The most important intermolecular interactions in the two indole derivatives described here are N—H⋯π bonds, which lead to chains in one case and inversion dimers in the other; C—H⋯π interactions appear to reinforce the N—H⋯π bonds in each case.
Keywords: crystal structure, indole, N—H⋯π interaction, chains, inversion dimers
Abstract
We describe the syntheses and crystal structures of two indole derivatives, namely 6-isopropyl-3-(2-nitro-1-phenylethyl)-1H-indole, C19H20N2O2, (I), and 2-(4-methoxyphenyl)-3-(2-nitro-1-phenylethyl)-1H-indole, C23H20N2O3, (II); the latter crystallizes with two molecules (A and B) with similar conformations (r.m.s. overlay fit = 0.139 Å) in the asymmetric unit. Despite the presence of O atoms as potential acceptors for classical hydrogen bonds, the dominant intermolecular interaction in each crystal is an N—H⋯π bond, which generates chains in (I) and A+A and B+B inversion dimers in (II). A different aromatic ring acts as the acceptor in each case. The packing is consolidated by C—H⋯π interactions in each case but aromatic π–π stacking interactions are absent.
Chemical context
N—H⋯π interactions are now a well-recognised type of ‘non-classical’ weak bond (Desiraju & Steiner, 1999 ▸). They are of special significance in biological systems (Burley & Petsko, 1986 ▸; Levitt & Perutz, 1998 ▸) and are thought to play an important role in establishing protein secondary structures (Lavanya et al., 2014 ▸). They may even influence the charge-transport properties of organic semiconductors (Zhao et al., 2009 ▸). The presence of N—H⋯π interactions in indole complexes with aromatic species has been investigated by IR spectroscopy (Muñoz et al., 2004 ▸), and such bonds have also been observed in many crystal structures of indole derivatives (e.g. Krishna et al., 1999 ▸; Cordes et al., 2011 ▸).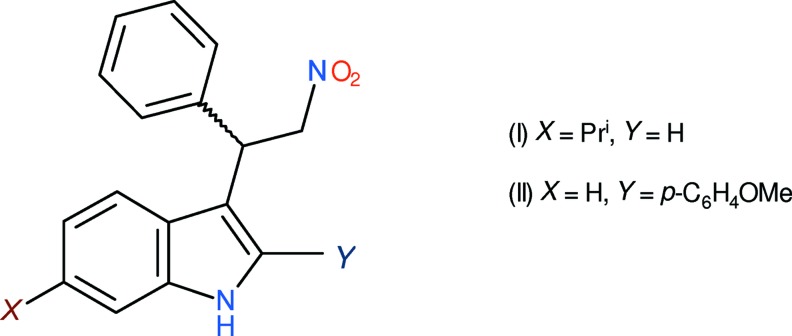
As part of our ongoing synthetic, biological (Kerr, 2013 ▸) and structural studies (Kerr et al., 2015 ▸, 2016 ▸) of variously substituted indole derivatives, we now report the syntheses and crystal structures of 6-isopropyl-3-(2-nitro-1-phenylethyl)-1H-indole, C19H20N2O2, (I), and 2-(4-methoxyphenyl)-3-(2-nitro-1-phenylethyl)-1H-indole, C23H20N2O3, (II), in which N—H⋯π bonds are the most important intermolecular interactions, but result in quite different structures.
Structural commentary
Compound (I) crystallizes in a Sohncke space group with one molecule in the asymmetric unit (Fig. 1 ▸). The absolute structure was indeterminate in the present study and C9 was assigned an arbitrary S configuration (given the synthesis, we presume that the bulk sample consists of a statistical mixture of enantiomers). The dihedral angle between the mean plane of the N1/C1–C8 indole ring system (r.m.s. deviation = 0.018 Å) and the C11–C16 phenyl ring is 83.59 (11)°. Atom C17 of the 6-isopropyl substituent deviates slightly from the indole plane, by −0.092 (6) Å. In terms of the terminal carbon atoms of this group, C18 and C19 deviate from the indole plane by −1.461 (6) and 1.030 (6) Å, respectively. Atom C9 shows a relatively large deviation from the indole plane of −0.084 (6) Å, perhaps because of steric crowding. In terms of the orientation of the substituents attached to C9, the C6—C7—C9—C10 torsion angle of 174.6 (5)° (anti about C7—C9) indicates that the C10 atom of the CH2NO2 group lies roughly in the plane of the indole ring, whereas the C6—C7—C9—C11 angle of −61.6 (7)° (gauche about C7—C9) indicates that the pendant ring lies to one side of the indole plane. Finally, the C7—C9—C10—N2 torsion angle of −176.5 (4)° indicates a near anti conformation about the C9—C10 bond.
Figure 1.
The molecular structure of (I), showing 50% probability displacement ellipsoids.
There are two molecules, A (Fig. 2 ▸) and B, in the asymmetric unit of (II). The space group for (II) is centrosymmetric and the stereogenic centres (C9 in molecule A and C32 in molecule B) were arbitrarily assigned an S configuration for ease of comparison with compound (I).
Figure 2.
The molecular structure of the N1 molecule in (II), showing 50% probability displacement ellipsoids. The molecular structure of the N3 molecule is very similar.
