Abstract
The marine bacterium Vibrio alginolyticus has four motor components, PomA, PomB, MotX, and MotY, responsible for its Na+-driven flagellar rotation. PomA and PomB are integral inner membrane proteins having four and one transmembrane segments (TMs), respectively, which are thought to form an ion channel complex. First, site-directed Cys mutagenesis was systematically performed from Asp-24 to Glu-41 of PomB, and the resulting mutant proteins were examined for susceptibility to a sulfhydryl reagent. Secondly, the Cys substitutions at the periplasmic boundaries of the PomB TM (Ser-38) and PomA TMs (Gly-23, Ser-34, Asp-170, and Ala-178) were combined. Cross-linked products were detected for the combination of PomB-S38C and PomA-D170C mutant proteins. The Cys substitutions in the periplasmic boundaries of PomA TM3 (from Met-169 to Asp-171) and the PomB TM (from Leu-37 to Ser-40) were combined to construct a series of double mutants. Most double mutations reduced the motility, whereas each single Cys substitution slightly affected it. Although the motility of the strain carrying PomA-D170C and PomB-S38C was significantly inhibited, it was recovered by reducing reagent. The strain with this combination showed a lower affinity for Na+ than the wild-type combination. PomA-D148C and PomB-P16C, which are located at the cytoplasmic boundaries of PomA TM3 and the PomB TM, also formed the cross-linked product. From these lines of evidence, we infer that TM3 of PomA and the TM of PomB are in close proximity over their entire length and that cooperation between these two TMs is required for coupling of Na+ conduction to flagellar rotation.
The chemotactic behavior of bacteria is a consequence of two swimming states, smooth swimming and tumbling (11, 33), and the alternative, which can be backward swimming (22), turns (40), or stopping (1). The flagellar filament is the propeller that drives the cell body, and the bacterium utilizes it to swim. It is driven by the rotation of the motor embedded in the cytoplasmic membrane, which is connected to the filament via the rod and hook structures. The basal body of the flagellum in gram-negative bacteria consists of the rod and several ring structures: the L ring, P ring, MS ring, and C ring, which reside in the outer membrane, peptidoglycan layer, cytoplasmic membrane, and cytoplasm, respectively (18). The MS and C rings are referred to as the rotor, and the motor protein complexes (see below) that surround the rotor are referred to as the stator (27).
The flagellar motor is energized by an electrochemical potential using either H+ or Na+ as the coupling ion (21, 34). The marine bacterium Vibrio alginolyticus and the pathogenic bacterium Vibrio parahaemolyticus have two different types of flagella, H+-driven lateral flagella and Na+-driven polar flagella (8, 26). The lateral flagella are useful for movement under viscous conditions, even on solid surfaces, but this motility is relatively slow. The study of the rotation mechanism of Na+-driven flagellar motors has two clear advantages: (i) it is easy to manipulate the ion-motive force by changing the Na+ concentration of the medium, and (ii) phenamil, which is a known inhibitor of Na+ channels, specifically inhibits Na+-driven flagella but not H+-driven flagella (7, 23, 28).
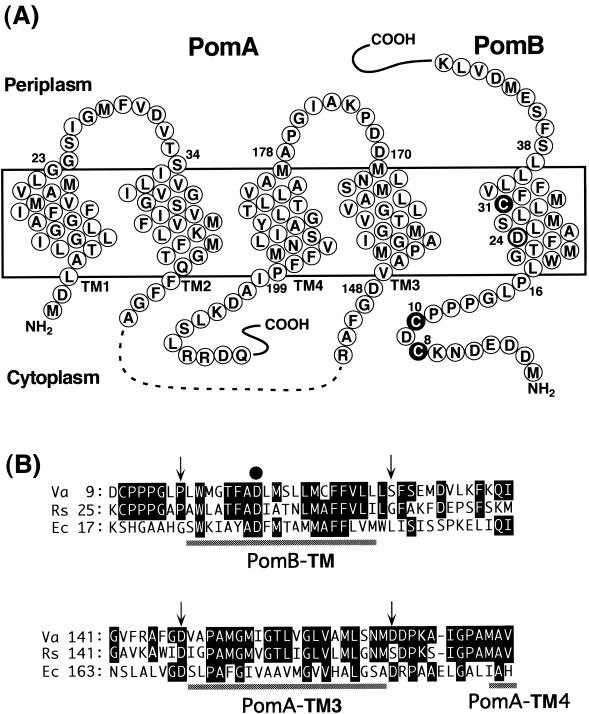
Four genes that are required for the function of the Na+-driven motor of V. alginolyticus have been identified: pomA, pomB, motX, and motY (2, 35, 37, 39). PomA and PomB are orthologs of MotA and MotB, respectively, which are the motor proteins of H+-driven flagella in Escherichia coli, Salmonella spp., and other species (33). PomA (MotA) and PomB (MotB) are integral cytoplasmic membrane proteins having four and one transmembrane segments (TMs), respectively. Figure 1 shows a model of the predicted membrane topology of PomA and PomB (5, 14, 19, 44, 45, 58). A multimeric complex composed of four PomA and two PomB proteins participates in the ion influx to drive the flagellar rotation (15, 30, 42, 43, 55). PomB (MotB) has a putative peptidoglycan-binding motif to attach the motor complex to the peptidoglycan layer in the periplasmic space (2, 17). MotX and MotY are exclusively found in Vibrio species, and they have been shown to be present in the outer membrane (38). It is not known how the outer membrane proteins function in the motor.
FIG. 1.
Putative membrane topology of PomA and PomB and sites of mutations used in this study. (A) The predicted structure of PomA (left) is taken from Asai et al. (5). PomA has four TMs, TM1 to TM4, and no Cys residues. PomA has a ca. 100-amino-acid cytoplasmic region (dashes) that plays a critical role in interactions with the rotor protein FliG. Wild-type PomB (right) has three Cys residues, Cys-8, Cys-10, and Cys-31 (white letters in filled circles). The predicted topology of PomB is from Asai et al. (2) and Braun and Blair (14) but modified as described in Discussion. PomB has an essential 24th Asp residue in its TM (bold circle) and a large (length, ∼270 amino acids) C-terminal periplasmic domain (not shown), which contains the putative peptidoglycan-binding region. (B) Alignments of the PomA TM3 and the PomB TM regions with the TM regions of MotA and MotB. Va, V. alginolyticus; Rs, Rhodobacter sphaeroides; Ec, E. coli. Residues that are identical to those of V. alginolyticus are indicated by white letters in black boxes. Arrows indicate cross-linked residues between PomA TM3 and PomB TM. The closed circle indicates the essential Asp residue, which is thought to bind protons or sodium ions.
The large (ca. 100-amino-acid) cytoplasmic region of PomA connecting the second TM (TM2) and the third TM (TM3) is thought to interact with the rotor protein FliG, which is associated with the MS ring (52). The 32nd Asp residue in MotB, which corresponds to Asp-24 of PomB, is located near the cytoplasmic end of its TM and is thought to bind ions (H+ or Na+) and play a critical role in ion flux and the energy conversion (57). It is thought that successive conformational changes in MotA, associated with the protonation and deprotonation of the MotB TM, drive the rotation of the FliG-MS ring complex (29). It is hypothesized that the cytoplasmic region of MotA interacts with the rotor component FliG, where electrostatic interactions between conserved charges on FliG of the rotor and on MotA are important for the rotation of the H+-driven motor (59). According to these hypotheses, it can be speculated that a conformational change in the MotB TM (PomB TM), which is evoked by binding of ions, is successively transferred to MotA (PomA), to its cytoplasmic region, and finally to the rotor protein FliG to drive the motor. Thus, the interactions between PomA and PomB are critical to understand the molecular mechanism of flagellar rotation. Several chimeric motors composed of both H+ and Na+ types of components are functional (3, 4, 6, 20), suggesting that a similar mechanism governs both types of motors. However, there are some inconsistencies because the conserved charges of PomA and FliG in the Na+-driven motor of V. alginolyticus are not particularly important (53, 54).
