The anion has an open-chain structure in which one of the oxygen atoms of the sulfate residue, the S atom, the C atoms of the sugar chain and the O atom of the hydroxymethyl group form an essentially planar zigzag chain. A three-dimensional bonding network exists in the crystal structure involving hexacoordination of sodium ions by O atoms, three of which are provided by a single d-lyxose–sulfonate unit and the other three by two sulfonate groups and one hydroxymethyl group, each from separate units of the adduct. Extensive intermolecular O—H⋯O hydrogen bonding supplements this bonding network.
Keywords: crystal structure, d-lyxose bisulfite adduct, sodium hydrogen sulfite, sodium metabisulfite
Abstract
The title compound, Na+·C5H11O8S− [systematic name: sodium (1S,2S,3S,4R)-1,2,3,4,5-pentahydroxypentane-1-sulfonate], is formed by reaction of d-lyxose with sodium bisulfite (sodium hydrogen sulfite) in water. The anion has an open-chain structure in which one of the oxygen atoms of the sulfonate residue, the S atom, the C atoms of the sugar chain and the O atom of the hydroxymethyl group form an essentially planar zigzag chain with the corresponding torsion angles lying between 179.80 (11) and 167.74 (14)°. A three-dimensional bonding network exists in the crystal structure involving hexacoordination of sodium ions by O atoms, three of which are provided by a single d-lyxose–sulfonate unit and the other three by two sulfonate groups and one hydroxymethyl group, each from separate units of the adduct. Extensive intermolecular O—H⋯O hydrogen bonding supplements this bonding network.
Chemical context
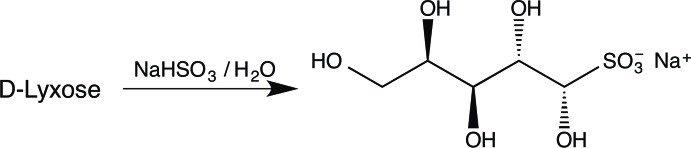
Bisulfite adducts of aldehydes are important compounds because, in many cases, they are crystalline and allow a means of purification and storage of those aldehydes which are liquids or which suffer from problems of instability. The importance of aldehydes in many synthetic processes for the production of commercially important compounds, including pharmaceuticals, means that there is continuing interest in these bisulfite adducts. A recent publication (Kissane et al., 2013 ▸) has focused on counter-ion effects in the preparation of aldehyde–bisulfite adducts. Of particular concern in that work was a comparison of the physical properties of sodium and potassium bisulfite adducts of a range of aldehydes, to include their hygroscopic nature and ease of filtration, in order to facilitate their preparation and storage on a large scale. Studies by X-ray crystallography on the bisulfite adducts of common aldehydo-sugars such as d-glucose (Cole et al., 2001 ▸) and our related work on d-galactose (Haines & Hughes, 2010 ▸), d-ribose (Haines & Hughes, 2014 ▸) and d-lyxose (Haines & Hughes, 2015 ▸) indicated the crystallinity and ease of isolation of such potassium adducts, and also, in the case of the sodium bisulfite adduct of d-glucose (Haines & Hughes, 2012 ▸), allowed a comparison of the potassium and sodium compounds. We now report the preparation (Fig. 1 ▸), properties, and crystal structure of the sodium bisulfite adduct of d-lyxose, which allows a further comparison of the influence of the two counter-ions in the properties of an adduct from the same substrate.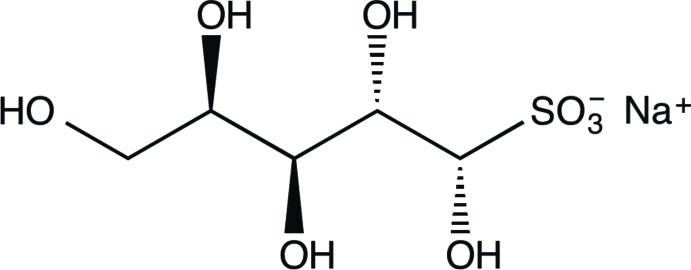
Figure 1.
Schematic representation of the preparation of the title compound.
Crystallization of the sodium bisulfite adduct of d-lyxose from water required a very concentrated solution from which highly crystalline material grew slowly on storage at room temperature. In contrast to the potassium adduct (Haines & Hughes, 2015 ▸), the crystals lacked water of crystallization but had the same S-configuration at C1, leading to a similar positive optical rotation for the two products. The melting points of the sodium and potassium adducts (417.6–420.1 K and 392–400 K, respectively, both with decomposition) were above that of d-lyxose (381–385 K). Both of the d-lyxose adducts were stable on storage in a sealed container at room temperature.