In molecule A, the dihedral angles between the indole (N1/C1–C8) mean plane (r.m.s. deviation = 0.012 Å) and the C11–C16 and C17–C22 rings are 65.49 (4) and 66.26 (4)°, respectively. The deviations of C9 and C17 from the indole plane are 0.017 (2) and 0.0168 (19) Å, respectively; C23 deviates from the C17–C22 plane by 0.322 (3) Å. The equivalent data for molecule B are 0.005 Å (N3/C24–C31 r.m.s. deviation), 64.92 (4)° (C34 ring), 58.31 (5)° (C40 ring), −0.071 (2) Å (C32), −0.014 (2) Å (C40), −0.214 (3) Å (C46). These data indicate that molecules A and B have similar but not quite identical conformations: the unweighted r.m.s. overlay fit for the 28 non-hydrogen atoms is 0.139 Å (Fig. 3 ▸).
Figure 3.
Overlay plot of the conformations of the N1 molecules (black) and N3 molecules (red) in the crystal of (II).
As just noted, molecules A and B in (II) have similar conformations, but the local geometry about the stereogenic atoms C9 and C32 are completely different from the corresponding local geometry about C9 in (I). This can be seen in the following data for the N1 molecule in (II): the C6—C7—C9—C10 torsion angle is −42.9 (2)° (compressed gauche about C7—C9) and the C6—C7—C9—C11 angle is 83.76 (19)° (expanded gauche about C7—C9); the C7—C9—C10—N2 torsion angle of −58.42 (17)° (gauche about C9—C10) is also completely different from the corresponding angle in (I). The corresponding torsion angles for the N3 molecule in (II) are −38.4 (2), 87.50 (19) and −56.24 (19)°, respectively. In essence, the 2-nitro 1-phenyl ethyl substituent has rotated around the C7—C9 bond, so that the H atom attached to C9 and C32 in (II) lies approximately above C8 whereas in (I) the CH2NO2 group takes on this role.
Supramolecular features
In the crystal of (I), the molecules are linked by N—H⋯π interactions (Table 1 ▸, Fig. 4 ▸) to generate [010] chains, in which adjacent molecules are related by the 21 screw axis. The acceptor ring is the C1–C6 benzene ring of the indole system; the dihedral angle between any adjacent pair of indole ring systems in the chain is 68.89 (8)°. The chain appears to be reinforced by a C—H⋯π bond from the C2—H2 group of the benzene ring syn to the N—H group to the five-membered ring of the same adjacent molecule; the H⋯π separation is actually marginally shorter for this bond than for the N—H⋯π bond. Two further C—H⋯π interactions (Fig. 5 ▸) also occur in the crystal of (I): based on their lengths, these are presumably significantly weaker than the C2—H2 bond. They arise from adjacent C—H groups on the pendant C11–C16 benzene ring with the acceptor rings being another C11–C16 ring and the C1–C6 indole ring of the same adjacent molecule. Taken together, the intermolecular interactions lead to (100) sheets in the crystal of (I).
Table 1. Hydrogen-bond geometry (Å, °) for (I) .
Cg1, Cg2 and Cg3 are the centroids of the N1/C1/C6–C8, C1–C6 and C11–C16 rings, respectively.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1⋯Cg2i | 0.84 (6) | 2.64 (6) | 3.386 (5) | 148 (6) |
| C2—H2⋯Cg1i | 0.95 | 2.63 | 3.468 (6) | 147 |
| C14—H14⋯Cg2ii | 0.95 | 2.79 | 3.638 (6) | 148 |
| C15—H15⋯Cg3ii | 0.95 | 2.87 | 3.551 (7) | 129 |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
Figure 4.
Partial packing diagram for (I), showing the formation of [010] chains linked by N—H⋯π and C—H⋯π interactions (double-dashed lines). [Symmetry codes: (i) 1 − x, y −  , 1 − z; (ii) 1 − x, y +
, 1 − z; (ii) 1 − x, y +  , 1 − z.] All H atoms, except H1 and H2, have been omitted for clarity. The orange circles indicate ring centroids.
, 1 − z.] All H atoms, except H1 and H2, have been omitted for clarity. The orange circles indicate ring centroids.
Figure 5.
Fragment of the packing for (I), showing C—H⋯π bonds arising from adjacent C—H groups of the pendant benzene ring. All H atoms, except H14 and H15, have been omitted for clarity. [Symmetry code: (i) 1 − x, y +  , −z.] The orange circles indicate ring centroids.
, −z.] The orange circles indicate ring centroids.
In the crystal of (II), inversion dimers linked by pairs of N—H⋯π interactions (Table 2 ▸, Fig. 6 ▸) occur for both independent molecules. In this case, the acceptor ring is the pendant C11–C16 or C34–C39 benzene ring for molecules A and B, respectively. This bonding mode possibly correlates with the different orientation of the substituents attached to C9 and C32, as described above. Again, the N—H⋯π bonds appear to be reinforced, but this time by two pairs of C—H⋯π interactions. As for (I), they arise from adjacent C—H groups in a benzene ring but this time they are part of the pendant 4-methoxybenzene ring at the indole 2-position. Further C—H⋯π bonds link the A+A and B+B dimers into a three-dimensional network in the crystal of (II).
Table 2. Hydrogen-bond geometry (Å, °) for (II) .
