 |
| O2—C7—O1 | 123.06 (15) | C6—C5—H5A | 119.2 |
| O2—C7—C1 | 119.38 (15) | C4—C5—C6 | 121.53 (18) |
| O1—C7—C1 | 117.56 (14) | C4—C5—H5A | 119.2 |
| C3—O3—H3 | 109.5 | N1—C12—H12A | 107.8 |
| O3—C3—C2 | 122.12 (17) | N1—C12—H12B | 107.8 |
| O3—C3—C4 | 118.51 (17) | N1—C12—C13 | 117.92 (17) |
| C4—C3—C2 | 119.37 (18) | H12A—C12—H12B | 107.2 |
| C3—C2—H2 | 119.5 | C13—C12—H12A | 107.8 |
| C3—C2—C1 | 120.92 (16) | C13—C12—H12B | 107.8 |
| C1—C2—H2 | 119.5 | O6—C13—C12 | 111.83 (19) |
| N1—C10—H10A | 109.1 | O6—C13—H13A | 109.2 |
| N1—C10—H10B | 109.1 | O6—C13—H13B | 109.2 |
| N1—C10—C11 | 112.46 (15) | C12—C13—H13A | 109.2 |
| H10A—C10—H10B | 107.8 | C12—C13—H13B | 109.2 |
| C11—C10—H10A | 109.1 | H13A—C13—H13B | 107.9 |
| C11—C10—H10B | 109.1 | H2WA—O2W—H2WB | 109.4 |
| C2—C1—C7 | 119.25 (14) | H1WA—O1W—H1WB | 97 (3) |
| Ni1—O4—C9—C8 | −22.1 (2) | C3—C2—C1—C7 | −179.82 (16) |
| Ni1—N1—C10—C11 | 31.50 (18) | C3—C2—C1—C6 | 0.8 (3) |
| Ni1—N1—C8—C9 | −40.0 (2) | C3—C4—C5—C6 | 0.9 (4) |
| Ni1—N1—C12—C13 | 177.98 (15) | C2—C3—C4—C5 | −0.2 (3) |
| O4—C9—C8—N1 | 42.1 (2) | C2—C1—C6—C5 | −0.1 (3) |
| O2—C7—C1—C2 | 166.38 (16) | C10—N1—C8—C9 | 76.4 (2) |
| O2—C7—C1—C6 | −14.2 (3) | C10—N1—C12—C13 | 60.5 (2) |
| N1—C10—C11—O5i | −46.7 (2) | C1—C6—C5—C4 | −0.7 (4) |
| N1—C12—C13—O6 | −79.9 (2) | C8—N1—C10—C11 | −84.60 (19) |
| O1—C7—C1—C2 | −13.7 (2) | C8—N1—C12—C13 | −66.2 (2) |
| O1—C7—C1—C6 | 165.70 (18) | C4—C3—C2—C1 | −0.6 (3) |
| C7—C1—C6—C5 | −179.51 (19) | C12—N1—C10—C11 | 150.73 (16) |
| O3—C3—C2—C1 | 180.00 (18) | C12—N1—C8—C9 | −157.89 (17) |
| O3—C3—C4—C5 | 179.2 (2) |
Symmetry code: (i) −x+1, −y, −z+1.
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1W—H1WA···O2ii | 0.83 (3) | 1.89 (3) | 2.711 (2) | 168 (3) |
| O1W—H1WB···O6iii | 1.06 (5) | 1.63 (3) | 2.674 (2) | 168 (4) |
| O2W—H2WA···O1i | 0.85 | 1.95 | 2.775 (2) | 165 |
| O2W—H2WB···O1Wi | 0.85 | 2.07 | 2.830 (2) | 149 |
| O3—H3···O1W | 0.82 | 1.96 | 2.775 (2) | 177 |
| O4—H4···O1i | 0.87 (2) | 1.72 (2) | 2.579 (2) | 169 (2) |
| O5—H5···O2i | 0.74 (3) | 1.90 (3) | 2.638 (2) | 175 (3) |
| O6—H6···O2Wi | 0.82 | 1.93 | 2.728 (3) | 165 |
Symmetry codes: (i) −x+1, −y, −z+1; (ii) x−1, y, z; (iii) −x, −y, −z+2.
References
- Agilent (2014). CrysAlis PRO. Agilent Technologies Ltd, Yarnton, England.
- Ashton Acton, Q. (2013). Editor. In Ethanolamines – Advances in Research and Application. Atlanta: Scholarly Editions.