Cys-scanning mutagenesis in PomA, especially in the periplasmic loop regions, suggests that the environments around loop1-2, which connects TM1 and TM2, and loop3-4, which connects TM3 and TM4, are very different from each other (5). Many Cys substitutions in loop3-4 impair motility, implying that this loop region or the connecting TM3 or TM4 have important roles. The 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) modification of PomA-D170C at the periplasmic boundary of TM3 was affected by NaCl concentrations, suggesting that Na+ competes with DTNB and that this region may be involved in ion conductance in the PomA/PomB complex (5). The mutations conferring phenamil resistance for Na+-driven flagellar rotation have been mapped near the cytoplasmic boundaries of PomA TM3 and PomB TM (23, 28). The charged residue Asp-24 of PomB TM, the hypothetical Na+-binding site, is fairly close to the position of the phenamil-resistant mutations. In the H+-driven motor, Trp scanning of MotA showed that several different amino acid substitutions in TM3 impaired motility (44). Furthermore, Pro-173 (TM3) of MotA is suggested to be close to Asp-32 of MotB (16).
In order to understand the molecular mechanisms that connect the ion conductance and the subsequent conformational change of PomA, it is important to examine the interactions between PomA, which interacts with FliG, and PomB, which has a putative Na+-binding site. In this study, we assessed the possibility of functional and physical interactions between PomA TM3 and PomB TM, because they appear to be important TMs in the PomA/PomB complex. Through biochemical and physiological analyses using combinations of Cys replacements of PomA and PomB, two major conclusions are described: (i) the PomA TM3 and PomB TM are associated at both the periplasmic and cytoplasmic faces of the membrane, and (ii) at the periplasmic side, PomA TM3 and PomB TM may comprise part of an entrance for Na+ into the PomA/PomB complex.
MATERIALS AND METHODS
Bacterial strains, plasmids, media and growth conditions.
E. coli strain JM109 [recA1 endA1 gyrA96 thi hsdR17 relA1 supE44 λ− Δ(lac-proAB) F′ traD36 proAB lacIqΔM15) was used for DNA manipulations (49). The strains of V. alginolyticus used in this study are listed in Table 1. V. alginolyticus strain NMB136, which is defective in chemotaxis (Che−), was isolated after ethyl methanesulfonate treatment (22) from the lateral-flagellar-defective strain VIO5. V. alginolyticus strain NMB195, which has a pomAB deletion and Che− phenotype, was constructed from NMB136 by using the suicide plasmid pYA802 carrying ΔpomAB as described previously (50). V. alginolyticus cells were cultured at 30°C on VC medium (0.5% tryptone, 0.5% yeast extract, 0.4% K2HPO4, 3% NaCl, 0.2% glucose) or VPG500 medium (1% tryptone, 0.4% K2HPO4, 500 mM NaCl, 0.5% glycerol). When necessary, kanamycin was added to a final concentration of 100 μg/ml (for V. alginolyticus) or 50 μg/ml (for E. coli). The plasmids used are listed in Table 1. The motor genes on the plasmids constructed in this study are expressed from the lac promoter-operator in pSU41 (10). Transformation of Vibrio cells by electroporation was carried out as described previously (25). DNA manipulations were carried out according to standard procedures (41). Amino acid substitution was carried out using a site-directed mutagenesis kit (Stratagene).
TABLE 1.
V. alginolyticus strains and plasmids
| Strain or plasmid | Genotype or descriptiona | Reference or source |
|---|---|---|
| V. alginolyticus strain | ||
| VIO5 | Rifr Laf− | 39 |
| NMB136 | Rifr Laf− Che− | This study |
| NMB192 | Rifr Laf− ΔpomB | This study |
| NMB191 | Rifr Laf− ΔpomAB | 50 |
| NMB195 | Rifr Laf− Che− ΔpomAB | This study |
| Plasmid | ||
| pSU41 | kan (Kmr) Plac lacZα | 10 |
| pHK4 | pSU41, 1.0-kb HindIII-SacI fragment (pomB+) | 28 |
| pYA303 | pSU41, 1.9-kb BamHI-SacI fragment (pomAB+) | 28 |
| pYA803 | pKY704, 1.2-kb XbaI-HpaI and 500-base SnaBI-SacI fragments (pomA-ΔpomB) | Y. Asai |
| pYA802 | pKY704, 250-base BamHI-DraI and 700-base HpaI-SacI fragments (ΔpomAB) | 50 |
Rifr, rifampin resistant; Kmr, kanamycin resistant; Laf−, defective in lateral flagellar formation; Che−, defective in chemotaxis; Plac, lac promoter-operator.
Swarming assay.
An aliquot (1 μl) of overnight culture in VC medium was spotted onto VPG500 plates containing 0.25% agar and 100 μg of kanamycin/ml and incubated at 30°C. Dithiothreitol (DTT) was added to final concentrations as specified. DTNB was dissolved in dimethyl sulfoxide at 100 mM prior to use.
Measurement of swimming speed and data processing.
An overnight culture in VC medium was inoculated into VPG500 medium at a 50-fold dilution and grown at 30°C to exponential growth phase. Cells were centrifuged at 3,500 × g for 3 min, and the sedimented cells were resuspended in TMN50 (50 mM Tris-HCl [pH 7.5], 5 mM MgSO4, 5 mM glucose, 50 mM NaCl, 450 mM KCl) supplemented with 1 mM DTT. Cell suspensions were diluted 20-fold into TMN containing 1 mM DTT and various concentrations of NaCl, and the motility of the cells was observed immediately under a dark-field microscope. The KCl concentration was also changed to hold the total ion concentration at a fixed level (500 mM). Swimming speeds were determined from at least 20 individual cells as described previously (9). The Na+ influx through the motor was estimated by the kinetic treatment as described previously (47). Na+ influx corresponds to the equation v2/(ɛ × Na+-motive force), where v is the swimming speed and ɛ is the efficiency. For this estimation, the intracellular Na+ concentration and membrane potential were assumed to be 30 mM and −150 mV, respectively. Apparent Km values for Na+ in the estimated Na+ influx were calculated from double-reciprocal plots of the kinetic treatment. The maximum velocity of Na+ influx through the motor cannot be determined, because the efficiency ɛ cannot be determined, whereas it is assumed to be constant.
Detection of PomA, PomB, and the cross-linked products.
Cells were cultured in VPG500 medium at the mid-log phase of growth (optical density at 660 nm, ∼1.0). An equal volume (100 μl) of 20% (wt/vol) trichloroacetic acid (TCA) was added to the culture. Alternatively, for a combination of PomA-D148C/PomBcl-P16C, cells were collected by centrifugation, disrupted by sonication, and incubated for 30 min at room temperature prior to TCA precipitation. TCA-insoluble materials were collected by centrifugation, washed with acetone and diethylether, and dried. The dried materials were dissolved in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer containing 2 mM N-ethylmaleimide or 5% (vol/vol) 2-mercaptoethanol and subjected to SDS-PAGE. Immunoblotting was performed using anti-PomA and -PomB antisera as described previously (50). The band densities were quantified by using the NIH Image program to view the captured gel images through a charge-coupled device camera.
RESULTS
Cys substitutions in the TM of PomB.
PomB has a single TM whose precise boundary has not yet been determined. Each amino acid in the single TM of PomB was systematically replaced with Cys (Asp-24 to Glu-41) to assess the effects of each substitution per se and, thereafter, the effects of treatment with the water-soluble sulfhydryl reagent DTNB.