Structural commentary
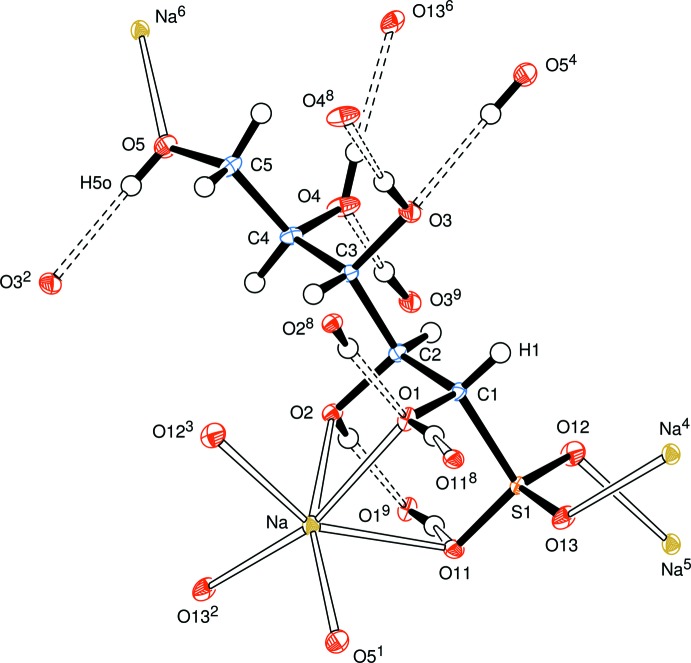
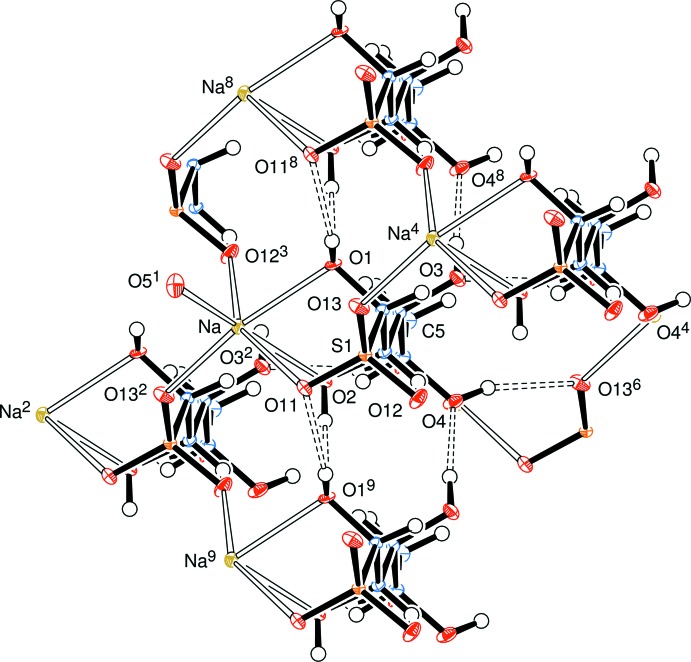
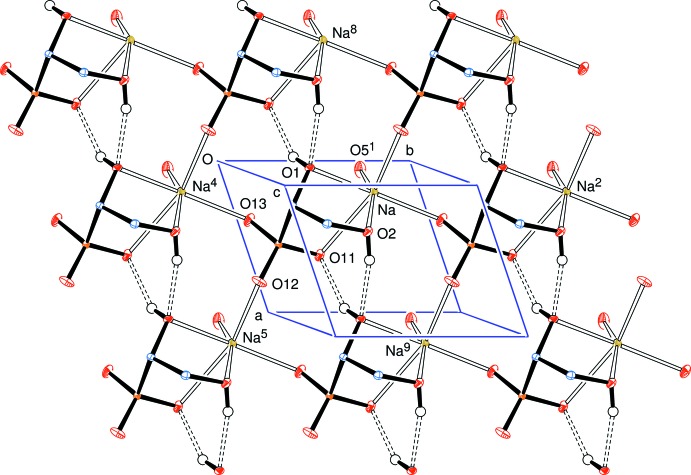
The newly formed chiral centre at C1 has the S-configuration (Fig. 2 ▸) and the systematic name for the salt is sodium (1S,2S,3S,4R)-1,2,3,4,5-pentahydroxypentane-1-sulfonate. The anion has an open-chain structure in which one of the oxygen atoms, O13, of the sulfonate residue, the S atom, the C atoms of the sugar chain and the oxygen atom, O5, of the terminal hydroxymethyl group form an essentially planar zigzag (all-trans) chain with the corresponding torsion angles lying between the absolute values of 179.80 (11) and 167.74 (14)°. The atoms O13–C4 form a plane, with C5 and O5 displaced 0.229 (3) and 0.525 (2) Å, respectively, from that mean plane. All of the hydroxyl groups form medium-strength to weak intermolecular hydrogen bonds which connect molecules in an extensive three-dimensional network (Fig. 3 ▸ and Table 1 ▸). This network is enhanced through complexation of the sodium atom which has a coordination sphere of six oxygen atoms with an approximately octahedral pattern in which three sites are occupied by oxygen atoms O1, O2, and O11 of one basic d-lyxose-sulfonate unit and the remaining three sites are occupied by oxygen atoms O12 and O13 arising from two different sulfonate groups, and O5 of another d-lyxose-sulfonate unit. The Na—O bond lengths lie in the range 2.2524 (16) to 2.5265 (16) Å. The sodium atoms are linked in planes parallel to the ab plane through coordinating sulfonate groups supported by H–O⋯Na coordination and hydrogen bonds (Fig. 4 ▸). There is no symmetry in this space group; all the molecules lie parallel and are arranged by translation parallel to the cell axes.
