In both structures, molecules are linked into hydrogen-bonded chains. In (Z)-5-[2-(benzo[b]thiophen-2-yl)-1-(3,5-dimethoxyphenyl)ethenyl]-1H-tetrazole methanol monosolvate, these chains involve both tetrazole and methanol, and are parallel to the b axis. In (Z)-5-[2-(benzo[b]thiophen-3-yl)-1-(3,4,5-trimethoxyphenyl)ethenyl]-1H-tetrazole, molecules are linked into chains parallel to the a axis by N—H⋯N hydrogen bonds between adjacent tetrazole rings.
Keywords: crystal structure, 5-substituted-1H-tetrazoles, tetrazole-tethered combretastatin A-4 analogs, anticancer agents, hydrogen bonding
Abstract
(Z)-5-[2-(Benzo[b]thiophen-2-yl)-1-(3,5-dimethoxyphenyl)ethenyl]-1H-tetrazole methanol monosolvate, C19H16N4O2S·CH3OH, (I), was prepared by the reaction of (Z)-3-(benzo[b]thiophen-2-yl)-2-(3,5-dimethoxyphenyl)acrylonitrile with tributyltin azide via a [3 + 2]cycloaddition azide condensation reaction. The structurally related compound (Z)-5-[2-(benzo[b]thiophen-3-yl)-1-(3,4,5-trimethoxyphenyl)ethenyl]-1H-tetrazole, C20H18N4O3S, (II), was prepared by the reaction of (Z)-3-(benzo[b]thiophen-3-yl)-2-(3,4,5-trimethoxyphenyl)acrylonitrile with tributyltin azide. Crystals of (I) have two molecules in the asymmetric unit (Z′ = 2), whereas crystals of (II) have Z′ = 1. The benzothiophene rings in (I) and (II) are almost planar, with r.m.s deviations from the mean plane of 0.0084 and 0.0037 Å in (I) and 0.0084 Å in (II). The tetrazole rings of (I) and (II) make dihedral angles with the mean planes of the benzothiophene rings of 88.81 (13) and 88.92 (13)° in (I), and 60.94 (6)° in (II). The dimethoxyphenyl and trimethoxyphenyl rings make dihedral angles with the benzothiophene rings of 23.91 (8) and 24.99 (8)° in (I) and 84.47 (3)° in (II). In both structures, molecules are linked into hydrogen-bonded chains. In (I), these chains involve both tetrazole and methanol, and are parallel to the b axis. In (II), molecules are linked into chains parallel to the a axis by N—H⋯N hydrogen bonds between adjacent tetrazole rings.
Chemical context
We have reported on benzothiophene cyanocombretastatin A-4 analogs (Penthala et al., 2013 ▸), and benzothiophene triazolylcombretastatin A-4 analogs as promising anti-cancer agents (Penthala et al., 2015 ▸). Previously, we published the synthesis of triazolylcombretastatin A-4 analogs utilizing a [3 + 2]cycloaddition azide condensation reaction with sodium azide in the presence of l-proline as catalyst (Penthala et al., 2014a ▸). In a continuation of our work on the chemical modification of the cyano group on the stilbene moiety of cyanocombretastatin A-4 analogs (Penthala et al., 2014a ▸), we have recently synthesized tetrazolylcombretastatin A-4 analogs as potential anti-cancer agents (Penthala et al., 2016 ▸).
Structural commentary
Single crystal X-ray analysis was carried out to obtain the structural conformations of the tetrazolylcombretastatin A-4 analogs (I) and (II) for the analysis of structure–activity relationships (SAR), the relevance of the geometry of the tetrazole ring on the stilbene scaffold and to confirm the position of the hydrogen atom in the tetrazole ring system. The single crystal X-ray structures of (I) and (II) are shown in Figs. 1 ▸ and 2 ▸, respectively.
Figure 1.
The molecular structure of (I), with displacement ellipsoids drawn at the 50% probability level.
Figure 2.
The molecular structure of (II), with displacement ellipsoids drawn at the 50% probability level.
The benzothiophene rings are almost planar with r.m.s. deviations from the mean plane of 0.0084 and 0.0037 Å in (I) and 0.0084 Å in (II), with bond distances and angles comparable with those reported for other benzothiophene derivatives (Sonar et al., 2007 ▸; Penthala et al., 2014b ▸). The tetrazole rings make dihedral angles with the mean plane of the benzothiophene rings of 88.81 (13) and 88.92 (13)° in (I), and 60.94 (6)° in (II). The dimethoxyphenyl ring in (I) and trimethoxyphenyl ring in (II) make dihedral angles with the benzothiophene rings of 23.91 (8) and 24.99 (8)° in (I) and 84.47 (3)° in (II). Bond lengths and angles in both (I) and (II) are, by and large, unremarkable.
Supramolecular features
Hydrogen bonding and the mode of packing of (I) is illustrated in Fig. 3 ▸, and the mode of packing of (II) is illustrated in Fig. 4 ▸. In the structure of (I), the molecules are linked into hydrogen-bonded (Table 1 ▸) chains parallel to the crystallographic b axis involving interaction between tetrazole–tetrazole (N—H⋯N), tetrazole–methanol (O—H⋯N and N—H⋯O), and methanol–methanol (O—H⋯O). These chains are bidirectional, as the hydrogen atoms on the tetrazole rings and the methanol oxygen atom appear to be disordered over two positions. In the structure of (II), the molecules are linked into chains parallel to the a axis by intermolecular N—H⋯N hydrogen bonds (Table 2 ▸) between adjacent tetrazole rings.
Figure 3.
Crystal packing of (I), viewed down the c axis.
Figure 4.
Crystal packing of (II), viewed down the c axis.
Table 1. Hydrogen-bond geometry (Å, °) for (I) .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1A—H1NA⋯N1A i | 0.88 | 1.91 | 2.787 (9) | 176 |
| N4A—H4NA⋯O1SB | 0.88 | 1.87 | 2.736 (6) | 168 |
| N1B—H1NB⋯N1B ii | 0.88 | 1.92 | 2.792 (9) | 174 |
| N4B—H4NB⋯O1SA | 0.88 | 1.91 | 2.769 (6) | 165 |
| O1SA—H1SA⋯N4B | 0.84 | 1.97 | 2.769 (6) | 158 |
| O1SA—H2SA⋯O1SA iii | 0.84 | 1.81 | 2.646 (7) | 177 |
| O1SB—H1SB⋯N4A | 0.84 | 1.90 | 2.736 (6) | 176 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  .
.
Table 2. Hydrogen-bond geometry (Å, °) for (II) .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1N⋯N3i | 0.91 (2) | 2.65 (2) | 3.3886 (19) | 138.5 (16) |
| N1—H1N⋯N4i | 0.91 (2) | 1.85 (2) | 2.7482 (19) | 167.1 (18) |
Symmetry code: (i) 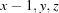 .
.
Database survey
A search of the 2015 Cambridge Structural Database (Groom & Allen, 2014 ▸) for tetrazole bonded via its carbon atom to another carbon atom yielded 255 hits. Of these, only two were bonded to an sp 2 carbon atom, namely 5-(2H-chromen-3-yl)-1H-tetrazole monohydrate (NEYCUR: Gawande et al., 2013 ▸) and (2Z,4E)-5-(dimethylamino)-2-(1H-tetrazol-5-yl)penta-2,4-dienenitrile methanol solvate (YUPPAB: Addicott et al., 2009 ▸). Neither NEYCUR nor YUPPAB have any particular similarity to compounds (I) and (II).
Synthesis and crystallization
The title compounds (I) and (II) were prepared by utilizing our recently reported literature procedure (Penthala et al., 2016 ▸). Recrystallization of the compounds from methanol afforded (I) and (II) as pale-yellow crystalline products which were suitable for X-ray analysis.
Refinement details
Crystal data, data collection and refinement details for both (I) and (II) are summarized in Table 3 ▸. H atoms were found in difference Fourier maps and subsequently placed at idealized positions with constrained distances of 0.95 Å (R 2Csp 2H), 0.98 Å (RCH3), 0.84 Å (OH), 0.88 Å (Nsp 2H). U iso(H) values were set to either 1.2U eq or 1.5U eq (RCH3, OH) of the attached atom. Final models were checked using PLATON (Spek, 2009 ▸), RT (Parkin, 2000 ▸), and by checkCIF.
Table 3. Experimental details.
| (I) | (II) | |
|---|---|---|
| Crystal data | ||
| Chemical formula | C19H16N4O2S·CH4O | C20H18N4O3S |
| M r | 396.46 | 394.44 |
| Crystal system, space group | Orthorhombic, P21212 | Monoclinic, P21/c |
| Temperature (K) | 90 | 90 |
| a, b, c (Å) | 18.2226 (4), 13.7954 (5), 15.5594 (5) | 4.8888 (1), 24.6650 (6), 15.5956 (4) |
| α, β, γ (°) | 90, 90, 90 | 90, 91.031 (1), 90 |
| V (Å3) | 3911.4 (2) | 1880.25 (8) |
| Z | 8 | 4 |
| Radiation type | Cu Kα | Cu Kα |
| μ (mm−1) | 1.72 | 1.78 |
| Crystal size (mm) | 0.21 × 0.15 × 0.12 | 0.10 × 0.08 × 0.02 |
| Data collection | ||
| Diffractometer | Bruker X8 Proteum | Bruker X8 Proteum |
| Absorption correction | Multi-scan (SADABS; Krause et al., 2015 ▸) | Multi-scan (SADABS; Krause et al., 2015 ▸) |
| T min, T max | 0.720, 0.915 | 0.693, 0.897 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 51755, 7112, 6916 | 23250, 3337, 3138 |
| R int | 0.038 | 0.037 |
| (sin θ/λ)max (Å−1) | 0.602 | 0.603 |
| Refinement | ||
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.042, 0.109, 1.10 | 0.034, 0.094, 1.13 |
| No. of reflections | 7112 | 3337 |
| No. of parameters | 514 | 259 |
| H-atom treatment | H-atom parameters constrained | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.33, −0.34 | 0.27, −0.31 |
| Absolute structure | Refined as an inversion twin | – |
| Absolute structure parameter | 0.50 (3) | – |
Refinement of (I) was hampered by the presence of pseudosymmetry. An alternative model using space group Pccn was also refined, but the overall quality of the refinement was not as good as the P21212 model given here. Indeed, the ADDSYM routine in PLATON (Spek, 2009 ▸) suggests a missing inversion centre and transformation to Pccn, but that model did not refine well (R 1 > 9%). Other alternatives using space groups Pcc2, Pban, and Pna21 were much less satisfactory. Not surprisingly, the P21212 model was twinned by inversion, which was dealt with using standard SHELXL methods (TWIN and BASF commands).