- Custelcean, R. & Jackson, J. E. (1998). J. Am. Chem. Soc. 120, 12935–12941.
- Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2009). J. Appl. Cryst. 42, 339–341.
- Etter, M. C. (1990). Acc. Chem. Res. 23, 120–126.
- Groom, C. R. & Allen, F. H. (2014). Angew. Chem. Int. Ed. 53, 662–671. [DOI] [PubMed]
- Haukka, M., Kirillov, A. M., Kopylovich, M. N. & Pombeiro, A. J. L. (2005). Acta Cryst. E61, m2746–m2748.
- Ilyukhin, A. B., Koroteev, P. S., Kiskin, M. A., Dobrokhotova, Z. V. & Novotortsev, V. M. (2013). J. Mol. Struct. 1033, 187–199.
- Koizumi, Y., Sawase, H., Suzuki, Y., Takeuchi, T., Shimoi, M. & Ouchi, A. (1984). Bull. Chem. Soc. Jpn, 57, 1809–1817.
- Köse, D. A., Necefoğlu, H., Şahin, O. & Büyükgüngör, O. (2012). J. Therm. Anal. Calorim. 110, 1233–1241.
- Koziol, A. E., Brzyska, W., Klimek, B., Kula, A., Palenik, G. J. & Stepniak, K. (1990). J. Coord. Chem. 21, 183–191.
- Kumar, R., Obrai, S., Kaur, A., Hundal, M. S., Meehnian, H. & Jana, A. K. (2014). New J. Chem. 38, 1186–1198.
- Langley, S. K., Chilton, N. F., Moubaraki, B. & Murray, K. S. (2011). Dalton Trans. 40, 12201–12209. [DOI] [PubMed]
- Li, J.-H., Nie, J.-J., Su, J.-R. & Xu, D.-J. (2008). Acta Cryst. E64, m382–m383. [DOI] [PMC free article] [PubMed]
- Loginov, S. V., Kuznetsov, B. A. & Petrichenko, B. A. (2012). Patent of the Russian Federation, No. 2450516.
- Long, D.-L., Abbas, H., Kögerler, P. & Cronin, L. (2004). J. Am. Chem. Soc. 126, 13880–13881. [DOI] [PubMed]
- Ma, Z., Lu, W., Liang, B. & Pombeiro, A. J. L. (2013). New J. Chem. 37, 1529–1537.
- Manos, M. J., Moushi, E. E., Papaefstathiou, G. S. & Tasiopoulos, A. J. (2012). Cryst. Growth Des. 12, 5471–5480.
- Mirskova, A. N., Adamovich, S. N., Mirskov, R. G. & Schilde, U. (2013). Chem. Cent. J. 7, 34–38. [DOI] [PMC free article] [PubMed]
- Müller-Schwarze, D. & Houlihan, P. W. (1991). J. Chem. Ecol. 17, 715–734. [DOI] [PubMed]
- Sharma, R. P., Saini, A., Venugopalan, P., Ferretti, V., Spizzo, F., Angeli, C. & Calzado, C. J. (2014). New J. Chem. 38, 574–583.
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
- Thompson, D. J., Paredes, J. E. B., Villalobos, L., Ciclosi, M., Elsby, R. J., Liu, B., Fanwick, P. E. & Ren, T. (2015). Inorg. Chim. Acta, 424, 150–155.
- Zaitsev, K. V., Churakov, A. V., Poleshchuk, O. Kh., Oprunenko, Y. F., Zaitseva, G. S. & Karlov, S. S. (2014). Dalton Trans. 43, 6605–6609. [DOI] [PubMed]
- Zaman, I. G., Çaylak Delibaş, N., Necefoğlu, H. & Hökelek, T. (2012). Acta Cryst. E68, m198–m199. [DOI] [PMC free article] [PubMed]
- Zaman, İ. G., Çaylak Delibaş, N., Necefoğlu, H. & Hökelek, T. (2013). Acta Cryst. E69, m198–m199. [DOI] [PMC free article] [PubMed]
- Zardini, H. Z., Davarpanah, M., Shanbedi, M., Amiri, A., Maghrebi, M. & Ebrahimi, L. J. (2014). J. Biomed. Mater. Res. 102, 1774–1781. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989016005521/wm5282sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989016005521/wm5282Isup2.hkl
CCDC reference: 1471925
Additional supporting information: crystallographic information; 3D view; checkCIF report