Figure 2.
View of the d-lyxose–NaHSO3 adduct, indicating the atom-numbering scheme, all sodium coordination contacts and hydrogen bonds involving the atoms of the basic adduct moieties. Displacement ellipsoids are drawn at the 50% probability level. [Symmetry codes: (1) x, y, z − 1; (2) x, y + 1, z; (3) x − 1, y + 1, z; (4) x, y − 1, z; (5) x + 1, y − 1, z; (6) x, y, z + 1; (7) x − 1, y, z + 1; (8) x − 1, y, z; (9) x + 1, y, z.]
Figure 3.
View approximately along the d-lyxose chain, showing the intermolecular hydrogen bonding and coordination links. Symmetry codes are as in Fig. 2 ▸.
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C5—H5B⋯O11i | 0.97 (4) | 2.48 (3) | 3.394 (2) | 157 (3) |
| O1—H1O⋯O11ii | 0.74 (4) | 2.00 (4) | 2.6813 (19) | 152 (4) |
| O2—H2O⋯O1iii | 0.88 (4) | 1.97 (4) | 2.8311 (19) | 164 (3) |
| O3—H3O⋯O4ii | 0.87 (3) | 1.79 (3) | 2.664 (2) | 173 (3) |
| O4—H4O⋯O13iv | 0.86 (4) | 2.11 (4) | 2.936 (2) | 162 (4) |
| O5—H5O⋯O3v | 0.80 (5) | 1.99 (5) | 2.782 (2) | 166 (5) |
Symmetry codes: (i) 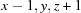 ; (ii)
; (ii) 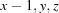 ; (iii)
; (iii) 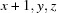 ; (iv)
; (iv) 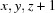 ; (v)
; (v) 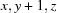 .
.
Figure 4.
View approximately onto the ab plane, showing the links between the sodium ions parallel to that plane. Symmetry codes are as in Fig. 2 ▸.
A comparison of the crystal structures of the sodium and potassium bisulfite adducts of d-lyxose illustrates the different coordination requirements of the two alkali metal cations. In the potassium salt hydrate (Haines & Hughes, 2015 ▸), two distinct environments for the cation are observed, involving both hexa- and octa-coordination of oxygen atoms, with each cation lying on a twofold symmetry axis. Oxygen atoms from the water of crystallization provide two of the coordination sites for the octa-coordinate potassium ion. In contrast, the sodium salt lacks water of crystallization and possesses a much simpler crystal structure having one environment only for the cation with hexa-coordination of oxygen atoms. However, in both cases the structures accommodate a nearly planar zigzag chain incorporating the sulfur atom, the five sugar carbon atoms and the oxygen of the terminal hydroxymethyl group, and both adducts crystallize with the same S-configuration at the newly formed chiral centre, despite evidence for the existence of the R-stereoisomer in solution.
Supramolecular features
A three-dimensional bonding network exists in the crystal structure through (i) hexa-coordination of a sodium cation with oxygens from four different lyxose bisulfite residues, three of those oxygens coming from one such residue, and (ii) intermolecular hydrogen bonds from each of the five hydroxyl groups to acceptor oxygens in four different residues.
Spectroscopic findings
High resolution mass spectrometry in negative ion mode showed no peak for ([C5H11O8S1]−) at m/z 231.0108 but a significant peak was observed at 213.0075 ([C5H11O8S1– H2O]−). The mono-anion of d-lyxose gave a peak at m/z 149.0458 ([C5H9O5]−) and the base peak of the spectrum, observed at m/z 299.0982 ([C10H19O10]−), was assigned to a dimer ion ([2M – H] −) produced by association of a d-lyxose molecule (M = C5H10O5) with the mono-anion of d-lyxose ([C5H9O5]−) under the electrospray ionization conditions of the mass spectrometric measurement.
The 1H NMR spectrum of the adduct in D2O indicated the presence of the α- and β-pyranose forms of d-lyxose and the major and minor forms of the acyclic sulfonate in the % ratios 11.62 : 5.47 : 74.78 : 8.14. Clearly, the R-stereoisomer at C1 is present in solution but only the S-isomer crystallizes. Further, some hydrolysis of the adduct to afford the parent sugar occurs during the NMR measurement. As expected, the NMR spectrum of the sodium bisulfite adduct is very similar to that of the related potassium sulfite adduct reported earlier (Haines & Hughes, 2015 ▸).
The 13C NMR spectrum showed signals for C1 nuclei at δC 94.81, 94.68, 84.21 and 82.17 arising, respectively, from the β- and α-pyranose forms of d-lyxose, the minor adduct and the major adduct, in the % ratios of 5.23 : 15.69 : 7.19 : 71.90.