The hydrogen on the tetrazole ring was initially placed solely on the atoms labelled N1A and N1B. This assignment results in impossible clashes with symmetry equivalents about the twofold axis. Since there were suitable small difference map peaks for hydrogen atoms attached to atoms N4A and N4B as well as N1A and N1B, these hydrogen atoms were included as split over the two sites at half occupancy. Disorder of the tetrazole ring hydrogen atoms in this way also requires that the hydroxyl hydrogen atoms of the methanol molecules are disordered. Again, suitable (albeit small) difference map peaks were apparent. Further evidence for the disorder is that the distances C11A—N1A, C11A—N4A and C11B—N1B, C11B—N4B are all very similar, indicating that the C=N double bond and C—N single bond in these rings are scrambled. Not surprisingly, convergence of the OH hydrogen-atom positions was rather problematic.
Supplementary Material
Crystal structure: contains datablock(s) global, I, II. DOI: 10.1107/S2056989016005600/hg5472sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989016005600/hg5472Isup2.hkl
Structure factors: contains datablock(s) II. DOI: 10.1107/S2056989016005600/hg5472IIsup3.hkl
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
The authors gratefully acknowledge the Arkansas Research Alliance for financial support, the UAMS sub award to PNR and SA from NIA Claude Pepper Center grant P30-AG028718 (J. Wei, P.I.) and the NIH/National Institute of General Medical Sciences (P20GM109005) for a COBRE award.
supplementary crystallographic information
(I) (Z)-5-[2-(Benzo[b]thiophen-2-yl)-1-(3,5-dimethoxyphenyl)ethenyl]-1H-tetrazole methanol monosolvate . Crystal data
| C19H16N4O2S·CH4O | Dx = 1.346 Mg m−3 |
| Mr = 396.46 | Cu Kα radiation, λ = 1.54178 Å |
| Orthorhombic, P21212 | Cell parameters from 9871 reflections |
| a = 18.2226 (4) Å | θ = 4.9–68.2° |
| b = 13.7954 (5) Å | µ = 1.72 mm−1 |
| c = 15.5594 (5) Å | T = 90 K |
| V = 3911.4 (2) Å3 | Irregular block, pale yellow |
| Z = 8 | 0.21 × 0.15 × 0.12 mm |
| F(000) = 1664 |
(I) (Z)-5-[2-(Benzo[b]thiophen-2-yl)-1-(3,5-dimethoxyphenyl)ethenyl]-1H-tetrazole methanol monosolvate . Data collection
| Bruker X8 Proteum diffractometer | 7112 independent reflections |
| Radiation source: fine-focus rotating anode | 6916 reflections with I > 2σ(I) |
| Detector resolution: 5.6 pixels mm-1 | Rint = 0.038 |
| φ and ω scans | θmax = 68.2°, θmin = 2.8° |
| Absorption correction: multi-scan (SADABS; Krause et al., 2015) | h = −21→17 |
| Tmin = 0.720, Tmax = 0.915 | k = −16→15 |
| 51755 measured reflections | l = −18→16 |
(I) (Z)-5-[2-(Benzo[b]thiophen-2-yl)-1-(3,5-dimethoxyphenyl)ethenyl]-1H-tetrazole methanol monosolvate . Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.042 | H-atom parameters constrained |
| wR(F2) = 0.109 | w = 1/[σ2(Fo2) + (0.0327P)2 + 4.5187P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.10 | (Δ/σ)max < 0.001 |
| 7112 reflections | Δρmax = 0.33 e Å−3 |
| 514 parameters | Δρmin = −0.34 e Å−3 |
| 0 restraints | Absolute structure: Refined as an inversion twin |
| Primary atom site location: structure-invariant direct methods | Absolute structure parameter: 0.50 (3) |
(I) (Z)-5-[2-(Benzo[b]thiophen-2-yl)-1-(3,5-dimethoxyphenyl)ethenyl]-1H-tetrazole methanol monosolvate . Special details
| Experimental. The crystal was mounted using polyisobutene oil on the tip of a fine glass fibre, which was fastened in a copper mounting pin with electrical solder. It was placed directly into the cold gas stream of a liquid-nitrogen based cryostat. Diffraction data were collected with the crystal at 90K, which is standard practice in this laboratory for the majority of flash-cooled crystals. |
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refined as a 2-component inversion twin. |
(I) (Z)-5-[2-(Benzo[b]thiophen-2-yl)-1-(3,5-dimethoxyphenyl)ethenyl]-1H-tetrazole methanol monosolvate . Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| S1A | −0.14677 (5) | 0.17392 (8) | 0.08523 (7) | 0.0251 (2) | |
| O1A | 0.27407 (16) | 0.0780 (3) | −0.00263 (19) | 0.0339 (8) | |
| O2A | 0.25387 (16) | 0.1201 (3) | 0.2996 (2) | 0.0318 (8) | |
| N1A | −0.00422 (19) | 0.1008 (3) | −0.0368 (2) | 0.0246 (8) | |
| H1NA | −0.0012 | 0.0373 | −0.0338 | 0.030* | 0.5 |
| N2A | −0.0252 (2) | 0.1531 (3) | −0.1071 (2) | 0.031 (1) | |
| N3A | −0.0231 (2) | 0.2448 (3) | −0.0848 (3) | 0.0349 (10) | |
| N4A | −0.00020 (19) | 0.2525 (3) | −0.0020 (3) | 0.0287 (8) | |
| H4NA | 0.0062 | 0.3063 | 0.0274 | 0.034* | 0.5 |
| C1A | −0.0953 (2) | 0.1363 (3) | 0.1756 (3) | 0.0223 (9) | |
| C2A | −0.1397 (2) | 0.1174 (3) | 0.2428 (3) | 0.0231 (9) | |
| H2A | −0.1214 | 0.0973 | 0.2971 | 0.028* | |
| C3A | −0.2158 (2) | 0.1295 (3) | 0.2268 (3) | 0.0239 (9) | |
| C4A | −0.2763 (2) | 0.1176 (3) | 0.2813 (3) | 0.0244 (9) | |
| H4A | −0.2688 | 0.0980 | 0.3391 | 0.029* | |
| C5A | −0.3458 (2) | 0.1339 (3) | 0.2521 (3) | 0.0277 (10) | |
| H5A | −0.3865 | 0.1249 | 0.2894 | 0.033* | |
| C6A | −0.3574 (2) | 0.1642 (3) | 0.1660 (3) | 0.0273 (10) | |
| H6A | −0.4060 | 0.1742 | 0.1457 | 0.033* | |
| C7A | −0.2992 (2) | 0.1793 (3) | 0.1117 (3) | 0.0270 (9) | |
| H7A | −0.3068 | 0.2022 | 0.0548 | 0.032* | |
| C8A | −0.2287 (2) | 0.1604 (3) | 0.1415 (3) | 0.0226 (9) | |
| C9A | −0.0161 (2) | 0.1270 (3) | 0.1772 (3) | 0.0204 (8) | |
| H9A | 0.0036 | 0.1098 | 0.2316 | 0.025* | |
| C10A | 0.0341 (2) | 0.1385 (3) | 0.1153 (3) | 0.0211 (9) | |
| C11A | 0.0106 (2) | 0.1620 (4) | 0.0259 (3) | 0.0225 (9) | |
| C12A | 0.1135 (2) | 0.1267 (3) | 0.1272 (3) | 0.0224 (9) | |
| C13A | 0.1443 (2) | 0.1325 (3) | 0.2116 (3) | 0.0234 (9) | |
| H13A | 0.1143 | 0.1463 | 0.2600 | 0.028* | |
| C14A | 0.2187 (2) | 0.1177 (3) | 0.2212 (3) | 0.0233 (9) | |