Cg1, Cg2, Cg3, Cg6, Cg7 and Cg8 are the centroids of the N1/C1/C6–C8, C1–C6, C11–C16, N3/C24/C29–C34--C31, C24–C29 and C34–C39 rings, respectively.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1⋯Cg3i | 0.886 (19) | 2.640 (19) | 3.3631 (15) | 139.6 (15) |
| N3—H3⋯Cg8ii | 0.875 (18) | 2.582 (19) | 3.3364 (15) | 144.9 (16) |
| C14—H14⋯Cg2iii | 0.95 | 2.58 | 3.4228 (18) | 149 |
| C21—H21⋯Cg2i | 0.95 | 2.69 | 3.4133 (17) | 134 |
| C22—H22⋯Cg1i | 0.95 | 2.68 | 3.4543 (17) | 138 |
| C23—H23B⋯Cg3iv | 0.98 | 2.79 | 3.6739 (18) | 150 |
| C37—H37⋯Cg6v | 0.95 | 2.86 | 3.7660 (18) | 160 |
| C41—H41⋯Cg6ii | 0.95 | 2.70 | 3.3793 (17) | 129 |
| C42—H42⋯Cg7ii | 0.95 | 2.67 | 3.3627 (17) | 130 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  ; (iv)
; (iv)  ; (v)
; (v)  .
.
Figure 6.
An inversion dimer of N1 molecules in the crystal of (II) linked by pairs of N—H⋯π and C—H⋯π interactions (double-dashed lines). [Symmetry code: (i) 1 − x, −y, −z.] The N3 molecules associate into similar dimers. The orange circles indicate ring centroids.
Database survey
There are over 7000 crystal structures of indole derivatives in the Cambridge Structural Database (CSD; Groom et al., 2016 ▸), but none of them have an iso-propyl group at the 6-position. Six structures contain a p-methoxybenzene grouping at the 2-position and four contain a 2-nitro-1-phenylethyl grouping at the 3-position; these latter structures are the ones recently described by us (Kerr et al., 2015 ▸).
Synthesis and crystallization
To prepare (I), 6-isopropylindole (452 mg, 2.84 mmol), trans-β-nitrostyrene (28, 429 mg, 2.88 mmol) and sulfamic acid (57 mg, 0.59 mmol) were stirred in EtOH (10 ml) at 323 K for 48 h. Evaporation of the solvent and flash chromatography (1:6 EtOAc, hexanes) gave 6-isopropyl-3-(2-nitro-1-phenylethyl)-1H-indole as an orange solid (550 mg, 63%). Red blades of (I) were recrystallized from methanol solution. δC (101 MHz; CDCl3) 144.0 (Cq), 139.3 (Cq), 136.9 (Cq), 127.9 (Cq), 127.8 (CH), 127.5 (CH), 124.3 (CH), 121.1 (CH), 119.4 (CH), 118.6 (CH), 114.3 (Cq), 108.5 (CH), 79.5 (CH2), 41.6 (CH), 34.3 (CH) and 24.4 (CH3); δH (400 MHz; CDCl3) 7.89 (1 H, br s), 7.30–7.21 (5 H, m), 7.18–7.15 (1 H, m), 7.12 (1 H, t, J 0.6), 6.90 (2 H, td, J, 1.5, 8.8), 5.08 (1 H, t, J 8.0), 4.97 (1 H, dd, J 7.4, 12.2), 4.85 (1 H, dd, J 8.4, 12.4), 2.91 (1 H, sp, J 6.9) and 1.20 (6 H, d, J 6.8); R f 0.16 (1:6 ethyl acetate, hexanes); m.p. 374–376 K; IR (KBr, cm−1) 3433, 3007, 2924,1550, 1429, 1377, 1089 and 750; HRMS (ESI) for C19H21N2O2 [M + H]+ calculated 309.1604, found 309.1619.