Synthesis and crystallization
d-Lyxose (1 g) was dissolved in water (2 ml) and sodium metabisulfite (0.633 g) was added, Fig. 1 ▸. Complete solution was achieved on warming (to ca 313 K). Crystallization did not occur on prolonged standing, so the solution was evaporated at ca 303 K until the volume was ca 1 ml. On further storage, crystals (0.313 g) were deposited, m.p. 417.6–420.1 K (decomp.) and after concentration of the mother liquors, a further crop (0.204 g) was obtained, m.p. 414–420 K; [α]D 21 +8.9 (12 min.) (c, 0.68 in 9 : 1 H2O : HOAc). 1H NMR (D2O, 400 MHz, reference Me 3COH at δH 1.24): δH 5.00 (d, J 1,2 = 4.6 Hz, H-1 of α-pyranose), 4.86 (d, J 1,2 = 1.4 Hz, H-1 of β-pyranose); signals for the major acyclic sulfonate: δH 4.71 (d, J 1,2 = 0.6 Hz, H-1), 4.19 (dd, J 2,3 = 9.5 Hz, H-2), 3.99 (td, J 3,4 = 6.4, J 4,5b = 6.4, J 4,5a =1.2 Hz, H-4), 3.63 (dd, J 5a,5b = − 9.4 Hz, H-5a); for the minor acyclic sulfonate: δH 4.63 (d, J 1,2 = 5.4 Hz, H-1); ratio of major to minor sulfonate = 9.2 : 1. 13C NMR (D2O, 100 MHz, reference Me3COH at δC 30.29): δC 94.81 (C1, β-pyranose), 94.68 (C1, α-pyranose); signals for the major acyclic sulfonate: δC 82.17 (C1), 70.43, 69.85, 69.32 (C2, C3, C4), 63.78 (C5); the minor acyclic sulfonate showed a peak at δC 84.21 (C1).
Integration of the various signals for H-1 in the 1H NMR spectrum indicated the species α-pyranose, β-pyranose, major acyclic sulfonate and minor acyclic sulfonate were present in the % ratios of 11.62 : 5.47 : 74.78 : 8.14. In the 13C NMR spectrum, based on peak heights, the corresponding ratios were: 15.69 : 5.23 : 71.90 : 7.19.
HRESMS (negative ion mode, measured in H2O/MeOH, solution) gave a peak at m/z 149.0458 ([C5H9O5]−), a significant peak at 213.0075 ([C5H11O8S1 – H2O]−), and the base peak at 299.0982 ([C10H19O10]−). No significant peak was observed for ([C5H11O8S1]−) at the calculated m/z of 231.0180.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. Hydrogen atoms were located in difference maps and were refined freely with isotropic displacement parameters, except for H1 and H3 for which the U iso values were set at the positive value of 0.010 (rather than refining to a very low or negative value).
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | Na+·C5H11O8S− |
| M r | 254.19 |
| Crystal system, space group | Triclinic, P1 |
| Temperature (K) | 140 |
| a, b, c (Å) | 4.8558 (7), 5.8496 (10), 8.7950 (13) |
| α, β, γ (°) | 76.517 (13), 81.528 (12), 71.392 (14) |
| V (Å3) | 229.51 (7) |
| Z | 1 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.42 |
| Crystal size (mm) | 0.37 × 0.22 × 0.15 |
| Data collection | |
| Diffractometer | Oxford Diffraction Xcalibur 3/Sapphire3 CCD |
| Absorption correction | Multi-scan (CrysAlis PRO; Agilent, 2014 ▸) |
| T min, T max | 0.608, 1.000 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 4256, 2668, 2637 |
| R int | 0.031 |
| (sin θ/λ)max (Å−1) | 0.703 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.023, 0.059, 1.09 |
| No. of reflections | 2668 |
| No. of parameters | 178 |
| No. of restraints | 3 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.25, −0.45 |
| Absolute structure | Flack x determined using 1289 quotients [(I +)−(I −)]/[(I +)+(I −)] (Parsons et al., 2013 ▸) |
| Absolute structure parameter | 0.03 (3) |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989016005375/sj5496sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989016005375/sj5496Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989016005375/sj5496Isup3.cml
CCDC reference: 1471425
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
We thank the EPSRC UK National Mass Spectrometry Facility (NMSF) at Swansea University for determination of the low and high resolution mass spectra and Dr Sergey Nepogodiev for measurement of the NMR spectra.