| C15A | 0.2651 (2) | 0.1003 (3) | 0.1514 (3) | 0.0271 (10) | |
| H15A | 0.3163 | 0.0915 | 0.1594 | 0.033* | |
| C16A | 0.2347 (2) | 0.0964 (3) | 0.0717 (3) | 0.0268 (10) | |
| C17A | 0.1596 (2) | 0.1100 (3) | 0.0598 (3) | 0.0264 (10) | |
| H17A | 0.1401 | 0.1077 | 0.0032 | 0.032* | |
| C18A | 0.3507 (2) | 0.0617 (4) | 0.0066 (3) | 0.0377 (12) | |
| H18A | 0.3733 | 0.1180 | 0.0344 | 0.057* | |
| H18B | 0.3728 | 0.0520 | −0.0501 | 0.057* | |
| H18C | 0.3588 | 0.0039 | 0.0420 | 0.057* | |
| C19A | 0.2093 (3) | 0.1288 (4) | 0.3753 (3) | 0.0335 (11) | |
| H19A | 0.1845 | 0.1919 | 0.3752 | 0.050* | |
| H19B | 0.2404 | 0.1236 | 0.4265 | 0.050* | |
| H19C | 0.1725 | 0.0769 | 0.3759 | 0.050* | |
| S1B | 0.63976 (5) | 0.33164 (8) | 0.41542 (7) | 0.0245 (2) | |
| O1B | 0.21860 (16) | 0.4196 (3) | 0.5019 (2) | 0.0393 (9) | |
| O2B | 0.23920 (16) | 0.3710 (3) | 0.2009 (2) | 0.0306 (7) | |
| N1B | 0.49906 (19) | 0.3988 (3) | 0.5359 (2) | 0.0250 (8) | |
| H1NB | 0.4969 | 0.4625 | 0.5332 | 0.030* | 0.5 |
| N2B | 0.5193 (2) | 0.3455 (3) | 0.6043 (2) | 0.0321 (10) | |
| N3B | 0.5154 (2) | 0.2549 (3) | 0.5866 (3) | 0.0332 (9) | |
| N4B | 0.49224 (19) | 0.2483 (3) | 0.5038 (3) | 0.0260 (8) | |
| H4NB | 0.4848 | 0.1940 | 0.4754 | 0.031* | 0.5 |
| C1B | 0.5881 (2) | 0.3683 (3) | 0.3263 (3) | 0.0218 (9) | |
| C2B | 0.6327 (2) | 0.3889 (3) | 0.2573 (3) | 0.0244 (9) | |
| H2B | 0.6144 | 0.4095 | 0.2031 | 0.029* | |
| C3B | 0.7093 (2) | 0.3762 (3) | 0.2746 (3) | 0.0215 (9) | |
| C4B | 0.7694 (2) | 0.3905 (3) | 0.2191 (3) | 0.0252 (9) | |
| H4B | 0.7625 | 0.4114 | 0.1615 | 0.030* | |
| C5B | 0.8392 (2) | 0.3730 (3) | 0.2517 (3) | 0.0247 (9) | |
| H5B | 0.8806 | 0.3819 | 0.2153 | 0.030* | |
| C6B | 0.8503 (2) | 0.3429 (3) | 0.3357 (3) | 0.0271 (10) | |
| H6B | 0.8989 | 0.3321 | 0.3556 | 0.032* | |
| C7B | 0.7919 (2) | 0.3283 (3) | 0.3910 (3) | 0.0261 (9) | |
| H7B | 0.7996 | 0.3076 | 0.4485 | 0.031* | |
| C8B | 0.7214 (2) | 0.3451 (3) | 0.3595 (3) | 0.0253 (9) | |
| C9B | 0.5091 (2) | 0.3757 (4) | 0.3228 (3) | 0.0233 (9) | |
| H9B | 0.4895 | 0.3923 | 0.2681 | 0.028* | |
| C10B | 0.4590 (2) | 0.3625 (3) | 0.3856 (3) | 0.0211 (9) | |
| C11B | 0.4828 (2) | 0.3364 (3) | 0.4728 (3) | 0.0221 (9) | |
| C12B | 0.3782 (2) | 0.3708 (3) | 0.3707 (3) | 0.0200 (8) | |
| C13B | 0.3489 (2) | 0.3638 (3) | 0.2897 (3) | 0.0224 (9) | |
| H13B | 0.3799 | 0.3513 | 0.2419 | 0.027* | |
| C14B | 0.2740 (2) | 0.3750 (3) | 0.2778 (3) | 0.0261 (10) | |
| C15B | 0.2278 (2) | 0.3939 (3) | 0.3476 (3) | 0.0249 (9) | |
| H15B | 0.1765 | 0.4019 | 0.3390 | 0.030* | |
| C16B | 0.2576 (2) | 0.4008 (3) | 0.4304 (3) | 0.0261 (10) | |
| C17B | 0.3329 (2) | 0.3884 (3) | 0.4430 (3) | 0.0233 (9) | |
| H17B | 0.3534 | 0.3918 | 0.4991 | 0.028* | |
| C18B | 0.1415 (2) | 0.4338 (4) | 0.4934 (3) | 0.0382 (12) | |
| H18D | 0.1322 | 0.4897 | 0.4560 | 0.057* | |
| H18E | 0.1200 | 0.4457 | 0.5501 | 0.057* | |
| H18F | 0.1192 | 0.3758 | 0.4681 | 0.057* | |
| C19B | 0.2836 (3) | 0.3658 (4) | 0.1264 (3) | 0.0357 (12) | |
| H19D | 0.3218 | 0.4157 | 0.1292 | 0.054* | |
| H19E | 0.2531 | 0.3764 | 0.0753 | 0.054* | |
| H19F | 0.3065 | 0.3017 | 0.1230 | 0.054* | |
| C1SA | 0.4356 (4) | 0.1141 (5) | 0.3215 (5) | 0.069 (2) | |
| H1S1 | 0.3850 | 0.1165 | 0.3428 | 0.103* | |
| H1S2 | 0.4399 | 0.0632 | 0.2779 | 0.103* | |
| H1S3 | 0.4486 | 0.1768 | 0.2961 | 0.103* | |
| O1SA | 0.4839 (2) | 0.0935 (2) | 0.3908 (3) | 0.0400 (9) | |
| H1SA | 0.4764 | 0.1330 | 0.4310 | 0.060* | 0.5 |
| H2SA | 0.4955 | 0.0346 | 0.3897 | 0.060* | 0.5 |
| C1SB | 0.0583 (3) | 0.3839 (4) | 0.1772 (5) | 0.065 (2) | |
| H1S4 | 0.0789 | 0.4438 | 0.2009 | 0.098* | |
| H1S5 | 0.0982 | 0.3422 | 0.1567 | 0.098* | |
| H1S6 | 0.0305 | 0.3499 | 0.2219 | 0.098* | |
| O1SB | 0.0113 (2) | 0.4064 (3) | 0.1081 (2) | 0.0391 (9) | |
| H1SB | 0.0067 | 0.3576 | 0.0763 | 0.059* | 0.5 |
| H2SB | 0.0327 | 0.4446 | 0.0743 | 0.059* | 0.5 |
(I) (Z)-5-[2-(Benzo[b]thiophen-2-yl)-1-(3,5-dimethoxyphenyl)ethenyl]-1H-tetrazole methanol monosolvate . Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1A | 0.0254 (4) | 0.0331 (6) | 0.0167 (5) | −0.0001 (4) | 0.0001 (4) | 0.0019 (5) |
| O1A | 0.0268 (15) | 0.061 (2) | 0.0140 (15) | 0.0078 (15) | 0.0072 (13) | 0.0040 (16) |
| O2A | 0.0265 (14) | 0.049 (2) | 0.0195 (15) | 0.0074 (14) | −0.0059 (13) | −0.0006 (15) |
| N1A | 0.0252 (17) | 0.036 (2) | 0.0126 (17) | 0.0037 (15) | −0.0012 (15) | 0.0035 (16) |
| N2A | 0.0258 (17) | 0.050 (3) | 0.0170 (19) | 0.0041 (18) | 0.0003 (14) | 0.0097 (18) |
| N3A | 0.0286 (18) | 0.051 (3) | 0.025 (2) | 0.0004 (18) | 0.0037 (17) | 0.016 (2) |
| N4A | 0.0268 (18) | 0.0291 (19) | 0.0302 (19) | 0.0012 (15) | 0.0000 (17) | 0.0062 (16) |
| C1A | 0.0254 (19) | 0.023 (2) | 0.019 (2) | −0.0022 (16) | 0.0013 (16) | 0.0030 (18) |
| C2A | 0.033 (2) | 0.022 (2) | 0.015 (2) | −0.0017 (18) | −0.0008 (17) | −0.0009 (17) |
| C3A | 0.027 (2) | 0.020 (2) | 0.024 (2) | −0.0053 (17) | −0.0029 (17) | −0.0007 (18) |
| C4A | 0.036 (2) | 0.022 (2) | 0.015 (2) | −0.0051 (18) | 0.0026 (18) | 0.0012 (18) |
| C5A | 0.024 (2) | 0.029 (2) | 0.030 (2) | −0.0031 (17) | 0.0077 (18) | −0.004 (2) |
| C6A | 0.026 (2) | 0.030 (2) | 0.026 (2) | 0.0005 (19) | −0.0021 (16) | −0.004 (2) |
| C7A | 0.029 (2) | 0.028 (2) | 0.024 (2) | 0.0006 (18) | −0.0023 (17) | −0.0014 (19) |
| C8A | 0.029 (2) | 0.021 (2) | 0.018 (2) | −0.0023 (17) | 0.0012 (16) | −0.0041 (18) |
| C9A | 0.029 (2) | 0.020 (2) | 0.0124 (19) | −0.0040 (18) | −0.0033 (15) | 0.0030 (17) |
| C10A | 0.0253 (19) | 0.018 (2) | 0.020 (2) | 0.0006 (16) | −0.0028 (17) | −0.0017 (18) |
| C11A | 0.0185 (18) | 0.033 (2) | 0.016 (2) | 0.0016 (18) | 0.0017 (15) | 0.0023 (19) |
| C12A | 0.0260 (19) | 0.021 (2) | 0.020 (2) | −0.0018 (17) | −0.0037 (16) | 0.0065 (19) |
| C13A | 0.0245 (19) | 0.023 (2) | 0.022 (2) | 0.0034 (16) | 0.0028 (17) | 0.0009 (19) |