To prepare (II), 2-bromo-3-(2-nitro-1-phenylethyl)-1H-indole (Kerr et al., 2015 ▸) (90 mg, 0.26 mmol), 4-methoxyphenylboronic acid (53 mg, 0.35 mmol), Na2CO3 (29 mg, 0.27 mmol), LiCl (22 mg, 0.52 mmol) and tetrakis(triphenylphosphine)palladium(0) (12 mg, 0.01 mmol) were placed in a microwave reactor vessel under argon. Degassed water (4 ml), toluene (6 ml) and ethanol (6 ml) were added and the reaction was heated to 373 K (high absorbance mode, 30 W, 8 bar) for 2 h. The mixture was acidified to pH 2 with 10% HCl(aq) then extracted into EtOAc (10 ml × 3). The combined organic phases were washed with water (10 ml) and saturated NaCl(aq) (10 ml) then dried (magnesium sulfate), filtered and evaporated under reduced pressure. Flash chromatography of the isolated solid (1:5 ethyl acetate, hexanes) afforded 2-(4-methoxyphenyl)-3-(2-nitro-1-phenylethyl)-1H-indole as a colourless solid (48 mg, 50%). Colourless chunks of (II) were recrystallized from methanol solution. δC (63 MHz; CDCl3) 159.9 (Cq), 140.0 (Cq), 136.9 (Cq), 135.9 (CH), 130.1 (CH), 128.9 (Cq), 127.1 (CH), 125.0 (CH), 124.5 (Cq), 122.2 (Cq), 120.2 (CH), 119.8 (CH), 114.4 (CH), 111.3 (CH), 110.0 (CH), 109.1 (Cq), 79.1 (CH2), 55.4 (CH3) and 40.9 (CH); δH (250 MHz; CDCl3) 8.08 (1 H, br s), 7.45–7.25 (9 H, m), 7.19–6.90 (4 H, m), 5.19 (1 H, t, J 6.9) 5.10–5.01 (2 H, m) and 3.76 (3 H, s); R f 0.09 (1:5 EtOAc, hexanes); m.p. 472 K (EtOH); IR (Nujol, cm−1) 3401, 3013, 2854, 1616, 1548, 1324, 1250, 1203, 1099, 870 and 746; HRMS (ESI) for C23H21N2O3 [M + H]+ calculated 373.1553, found 373.1546.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 3 ▸. The N-bound H atoms were located in difference maps and their positions were freely refined. The C-bound H atoms were geometrically placed (C—H = 0.93–0.98 Å) and refined as riding atoms. The constraint U iso(H) = 1.2U eq(C, N carrier) or 1.5U eq(methyl carrier) was applied in all cases. The –CH3 groups were allowed to rotate, but not to tip, to best fit the electron density. Due to the similarity in the a and c unit-cell parameters for (I), twinning models were applied, but no improvement in fit resulted.
Table 3. Experimental details.
| (I) | (II) | |
|---|---|---|
| Crystal data | ||
| Chemical formula | C19H20N2O2 | C23H20N2O3 |
| M r | 308.37 | 372.41 |
| Crystal system, space group | Monoclinic, P21 | Triclinic, P

|
| Temperature (K) | 100 | 100 |
| a, b, c (Å) | 12.4525 (9), 5.7360 (4), 12.5896 (9) | 9.2014 (5), 9.4543 (7), 21.6201 (14) |
| α, β, γ (°) | 90, 116.081 (6), 90 | 98.563 (4), 93.416 (4), 98.354 (4) |
| V (Å3) | 807.68 (11) | 1833.7 (2) |
| Z | 2 | 4 |
| Radiation type | Mo Kα | Mo Kα |
| μ (mm−1) | 0.08 | 0.09 |
| Crystal size (mm) | 0.28 × 0.05 × 0.01 | 0.10 × 0.06 × 0.06 |
| Data collection | ||
| Diffractometer | Rigaku Mercury CCD | Rigaku Mercury CCD |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 7830, 3498, 2259 | 24774, 8621, 6769 |
| R int | 0.107 | 0.038 |
| (sin θ/λ)max (Å−1) | 0.649 | 0.668 |
| Refinement | ||
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.081, 0.164, 1.12 | 0.046, 0.124, 1.03 |
| No. of reflections | 3498 | 8621 |
| No. of parameters | 213 | 513 |
| No. of restraints | 1 | 0 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.28, −0.24 | 0.62, −0.31 |
Supplementary Material
Crystal structure: contains datablock(s) I, II, global. DOI: 10.1107/S2056989016006162/pk2578sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989016006162/pk2578Isup2.hkl