supplementary crystallographic information
Crystal data
| Na+·C5H11O8S− | Z = 1 |
| Mr = 254.19 | F(000) = 132 |
| Triclinic, P1 | Dx = 1.839 Mg m−3 |
| a = 4.8558 (7) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 5.8496 (10) Å | Cell parameters from 3183 reflections |
| c = 8.7950 (13) Å | θ = 4.0–32.9° |
| α = 76.517 (13)° | µ = 0.42 mm−1 |
| β = 81.528 (12)° | T = 140 K |
| γ = 71.392 (14)° | Block, colourless |
| V = 229.51 (7) Å3 | 0.37 × 0.22 × 0.15 mm |
Data collection
| Oxford Diffraction Xcalibur 3/Sapphire3 CCD diffractometer | 2668 independent reflections |
| Radiation source: Enhance (Mo) X-ray Source | 2637 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.031 |
| Detector resolution: 16.0050 pixels mm-1 | θmax = 30.0°, θmin = 3.8° |
| Thin–slice φ and ω scans | h = −6→6 |
| Absorption correction: multi-scan (CrysAlis PRO; Agilent, 2014) | k = −8→8 |
| Tmin = 0.608, Tmax = 1.000 | l = −12→12 |
| 4256 measured reflections |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.023 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.059 | w = 1/[σ2(Fo2) + (0.039P)2 + 0.0108P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.09 | (Δ/σ)max = 0.001 |
| 2668 reflections | Δρmax = 0.25 e Å−3 |
| 178 parameters | Δρmin = −0.45 e Å−3 |
| 3 restraints | Absolute structure: Flack x determined using 1289 quotients [(I+)-(I-)]/[(I+)+(I-)] (Parsons et al., 2013) |
| Primary atom site location: structure-invariant direct methods | Absolute structure parameter: 0.03 (3) |
Special details
| Experimental. Absorption correction: CrysAlisPro (Agilent 2014). Empirical absorption correction using spherical harmonics, implemented in SCALE3 ABSPACK scaling algorithm. |
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Na | 0.14247 (15) | 0.62758 (13) | 0.42059 (8) | 0.00942 (15) | |
| S1 | 0.47392 (5) | 0.02789 (4) | 0.47548 (4) | 0.00670 (10) | |
| O11 | 0.5597 (3) | 0.2447 (2) | 0.39414 (15) | 0.0105 (2) | |
| O12 | 0.7080 (3) | −0.1631 (3) | 0.55456 (16) | 0.0152 (3) | |
| O13 | 0.3305 (3) | −0.0520 (3) | 0.37321 (16) | 0.0132 (3) | |
| C1 | 0.1957 (3) | 0.1206 (3) | 0.62783 (19) | 0.0074 (3) | |
| C2 | 0.2966 (4) | 0.2336 (3) | 0.7408 (2) | 0.0080 (3) | |
| C3 | 0.0562 (4) | 0.3094 (3) | 0.86872 (19) | 0.0080 (3) | |
| C4 | 0.1694 (4) | 0.4162 (3) | 0.9803 (2) | 0.0098 (3) | |
| C5 | −0.0682 (4) | 0.5331 (4) | 1.0947 (2) | 0.0133 (3) | |
| O1 | −0.0431 (3) | 0.2976 (2) | 0.55444 (15) | 0.0090 (2) | |
| O2 | 0.3613 (3) | 0.4537 (2) | 0.66271 (15) | 0.0093 (2) | |
| O3 | −0.0376 (3) | 0.1035 (3) | 0.94619 (16) | 0.0123 (3) | |
| O4 | 0.4056 (3) | 0.2355 (3) | 1.05675 (17) | 0.0154 (3) | |
| O5 | 0.0303 (4) | 0.6739 (3) | 1.17348 (17) | 0.0163 (3) | |
| H1 | 0.158 (6) | −0.022 (5) | 0.679 (3) | 0.010* | |
| H2 | 0.450 (6) | 0.117 (5) | 0.794 (3) | 0.010 (6)* | |
| H3 | −0.096 (6) | 0.433 (5) | 0.816 (3) | 0.010* | |
| H4 | 0.246 (6) | 0.548 (5) | 0.919 (3) | 0.007 (5)* | |
| H5A | −0.236 (6) | 0.632 (5) | 1.037 (4) | 0.013 (7)* | |
| H5B | −0.133 (7) | 0.409 (6) | 1.171 (4) | 0.021 (7)* | |