| C14A | 0.034 (2) | 0.026 (2) | 0.0104 (19) | 0.0022 (18) | −0.0048 (16) | 0.0036 (18) |
| C15A | 0.023 (2) | 0.028 (2) | 0.030 (2) | 0.0047 (19) | 0.0015 (18) | 0.005 (2) |
| C16A | 0.031 (2) | 0.033 (2) | 0.017 (2) | 0.0018 (19) | 0.0050 (18) | 0.0025 (19) |
| C17A | 0.027 (2) | 0.027 (2) | 0.025 (2) | 0.0001 (18) | 0.0028 (17) | 0.0067 (19) |
| C18A | 0.028 (2) | 0.053 (3) | 0.032 (3) | 0.007 (2) | 0.008 (2) | 0.016 (2) |
| C19A | 0.037 (2) | 0.050 (3) | 0.013 (2) | 0.008 (2) | −0.0022 (18) | −0.004 (2) |
| S1B | 0.0225 (4) | 0.0342 (6) | 0.0167 (5) | −0.0002 (4) | −0.0002 (4) | 0.0009 (5) |
| O1B | 0.0211 (14) | 0.065 (2) | 0.0322 (19) | 0.0067 (16) | 0.0042 (14) | 0.0094 (18) |
| O2B | 0.0268 (14) | 0.0429 (19) | 0.0220 (15) | 0.0032 (14) | −0.0041 (13) | −0.0040 (15) |
| N1B | 0.0236 (17) | 0.031 (2) | 0.0200 (19) | 0.0006 (14) | 0.0013 (16) | −0.0029 (17) |
| N2B | 0.0260 (18) | 0.052 (3) | 0.0183 (19) | 0.0023 (18) | 0.0002 (14) | 0.0063 (19) |
| N3B | 0.0288 (18) | 0.045 (2) | 0.025 (2) | 0.0006 (17) | −0.0004 (17) | 0.012 (2) |
| N4B | 0.0288 (18) | 0.0294 (19) | 0.0198 (17) | −0.0002 (15) | −0.0006 (15) | 0.0044 (15) |
| C1B | 0.026 (2) | 0.023 (2) | 0.016 (2) | 0.0008 (17) | −0.0020 (15) | −0.0066 (17) |
| C2B | 0.0201 (19) | 0.025 (2) | 0.028 (2) | −0.0035 (17) | 0.0008 (17) | 0.0008 (19) |
| C3B | 0.027 (2) | 0.022 (2) | 0.016 (2) | −0.0023 (17) | 0.0010 (16) | −0.0013 (18) |
| C4B | 0.026 (2) | 0.022 (2) | 0.028 (2) | −0.0029 (17) | 0.0007 (18) | 0.0005 (19) |
| C5B | 0.025 (2) | 0.028 (2) | 0.021 (2) | −0.0074 (17) | 0.0018 (17) | −0.0008 (19) |
| C6B | 0.0210 (18) | 0.029 (2) | 0.031 (2) | −0.0004 (18) | −0.0029 (16) | −0.002 (2) |
| C7B | 0.029 (2) | 0.030 (2) | 0.019 (2) | −0.0010 (19) | −0.0023 (16) | −0.0011 (19) |
| C8B | 0.0221 (19) | 0.029 (2) | 0.024 (2) | −0.0014 (17) | −0.0006 (16) | 0.0004 (19) |
| C9B | 0.025 (2) | 0.024 (2) | 0.020 (2) | 0.0000 (18) | −0.0028 (17) | −0.0047 (19) |
| C10B | 0.025 (2) | 0.022 (2) | 0.017 (2) | 0.0004 (16) | 0.0008 (16) | 0.0007 (17) |
| C11B | 0.0200 (18) | 0.024 (2) | 0.022 (2) | −0.0014 (18) | 0.0021 (15) | 0.0006 (18) |
| C12B | 0.0215 (18) | 0.021 (2) | 0.017 (2) | 0.0015 (16) | 0.0018 (16) | −0.0012 (18) |
| C13B | 0.0259 (19) | 0.024 (2) | 0.017 (2) | 0.0009 (16) | −0.0008 (17) | 0.0019 (18) |
| C14B | 0.024 (2) | 0.0194 (19) | 0.035 (3) | −0.0005 (17) | −0.0026 (18) | −0.001 (2) |
| C15B | 0.0230 (19) | 0.030 (2) | 0.021 (2) | 0.0012 (18) | −0.0025 (17) | 0.0029 (19) |
| C16B | 0.0230 (19) | 0.028 (2) | 0.028 (2) | 0.0013 (17) | 0.0040 (17) | 0.0082 (19) |
| C17B | 0.026 (2) | 0.031 (2) | 0.0124 (18) | 0.0000 (18) | 0.0008 (15) | 0.0010 (17) |
| C18B | 0.025 (2) | 0.061 (3) | 0.029 (2) | 0.009 (2) | 0.0073 (19) | 0.011 (2) |
| C19B | 0.029 (2) | 0.049 (3) | 0.029 (3) | 0.006 (2) | −0.0046 (19) | −0.007 (2) |
| C1SA | 0.070 (4) | 0.045 (4) | 0.091 (6) | 0.008 (3) | −0.055 (4) | −0.004 (4) |
| O1SA | 0.0391 (17) | 0.0270 (18) | 0.054 (2) | 0.0036 (15) | −0.0169 (17) | −0.0028 (17) |
| C1SB | 0.062 (4) | 0.033 (3) | 0.101 (6) | 0.001 (3) | −0.051 (4) | −0.002 (4) |
| O1SB | 0.0427 (18) | 0.0318 (19) | 0.043 (2) | 0.0008 (16) | −0.0132 (16) | −0.0011 (17) |
(I) (Z)-5-[2-(Benzo[b]thiophen-2-yl)-1-(3,5-dimethoxyphenyl)ethenyl]-1H-tetrazole methanol monosolvate . Geometric parameters (Å, º)
| S1A—C8A | 1.742 (4) | N1B—C11B | 1.339 (6) |
| S1A—C1A | 1.768 (4) | N1B—N2B | 1.346 (5) |
| O1A—C16A | 1.385 (5) | N1B—H1NB | 0.8800 |
| O1A—C18A | 1.422 (5) | N2B—N3B | 1.281 (6) |
| O2A—C14A | 1.379 (5) | N3B—N4B | 1.360 (6) |
| O2A—C19A | 1.437 (5) | N4B—C11B | 1.319 (6) |
| N1A—C11A | 1.317 (6) | N4B—H4NB | 0.8800 |
| N1A—N2A | 1.365 (5) | C1B—C2B | 1.376 (6) |
| N1A—H1NA | 0.8800 | C1B—C9B | 1.444 (6) |
| N2A—N3A | 1.313 (7) | C2B—C3B | 1.433 (6) |
| N3A—N4A | 1.358 (6) | C2B—H2B | 0.9500 |
| N4A—C11A | 1.337 (7) | C3B—C8B | 1.406 (6) |
| N4A—H4NA | 0.8800 | C3B—C4B | 1.408 (6) |
| C1A—C2A | 1.348 (6) | C4B—C5B | 1.391 (6) |
| C1A—C9A | 1.449 (6) | C4B—H4B | 0.9500 |
| C2A—C3A | 1.418 (6) | C5B—C6B | 1.386 (6) |
| C2A—H2A | 0.9500 | C5B—H5B | 0.9500 |
| C3A—C4A | 1.400 (7) | C6B—C7B | 1.383 (6) |
| C3A—C8A | 1.413 (6) | C6B—H6B | 0.9500 |
| C4A—C5A | 1.364 (6) | C7B—C8B | 1.395 (6) |
| C4A—H4A | 0.9500 | C7B—H7B | 0.9500 |
| C5A—C6A | 1.419 (7) | C9B—C10B | 1.351 (6) |
| C5A—H5A | 0.9500 | C9B—H9B | 0.9500 |
| C6A—C7A | 1.372 (6) | C10B—C11B | 1.469 (6) |
| C6A—H6A | 0.9500 | C10B—C12B | 1.494 (5) |
| C7A—C8A | 1.389 (6) | C12B—C13B | 1.372 (6) |
| C7A—H7A | 0.9500 | C12B—C17B | 1.416 (6) |
| C9A—C10A | 1.338 (6) | C13B—C14B | 1.386 (6) |
| C9A—H9A | 0.9500 | C13B—H13B | 0.9500 |
| C10A—C12A | 1.469 (6) | C14B—C15B | 1.400 (7) |
| C10A—C11A | 1.491 (6) | C15B—C16B | 1.400 (6) |
| C12A—C17A | 1.363 (6) | C15B—H15B | 0.9500 |
| C12A—C13A | 1.431 (6) | C16B—C17B | 1.398 (6) |
| C13A—C14A | 1.378 (6) | C17B—H17B | 0.9500 |
| C13A—H13A | 0.9500 | C18B—H18D | 0.9800 |
| C14A—C15A | 1.396 (6) | C18B—H18E | 0.9800 |
| C15A—C16A | 1.359 (6) | C18B—H18F | 0.9800 |
| C15A—H15A | 0.9500 | C19B—H19D | 0.9800 |
| C16A—C17A | 1.394 (6) | C19B—H19E | 0.9800 |
| C17A—H17A | 0.9500 | C19B—H19F | 0.9800 |
| C18A—H18A | 0.9800 | C1SA—O1SA | 1.421 (7) |
| C18A—H18B | 0.9800 | C1SA—H1S1 | 0.9800 |
| C18A—H18C | 0.9800 | C1SA—H1S2 | 0.9800 |
| C19A—H19A | 0.9800 | C1SA—H1S3 | 0.9800 |
| C19A—H19B | 0.9800 | O1SA—H1SA | 0.8400 |
| C19A—H19C | 0.9800 | O1SA—H2SA | 0.8400 |
| S1B—C8B | 1.733 (4) | C1SB—O1SB | 1.409 (6) |
| S1B—C1B | 1.750 (4) | C1SB—H1S4 | 0.9800 |
| O1B—C16B | 1.345 (5) | C1SB—H1S5 | 0.9800 |
| O1B—C18B | 1.425 (5) | C1SB—H1S6 | 0.9800 |
| O2B—C14B | 1.355 (6) | O1SB—H1SB | 0.8400 |
| O2B—C19B | 1.415 (6) | O1SB—H2SB | 0.8400 |
| C8A—S1A—C1A | 91.3 (2) | C11B—N4B—N3B | 108.9 (4) |