Structure factors: contains datablock(s) II. DOI: 10.1107/S2056989016006162/pk2578IIsup3.hkl
Supporting information file. DOI: 10.1107/S2056989016006162/pk2578Isup4.cml
Supporting information file. DOI: 10.1107/S2056989016006162/pk2578IIsup5.cml
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
We thank the EPSRC National Crystallography Service (University of Southampton) for the data collections and the EPSRC National Mass Spectrometry Service (University of Swansea) for the HRMS data.
supplementary crystallographic information
(I) 6-Isopropyl-3-(2-nitro-1-phenylethyl)-1H-indole . Crystal data
| C19H20N2O2 | F(000) = 328 |
| Mr = 308.37 | Dx = 1.268 Mg m−3 |
| Monoclinic, P21 | Mo Kα radiation, λ = 0.71073 Å |
| a = 12.4525 (9) Å | Cell parameters from 6780 reflections |
| b = 5.7360 (4) Å | θ = 3.1–27.5° |
| c = 12.5896 (9) Å | µ = 0.08 mm−1 |
| β = 116.081 (6)° | T = 100 K |
| V = 807.68 (11) Å3 | Blade, light red |
| Z = 2 | 0.28 × 0.05 × 0.01 mm |
(I) 6-Isopropyl-3-(2-nitro-1-phenylethyl)-1H-indole . Data collection
| Rigaku Mercury CCD diffractometer | Rint = 0.107 |
| ω scans | θmax = 27.5°, θmin = 3.1° |
| 7830 measured reflections | h = −13→16 |
| 3498 independent reflections | k = −7→7 |
| 2259 reflections with I > 2σ(I) | l = −16→13 |
(I) 6-Isopropyl-3-(2-nitro-1-phenylethyl)-1H-indole . Refinement
| Refinement on F2 | 1 restraint |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.081 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.164 | w = 1/[σ2(Fo2) + (0.0357P)2 + 0.4188P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.12 | (Δ/σ)max < 0.001 |
| 3498 reflections | Δρmax = 0.28 e Å−3 |
| 213 parameters | Δρmin = −0.24 e Å−3 |
(I) 6-Isopropyl-3-(2-nitro-1-phenylethyl)-1H-indole . Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
(I) 6-Isopropyl-3-(2-nitro-1-phenylethyl)-1H-indole . Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.5003 (5) | 0.1427 (9) | 0.3881 (4) | 0.0226 (13) | |
| C2 | 0.3850 (5) | 0.0617 (12) | 0.3609 (4) | 0.0254 (13) | |
| H2 | 0.3737 | −0.0752 | 0.3972 | 0.031* | |
| C3 | 0.2883 (5) | 0.1850 (11) | 0.2804 (5) | 0.0264 (13) | |
| C4 | 0.3092 (5) | 0.3871 (10) | 0.2291 (4) | 0.0257 (14) | |
| H4 | 0.2422 | 0.4726 | 0.1747 | 0.031* | |
| C5 | 0.4222 (5) | 0.4675 (10) | 0.2538 (4) | 0.0241 (13) | |
| H5 | 0.4329 | 0.6061 | 0.2183 | 0.029* | |
| C6 | 0.5208 (5) | 0.3389 (10) | 0.3329 (4) | 0.0239 (12) | |
| C7 | 0.6491 (5) | 0.3646 (10) | 0.3796 (4) | 0.0245 (13) | |
| C8 | 0.6977 (5) | 0.1894 (10) | 0.4586 (5) | 0.0256 (13) | |
| H8 | 0.7811 | 0.1642 | 0.5038 | 0.031* | |
| C9 | 0.7126 (5) | 0.5512 (11) | 0.3434 (4) | 0.0232 (12) | |
| H9 | 0.6848 | 0.7063 | 0.3577 | 0.028* | |
| C10 | 0.8468 (5) | 0.5371 (11) | 0.4194 (4) | 0.0263 (13) | |
| H10A | 0.8643 | 0.5484 | 0.5040 | 0.032* | |
| H10B | 0.8771 | 0.3851 | 0.4067 | 0.032* | |
| C11 | 0.6842 (4) | 0.5377 (10) | 0.2123 (4) | 0.0222 (12) | |
| C12 | 0.7189 (5) | 0.3446 (10) | 0.1683 (4) | 0.0264 (13) | |
| H12 | 0.7558 | 0.2160 | 0.2187 | 0.032* | |
| C13 | 0.6998 (5) | 0.3383 (11) | 0.0510 (5) | 0.0293 (14) | |
| H13 | 0.7245 | 0.2066 | 0.0216 | 0.035* | |
| C14 | 0.6451 (5) | 0.5229 (11) | −0.0224 (5) | 0.0288 (14) | |
| H14 | 0.6323 | 0.5190 | −0.1025 | 0.035* | |
| C15 | 0.6086 (5) | 0.7141 (11) | 0.0200 (5) | 0.0280 (13) | |
| H15 | 0.5692 | 0.8400 | −0.0315 | 0.034* | |
| C16 | 0.6298 (5) | 0.7225 (11) | 0.1384 (5) | 0.0258 (13) | |
| H16 | 0.6065 | 0.8558 | 0.1680 | 0.031* | |
| C17 | 0.1618 (5) | 0.0955 (10) | 0.2462 (5) | 0.0294 (14) | |
| H17 | 0.1681 | −0.0383 | 0.2995 | 0.035* | |