| H1O | −0.107 (8) | 0.248 (6) | 0.503 (4) | 0.023 (7)* | |
| H2O | 0.552 (8) | 0.420 (7) | 0.642 (4) | 0.032 (9)* | |
| H3O | −0.219 (7) | 0.158 (6) | 0.982 (3) | 0.019 (7)* | |
| H4O | 0.345 (9) | 0.159 (8) | 1.144 (5) | 0.034 (9)* | |
| H5O | 0.035 (10) | 0.799 (9) | 1.113 (6) | 0.055 (13)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Na | 0.0114 (3) | 0.0081 (3) | 0.0094 (3) | −0.0030 (2) | −0.0021 (2) | −0.0021 (2) |
| S1 | 0.00646 (16) | 0.00679 (16) | 0.00738 (16) | −0.00169 (12) | −0.00029 (12) | −0.00294 (11) |
| O11 | 0.0130 (6) | 0.0106 (6) | 0.0095 (5) | −0.0065 (4) | 0.0013 (4) | −0.0021 (4) |
| O12 | 0.0110 (6) | 0.0144 (6) | 0.0138 (6) | 0.0048 (5) | −0.0017 (5) | −0.0021 (5) |
| O13 | 0.0161 (6) | 0.0170 (6) | 0.0123 (6) | −0.0098 (5) | 0.0009 (5) | −0.0083 (5) |
| C1 | 0.0065 (7) | 0.0075 (7) | 0.0082 (7) | −0.0017 (5) | 0.0005 (5) | −0.0025 (5) |
| C2 | 0.0074 (6) | 0.0093 (7) | 0.0074 (7) | −0.0020 (5) | −0.0009 (5) | −0.0021 (5) |
| C3 | 0.0076 (6) | 0.0090 (7) | 0.0069 (7) | −0.0020 (6) | −0.0001 (5) | −0.0018 (5) |
| C4 | 0.0098 (7) | 0.0135 (8) | 0.0068 (7) | −0.0042 (6) | −0.0006 (5) | −0.0025 (6) |
| C5 | 0.0147 (8) | 0.0166 (8) | 0.0099 (7) | −0.0043 (6) | 0.0009 (6) | −0.0069 (6) |
| O1 | 0.0063 (5) | 0.0103 (6) | 0.0116 (6) | −0.0013 (4) | −0.0036 (4) | −0.0045 (4) |
| O2 | 0.0094 (5) | 0.0113 (6) | 0.0090 (5) | −0.0054 (4) | 0.0012 (4) | −0.0035 (4) |
| O3 | 0.0115 (6) | 0.0111 (6) | 0.0139 (6) | −0.0051 (5) | 0.0029 (5) | −0.0017 (4) |
| O4 | 0.0091 (5) | 0.0267 (7) | 0.0081 (6) | −0.0025 (5) | −0.0021 (4) | −0.0020 (5) |
| O5 | 0.0276 (7) | 0.0142 (7) | 0.0093 (6) | −0.0067 (6) | −0.0040 (5) | −0.0041 (5) |
Geometric parameters (Å, º)
| Na—O5i | 2.2524 (16) | C1—H1 | 0.91 (3) |
| Na—O13ii | 2.2661 (16) | C2—O2 | 1.417 (2) |
| Na—O12iii | 2.3728 (15) | C2—C3 | 1.530 (2) |
| Na—O1 | 2.3791 (16) | C2—H2 | 0.93 (3) |
| Na—O2 | 2.3800 (15) | C3—O3 | 1.4136 (19) |
| Na—O11 | 2.5265 (16) | C3—C4 | 1.523 (2) |
| Na—C1 | 3.0515 (19) | C3—H3 | 0.94 (3) |
| Na—S1 | 3.3114 (10) | C4—O4 | 1.417 (2) |
| Na—S1ii | 3.3864 (9) | C4—C5 | 1.512 (2) |
| Na—S1iii | 3.3866 (10) | C4—H4 | 0.98 (3) |
| S1—O12 | 1.4435 (14) | C5—O5 | 1.415 (2) |
| S1—O13 | 1.4496 (13) | C5—H5A | 0.97 (3) |
| S1—O11 | 1.4562 (13) | C5—H5B | 0.97 (4) |
| S1—C1 | 1.8034 (17) | O1—H1O | 0.74 (4) |
| S1—Naiv | 3.3865 (9) | O2—H2O | 0.88 (4) |
| S1—Nav | 3.3866 (10) | O3—H3O | 0.87 (3) |
| O12—Nav | 2.3728 (15) | O4—H4O | 0.86 (4) |
| O13—Naiv | 2.2661 (16) | O5—Navi | 2.2524 (16) |
| C1—O1 | 1.406 (2) | O5—H5O | 0.80 (5) |
| C1—C2 | 1.523 (2) | ||
| O5i—Na—O13ii | 96.36 (6) | O11—S1—Naiv | 141.71 (5) |
| O5i—Na—O12iii | 105.63 (6) | C1—S1—Naiv | 90.25 (6) |
| O13ii—Na—O12iii | 93.66 (6) | Na—S1—Naiv | 121.70 (3) |
| O5i—Na—O1 | 101.70 (6) | O12—S1—Nav | 35.88 (6) |
| O13ii—Na—O1 | 161.60 (6) | O13—S1—Nav | 96.24 (6) |
| O12iii—Na—O1 | 78.19 (5) | O11—S1—Nav | 94.58 (6) |
| O5i—Na—O2 | 161.20 (6) | C1—S1—Nav | 141.61 (6) |
| O13ii—Na—O2 | 92.49 (6) | Na—S1—Nav | 140.21 (3) |
| O12iii—Na—O2 | 90.27 (5) | Naiv—S1—Nav | 91.60 (2) |