| C16A—O1A—C18A | 116.9 (4) | C11B—N4B—H4NB | 125.5 |
| C14A—O2A—C19A | 117.7 (3) | N3B—N4B—H4NB | 125.5 |
| C11A—N1A—N2A | 108.2 (4) | C2B—C1B—C9B | 122.9 (4) |
| C11A—N1A—H1NA | 125.9 | C2B—C1B—S1B | 111.2 (3) |
| N2A—N1A—H1NA | 125.9 | C9B—C1B—S1B | 125.9 (3) |
| N3A—N2A—N1A | 106.8 (4) | C1B—C2B—C3B | 113.7 (4) |
| N2A—N3A—N4A | 109.6 (4) | C1B—C2B—H2B | 123.1 |
| C11A—N4A—N3A | 106.2 (4) | C3B—C2B—H2B | 123.1 |
| C11A—N4A—H4NA | 126.9 | C8B—C3B—C4B | 119.8 (4) |
| N3A—N4A—H4NA | 126.9 | C8B—C3B—C2B | 111.5 (4) |
| C2A—C1A—C9A | 124.6 (4) | C4B—C3B—C2B | 128.6 (4) |
| C2A—C1A—S1A | 110.8 (3) | C5B—C4B—C3B | 117.6 (4) |
| C9A—C1A—S1A | 124.6 (3) | C5B—C4B—H4B | 121.2 |
| C1A—C2A—C3A | 115.3 (4) | C3B—C4B—H4B | 121.2 |
| C1A—C2A—H2A | 122.4 | C6B—C5B—C4B | 121.9 (4) |
| C3A—C2A—H2A | 122.4 | C6B—C5B—H5B | 119.1 |
| C4A—C3A—C8A | 118.2 (4) | C4B—C5B—H5B | 119.1 |
| C4A—C3A—C2A | 130.4 (4) | C7B—C6B—C5B | 121.3 (4) |
| C8A—C3A—C2A | 111.4 (4) | C7B—C6B—H6B | 119.4 |
| C5A—C4A—C3A | 120.6 (4) | C5B—C6B—H6B | 119.4 |
| C5A—C4A—H4A | 119.7 | C6B—C7B—C8B | 117.8 (4) |
| C3A—C4A—H4A | 119.7 | C6B—C7B—H7B | 121.1 |
| C4A—C5A—C6A | 120.1 (4) | C8B—C7B—H7B | 121.1 |
| C4A—C5A—H5A | 120.0 | C7B—C8B—C3B | 121.6 (4) |
| C6A—C5A—H5A | 120.0 | C7B—C8B—S1B | 126.7 (4) |
| C7A—C6A—C5A | 120.8 (4) | C3B—C8B—S1B | 111.7 (3) |
| C7A—C6A—H6A | 119.6 | C10B—C9B—C1B | 129.6 (4) |
| C5A—C6A—H6A | 119.6 | C10B—C9B—H9B | 115.2 |
| C6A—C7A—C8A | 118.6 (4) | C1B—C9B—H9B | 115.2 |
| C6A—C7A—H7A | 120.7 | C9B—C10B—C11B | 120.1 (4) |
| C8A—C7A—H7A | 120.7 | C9B—C10B—C12B | 123.0 (4) |
| C7A—C8A—C3A | 121.7 (4) | C11B—C10B—C12B | 116.9 (4) |
| C7A—C8A—S1A | 127.1 (3) | N4B—C11B—N1B | 107.2 (4) |
| C3A—C8A—S1A | 111.2 (3) | N4B—C11B—C10B | 127.0 (4) |
| C10A—C9A—C1A | 131.2 (4) | N1B—C11B—C10B | 125.8 (4) |
| C10A—C9A—H9A | 114.4 | C13B—C12B—C17B | 120.9 (4) |
| C1A—C9A—H9A | 114.4 | C13B—C12B—C10B | 121.3 (4) |
| C9A—C10A—C12A | 124.7 (4) | C17B—C12B—C10B | 117.7 (4) |
| C9A—C10A—C11A | 120.1 (4) | C12B—C13B—C14B | 119.9 (4) |
| C12A—C10A—C11A | 115.1 (4) | C12B—C13B—H13B | 120.0 |
| N1A—C11A—N4A | 109.2 (4) | C14B—C13B—H13B | 120.0 |
| N1A—C11A—C10A | 127.6 (4) | O2B—C14B—C13B | 125.1 (4) |
| N4A—C11A—C10A | 123.2 (4) | O2B—C14B—C15B | 114.3 (4) |
| C17A—C12A—C13A | 118.3 (4) | C13B—C14B—C15B | 120.6 (5) |
| C17A—C12A—C10A | 121.9 (4) | C14B—C15B—C16B | 119.6 (4) |
| C13A—C12A—C10A | 119.8 (4) | C14B—C15B—H15B | 120.2 |
| C14A—C13A—C12A | 118.5 (4) | C16B—C15B—H15B | 120.2 |
| C14A—C13A—H13A | 120.8 | O1B—C16B—C17B | 115.2 (4) |
| C12A—C13A—H13A | 120.8 | O1B—C16B—C15B | 124.7 (4) |
| C13A—C14A—O2A | 123.3 (4) | C17B—C16B—C15B | 120.1 (4) |
| C13A—C14A—C15A | 122.5 (4) | C16B—C17B—C12B | 118.8 (4) |
| O2A—C14A—C15A | 114.2 (4) | C16B—C17B—H17B | 120.6 |
| C16A—C15A—C14A | 118.0 (4) | C12B—C17B—H17B | 120.6 |
| C16A—C15A—H15A | 121.0 | O1B—C18B—H18D | 109.5 |
| C14A—C15A—H15A | 121.0 | O1B—C18B—H18E | 109.5 |
| C15A—C16A—O1A | 124.0 (4) | H18D—C18B—H18E | 109.5 |
| C15A—C16A—C17A | 121.1 (4) | O1B—C18B—H18F | 109.5 |
| O1A—C16A—C17A | 114.9 (4) | H18D—C18B—H18F | 109.5 |
| C12A—C17A—C16A | 121.7 (4) | H18E—C18B—H18F | 109.5 |
| C12A—C17A—H17A | 119.2 | O2B—C19B—H19D | 109.5 |
| C16A—C17A—H17A | 119.2 | O2B—C19B—H19E | 109.5 |
| O1A—C18A—H18A | 109.5 | H19D—C19B—H19E | 109.5 |
| O1A—C18A—H18B | 109.5 | O2B—C19B—H19F | 109.5 |
| H18A—C18A—H18B | 109.5 | H19D—C19B—H19F | 109.5 |
| O1A—C18A—H18C | 109.5 | H19E—C19B—H19F | 109.5 |
| H18A—C18A—H18C | 109.5 | O1SA—C1SA—H1S1 | 109.5 |
| H18B—C18A—H18C | 109.5 | O1SA—C1SA—H1S2 | 109.5 |
| O2A—C19A—H19A | 109.5 | H1S1—C1SA—H1S2 | 109.5 |
| O2A—C19A—H19B | 109.5 | O1SA—C1SA—H1S3 | 109.5 |
| H19A—C19A—H19B | 109.5 | H1S1—C1SA—H1S3 | 109.5 |
| O2A—C19A—H19C | 109.5 | H1S2—C1SA—H1S3 | 109.5 |
| H19A—C19A—H19C | 109.5 | C1SA—O1SA—H1SA | 109.5 |
| H19B—C19A—H19C | 109.5 | C1SA—O1SA—H2SA | 109.5 |
| C8B—S1B—C1B | 91.9 (2) | O1SB—C1SB—H1S4 | 109.5 |
| C16B—O1B—C18B | 118.0 (4) | O1SB—C1SB—H1S5 | 109.5 |
| C14B—O2B—C19B | 117.2 (3) | H1S4—C1SB—H1S5 | 109.5 |
| C11B—N1B—N2B | 106.8 (4) | O1SB—C1SB—H1S6 | 109.5 |
| C11B—N1B—H1NB | 126.6 | H1S4—C1SB—H1S6 | 109.5 |
| N2B—N1B—H1NB | 126.6 | H1S5—C1SB—H1S6 | 109.5 |
| N3B—N2B—N1B | 110.4 (4) | C1SB—O1SB—H1SB | 109.5 |
| N2B—N3B—N4B | 106.7 (4) | C1SB—O1SB—H2SB | 109.5 |
| C11A—N1A—N2A—N3A | −0.6 (4) | C11B—N1B—N2B—N3B | −0.8 (5) |
| N1A—N2A—N3A—N4A | 0.9 (5) | N1B—N2B—N3B—N4B | 0.4 (5) |
| N2A—N3A—N4A—C11A | −0.8 (5) | N2B—N3B—N4B—C11B | 0.2 (5) |
| C8A—S1A—C1A—C2A | −1.1 (4) | C8B—S1B—C1B—C2B | 0.5 (4) |
| C8A—S1A—C1A—C9A | 178.9 (4) | C8B—S1B—C1B—C9B | 179.6 (4) |
| C9A—C1A—C2A—C3A | −178.9 (4) | C9B—C1B—C2B—C3B | −179.6 (4) |
| S1A—C1A—C2A—C3A | 1.1 (5) | S1B—C1B—C2B—C3B | −0.4 (5) |
| C1A—C2A—C3A—C4A | −179.1 (5) | C1B—C2B—C3B—C8B | 0.1 (6) |
| C1A—C2A—C3A—C8A | −0.4 (6) | C1B—C2B—C3B—C4B | 179.9 (4) |
| C8A—C3A—C4A—C5A | 1.1 (7) | C8B—C3B—C4B—C5B | −0.1 (7) |
| C2A—C3A—C4A—C5A | 179.7 (5) | C2B—C3B—C4B—C5B | −180.0 (5) |
| C3A—C4A—C5A—C6A | −0.7 (7) | C3B—C4B—C5B—C6B | −0.3 (7) |
| C4A—C5A—C6A—C7A | −1.2 (7) | C4B—C5B—C6B—C7B | 0.4 (8) |
| C5A—C6A—C7A—C8A | 2.6 (7) | C5B—C6B—C7B—C8B | −0.1 (7) |
| C6A—C7A—C8A—C3A | −2.2 (7) | C6B—C7B—C8B—C3B | −0.3 (7) |
| C6A—C7A—C8A—S1A | −180.0 (4) | C6B—C7B—C8B—S1B | 179.7 (4) |
| C4A—C3A—C8A—C7A | 0.4 (7) | C4B—C3B—C8B—C7B | 0.4 (7) |
| C2A—C3A—C8A—C7A | −178.5 (4) | C2B—C3B—C8B—C7B | −179.7 (4) |
| C4A—C3A—C8A—S1A | 178.4 (3) | C4B—C3B—C8B—S1B | −179.6 (4) |
| C2A—C3A—C8A—S1A | −0.4 (5) | C2B—C3B—C8B—S1B | 0.3 (5) |
| C1A—S1A—C8A—C7A | 178.8 (5) | C1B—S1B—C8B—C7B | 179.6 (5) |
| C1A—S1A—C8A—C3A | 0.8 (3) | C1B—S1B—C8B—C3B | −0.4 (4) |
| C2A—C1A—C9A—C10A | 176.3 (5) | C2B—C1B—C9B—C10B | −176.6 (5) |
| S1A—C1A—C9A—C10A | −3.6 (8) | S1B—C1B—C9B—C10B | 4.3 (8) |