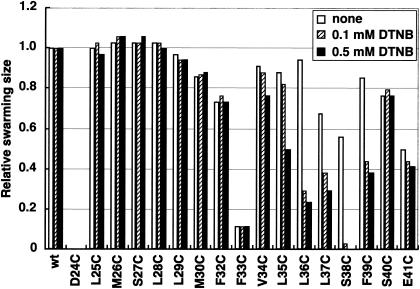
The Cys derivatives were expressed in a ΔpomB strain, and the motor function was assayed by monitoring swarming behavior. The swarming ability of all mutants except two, D24C and F33C, was comparable to that of the wild-type strain (Fig. 2). When cells were observed under the microscope, D24C was completely nonmotile and F33C showed very slow swimming (data not shown). The swarming diameter is assumed to reflect the flagellar function under these conditions. Since all of the PomB mutant proteins were detected at almost the same level by immunoblotting (data not shown), any loss or decrease of function must reflect the effects of the amino acid substitution rather than protein stability or expression levels. Asp-24 of PomB is an essential negatively charged residue, as has been demonstrated in MotB of the H+-type motor, where it corresponds to Asp-32 (57). Among the Cys substitutions, PomB-S38C was the most susceptible to DTNB treatment: in the presence of 0.1 mM DTNB, the swarm size of the PomB-S38C mutant was reduced to ∼5% of that in the control experiment. Swimming cells were almost nonmotile in the presence of 0.1 mM DTNB, but motility could be restored by addition of the reducing reagent DTT (data not shown). Around Ser-38 of PomB, Cys substitutions at positions 35 to 39 were sensitive to DTNB. This suggests that the environment around Ser-38 is exposed to the solvent, i.e., to the periplasmic space.
FIG. 2.
Effect of the sulfhydryl reagent DTNB on swarming of cells with Cys-substituted variants of PomB. Vibrio strain NMB192 (ΔpomB) harboring derivatives of pHK4 (pomB+) with Cys-substituted variants of pomB were spotted on semisolid plates containing no DTNB, 0.1 mM DTNB, and 0.5 mM DTNB and photographed after 4 h of incubation at 30°C. Relative swarm sizes were normalized to that of the wild type under the same conditions. The 31st residue is Cys in the wild-type PomB.
Combinations of Cys substitutions in the periplasmic boundaries of TMs.
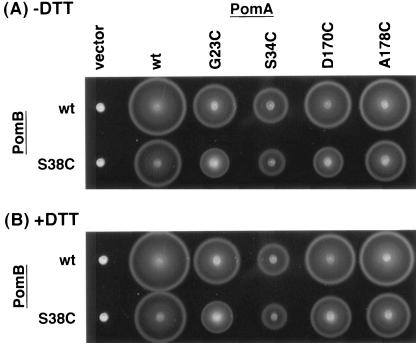
We tried to acquire topological information about the periplasmic regions of the PomA TMs and PomB TM. Cys substitutions were performed at Gly-23, Ser-34, Asp-170, and Ala-178, which correspond to the putative periplasmic boundaries of TM1, TM2, TM3, and TM4 in PomA, respectively (5). These mutants were combined with PomB-S38C, which is the most susceptible to DTNB treatment. The swimming and swarming assays were done without addition of oxidants. The periplasm is generally an oxidative environment, so a disulfide bond is formed spontaneously if two Cys residues are close to each other.
The swarming ability of the combined mutants was investigated in the presence or absence of the reductant DTT (Fig. 3). An S34C mutation in PomA significantly decreased the swarming ability, both with the wild type and with PomB-S38C. Swarming by the PomA-A178C or -G23C mutants was affected by the Na+ concentration, especially when combined with PomB-S38C (particularly PomA-G23C) (data not shown). No significant restoration of swarming was observed upon addition of DTT to the A178C and G23C mutants. On the other hand, addition of DTT restored the swarming ability in the combination mutant carrying PomA-D170C and PomB-S38C.
FIG. 3.
Combinations between Cys mutations in PomA TMs and PomB TM. Vibrio strain NMB191 (ΔpomAB) harboring derivatives of pYA303 (pomAB+) with Cys-substituted variants of pomA and pomB were spotted on semisolid plates containing 500 mM NaCl and 0 mM (A) or 2 mM (B) DTT and photographed after 4 h of incubation at 30°C. wt, wild type.
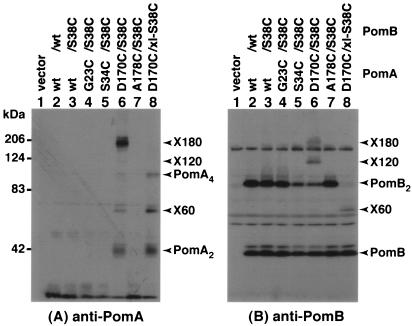
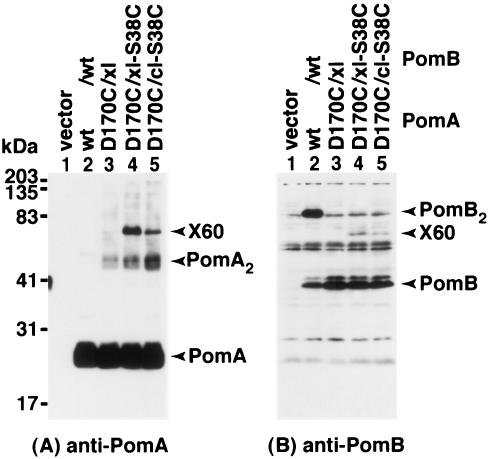
Cross-link formation between PomA and PomB was assessed with the double Cys mutants. PomB has three Cys residues, Cys-8 and Cys-10, which are predicted to be in the cytoplasm, and Cys-31, which is in the TM (Fig. 1) (2). PomB forms a cross-linked dimer (PomB2) in the absence of reducing agent but not in its presence (55). It has been shown that either Cys-8 or Cys-10 is responsible for dimer formation, but Cys-31 is not involved. Thus, the cross-link-less PomB (PomB-C8A/C10A [PomBxl]) and the Cys-less PomB (PomB-C8A/C10A/C31A [PomBcl]) were used to eliminate the complications in some experiments. Cross-linked products of PomA and PomB were not detected in the G23C, S34C, and A178C mutant forms of PomA with PomB-S38C but were found with the PomA D170C mutant protein (Fig. 4). In the combination of PomA-D170C/PomB-S38C, three cross-linked products could be detected without oxidant: a ca. 180-kDa product (X180) and a ca. 60-kDa product (X60), which were recognized by both anti-PomA and anti-PomB antisera, and a ca.120-kDa product (X120), which was recognized by anti-PomB antiserum. The X180 and X60 products could not be detected when a reducing agent was added or when either PomA or PomB was wild type (data not shown). The apparent molecular size of X60 is consistent with the sum of those of PomA (25 kDa) and PomB (37 kDa). X180 can be assignable to linear cross-linkings of PomA-PomB-PomB-PomA. On the other hand, the X120 product is not readily known, because X120 is rarely detected by anti-PomA antiserum. We suggest that the X120 product is assignable to an entangled complex of PomA-PomA and PomB-PomB disulfides due to the following observations: (i) X120 was not detected in the absence of the D170C mutation (Fig. 4B, lane 3), and (ii) the band equivalent to X120 was detected with the combination of PomA-D170C and wild-type PomB, although the amount was small (data not shown). Presumably because PomB has two endogenous Cys residues, Cys-8 and Cys-10, forming the cross-linked homodimer, several kinds of cross-linked products of PomA and PomB would be formed. We constructed the combination of PomA-D170C/PomBxl-S38C to interpret the results of cross-linking between PomA and PomB readily. Both X120 and X180 products were eliminated in this combination (Fig. 4A and B, lane 8). Among the Cys mutants of PomA tested in Fig. 4, a PomA dimer (PomA2) was only observed with D170C, which is consistent with results of a previous study (51). A PomA tetramer (PomA4), the ca.100-kDa band, was also detected with PomA-D170C (Fig. 4A), because a small proportion of the wild-type PomA runs as an SDS-resistant dimer, presumably because of the strong hydrophobic property (31). The PomA4 band otherwise may be an intertwining of cross-linked PomA dimers.