| O1—Na—O2 | 71.30 (5) | S1—O11—Na | 109.54 (7) |
| O5i—Na—O11 | 91.52 (6) | S1—O12—Nav | 123.24 (8) |
| O13ii—Na—O11 | 107.60 (5) | S1—O13—Naiv | 130.10 (9) |
| O12iii—Na—O11 | 151.12 (6) | O1—C1—C2 | 108.62 (13) |
| O1—Na—O11 | 75.63 (5) | O1—C1—S1 | 107.39 (11) |
| O2—Na—O11 | 69.96 (5) | C2—C1—S1 | 112.68 (11) |
| O5i—Na—C1 | 115.69 (6) | O1—C1—Na | 49.01 (8) |
| O13ii—Na—C1 | 142.37 (6) | C2—C1—Na | 81.73 (9) |
| O12iii—Na—C1 | 95.93 (5) | S1—C1—Na | 81.65 (6) |
| O1—Na—C1 | 26.49 (4) | O1—C1—H1 | 113.8 (17) |
| O2—Na—C1 | 51.26 (5) | C2—C1—H1 | 110.4 (17) |
| O11—Na—C1 | 55.35 (5) | S1—C1—H1 | 104.0 (16) |
| O5i—Na—S1 | 96.73 (5) | Na—C1—H1 | 162.5 (17) |
| O13ii—Na—S1 | 130.25 (5) | O2—C2—C1 | 111.14 (13) |
| O12iii—Na—S1 | 127.92 (5) | O2—C2—C3 | 105.16 (13) |
| O1—Na—S1 | 51.16 (3) | C1—C2—C3 | 111.34 (13) |
| O2—Na—S1 | 65.10 (4) | O2—C2—H2 | 113.6 (17) |
| O11—Na—S1 | 24.48 (3) | C1—C2—H2 | 109.9 (17) |
| C1—Na—S1 | 32.60 (3) | C3—C2—H2 | 105.4 (16) |
| O5i—Na—S1ii | 115.37 (5) | O3—C3—C4 | 112.49 (13) |
| O13ii—Na—S1ii | 19.11 (4) | O3—C3—C2 | 109.19 (13) |
| O12iii—Na—S1ii | 89.97 (4) | C4—C3—C2 | 109.33 (13) |
| O1—Na—S1ii | 142.89 (4) | O3—C3—H3 | 110.7 (18) |
| O2—Na—S1ii | 73.74 (4) | C4—C3—H3 | 109.1 (17) |
| O11—Na—S1ii | 103.67 (4) | C2—C3—H3 | 105.7 (17) |
| C1—Na—S1ii | 124.58 (4) | O4—C4—C5 | 112.34 (15) |
| S1—Na—S1ii | 121.70 (3) | O4—C4—C3 | 110.35 (14) |
| O5i—Na—S1iii | 86.16 (5) | C5—C4—C3 | 112.70 (14) |
| O13ii—Na—S1iii | 88.41 (4) | O4—C4—H4 | 106.7 (16) |
| O12iii—Na—S1iii | 20.89 (4) | C5—C4—H4 | 105.9 (15) |
| O1—Na—S1iii | 89.30 (4) | C3—C4—H4 | 108.5 (16) |
| O2—Na—S1iii | 110.71 (4) | O5—C5—C4 | 111.02 (15) |
| O11—Na—S1iii | 163.98 (4) | O5—C5—H5A | 112.1 (17) |
| C1—Na—S1iii | 111.76 (4) | C4—C5—H5A | 108.1 (17) |
| S1—Na—S1iii | 140.21 (3) | O5—C5—H5B | 109 (2) |
| S1ii—Na—S1iii | 91.60 (2) | C4—C5—H5B | 110.9 (19) |
| O12—S1—O13 | 114.87 (9) | H5A—C5—H5B | 106 (3) |
| O12—S1—O11 | 112.80 (8) | C1—O1—Na | 104.51 (10) |
| O13—S1—O11 | 110.92 (8) | C1—O1—H1O | 112 (3) |
| O12—S1—C1 | 105.74 (8) | Na—O1—H1O | 114 (3) |
| O13—S1—C1 | 104.52 (8) | C2—O2—Na | 112.86 (10) |
| O11—S1—C1 | 107.22 (7) | C2—O2—H2O | 109 (2) |
| O12—S1—Na | 142.43 (6) | Na—O2—H2O | 108 (2) |
| O13—S1—Na | 102.56 (6) | C3—O3—H3O | 108 (2) |
| O11—S1—Na | 45.97 (6) | C4—O4—H4O | 111 (3) |
| C1—S1—Na | 65.75 (6) | C5—O5—Navi | 134.32 (12) |
| O12—S1—Naiv | 93.81 (6) | C5—O5—H5O | 108 (3) |
| O13—S1—Naiv | 30.79 (6) | Navi—O5—H5O | 118 (3) |
| O12—S1—O11—Na | −142.14 (7) | O12—S1—C1—Na | 140.92 (7) |
| O13—S1—O11—Na | 87.39 (8) | O13—S1—C1—Na | −97.46 (7) |
| C1—S1—O11—Na | −26.15 (9) | O11—S1—C1—Na | 20.34 (7) |
| Naiv—S1—O11—Na | 87.43 (9) | Naiv—S1—C1—Na | −125.05 (4) |
| Nav—S1—O11—Na | −174.14 (5) | Nav—S1—C1—Na | 142.05 (6) |
| O13—S1—O12—Nav | 64.12 (12) | O1—C1—C2—O2 | −55.53 (17) |
| O11—S1—O12—Nav | −64.32 (11) | S1—C1—C2—O2 | 63.32 (15) |
| C1—S1—O12—Nav | 178.80 (9) | Na—C1—C2—O2 | −13.98 (11) |
| Na—S1—O12—Nav | −110.68 (9) | O1—C1—C2—C3 | 61.35 (16) |