| C1A—C9A—C10A—C12A | 179.7 (5) | C1B—C9B—C10B—C11B | −0.1 (8) |
| C1A—C9A—C10A—C11A | −2.2 (8) | C1B—C9B—C10B—C12B | −178.6 (5) |
| N2A—N1A—C11A—N4A | 0.0 (4) | N3B—N4B—C11B—N1B | −0.7 (5) |
| N2A—N1A—C11A—C10A | 178.6 (4) | N3B—N4B—C11B—C10B | 179.4 (4) |
| N3A—N4A—C11A—N1A | 0.5 (4) | N2B—N1B—C11B—N4B | 0.9 (4) |
| N3A—N4A—C11A—C10A | −178.2 (3) | N2B—N1B—C11B—C10B | −179.2 (4) |
| C9A—C10A—C11A—N1A | −90.9 (5) | C9B—C10B—C11B—N4B | −90.0 (6) |
| C12A—C10A—C11A—N1A | 87.3 (5) | C12B—C10B—C11B—N4B | 88.6 (5) |
| C9A—C10A—C11A—N4A | 87.5 (5) | C9B—C10B—C11B—N1B | 90.1 (5) |
| C12A—C10A—C11A—N4A | −94.3 (5) | C12B—C10B—C11B—N1B | −91.3 (5) |
| C9A—C10A—C12A—C17A | 159.7 (5) | C9B—C10B—C12B—C13B | 19.7 (7) |
| C11A—C10A—C12A—C17A | −18.4 (6) | C11B—C10B—C12B—C13B | −158.9 (4) |
| C9A—C10A—C12A—C13A | −20.2 (7) | C9B—C10B—C12B—C17B | −159.0 (5) |
| C11A—C10A—C12A—C13A | 161.6 (4) | C11B—C10B—C12B—C17B | 22.5 (6) |
| C17A—C12A—C13A—C14A | −2.2 (6) | C17B—C12B—C13B—C14B | 0.7 (7) |
| C10A—C12A—C13A—C14A | 177.7 (4) | C10B—C12B—C13B—C14B | −177.9 (4) |
| C12A—C13A—C14A—O2A | −178.9 (4) | C19B—O2B—C14B—C13B | −7.5 (7) |
| C12A—C13A—C14A—C15A | 2.0 (7) | C19B—O2B—C14B—C15B | 171.4 (5) |
| C19A—O2A—C14A—C13A | 6.4 (7) | C12B—C13B—C14B—O2B | 179.0 (4) |
| C19A—O2A—C14A—C15A | −174.5 (4) | C12B—C13B—C14B—C15B | 0.3 (7) |
| C13A—C14A—C15A—C16A | −1.2 (7) | O2B—C14B—C15B—C16B | −179.3 (4) |
| O2A—C14A—C15A—C16A | 179.7 (4) | C13B—C14B—C15B—C16B | −0.4 (7) |
| C14A—C15A—C16A—O1A | −178.8 (4) | C18B—O1B—C16B—C17B | 179.5 (5) |
| C14A—C15A—C16A—C17A | 0.5 (7) | C18B—O1B—C16B—C15B | −0.2 (7) |
| C18A—O1A—C16A—C15A | 0.5 (7) | C14B—C15B—C16B—O1B | 179.3 (4) |
| C18A—O1A—C16A—C17A | −178.9 (4) | C14B—C15B—C16B—C17B | −0.4 (7) |
| C13A—C12A—C17A—C16A | 1.7 (7) | O1B—C16B—C17B—C12B | −178.4 (4) |
| C10A—C12A—C17A—C16A | −178.3 (4) | C15B—C16B—C17B—C12B | 1.3 (7) |
| C15A—C16A—C17A—C12A | −0.8 (8) | C13B—C12B—C17B—C16B | −1.4 (7) |
| O1A—C16A—C17A—C12A | 178.6 (4) | C10B—C12B—C17B—C16B | 177.2 (4) |
(I) (Z)-5-[2-(Benzo[b]thiophen-2-yl)-1-(3,5-dimethoxyphenyl)ethenyl]-1H-tetrazole methanol monosolvate . Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1A—H1NA···N1Ai | 0.88 | 1.91 | 2.787 (9) | 176 |
| N4A—H4NA···O1SB | 0.88 | 1.87 | 2.736 (6) | 168 |
| N1B—H1NB···N1Bii | 0.88 | 1.92 | 2.792 (9) | 174 |
| N4B—H4NB···O1SA | 0.88 | 1.91 | 2.769 (6) | 165 |
| O1SA—H1SA···N4B | 0.84 | 1.97 | 2.769 (6) | 158 |
| O1SA—H2SA···O1SAiii | 0.84 | 1.81 | 2.646 (7) | 177 |
| O1SB—H1SB···N4A | 0.84 | 1.90 | 2.736 (6) | 176 |
Symmetry codes: (i) −x, −y, z; (ii) −x+1, −y+1, z; (iii) −x+1, −y, z.
(II) (Z)-5-[2-(Benzo[b]thiophen-3-yl)-1-(3,4,5-trimethoxyphenyl)ethenyl]-1H-tetrazole . Crystal data
| C20H18N4O3S | F(000) = 824 |
| Mr = 394.44 | Dx = 1.393 Mg m−3 |
| Monoclinic, P21/c | Cu Kα radiation, λ = 1.54178 Å |
| a = 4.8888 (1) Å | Cell parameters from 9924 reflections |
| b = 24.6650 (6) Å | θ = 3.4–68.3° |
| c = 15.5956 (4) Å | µ = 1.78 mm−1 |
| β = 91.031 (1)° | T = 90 K |
| V = 1880.25 (8) Å3 | Plate, colourless |
| Z = 4 | 0.10 × 0.08 × 0.02 mm |
(II) (Z)-5-[2-(Benzo[b]thiophen-3-yl)-1-(3,4,5-trimethoxyphenyl)ethenyl]-1H-tetrazole . Data collection
| Bruker X8 Proteum diffractometer | 3337 independent reflections |
| Radiation source: fine-focus rotating anode | 3138 reflections with I > 2σ(I) |
| Detector resolution: 5.6 pixels mm-1 | Rint = 0.037 |
| φ and ω scans | θmax = 68.5°, θmin = 3.4° |
| Absorption correction: multi-scan (SADABS; Krause et al., 2015) | h = −5→2 |
| Tmin = 0.693, Tmax = 0.897 | k = −29→29 |
| 23250 measured reflections | l = −18→18 |
(II) (Z)-5-[2-(Benzo[b]thiophen-3-yl)-1-(3,4,5-trimethoxyphenyl)ethenyl]-1H-tetrazole . Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.034 | Hydrogen site location: mixed |
| wR(F2) = 0.094 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.13 | w = 1/[σ2(Fo2) + (0.0391P)2 + 1.3628P] where P = (Fo2 + 2Fc2)/3 |
| 3337 reflections | (Δ/σ)max = 0.005 |
| 259 parameters | Δρmax = 0.27 e Å−3 |
| 0 restraints | Δρmin = −0.31 e Å−3 |
(II) (Z)-5-[2-(Benzo[b]thiophen-3-yl)-1-(3,4,5-trimethoxyphenyl)ethenyl]-1H-tetrazole . Special details
| Experimental. The crystal was mounted using polyisobutene oil on the tip of a fine glass fibre, which was fastened in a copper mounting pin with electrical solder. It was placed directly into the cold gas stream of a liquid-nitrogen based cryostat. Diffraction data were collected with the crystal at 90K, which is standard practice in this laboratory for the majority of flash-cooled crystals. |
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refinement progress was checked using Platon (Spek, 2009) and by an R-tensor (Parkin, 2000). The final model was further checked with the IUCr utility checkCIF. |
(II) (Z)-5-[2-(Benzo[b]thiophen-3-yl)-1-(3,4,5-trimethoxyphenyl)ethenyl]-1H-tetrazole . Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | 0.58440 (8) | 0.92397 (2) | 0.22030 (3) | 0.02003 (13) | |
| N1 | 0.5809 (3) | 0.76447 (5) | 0.36657 (9) | 0.0158 (3) | |
| H1N | 0.397 (4) | 0.7636 (8) | 0.3563 (12) | 0.019* | |
| N2 | 0.6748 (3) | 0.78140 (6) | 0.44376 (9) | 0.0189 (3) | |
| N3 | 0.9385 (3) | 0.78051 (6) | 0.44059 (9) | 0.0188 (3) | |
| N4 | 1.0187 (3) | 0.76305 (5) | 0.36194 (9) | 0.0167 (3) | |
| C1 | 0.7332 (3) | 0.86078 (7) | 0.22620 (11) | 0.0195 (3) | |
| H1 | 0.8771 | 0.8522 | 0.2657 | 0.023* | |
| C2 | 0.6269 (3) | 0.82413 (7) | 0.1694 (1) | 0.0169 (3) | |
| C3 | 0.4172 (3) | 0.84839 (7) | 0.11505 (10) | 0.0162 (3) | |
| C4 | 0.2612 (3) | 0.82496 (7) | 0.04825 (10) | 0.0185 (3) | |
| H4 | 0.2885 | 0.7881 | 0.0327 | 0.022* | |
| C5 | 0.0676 (3) | 0.85608 (7) | 0.00545 (11) | 0.0225 (4) | |
| H5 | −0.0373 | 0.8405 | −0.0401 | 0.027* | |