FIG. 4.
Cross-linking between PomA TM3 and PomB TM. Anti-PomA (A) and anti-PomB (B) immunoblots of whole-cell proteins of Vibrio strain NMB191 (ΔpomAB) harboring plasmids producing various combinations of Cys mutations of PomA and PomB as indicated above each lane. The whole-cell proteins were prepared as described in Materials and Methods. The details of X180, X120, and X60 are described in the text. wt, wild type.
Decrease in cross-linking of PomA TM3 and PomB TM by the C31A mutation in PomB.
The cells with PomB-C31A show a lesser swarm size (55) and swim more slowly (T. Yakushi and M. Homma, unpublished data) than those with wild-type PomB. Furthermore, wild-type PomA seems to be dissociated more readily from PomB with the C31A mutation than the wild-type protein under detergent-solubilized conditions (Yakushi and Homma, unpublished). When the C31A mutation was further introduced into PomBxl-S38C to construct PomBcl-S38C, the amount of X60 significantly decreased (Fig. 5). Cross-linking between Cys-170 in PomA-D170C and Cys-31 in PomBxl was not observed (Fig. 5A and B, lane 3). These results suggest that the formation of cross-linked PomA and PomB requires interactions between PomA and PomB.
FIG. 5.
Cross-linking between PomA TM3 and PomB requires their interaction. Whole-cell proteins of V. alginolyticus NMB191 (ΔpomAB) harboring plasmids producing various combinations of Cys mutations of PomA and PomB as indicated above each lane were separated by SDS-PAGE and immunoblotted with anti-PomA antibody (A) and anti-PomB antibody (B). wt, wild type.
Effects of double Cys mutations at the periplasmic boundaries of PomA TM3 and PomB TM.
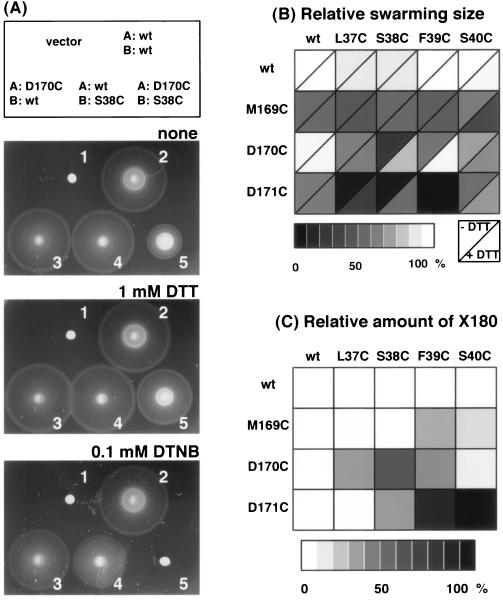
To further investigate the periplasmic boundaries of the PomA TM3 and PomB TM interface, Cys substitutions around the putative periplasmic boundary of PomA TM3 (M169-D171) and PomB TM (L37-S40) were prepared, and the combinations substituted in these sites were constructed in this study (Fig. 6).
FIG. 6.
Functional analysis and cross-link formation by the combinations of the PomA TM3 and PomB TM mutants in which Cys was introduced in the periplasmic boundaries. (A) Swarming ability of Vibrio strain NMB191 (ΔpomAB) harboring plasmids producing the PomA/PomB combinations as indicated in the top square of the photographs: 2, PomA and PomB; 3, PomA-D170C and PomB; 4, PomA and PomB-S38C; 5, PomA-D170C and PomB-S38C; or 1, vector plasmid. Semisolid agar plates contained no additions (top), 1 mM DTT (middle), or 0.1 mM DTNB (bottom). (B) Summary of relative swarm sizes of cells producing various PomA/PomB combinations. Each square represents a combination of different versions of PomA and PomB and is split into two triangles, according to whether DTT was absent or present at 2 mM. The cells are shaded according to the scale shown below. (C) Summary of cross-linking as represented by formation of X180 in each PomA/PomB combination (see the text). Amounts of X180 in each combination were measured, and the relative amounts were normalized to that of D171C/S40C as 100%. Each cell shows the results in the format from panel B and is shaded according to the scale shown below. wt, wild type.
For example, the swarming ability of the combined mutants PomA-D170C and PomB-S38C was shown in the presence or absence of the reductant DTT and the sulfhydryl reagent DTNB (Fig. 6A). The swarming was reduced to about 20 ± 6% (n = 4) of that of the wild type in the absence of DTT, but in the presence of DTT, it recovered to 65 ± 11% (n = 4) of the wild type level. The sulfhydryl reagent DTNB completely abolished the motility of a PomA-D170C/PomB-S38C mutant. On the other hand, the combinations of PomA-D170C/PomB wild type and PomA wild type/PomB-S38C are resistant to DTNB treatment. The other combinations are shown in Fig. 6B. Most combinations impaired the swarming ability, although single mutations in PomA or PomB did not significantly affect the swarming ability. The swarming ability was improved by the addition of DTT in D170C/F39C, D171C/S37C, and D171C/S38C as well as D170C/S38C. PomA-D171C/PomB-F39C abolished the swarming ability completely with or without DTT.
The swarming ability of all combined mutants other than combinations with PomB-S40C was impaired (>90% decrease) in the presence of 0.1 mM DTNB, although all combinations containing either wild-type PomA or PomB were resistant to this treatment (data not shown).
A cross-linked product comprised of PomA and PomB was detected as the X180 band (Fig. 6C). Asp-171 of PomA and Ser-40 of PomB are presumably very close and in easy access of each other because the highest amount of X180 was observed. However, the swarming ability was not lost in the presence of the combined mutations nor was it significantly affected by DTT (Fig. 6B). On the other hand, the cells with the combination of PomA-D171C and PomB-F39C, which showed a relatively high amount of X180, had severely decreased motility even in the presence of DTT, although each single mutation did not significantly affect motility.
Alteration of the Na+ requirement for the motility of PomA-D170C/PomB-S38C.