| Naiv—S1—O12—Nav | 87.47 (9) | S1—C1—C2—C3 | −179.80 (11) |
| O12—S1—O13—Naiv | 50.59 (13) | Na—C1—C2—C3 | 102.90 (12) |
| O11—S1—O13—Naiv | 179.96 (9) | O2—C2—C3—O3 | 175.85 (13) |
| C1—S1—O13—Naiv | −64.81 (12) | C1—C2—C3—O3 | 55.39 (16) |
| Na—S1—O13—Naiv | −132.66 (9) | O2—C2—C3—C4 | −60.69 (16) |
| Nav—S1—O13—Naiv | 82.62 (10) | C1—C2—C3—C4 | 178.85 (14) |
| O12—S1—C1—O1 | −176.87 (11) | O3—C3—C4—O4 | 58.72 (17) |
| O13—S1—C1—O1 | −55.25 (13) | C2—C3—C4—O4 | −62.77 (18) |
| O11—S1—C1—O1 | 62.54 (12) | O3—C3—C4—C5 | −67.77 (18) |
| Na—S1—C1—O1 | 42.21 (9) | C2—C3—C4—C5 | 170.74 (14) |
| Naiv—S1—C1—O1 | −82.85 (10) | O4—C4—C5—O5 | 66.8 (2) |
| Nav—S1—C1—O1 | −175.75 (7) | C3—C4—C5—O5 | −167.74 (14) |
| O12—S1—C1—C2 | 63.56 (13) | C2—C1—O1—Na | 60.41 (13) |
| O13—S1—C1—C2 | −174.82 (12) | S1—C1—O1—Na | −61.72 (10) |
| O11—S1—C1—C2 | −57.02 (13) | C1—C2—O2—Na | 19.43 (15) |
| Na—S1—C1—C2 | −77.36 (11) | C3—C2—O2—Na | −101.16 (12) |
| Naiv—S1—C1—C2 | 157.59 (11) | C4—C5—O5—Navi | −108.80 (17) |
| Nav—S1—C1—C2 | 64.69 (15) |
Symmetry codes: (i) x, y, z−1; (ii) x, y+1, z; (iii) x−1, y+1, z; (iv) x, y−1, z; (v) x+1, y−1, z; (vi) x, y, z+1.
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C5—H5B···O11vii | 0.97 (4) | 2.48 (3) | 3.394 (2) | 157 (3) |
| O1—H1O···O11viii | 0.74 (4) | 2.00 (4) | 2.6813 (19) | 152 (4) |
| O2—H2O···O1ix | 0.88 (4) | 1.97 (4) | 2.8311 (19) | 164 (3) |
| O3—H3O···O4viii | 0.87 (3) | 1.79 (3) | 2.664 (2) | 173 (3) |
| O4—H4O···O13vi | 0.86 (4) | 2.11 (4) | 2.936 (2) | 162 (4) |
| O5—H5O···O3ii | 0.80 (5) | 1.99 (5) | 2.782 (2) | 166 (5) |
Symmetry codes: (ii) x, y+1, z; (vi) x, y, z+1; (vii) x−1, y, z+1; (viii) x−1, y, z; (ix) x+1, y, z.
References
- Agilent (2014). CrysAlis PRO. Agilent Technologies Ltd, Yarnton, England.
- Cole, E. R., Craig, D. C., Fitzpatrick, L. J., Hibbert, D. B. & Stevens, J. D. (2001). Carbohydr. Res. 335, 1–10. [DOI] [PubMed]
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Haines, A. H. & Hughes, D. L. (2010). Carbohydr. Res. 345, 2705–2708. [DOI] [PubMed]
- Haines, A. H. & Hughes, D. L. (2012). Acta Cryst. E68, m377–m378. [DOI] [PMC free article] [PubMed]
- Haines, A. H. & Hughes, D. L. (2014). Acta Cryst. E70, 406–409. [DOI] [PMC free article] [PubMed]
- Haines, A. H. & Hughes, D. L. (2015). Acta Cryst. E71, 993–996. [DOI] [PMC free article] [PubMed]
- Johnson, C. K. (1976). ORTEPII. Report ORNL-5138. Oak Ridge National Laboratory, Tennessee, USA.
- Kissane, M. G., Frank, S. A., Rener, G. A., Ley, C. P., Alt, C. A., Stroud, P. A., Vaid, R. K., Boini, S. K., McKee, L. A., Vicenzi, J. T. & Stephenson, G. A. (2013). Tetrahedron Lett. 54, 6587–6591.
- Parsons, S., Flack, H. D. & Wagner, T. (2013). Acta Cryst. B69, 249–259. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989016005375/sj5496sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989016005375/sj5496Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989016005375/sj5496Isup3.cml
CCDC reference: 1471425
Additional supporting information: crystallographic information; 3D view; checkCIF report