| C6 | 0.0238 (3) | 0.91033 (7) | 0.02841 (12) | 0.0230 (4) | |
| H6 | −0.1108 | 0.9310 | −0.0018 | 0.028* | |
| C7 | 0.1728 (3) | 0.93419 (7) | 0.09405 (11) | 0.0207 (4) | |
| H7 | 0.1422 | 0.9709 | 0.1098 | 0.025* | |
| C8 | 0.3703 (3) | 0.90284 (7) | 0.13672 (11) | 0.0178 (3) | |
| C9 | 0.7039 (3) | 0.76681 (6) | 0.1624 (1) | 0.0165 (3) | |
| H9 | 0.7003 | 0.7515 | 0.1065 | 0.020* | |
| C10 | 0.7788 (3) | 0.73397 (6) | 0.22705 (10) | 0.0148 (3) | |
| C11 | 0.7926 (3) | 0.75371 (6) | 0.31634 (10) | 0.0134 (3) | |
| C12 | 0.8405 (3) | 0.67535 (6) | 0.21635 (10) | 0.0154 (3) | |
| C13 | 1.0177 (3) | 0.65742 (6) | 0.15372 (10) | 0.0161 (3) | |
| H13 | 1.1099 | 0.6827 | 0.1186 | 0.019* | |
| C14 | 1.0588 (3) | 0.60179 (7) | 0.14293 (10) | 0.0167 (3) | |
| C15 | 0.9205 (3) | 0.56460 (7) | 0.19391 (11) | 0.0175 (3) | |
| C16 | 0.7445 (3) | 0.58324 (7) | 0.25707 (11) | 0.0170 (3) | |
| C17 | 0.7064 (3) | 0.63845 (7) | 0.26888 (10) | 0.0166 (3) | |
| H17 | 0.5894 | 0.6511 | 0.3126 | 0.020* | |
| O1 | 1.2285 (2) | 0.57947 (5) | 0.08429 (8) | 0.0210 (3) | |
| C18 | 1.3890 (3) | 0.61614 (7) | 0.03497 (11) | 0.0211 (4) | |
| H18A | 1.2677 | 0.6393 | 0.0002 | 0.032* | |
| H18B | 1.5088 | 0.5955 | −0.0027 | 0.032* | |
| H18C | 1.5006 | 0.6387 | 0.0736 | 0.032* | |
| O2 | 0.9650 (2) | 0.50992 (5) | 0.18551 (8) | 0.0236 (3) | |
| C19 | 0.8111 (4) | 0.48639 (8) | 0.11584 (12) | 0.0308 (4) | |
| H19A | 0.6154 | 0.4924 | 0.1246 | 0.046* | |
| H19B | 0.8476 | 0.4474 | 0.1134 | 0.046* | |
| H19C | 0.8650 | 0.5033 | 0.0619 | 0.046* | |
| O3 | 0.6211 (2) | 0.54394 (5) | 0.30402 (8) | 0.0226 (3) | |
| C20 | 0.4227 (3) | 0.56095 (7) | 0.36440 (11) | 0.0220 (4) | |
| H20A | 0.5106 | 0.5841 | 0.4079 | 0.033* | |
| H20B | 0.3436 | 0.5291 | 0.3921 | 0.033* | |
| H20C | 0.2776 | 0.5814 | 0.3346 | 0.033* |
(II) (Z)-5-[2-(Benzo[b]thiophen-3-yl)-1-(3,4,5-trimethoxyphenyl)ethenyl]-1H-tetrazole . Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.0226 (2) | 0.0163 (2) | 0.0211 (2) | 0.00207 (14) | −0.00164 (16) | −0.00240 (15) |
| N1 | 0.0109 (7) | 0.0222 (7) | 0.0142 (7) | 0.0006 (5) | 0.0001 (5) | −0.0018 (5) |
| N2 | 0.0157 (7) | 0.0246 (7) | 0.0163 (7) | 0.0013 (5) | 0.0004 (5) | −0.0026 (6) |
| N3 | 0.0158 (7) | 0.0242 (7) | 0.0163 (7) | 0.0007 (5) | −0.0002 (5) | −0.0029 (6) |
| N4 | 0.0143 (7) | 0.0214 (7) | 0.0143 (7) | 0.0004 (5) | −0.0001 (5) | −0.0016 (5) |
| C1 | 0.0197 (8) | 0.0197 (8) | 0.0189 (9) | 0.0015 (6) | −0.0017 (6) | 0.0008 (6) |
| C2 | 0.0166 (8) | 0.0197 (8) | 0.0146 (8) | 0.0012 (6) | 0.0033 (6) | 0.0019 (6) |
| C3 | 0.0144 (7) | 0.0191 (8) | 0.0153 (8) | 0.0000 (6) | 0.0042 (6) | 0.0028 (6) |
| C4 | 0.0181 (8) | 0.0199 (8) | 0.0176 (8) | −0.0011 (6) | 0.0023 (6) | 0.0000 (6) |
| C5 | 0.0184 (8) | 0.0282 (9) | 0.0207 (9) | −0.0025 (7) | −0.0022 (6) | 0.0010 (7) |
| C6 | 0.0154 (8) | 0.0263 (9) | 0.0271 (10) | 0.0026 (7) | −0.0013 (7) | 0.0054 (7) |
| C7 | 0.0174 (8) | 0.0177 (8) | 0.0272 (9) | 0.0020 (6) | 0.0035 (6) | 0.0025 (7) |
| C8 | 0.0163 (8) | 0.0193 (8) | 0.0180 (8) | −0.0004 (6) | 0.0038 (6) | 0.0007 (6) |
| C9 | 0.0163 (8) | 0.0189 (8) | 0.0142 (8) | −0.0003 (6) | 0.0014 (6) | −0.0019 (6) |
| C10 | 0.0104 (7) | 0.0184 (8) | 0.0157 (8) | −0.0010 (6) | 0.0015 (6) | −0.0016 (6) |
| C11 | 0.0124 (7) | 0.0135 (7) | 0.0144 (8) | 0.0004 (5) | 0.0005 (6) | 0.0007 (6) |
| C12 | 0.0125 (7) | 0.0178 (8) | 0.0158 (8) | 0.0000 (6) | −0.0034 (6) | −0.0011 (6) |
| C13 | 0.0143 (7) | 0.0179 (8) | 0.0162 (8) | −0.0005 (6) | −0.0012 (6) | 0.0014 (6) |
| C14 | 0.0138 (7) | 0.0207 (8) | 0.0155 (8) | 0.0020 (6) | −0.0012 (6) | −0.0026 (6) |
| C15 | 0.0174 (8) | 0.0165 (8) | 0.0186 (8) | 0.0020 (6) | −0.0026 (6) | −0.0011 (6) |
| C16 | 0.0158 (8) | 0.0185 (8) | 0.0168 (8) | −0.0023 (6) | −0.0015 (6) | 0.0021 (6) |
| C17 | 0.0150 (7) | 0.0205 (8) | 0.0142 (8) | 0.0010 (6) | −0.0001 (6) | −0.0015 (6) |
| O1 | 0.0206 (6) | 0.0196 (6) | 0.0231 (7) | 0.0018 (4) | 0.0073 (5) | −0.0023 (5) |
| C18 | 0.0174 (8) | 0.0254 (9) | 0.0206 (9) | −0.0004 (6) | 0.0047 (6) | −0.0008 (7) |
| O2 | 0.0285 (6) | 0.0152 (6) | 0.0270 (7) | 0.0027 (5) | 0.0005 (5) | −0.0012 (5) |
| C19 | 0.0457 (11) | 0.0204 (9) | 0.0266 (10) | −0.0074 (8) | 0.0069 (8) | −0.0056 (7) |
| O3 | 0.0267 (6) | 0.0173 (6) | 0.0242 (7) | −0.0021 (5) | 0.0084 (5) | 0.0021 (5) |
| C20 | 0.0212 (8) | 0.0234 (9) | 0.0216 (9) | −0.0028 (7) | 0.0049 (7) | 0.0010 (7) |
(II) (Z)-5-[2-(Benzo[b]thiophen-3-yl)-1-(3,4,5-trimethoxyphenyl)ethenyl]-1H-tetrazole . Geometric parameters (Å, º)
| S1—C1 | 1.7218 (17) | C10—C12 | 1.487 (2) |
| S1—C8 | 1.7371 (17) | C12—C13 | 1.389 (2) |
| N1—C11 | 1.336 (2) | C12—C17 | 1.396 (2) |
| N1—N2 | 1.3470 (19) | C13—C14 | 1.397 (2) |
| N1—H1N | 0.91 (2) | C13—H13 | 0.9500 |
| N2—N3 | 1.2911 (19) | C14—O1 | 1.3622 (19) |
| N3—N4 | 1.3641 (19) | C14—C15 | 1.396 (2) |
| N4—C11 | 1.324 (2) | C15—O2 | 1.373 (2) |
| C1—C2 | 1.362 (2) | C15—C16 | 1.397 (2) |
| C1—H1 | 0.9500 | C16—O3 | 1.362 (2) |
| C2—C3 | 1.448 (2) | C16—C17 | 1.387 (2) |
| C2—C9 | 1.467 (2) | C17—H17 | 0.9500 |
| C3—C4 | 1.404 (2) | O1—C18 | 1.431 (2) |
| C3—C8 | 1.405 (2) | C18—H18A | 0.9800 |
| C4—C5 | 1.381 (2) | C18—H18B | 0.9800 |
| C4—H4 | 0.9500 | C18—H18C | 0.9800 |
| C5—C6 | 1.403 (3) | O2—C19 | 1.433 (2) |
| C5—H5 | 0.9500 | C19—H19A | 0.9800 |
| C6—C7 | 1.378 (3) | C19—H19B | 0.9800 |
| C6—H6 | 0.9500 | C19—H19C | 0.9800 |
| C7—C8 | 1.396 (2) | O3—C20 | 1.427 (2) |
| C7—H7 | 0.9500 | C20—H20A | 0.9800 |
| C9—C10 | 1.339 (2) | C20—H20B | 0.9800 |
| C9—H9 | 0.9500 | C20—H20C | 0.9800 |
| C10—C11 | 1.476 (2) | ||
| C1—S1—C8 | 90.97 (8) | C13—C12—C17 | 120.70 (15) |
| C11—N1—N2 | 109.29 (13) | C13—C12—C10 | 121.24 (14) |
| C11—N1—H1N | 131.7 (12) | C17—C12—C10 | 118.02 (14) |
| N2—N1—H1N | 119.0 (12) | C12—C13—C14 | 119.34 (15) |