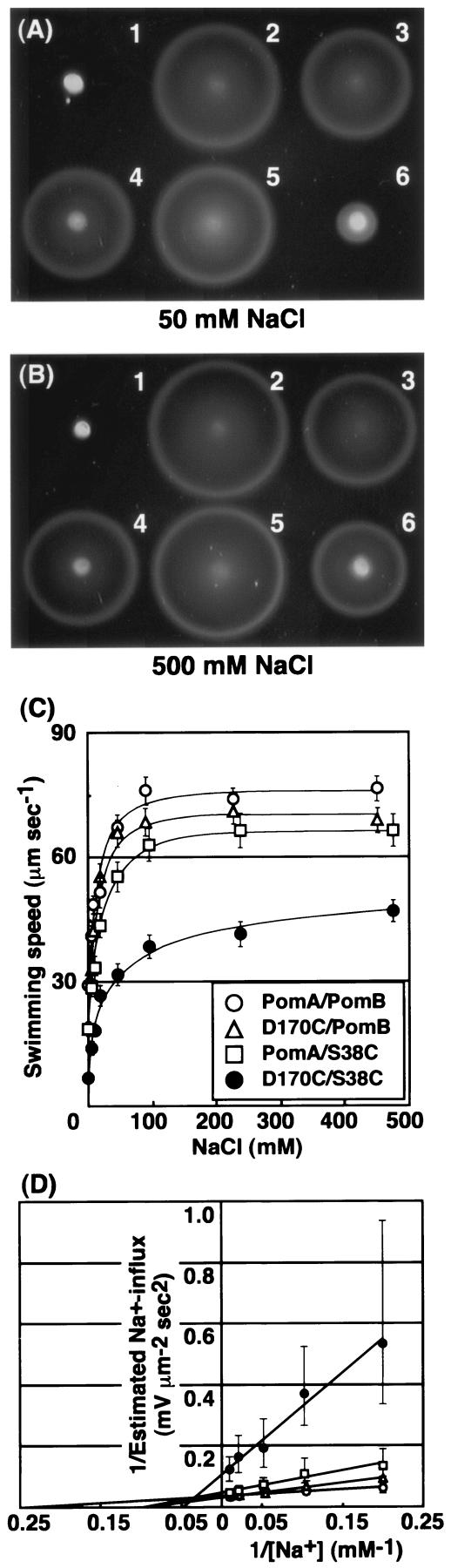
The swarming ability of the combination mutant PomA-D170C/PomB-S38C was examined at two different NaCl concentrations, 50 and 500 mM in the presence of DTT (Fig. 7A and B). As described above, the impaired swarming ability of the PomA-D170C/PomB-S38C mutant is significantly relieved by DTT (Fig. 6). The corresponding single mutants had almost the same swarming ability under the two salt conditions. However, swarming of the PomA-D170C/PomB-S38C mutant was severely affected at the lower NaCl concentration even in the presence of DTT. Swimming speeds were measured as a function of NaCl concentration in the presence of DTT for the PomA-D170C/PomB-S38C mutant and the corresponding single mutants (Fig. 7C). If DTT was omitted, cells having the PomA-D170C/PomB-S38C mutation could swim slightly (data not shown). Table 2 shows kinetic parameters of the single and the double mutants, as well as the swimming speeds at an NaCl concentration of 500 mM. The Na+ influx through the motor can be estimated as described previously (47). Apparent Km values for Na+ in the estimated Na+ influx were calculated for each PomA and PomB mutation (Fig. 7D; Table 2). Each single mutation showed a lower affinity for Na+ (∼twofold-higher Km value) than wild-type PomA/PomB. The affinity for Na+ in the combination mutant D170C/S38C was affected further (∼fivefold-higher Km value). The cells required significantly higher Na+ concentrations to achieve their highest swimming speeds and therefore presumably to achieve maximal rates of Na+ influx. Such swimming behaviors seem to correlate with the swarming results. These results may suggest that Asp-170 of PomA and Ser-38 of PomB functionally interact with each other and the interface is involved in the Na+ flux.
FIG. 7.
Effect of NaCl concentration on the motility of the mutant cells containing the combination of PomA-D170C and PomB-S38C. Swarming behavior in the presence of 50 mM NaCl (A) or 500 mM NaCl (B) of Vibrio strain NMB191 (ΔpomAB) harboring plasmids producing combinations of the following: 2, PomA and PomB; 3, PomA and PomBxl; 4, PomA-D170C and PomBxl; 5, PomA and PomBxl-S38C; 6, PomA-D170C and PomBxl-S38C; or 1, vector plasmid. Cells were incubated on semisolid agar plates for 5 h at 30°C. Both plates contained 2 mM DTT. (C) Swimming speed of Vibrio strain NMB195 (Che− ΔpomAB) harboring PomA and PomB, PomA-D170C and PomB, PomA and PomB-S38C, and PomA-D170C and PomB-S38C as a function of NaCl concentration. Symbols and error bars indicate the average values and standard deviations, respectively. (D) The double-reciprocal plots of the data shown in panel C after the kinetic treatment as described in Materials and Methods. Symbols and error bars are as described for panel C.
TABLE 2.
Swimming speeds and apparent Km values for estimated Na+ influx through the motor in the swimming cells
| PomA/PomBa | Swimming speedsb (μm/sec) | Kmc (mM) |
|---|---|---|
| wt/wt | 77 ± 6.2 | 4.1 |
| D170C/wt | 69 ± 6.3 | 9.7 |
| wt/S38C | 66 ± 8.0 | 10.4 |
| D170C/S38C | 47 ± 5.5 | 20.7 |
wt, wild type.
Swimming speeds were measured at an NaCl concentration of 500 mM.
Swimming speeds of Vibrio strain NMB195 (Che− ΔpomAB) producing PomA and PomB. PomA-D170C and PomB, PomA and PomB-S38C, and PomA-D170C and PomB-S38C were measured under different NaCl concentrations. Apparent Km values for Na+ in the estimated Na+ influx were calculated from the double-reciprocal plots (Fig. 7D) as described in Materials and Methods.
Cross-linking at the cytoplasmic boundaries of PomA TM3 and PomB TM.
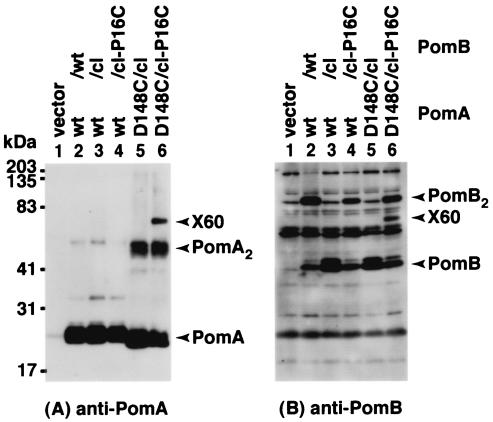
Phenamil-resistant mutations have been mapped at both Asp-148 of PomA and Pro-16 of PomB, suggesting that they form a phenamil high-affinity site and are in close proximity (28). The double mutant PomA-D148C/PomBcl-P16C was thus constructed to assess the physical interactions between the two proteins. A 60-kDa cross-linked product (X60), whose size corresponds to the sum of those of PomA and PomB, could be detected by both anti-PomA and anti-PomB antisera (Fig. 8A and B, lane 6). X60 was not detected in the singly substituted mutants (Fig. 8) and was lost upon addition of reducing reagents such as 2-mercaptoethanol (data not shown). Cells expressing either PomA-D148C or PomBcl-P16C were motile but less so than the wild type, as reported previously (28). Cells having both mutations were not motile even in the presence of DTT (data not shown). This is probably a result of a synergistic effect between the two mutations. These results directly support the proposal that Asp-148 of PomA and Pro-16 of PomB are close to each other.
FIG. 8.
Cross-linking between PomA-D148C and PomBcl-P16C. A P16C mutation was introduced into Cys-less PomB (PomBcl). Anti-PomA (A) and anti-PomB (B) immunoblots of whole-cell proteins of V. alginolyticus NMB191 (ΔpomAB) harboring plasmids producing various combinations of wild-type PomA or PomA-D148C and wild-type PomB or PomBcl-P16C, as indicated above each lane. The whole-cell proteins were derived from the cell lysates prepared by disruption using sonicator prior to TCA precipitation as described in Materials and Methods. wt, wild type.
DISCUSSION
The flagellar motor proteins participate in the energy transduction from the electrochemical potential to a mechanical output, the rotation of the flagellum. The PomA/PomB complex and MotA/MotB complex function as ion conductors in the Na+-driven and H+-driven flagella, respectively (13, 43). The interactions between PomA and PomB and the Na+ conductance are likely to be critical factors for the flagellar rotation. Cys-scanning mutagenesis is useful for studying the structure and function of membrane proteins for which the stereostructure has not been solved (24). In order to understand the interaction between PomA and PomB, site-directed Cys mutagenesis was performed on the PomB TM, and the Cys mutants of PomA TM3 and PomB TM were combined. Our proposals are consistent with those for the H+-driven motor, where it has been proposed that the essential Asp-32 of MotB (corresponding to Asp-24 of PomB) faces MotA to make the H+-conducting channel because Asp-32 was presumed to point away from the interface of the MotB dimer. Recently, a more detailed hypothetical arrangement of the TMs of MotA/MotB complex was proposed (12, 15).