| C18 | 0.1043 (5) | 0.0033 (12) | 0.1182 (5) | 0.0405 (17) | |
| H18A | 0.1532 | −0.1237 | 0.1106 | 0.061* | |
| H18B | 0.0994 | 0.1298 | 0.0639 | 0.061* | |
| H18C | 0.0238 | −0.0547 | 0.0986 | 0.061* | |
| C19 | 0.0831 (6) | 0.2786 (12) | 0.2629 (6) | 0.0420 (18) | |
| H19A | 0.0047 | 0.2109 | 0.2449 | 0.063* | |
| H19B | 0.0732 | 0.4100 | 0.2096 | 0.063* | |
| H19C | 0.1205 | 0.3334 | 0.3450 | 0.063* | |
| N1 | 0.6094 (4) | 0.0531 (9) | 0.4643 (4) | 0.0260 (11) | |
| N2 | 0.9084 (4) | 0.7309 (9) | 0.3883 (4) | 0.0267 (11) | |
| H1 | 0.625 (5) | −0.056 (11) | 0.514 (5) | 0.032* | |
| O1 | 0.9829 (4) | 0.6817 (8) | 0.3538 (4) | 0.0414 (11) | |
| O2 | 0.8825 (4) | 0.9308 (7) | 0.4015 (4) | 0.0376 (11) |
(I) 6-Isopropyl-3-(2-nitro-1-phenylethyl)-1H-indole . Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.035 (3) | 0.016 (3) | 0.020 (3) | 0.006 (2) | 0.015 (2) | 0.000 (2) |
| C2 | 0.035 (3) | 0.022 (3) | 0.026 (3) | −0.003 (3) | 0.020 (3) | −0.002 (3) |
| C3 | 0.033 (3) | 0.026 (4) | 0.023 (3) | −0.003 (3) | 0.015 (2) | −0.005 (3) |
| C4 | 0.033 (3) | 0.023 (4) | 0.022 (3) | 0.002 (3) | 0.013 (2) | −0.002 (2) |
| C5 | 0.037 (3) | 0.017 (3) | 0.022 (3) | 0.001 (3) | 0.017 (3) | 0.000 (2) |
| C6 | 0.033 (3) | 0.019 (3) | 0.023 (3) | 0.002 (3) | 0.016 (2) | 0.000 (3) |
| C7 | 0.034 (3) | 0.022 (3) | 0.020 (3) | 0.002 (3) | 0.015 (2) | −0.002 (2) |
| C8 | 0.025 (3) | 0.027 (3) | 0.028 (3) | 0.004 (3) | 0.015 (2) | −0.002 (3) |
| C9 | 0.031 (3) | 0.016 (3) | 0.025 (3) | 0.000 (3) | 0.015 (2) | 0.003 (3) |
| C10 | 0.032 (3) | 0.023 (3) | 0.028 (3) | 0.000 (3) | 0.016 (2) | 0.003 (3) |
| C11 | 0.027 (3) | 0.017 (3) | 0.023 (3) | −0.003 (3) | 0.013 (2) | −0.001 (3) |
| C12 | 0.035 (3) | 0.019 (3) | 0.024 (3) | −0.004 (3) | 0.012 (2) | 0.003 (3) |
| C13 | 0.040 (3) | 0.023 (3) | 0.029 (3) | −0.003 (3) | 0.019 (3) | −0.004 (3) |
| C14 | 0.042 (4) | 0.024 (3) | 0.025 (3) | −0.007 (3) | 0.019 (3) | −0.005 (3) |
| C15 | 0.033 (3) | 0.024 (3) | 0.027 (3) | −0.001 (3) | 0.013 (2) | 0.005 (3) |
| C16 | 0.032 (3) | 0.021 (3) | 0.028 (3) | 0.000 (3) | 0.016 (2) | −0.001 (3) |
| C17 | 0.034 (3) | 0.026 (4) | 0.029 (3) | −0.003 (3) | 0.015 (3) | 0.003 (3) |
| C18 | 0.039 (4) | 0.038 (5) | 0.044 (4) | −0.005 (3) | 0.018 (3) | −0.005 (3) |
| C19 | 0.036 (4) | 0.041 (5) | 0.057 (4) | −0.001 (3) | 0.027 (3) | −0.009 (3) |
| N1 | 0.033 (3) | 0.024 (3) | 0.026 (2) | 0.007 (2) | 0.017 (2) | 0.006 (2) |
| N2 | 0.028 (3) | 0.024 (3) | 0.029 (3) | 0.000 (2) | 0.013 (2) | −0.004 (2) |
| O1 | 0.048 (3) | 0.033 (3) | 0.057 (3) | −0.002 (2) | 0.036 (2) | −0.010 (2) |
| O2 | 0.042 (3) | 0.018 (2) | 0.057 (3) | 0.002 (2) | 0.025 (2) | −0.006 (2) |
(I) 6-Isopropyl-3-(2-nitro-1-phenylethyl)-1H-indole . Geometric parameters (Å, º)
| C1—N1 | 1.372 (7) | C11—C12 | 1.389 (8) |
| C1—C2 | 1.400 (7) | C12—C13 | 1.389 (7) |
| C1—C6 | 1.404 (7) | C12—H12 | 0.9500 |
| C2—C3 | 1.380 (8) | C13—C14 | 1.373 (8) |
| C2—H2 | 0.9500 | C13—H13 | 0.9500 |
| C3—C4 | 1.406 (8) | C14—C15 | 1.381 (8) |
| C3—C17 | 1.527 (8) | C14—H14 | 0.9500 |
| C4—C5 | 1.380 (7) | C15—C16 | 1.397 (7) |
| C4—H4 | 0.9500 | C15—H15 | 0.9500 |
| C5—C6 | 1.403 (7) | C16—H16 | 0.9500 |
| C5—H5 | 0.9500 | C17—C19 | 1.513 (8) |
| C6—C7 | 1.447 (7) | C17—C18 | 1.541 (8) |
| C7—C8 | 1.355 (8) | C17—H17 | 1.0000 |
| C7—C9 | 1.516 (8) | C18—H18A | 0.9800 |
| C8—N1 | 1.377 (7) | C18—H18B | 0.9800 |
| C8—H8 | 0.9500 | C18—H18C | 0.9800 |
| C9—C10 | 1.519 (7) | C19—H19A | 0.9800 |
| C9—C11 | 1.530 (7) | C19—H19B | 0.9800 |