| N3—N2—N1 | 106.55 (12) | C12—C13—H13 | 120.3 |
| N2—N3—N4 | 110.08 (12) | C14—C13—H13 | 120.3 |
| C11—N4—N3 | 106.69 (13) | O1—C14—C15 | 115.07 (14) |
| C2—C1—S1 | 114.20 (13) | O1—C14—C13 | 124.64 (15) |
| C2—C1—H1 | 122.9 | C15—C14—C13 | 120.29 (15) |
| S1—C1—H1 | 122.9 | O2—C15—C14 | 120.79 (15) |
| C1—C2—C3 | 111.37 (15) | O2—C15—C16 | 119.41 (15) |
| C1—C2—C9 | 126.31 (15) | C14—C15—C16 | 119.73 (15) |
| C3—C2—C9 | 122.31 (15) | O3—C16—C17 | 124.42 (15) |
| C4—C3—C8 | 118.88 (15) | O3—C16—C15 | 115.42 (14) |
| C4—C3—C2 | 129.32 (15) | C17—C16—C15 | 120.16 (15) |
| C8—C3—C2 | 111.79 (15) | C16—C17—C12 | 119.76 (15) |
| C5—C4—C3 | 119.22 (16) | C16—C17—H17 | 120.1 |
| C5—C4—H4 | 120.4 | C12—C17—H17 | 120.1 |
| C3—C4—H4 | 120.4 | C14—O1—C18 | 116.88 (13) |
| C4—C5—C6 | 120.87 (16) | O1—C18—H18A | 109.5 |
| C4—C5—H5 | 119.6 | O1—C18—H18B | 109.5 |
| C6—C5—H5 | 119.6 | H18A—C18—H18B | 109.5 |
| C7—C6—C5 | 121.07 (16) | O1—C18—H18C | 109.5 |
| C7—C6—H6 | 119.5 | H18A—C18—H18C | 109.5 |
| C5—C6—H6 | 119.5 | H18B—C18—H18C | 109.5 |
| C6—C7—C8 | 118.00 (16) | C15—O2—C19 | 112.84 (13) |
| C6—C7—H7 | 121.0 | O2—C19—H19A | 109.5 |
| C8—C7—H7 | 121.0 | O2—C19—H19B | 109.5 |
| C7—C8—C3 | 121.96 (16) | H19A—C19—H19B | 109.5 |
| C7—C8—S1 | 126.42 (13) | O2—C19—H19C | 109.5 |
| C3—C8—S1 | 111.62 (12) | H19A—C19—H19C | 109.5 |
| C10—C9—C2 | 126.42 (15) | H19B—C19—H19C | 109.5 |
| C10—C9—H9 | 116.8 | C16—O3—C20 | 117.28 (13) |
| C2—C9—H9 | 116.8 | O3—C20—H20A | 109.5 |
| C9—C10—C11 | 121.21 (14) | O3—C20—H20B | 109.5 |
| C9—C10—C12 | 123.84 (15) | H20A—C20—H20B | 109.5 |
| C11—C10—C12 | 114.88 (13) | O3—C20—H20C | 109.5 |
| N4—C11—N1 | 107.39 (14) | H20A—C20—H20C | 109.5 |
| N4—C11—C10 | 126.03 (14) | H20B—C20—H20C | 109.5 |
| N1—C11—C10 | 126.58 (14) | ||
| C11—N1—N2—N3 | −0.48 (17) | C9—C10—C11—N4 | −107.56 (19) |
| N1—N2—N3—N4 | −0.08 (17) | C12—C10—C11—N4 | 75.24 (19) |
| N2—N3—N4—C11 | 0.61 (17) | C9—C10—C11—N1 | 73.8 (2) |
| C8—S1—C1—C2 | −1.09 (14) | C12—C10—C11—N1 | −103.42 (18) |
| S1—C1—C2—C3 | 2.01 (18) | C9—C10—C12—C13 | 49.6 (2) |
| S1—C1—C2—C9 | −177.01 (13) | C11—C10—C12—C13 | −133.26 (15) |
| C1—C2—C3—C4 | 179.11 (16) | C9—C10—C12—C17 | −127.84 (17) |
| C9—C2—C3—C4 | −1.8 (3) | C11—C10—C12—C17 | 49.29 (19) |
| C1—C2—C3—C8 | −2.11 (19) | C17—C12—C13—C14 | 0.7 (2) |
| C9—C2—C3—C8 | 176.95 (14) | C10—C12—C13—C14 | −176.67 (14) |
| C8—C3—C4—C5 | 0.4 (2) | C12—C13—C14—O1 | −179.49 (14) |
| C2—C3—C4—C5 | 179.09 (15) | C12—C13—C14—C15 | 0.8 (2) |
| C3—C4—C5—C6 | −0.6 (2) | O1—C14—C15—O2 | 2.1 (2) |
| C4—C5—C6—C7 | 0.2 (3) | C13—C14—C15—O2 | −178.17 (14) |
| C5—C6—C7—C8 | 0.4 (3) | O1—C14—C15—C16 | 178.98 (14) |
| C6—C7—C8—C3 | −0.6 (2) | C13—C14—C15—C16 | −1.3 (2) |
| C6—C7—C8—S1 | 179.37 (13) | O2—C15—C16—O3 | −2.4 (2) |
| C4—C3—C8—C7 | 0.2 (2) | C14—C15—C16—O3 | −179.38 (14) |
| C2—C3—C8—C7 | −178.70 (14) | O2—C15—C16—C17 | 177.21 (14) |
| C4—C3—C8—S1 | −179.77 (12) | C14—C15—C16—C17 | 0.3 (2) |
| C2—C3—C8—S1 | 1.32 (17) | O3—C16—C17—C12 | −179.17 (14) |
| C1—S1—C8—C7 | 179.83 (15) | C15—C16—C17—C12 | 1.2 (2) |
| C1—S1—C8—C3 | −0.18 (12) | C13—C12—C17—C16 | −1.7 (2) |
| C1—C2—C9—C10 | 34.3 (3) | C10—C12—C17—C16 | 175.75 (14) |
| C3—C2—C9—C10 | −144.62 (16) | C15—C14—O1—C18 | −175.64 (14) |
| C2—C9—C10—C11 | −0.3 (2) | C13—C14—O1—C18 | 4.6 (2) |
| C2—C9—C10—C12 | 176.63 (14) | C14—C15—O2—C19 | −81.92 (19) |
| N3—N4—C11—N1 | −0.88 (17) | C16—C15—O2—C19 | 101.17 (17) |
| N3—N4—C11—C10 | −179.75 (14) | C17—C16—O3—C20 | 5.4 (2) |
| N2—N1—C11—N4 | 0.86 (17) | C15—C16—O3—C20 | −174.98 (14) |
| N2—N1—C11—C10 | 179.73 (14) |
(II) (Z)-5-[2-(Benzo[b]thiophen-3-yl)-1-(3,4,5-trimethoxyphenyl)ethenyl]-1H-tetrazole . Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1N···N3i | 0.91 (2) | 2.65 (2) | 3.3886 (19) | 138.5 (16) |
| N1—H1N···N4i | 0.91 (2) | 1.85 (2) | 2.7482 (19) | 167.1 (18) |
Symmetry code: (i) x−1, y, z.
References
- Addicott, C., Bernhardt, P. V. & Wentrup, C. (2009). Arkivoc, 10, 30–36.
- Bruker (2006). APEX2 and SAINT. Bruker AXS Inc., Madison, Wisconsin, USA.
- Gawande, S. D., Raihan, M. J., Zanwar, M. R., Kavala, V., Janreddy, D., Kuo, C.-W., Chen, M.-L., Kuo, T.-S. & Yao, C.-F. (2013). Tetrahedron, 69, 1841–1848.
- Groom, C. & Allen, F. H. (2014). Angew. Chem. Int. Ed. 53, 662–671. [DOI] [PubMed]
- Krause, L., Herbst-Irmer, R., Sheldrick, G. M. & Stalke, D. (2015). J. Appl. Cryst. 48, 3–10. [DOI] [PMC free article] [PubMed]
- Parkin, S. (2000). Acta Cryst. A56, 157–162. [DOI] [PubMed]
- Parkin, S. (2013). CIFFIX. http://xray.uky.edu/people/parkin/programs/ciffix.
- Penthala, N. R., Bommagani, S., Yadlapalli, J. & Crooks, P. A. (2016). Tetrahedron Lett. 57, 1807–1810.
- Penthala, N. R., Madadi, N. R., Bommagani, S., Parkin, S. & Crooks, P. A. (2014b). Acta Cryst. E70, 392–395. [DOI] [PMC free article] [PubMed]
- Penthala, N. R., Madadi, N. R., Janganati, V. & Crooks, P. A. (2014a). Tetrahedron Lett. 55, 5562–5565. [DOI] [PMC free article] [PubMed]
- Penthala, N. R., Madhukuri, L., Thakkar, S., Madadi, N. R., Lamture, G., Eoff, R. L. & Crooks, P. A. (2015). Med. Chem. Commun. 6, 1535–1543. [DOI] [PMC free article] [PubMed]
- Penthala, N. R., Sonar, V. N., Horn, J., Leggas, M., Yadlapalli, K. B. J. S. & Crooks, P. A. (2013). Med. Chem. Commun. 4, 1073–1078. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Sonar, V. N., Parkin, S. & Crooks, P. A. (2007). Acta Cryst. C63, o743–o745. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I, II. DOI: 10.1107/S2056989016005600/hg5472sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989016005600/hg5472Isup2.hkl
Structure factors: contains datablock(s) II. DOI: 10.1107/S2056989016005600/hg5472IIsup3.hkl
Additional supporting information: crystallographic information; 3D view; checkCIF report