Systematic substitutions in the TM of PomB showed that the putative Na+-binding site Asp-24 is essential for PomB function, consistent with data on Asp-32 of E. coli MotB (57). The F33C mutation also significantly impaired motility. This phenylalanine, which has a bulky side chain, might play an important role in maintaining an interaction among the TMs. In E. coli, MotB-F40C and MotB-W45C resulted in more severe effects on motility than the other Cys mutations of the TM region (14). Treatment with DTNB had the greatest effect on the Cys mutant, PomB-S38C, and also had an effect on residues from L35 to F39, suggesting that this region may have high solvent accessibility. The TM of PomB has been tentatively defined by hydrophobicity analysis from Gly-20 to Phe-39 (2); the precise periplasmic and cytoplasmic boundaries have not yet been determined. According to the previous prediction, Ser-38 is buried under the membrane, which is inconsistent with the assumption in this study. Therefore, assignment of the TM of PomB may be shifted from that of the previous study to give a tentative model as shown in Fig. 1. Plasmid-borne PomB-S38C expressed with the chromosomal PomA was sensitive to DTNB (Fig. 2). However, the DTNB sensitivity of PomB-S38C was repressed by coexpression of wild-type PomA (Fig. 6A). The PomB protein should be in excess of the PomA protein in the cells used in Fig. 2, whereas both PomA and PomB proteins are overproduced from a plasmid in the cells used in Fig. 6. An increase in the amount of PomA probably facilitates the formation of the PomA/PomB complex. If the PomA/PomB-S38C complex was influenced by the DTNB treatment within the flagellar motor, we expect that the motility would be inhibited irrespective of coexpression of PomA. It is likely that PomA affects the structure around the TM region of PomB, resulting in an alteration of the sensitivity to DTNB treatment. Therefore, we speculate that DTNB would affect the PomB-S38C protein that does not associate with PomA but it would no longer affect the PomB-S38C protein once the PomA/PomB-S38C complex is formed.
In this study, cross-link formation between PomA TM3 and PomB TM was detected. We suggest that the cross-linking between PomA TM3 and PomB TM reflects the association in the native PomA/PomB complex rather than aggregation due to overproduction because of the following observations: (i) cross-links were not detected in cells with single Cys mutations in either PomA or PomB and were specifically formed to the residues where Cys was introduced, (ii) the C31A mutation in PomB decreased the cross-linking between PomA-D170C and PomB-S38C, and moreover, (iii) the cross-linked products corresponding to the X180, X120, and X60 were detected in the same fraction where the wild-type PomA/PomB complex eluted, when detergent-solubilized cross-linked and un-cross-linked PomA/PomB complexes were separated by gel filtration (D. Yoshimura, T. Yakushi, and M. Homma, unpublished results). Treatment with DTNB did little to enhance the cross-link formation (data not shown), whereas it significantly enhanced the inhibition of swarming ability (Fig. 6A). Asp-171 of PomA and Ser-40 of PomB are presumed to be the closest residues of the combinations tested in this study because the highest amounts of cross-linked product were observed (Fig. 6C).
By using the wild-type PomB protein that has three endogenous Cys residues, the periplasmic substitutions resulted in the formation of X60 and X180, which reacted with both anti-PomA and anti-PomB antisera. PomA-D170C has a single Cys residue, so the sulfhydryl group of D170C can react with either that of another PomA-D170C or PomB-S38C. Two of the three endogenous Cys residues of PomB, Cys-8 and Cys-10, are responsible for the homotypic cross-link formation (55). Thus, it is likely that X180 is a heterotetramer, PomA-PomB-PomB-PomA: the cross-linked products of PomA-D170C and PomB-S38C are cross-linked through the cytoplasmic Cys residues of PomB to form the linear cross-linked product. The heterotetramer X180 seems to reflect a part of the complex of four PomA and two PomB proteins, (PomA2/PomB)2 (43, 55). On the other hand, X120 was clearly detected by anti-PomB antiserum, but just a faint band can be detected by anti-PomA antiserum (Fig. 4). The D170C mutation in PomA and three endogenous Cys of PomB are sufficient for the X120 formation (Fig. 4; data not shown). Because X120 are readily detected with anti-PomA antiserum through purification of the cross-linked products (Yakushi and Homma, unpublished), it is likely that X120 consists of PomA and PomB but there is some steric hindrance from the antibody recognition. Therefore, from the mobility of the X120 band on SDS-PAGE, we suggest that X120 is an entangled complex of the homotypic cross-linked products of PomA-PomA and PomB-PomB, being masked against anti-PomA antibody. The nature of PomA forming an SDS-resistant dimer would not be involved in the formation of the two different types of the larger cross-linked products, X120 and X180, because the combination of PomA-D170C and PomBxl-S38C showed a single cross-linked product, X60 (Fig. 4 and 5).
As PomA-D170C/PomB-S38C, the combinations of a series of the Cys substitutions in PomA TM3 and PomB TM synergistically affected the swarming ability and formed cross-linked products of PomA and PomB (Fig. 6). Furthermore, it has been proposed that Asp-148 in TM3 of PomA and Pro-16 of PomB may be close to each other and form a phenamil high-affinity site (28), results consistent with this study showing that substituted Cys residues at these positions are actually cross-linked (Fig. 8). Since PomA TM3 and PomB TM are close to each other on both faces of the membrane, it is reasonable to infer that they remain in intimate contact throughout the entire TM region from the periplasmic to the cytoplasmic side.
It has been shown that the sensitivity of PomA-D170C to DTNB treatment is affected by the NaCl concentration (5). Asp-170 is predicted to face the pore of the channel and to interact with Na+. The present study supports this possibility because of the higher apparent Km value for Na+. The swarming ability of cells with PomA-D170C/PomB-S38C was significantly decreased at low NaCl concentrations even in the presence of DTT (Fig. 7), although the swarming ability of the single mutants was only slightly affected by the NaCl concentration. The kinetic analysis of swimming confirms that the double mutant requires a higher concentration of Na+. Taken together, it is suggested that an interface around Asp-170 of PomA and Ser-38 of PomB participates in Na+ conductance to drive the motor. A working hypothesis has been proposed such that the motor has two ion-binding sites, extracellular and intracellular, for the ion flux (36, 46, 56). It is plausible that Asp-170 of PomA and Ser-38 of PomB may participate as an entrance for extracellular Na+. In this work, we found that the mutation of Gly-23 in PomA TM1 affects the Na+ requirement for motility and shows synergism with the PomB-S38C mutation (Fig. 3; data not shown). It has been shown that the strains with PomA substitutions at Asp-31 in periplasmic loop1-2 had a slow-motility phenotype and required higher Na+ concentrations to start swimming than those with wild-type PomA (32). This may suggest that the periplasmic loop1-2 or the boundary also contribute to an entrance for the Na+ translocation.
The cytoplasmic region of PomA, which connects TM2 and TM3, is involved in flagellar rotation and interacts with a rotor protein, FliG (52). As PomB TM has a central role in Na+ flux, we speculate that the Na+ flux in the interface between PomA TM3 and PomB TM evoke conformational changes of the cytoplasmic region of PomA to drive the flagellar rotation. In the H+-type motor of E. coli, it was shown that the protonation of Asp-32 in the MotB TM induces a conformational change in the cytoplasmic region between TM2 and TM3 (29). The stator complex is apparently composed of four PomA (MotA) and two PomB (MotB) molecules or a larger complex than the 4:2 complex (48). At least 18 TMs of the complex are arranged in the cytoplasmic membrane to form an ion channel or channels. Based on the recent cross-linking data, a model for the arrangements of 10 TMs in the MotA/MotB complex of E. coli has been proposed (15). The model is consistent with our results demonstrating that the PomB TM and the PomA TM3 regions are in closest association to each other in the TMs. To understand the mechanism of force generation by ion flux, the arrangement and structure of the TM regions must be clarified.