| C9—H9 | 1.0000 | C19—H19C | 0.9800 |
| C10—N2 | 1.497 (7) | N1—H1 | 0.84 (6) |
| C10—H10A | 0.9900 | N2—O1 | 1.217 (6) |
| C10—H10B | 0.9900 | N2—O2 | 1.222 (6) |
| C11—C16 | 1.376 (8) | ||
| N1—C1—C2 | 129.8 (5) | C13—C12—C11 | 120.4 (5) |
| N1—C1—C6 | 107.9 (5) | C13—C12—H12 | 119.8 |
| C2—C1—C6 | 122.3 (5) | C11—C12—H12 | 119.8 |
| C3—C2—C1 | 118.7 (5) | C14—C13—C12 | 119.9 (6) |
| C3—C2—H2 | 120.7 | C14—C13—H13 | 120.1 |
| C1—C2—H2 | 120.7 | C12—C13—H13 | 120.1 |
| C2—C3—C4 | 118.8 (5) | C13—C14—C15 | 120.2 (5) |
| C2—C3—C17 | 119.6 (5) | C13—C14—H14 | 119.9 |
| C4—C3—C17 | 121.6 (5) | C15—C14—H14 | 119.9 |
| C5—C4—C3 | 123.2 (5) | C14—C15—C16 | 120.0 (5) |
| C5—C4—H4 | 118.4 | C14—C15—H15 | 120.0 |
| C3—C4—H4 | 118.4 | C16—C15—H15 | 120.0 |
| C4—C5—C6 | 118.2 (5) | C11—C16—C15 | 120.0 (5) |
| C4—C5—H5 | 120.9 | C11—C16—H16 | 120.0 |
| C6—C5—H5 | 120.9 | C15—C16—H16 | 120.0 |
| C5—C6—C1 | 118.7 (5) | C19—C17—C3 | 112.2 (5) |
| C5—C6—C7 | 134.5 (5) | C19—C17—C18 | 110.6 (5) |
| C1—C6—C7 | 106.8 (5) | C3—C17—C18 | 111.0 (4) |
| C8—C7—C6 | 106.3 (5) | C19—C17—H17 | 107.6 |
| C8—C7—C9 | 128.4 (5) | C3—C17—H17 | 107.6 |
| C6—C7—C9 | 125.4 (5) | C18—C17—H17 | 107.6 |
| C7—C8—N1 | 110.5 (5) | C17—C18—H18A | 109.5 |
| C7—C8—H8 | 124.8 | C17—C18—H18B | 109.5 |
| N1—C8—H8 | 124.8 | H18A—C18—H18B | 109.5 |
| C7—C9—C10 | 110.3 (5) | C17—C18—H18C | 109.5 |
| C7—C9—C11 | 112.7 (5) | H18A—C18—H18C | 109.5 |
| C10—C9—C11 | 110.3 (4) | H18B—C18—H18C | 109.5 |
| C7—C9—H9 | 107.8 | C17—C19—H19A | 109.5 |
| C10—C9—H9 | 107.8 | C17—C19—H19B | 109.5 |
| C11—C9—H9 | 107.8 | H19A—C19—H19B | 109.5 |
| N2—C10—C9 | 110.1 (4) | C17—C19—H19C | 109.5 |
| N2—C10—H10A | 109.6 | H19A—C19—H19C | 109.5 |
| C9—C10—H10A | 109.6 | H19B—C19—H19C | 109.5 |
| N2—C10—H10B | 109.6 | C1—N1—C8 | 108.6 (5) |
| C9—C10—H10B | 109.6 | C1—N1—H1 | 129 (4) |
| H10A—C10—H10B | 108.2 | C8—N1—H1 | 122 (4) |
| C16—C11—C12 | 119.5 (5) | O1—N2—O2 | 123.6 (5) |
| C16—C11—C9 | 120.1 (5) | O1—N2—C10 | 118.7 (5) |
| C12—C11—C9 | 120.3 (5) | O2—N2—C10 | 117.7 (5) |
| N1—C1—C2—C3 | 179.9 (5) | C7—C9—C10—N2 | −176.5 (4) |
| C6—C1—C2—C3 | −2.6 (8) | C11—C9—C10—N2 | 58.4 (6) |
| C1—C2—C3—C4 | −0.2 (7) | C7—C9—C11—C16 | 118.9 (6) |
| C1—C2—C3—C17 | 177.9 (5) | C10—C9—C11—C16 | −117.3 (6) |
| C2—C3—C4—C5 | 1.0 (8) | C7—C9—C11—C12 | −64.5 (6) |
| C17—C3—C4—C5 | −177.0 (5) | C10—C9—C11—C12 | 59.3 (7) |
| C3—C4—C5—C6 | 0.9 (7) | C16—C11—C12—C13 | 0.6 (8) |
| C4—C5—C6—C1 | −3.5 (7) | C9—C11—C12—C13 | −176.0 (5) |
| C4—C5—C6—C7 | 179.8 (5) | C11—C12—C13—C14 | −0.8 (8) |
| N1—C1—C6—C5 | −177.5 (5) | C12—C13—C14—C15 | −0.2 (8) |
| C2—C1—C6—C5 | 4.5 (8) | C13—C14—C15—C16 | 1.4 (8) |
| N1—C1—C6—C7 | 0.0 (5) | C12—C11—C16—C15 | 0.6 (8) |
| C2—C1—C6—C7 | −178.0 (5) | C9—C11—C16—C15 | 177.2 (5) |
| C5—C6—C7—C8 | 176.5 (6) | C14—C15—C16—C11 | −1.6 (8) |
| C1—C6—C7—C8 | −0.4 (6) | C2—C3—C17—C19 | 126.2 (6) |
| C5—C6—C7—C9 | −4.3 (9) | C4—C3—C17—C19 | −55.7 (7) |
| C1—C6—C7—C9 | 178.7 (5) | C2—C3—C17—C18 | −109.4 (6) |
| C6—C7—C8—N1 | 0.7 (6) | C4—C3—C17—C18 | 68.6 (7) |
| C9—C7—C8—N1 | −178.4 (5) | C2—C1—N1—C8 | 178.2 (5) |
| C8—C7—C9—C10 | −6.4 (8) | C6—C1—N1—C8 | 0.4 (5) |
| C6—C7—C9—C10 | 174.6 (5) | C7—C8—N1—C1 | −0.7 (6) |
| C8—C7—C9—C11 | 117.3 (6) | C9—C10—N2—O1 | −120.8 (5) |
| C6—C7—C9—C11 | −61.6 (7) | C9—C10—N2—O2 | 60.7 (6) |
(I) 6-Isopropyl-3-(2-nitro-1-phenylethyl)-1H-indole . Hydrogen-bond geometry (Å, º)
Cg1, Cg2 and Cg3 are the centroids of the N1/C1/C6–C8, C1–C6 and C11–C16 rings, respectively.