Acknowledgments
We thank Noriko Ui and Emi Mitsuyama for technical assistance and Ikuro Kawagishi, Yoshiyuki Sowa, and Tomohiro Yorimitsu for stimulating discussions. We also thank Yukako Asai and Seiji Kojima for construction of bacterial strains. We are grateful to the late Robert M. Macnab for invaluable discussions and critically reading the manuscript.
This work was supported in part by grants-in-aid for scientific research from the Ministry of Education, Science, and Culture of Japan; the Japan Science and Technology Corporation (to M.H. and T.Y.); and from the Soft Nano-Machine Project of Japan Science and Technology Agency (to T.Y. and M.H.).
REFERENCES
- 1.Armitage, J. P., and R. M. Macnab. 1987. Unidirectional, intermittent rotation of the flagellum of Rhodobacter sphaeroides. J. Bacteriol. 169:514-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asai, Y., S. Kojima, H. Kato, N. Nishioka, I. Kawagishi, and M. Homma. 1997. Putative channel components for the fast-rotating sodium-driven flagellar motor of a marine bacterium. J. Bacteriol. 179:5104-5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asai, Y., I. Kawagishi, E. Sockett, and M. Homma. 1999. Hybrid motor with the H+-and Na+-driven components can rotate Vibrio polar flagella by using sodium ions. J. Bacteriol. 181:6322-6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asai, Y., R. E. Sockett, I. Kawagishi, and M. Homma. 2000. Coupling ion specificity of chimeras between H+- and Na+-driven motor proteins, MotB and PomB, in Vibrio polar flagella. EMBO J. 19:3639-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asai, Y., T. Shoji, I. Kawagishi, and M. Homma. 2000. Cysteine-scanning mutagenesis of the periplasmic loop regions of PomA, a putative channel component of the sodium-driven flagellar motor in Vibrio alginolyticus. J. Bacteriol. 182:1001-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asai, Y., T. Yakushi, I. Kawagishi, and M. Homma. 2003. Ion-coupling determinants of Na+-driven and H+-driven flagellar motors. J. Mol. Biol. 327:453-463. [DOI] [PubMed] [Google Scholar]
- 7.Atsumi, T., S. Sugiyama, E. J. Cragoe, Jr., and Y. Imae. 1990. Specific inhibition of the Na+-driven flagellar motors of alkalophilic Bacillus strains by the amiloride analog phenamil. J. Bacteriol. 172:1634-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Atsumi, T., L. McCarter, and Y. Imae. 1992. Polar and lateral flagellar motors of marine Vibrio are driven by different ion-motive forces. Nature 355:182-184. [DOI] [PubMed] [Google Scholar]
- 9.Atsumi, T., Y. Maekawa, T. Yamada, I. Kawagishi, Y. Imae, and M. Homma. 1996. Effect of viscosity on swimming by the lateral and polar flagella of Vibrio alginolyticus. J. Bacteriol. 178:5024-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartolome, B., Y. Jubete, E. Martinez, and F. de la Cruz. 1991. Construction and properties of a family of pACY184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 102:75-78. [DOI] [PubMed] [Google Scholar]
- 11.Blair, D. F. 1995. How bacteria sense and swim. Annu. Rev. Microbiol. 49:489-522. [DOI] [PubMed] [Google Scholar]
- 12.Blair, D. F. 2003. Flagellar movement driven by proton translocation. FEBS Lett. 545:86-95. [DOI] [PubMed] [Google Scholar]
- 13.Blair, D. F., and H. C. Berg. 1990. The MotA protein of E. coli is a proton-conducting component of the flagellar motor. Cell 60:439-449. [DOI] [PubMed] [Google Scholar]
- 14.Braun, T., and D. Blair. 2001. Targeted disulfide cross-linking of the MotB protein of Escherichia coli: evidence for two H+ channels in the stator complex. Biochemistry 40:13051-13059. [DOI] [PubMed] [Google Scholar]
- 15.Braun, T. F., L. Q. Al-Mawsawi, S. Kojima, and D. F. Blair. 2004. Arrangement of core membrane segments in the MotA/MotB proton-channel complex of Escherichia coli. Biochemistry 43:35-45. [DOI] [PubMed] [Google Scholar]
- 16.Braun, T. F., S. Poulson, J. B. Gully, J. C. Empey, S. Van Way, A. Putnam, and D. F. Blair. 1999. Function of proline residues of MotA in torque generation by the flagellar motor of Escherichia coli. J. Bacteriol. 181:3542-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Mot, R., and J. Vanderleyden. 1994. The C-terminal sequence conservation between OmpA-related outer membrane proteins and MotB suggests a common function in both gram-positive and gram-negative bacteria, possibly in the interaction of these domains with peptidoglycan. Mol. Microbiol. 12:333-334. [DOI] [PubMed] [Google Scholar]
- 18.Derosier, D. J. 1998. The turn of the screw: the bacterial flagellar motor. Cell 93:17-20. [DOI] [PubMed] [Google Scholar]
- 19.Garza, A., L. Harris-Haller, R. Stoebner, and M. Manson. 1995. Motility protein interactions in the bacterial flagellar motor. Proc. Natl. Acad. Sci. USA 92:1970-1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gosink, K. K., and C. C. Häse. 2000. Requirements for conversion of the Na+-driven flagellar motor of Vibrio cholerae to the H+-driven motor of Escherichia coli. J. Bacteriol. 182:4234-4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirota, N., M. Kitada, and Y. Imae. 1981. Flagellar motors of alkalophilic Bacillus are powered by an electrochemical potential gradient of Na+. FEBS Lett. 132:278-280. [Google Scholar]
- 22.Homma, M., H. Oota, S. Kojima, I. Kawagishi, and Y. Imae. 1996. Chemotactic responses to an attractant and a repellent in the flagellar systems of Vibrio alginolyticus. Microbiology 142:2777-2783. [DOI] [PubMed] [Google Scholar]
- 23.Jaques, S., Y.-K. Kim, and L. McCarter. 1999. Mutations conferring resistance to phenamil and amiloride, inhibitors of sodium-driven motility of Vibrio parahaemolyticus. Proc. Natl. Acad. Sci. USA 96:5740-5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaback, H. R., M. Sahin-Toth, and A. B. Weinglass. 2001. The kamikaze approach to membrane transport. Nat. Rev. Mol. Cell. Biol. 2:610-620. [DOI] [PubMed] [Google Scholar]
- 25.Kawagishi, I., I. Okunishi, M. Homma, and Y. Imae. 1994. Removal of the periplasmic DNase before electroporation enhances efficiency of transformation in a marine bacterium Vibrio alginolyticus. Microbiology 140:2355-2361. [Google Scholar]
- 26.Kawagishi, I., Y. Maekawa, T. Atsumi, M. Homma, and Y. Imae. 1995. Isolation of the polar and lateral flagellum-defective mutants in Vibrio alginolyticus and identification of their flagellar driving energy sources. J. Bacteriol. 177:5158-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khan, S., M. Dapice, and T. S. Reese. 1988. Effects of mot gene expression on the structure of the flagellar motor. J. Mol. Biol. 202:575-584. [DOI] [PubMed] [Google Scholar]
- 28.Kojima, S., Y. Asai, T. Atsumi, I. Kawagishi, and M. Homma. 1999. Na+-driven flagellar motor resistant to phenamil, an amiloride analog, caused by mutations of putative channel components. J. Mol. Biol. 285:1537-1547. [DOI] [PubMed] [Google Scholar]
- 29.Kojima, S., and D. F. Blair. 2001. Conformational change in the stator of the bacterial flagellar motor. Biochemistry 40:13041-13050. [DOI] [PubMed] [Google Scholar]
- 30.Kojima, S., and D. F. Blair. 2004. Solubilization and purification of the MotA/MotB complex of Escherichia coli. Biochemistry 43:26-34. [DOI] [PubMed] [Google Scholar]
- 31.Kojima, S., M. Kuroda, I. Kawagishi, and M. Homma. 1999. Random mutagenesis of the pomA gene encoding the putative channel component of the Na+-driven polar flagellar motor of Vibrio alginolyticus. Microbiology 145:1759-1767. [DOI] [PubMed] [Google Scholar]
- 32.Kojima, S., T. Shoji, Y. Asai, I. Kawagishi, and M. Homma. 2000. A slow-motility phenotype caused by substitutions at residue Asp31 in the PomA channel component of a sodium-driven flagellar motor. J. Bacteriol. 182:3314-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Macnab, R. 1996. Flagella and motility, p. 123-145. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella, 2nd ed., vol. 2. American Society for Microbiology, Washington, D.C.
- 34.Manson, M., P. Tedesco, H. Berg, F. Harold, and C. Van der Drift. 1977. A protonmotive force drives bacterial flagella. Proc. Natl. Acad. Sci. USA 74:3060-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCarter, L. L. 2001. Polar flagellar motility of the Vibrionaceae. Microbiol. Mol. Biol. Rev. 65:445-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Minamino, T., Y. Imae, F. Oosawa, Y. Kobayashi, and K. Oosawa. 2003. Effect of intracellular pH on rotational speed of bacterial flagellar motors. J. Bacteriol. 185:1190-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okabe, M., T. Yakushi, Y. Asai, and M. Homma. 2001. Cloning and characterization of motX, a Vibrio alginolyticus sodium-driven flagellar motor gene. J. Biochem. 130:879-884. [DOI] [PubMed] [Google Scholar]
- 38.Okabe, M., T. Yakushi, M. Kojima, and M. Homma. 2002. MotX and MotY, specific components of the sodium-driven flagellar motor, co-localize to the outer membrane in Vibrio alginolyticus. Mol. Microbiol. 46:125-134. [DOI] [PubMed] [Google Scholar]
- 39.Okunishi, I., I. Kawagishi, and M. Homma. 1996. Cloning and characterization of motY, a gene coding for a component of the sodium-driven flagellar motor in Vibrio alginolyticus. J. Bacteriol. 178:2409-2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Platzer, J., W. Sterr, M. Hausmann, and R. Schmitt. 1997. Three genes of a motility operon and their role in flagellar rotary speed variation in Rhizobium meliloti. J. Bacteriol. 179:6391-6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook, J., E. F. Fritsch, and T. Maniatis (ed.). 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 42.Sato, K., and M. Homma. 2000. Multimeric structure of PomA, the Na+-driven polar flagellar motor component of Vibrio alginolyticus. J. Biol. Chem. 275:20223-20228. [DOI] [PubMed] [Google Scholar]
- 43.Sato, K., and M. Homma. 2000. Functional reconstitution of the Na+-driven polar flagellar motor component of Vibrio alginolyticus. J. Biol. Chem. 275:5718-5722. [DOI] [PubMed] [Google Scholar]
- 44.Sharp, L. L., J. D. Zhou, and D. F. Blair. 1995. Features of MotA proton channel structure revealed by tryptophan-scanning mutagenesis. Proc. Natl. Acad. Sci. USA 92:7946-7950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharp, L. L., J. D. Zhou, and D. F. Blair. 1995. Tryptophan-scanning mutagenesis of MotB, an integral membrane protein essential for flagellar rotation in Escherichia coli. Biochemistry 34:9166-9171. [DOI] [PubMed] [Google Scholar]
- 46.Sowa, Y., H. Hotta, M. Homma, and A. Ishijima. 2003. Torque-speed relationship of the Na+-driven flagellar motor of Vibrio alginolyticus. J. Mol. Biol. 327:1043-1051. [DOI] [PubMed] [Google Scholar]
- 47.Sugiyama, S., E. J. Cragoe, Jr., and Y. Imae. 1988. Amiloride, a specific inhibitor for the Na+-driven flagellar motors of alkalophilic Bacillus. J. Biol. Chem. 263:8215-8219. [PubMed] [Google Scholar]
- 48.Yakushi, T., M. Kojima, and M. Homma. 2004. Isolation of Vibrio alginolyticus sodium-driven flagellar motor complex composed of PomA and PomB solubilized by sucrose monocaprate. Microbiology 150:911-920. [DOI] [PubMed] [Google Scholar]
- 49.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 50.Yorimitsu, T., K. Sato, Y. Asai, I. Kawagishi, and M. Homma. 1999. Functional interaction between PomA and PomB, the Na+-driven flagellar motor components of Vibrio alginolyticus. J. Bacteriol. 181:5103-5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yorimitsu, T., K. Sato, Y. Asai, and M. Homma. 2000. Intermolecular cross-linking between the periplasmic loop3-4 regions of PomA, a component of the Na+-driven flagellar motor of Vibrio alginolyticus. J. Biol. Chem. 275:31387-31391. [DOI] [PubMed] [Google Scholar]
- 52.Yorimitsu, T., and M. Homma. 2001. Na+-driven flagellar motor of Vibrio. Biochim. Biophys. Acta 1505:82-93. [DOI] [PubMed] [Google Scholar]
- 53.Yorimitsu, T., Y. Sowa, A. Ishijima, T. Yakushi, and M. Homma. 2002. The systematic substitutions around the conserved charged residues of the cytoplasmic loop of Na+-driven flagellar motor component PomA. J. Mol. Biol. 320:403-413. [DOI] [PubMed] [Google Scholar]
- 54.Yorimitsu, T., A. Mimaki, T. Yakushi, and M. Homma. 2003. The conserved charged residues of the C-terminal region of FliG, a rotor component of Na+-driven flagellar motor. J. Mol. Biol. 334:567-583. [DOI] [PubMed] [Google Scholar]
- 55.Yorimitsu, T., M. Kojima, T. Yakushi, and M. Homma. 2004. Multimeric structure of the PomA/PomB complex, the channel component of the Na+-driven flagellar motor of Vibrio alginolyticus. J. Biochem. 135:43-51. [DOI] [PubMed] [Google Scholar]
- 56.Yoshida, S., S. Sugiyama, Y. Hojo, H. Tokuda, and Y. Imae. 1990. Intracellular Na+ kinetically interferes with the rotation of the Na+-driven flagellar motors of Vibrio alginolyticus. J. Biol. Chem. 265:20346-20350. [PubMed] [Google Scholar]
- 57.Zhou, J., L. L. Sharp, H. L. Tang, S. A. Lloyd, S. Billings, T. F. Braun, and D. F. Blair. 1998. Function of protonatable residues in the flagellar motor of Escherichia coli: a critical role for Asp 32 of MotB. J. Bacteriol. 180:2729-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou, J. D., R. T. Fazzio, and D. F. Blair. 1995. Membrane topology of the MotA protein of Escherichia coli. J. Mol. Biol. 251:237-242. [DOI] [PubMed] [Google Scholar]
- 59.Zhou, J. D., S. A. Lloyd, and D. F. Blair. 1998. Electrostatic interactions between rotor and stator in the bacterial flagellar motor. Proc. Natl. Acad. Sci. USA 95:6436-6441. [DOI] [PMC free article] [PubMed] [Google Scholar]