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···Cg2i | 0.84 (6) | 2.64 (6) | 3.386 (5) | 148 (6) |
| C2—H2···Cg1i | 0.95 | 2.63 | 3.468 (6) | 147 |
| C14—H14···Cg2ii | 0.95 | 2.79 | 3.638 (6) | 148 |
| C15—H15···Cg3ii | 0.95 | 2.87 | 3.551 (7) | 129 |
Symmetry codes: (i) −x+1, y−1/2, −z+1; (ii) −x+1, y+1/2, −z.
(II) 2-(4-Methoxyphenyl)-3-(2-nitro-1-phenylethyl)-1H-indole . Crystal data
| C23H20N2O3 | Z = 4 |
| Mr = 372.41 | F(000) = 784 |
| Triclinic, P1 | Dx = 1.349 Mg m−3 |
| a = 9.2014 (5) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 9.4543 (7) Å | Cell parameters from 22268 reflections |
| c = 21.6201 (14) Å | θ = 2.2–27.5° |
| α = 98.563 (4)° | µ = 0.09 mm−1 |
| β = 93.416 (4)° | T = 100 K |
| γ = 98.354 (4)° | Plate, colourless |
| V = 1833.7 (2) Å3 | 0.10 × 0.06 × 0.06 mm |
(II) 2-(4-Methoxyphenyl)-3-(2-nitro-1-phenylethyl)-1H-indole . Data collection
| Rigaku Mercury CCD diffractometer | Rint = 0.038 |
| ω scans | θmax = 28.4°, θmin = 2.2° |
| 24774 measured reflections | h = −11→11 |
| 8621 independent reflections | k = −12→12 |
| 6769 reflections with I > 2σ(I) | l = −28→28 |
(II) 2-(4-Methoxyphenyl)-3-(2-nitro-1-phenylethyl)-1H-indole . Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.046 | Hydrogen site location: mixed |
| wR(F2) = 0.124 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.03 | w = 1/[σ2(Fo2) + (0.0548P)2 + 0.8021P] where P = (Fo2 + 2Fc2)/3 |
| 8621 reflections | (Δ/σ)max < 0.001 |
| 513 parameters | Δρmax = 0.62 e Å−3 |
| 0 restraints | Δρmin = −0.31 e Å−3 |
(II) 2-(4-Methoxyphenyl)-3-(2-nitro-1-phenylethyl)-1H-indole . Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
(II) 2-(4-Methoxyphenyl)-3-(2-nitro-1-phenylethyl)-1H-indole . Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.49618 (16) | 0.34164 (15) | 0.06414 (6) | 0.0170 (3) | |
| C2 | 0.38705 (17) | 0.42054 (15) | 0.08413 (7) | 0.0189 (3) | |
| H2A | 0.3201 | 0.4495 | 0.0551 | 0.023* | |
| C3 | 0.38042 (17) | 0.45411 (16) | 0.14638 (7) | 0.0213 (3) | |
| H3A | 0.3087 | 0.5094 | 0.1624 | 0.026* | |
| C4 | 0.47956 (18) | 0.40736 (16) | 0.18735 (7) | 0.0222 (3) | |
| H4 | 0.4737 | 0.4329 | 0.2312 | 0.027* | |
| C5 | 0.58536 (17) | 0.32607 (16) | 0.16733 (7) | 0.0199 (3) | |
| H5 | 0.6497 | 0.2950 | 0.1968 | 0.024* | |
| C6 | 0.59606 (16) | 0.29082 (15) | 0.10432 (7) | 0.0171 (3) | |
| C7 | 0.68766 (16) | 0.21018 (15) | 0.06651 (7) | 0.0177 (3) | |
| C8 | 0.64249 (16) | 0.21756 (15) | 0.00713 (7) | 0.0173 (3) | |
| C9 | 0.81582 (17) | 0.13561 (16) | 0.08290 (7) | 0.0217 (3) | |
| H9 | 0.8720 | 0.1249 | 0.0447 | 0.026* | |
| C10 | 0.92103 (18) | 0.22377 (17) | 0.13441 (8) | 0.0245 (3) | |
| H10A | 1.0027 | 0.1698 | 0.1428 | 0.029* | |
| H10B | 0.8695 | 0.2389 | 0.1732 | 0.029* | |
| C11 | 0.77242 (16) | −0.01722 (16) | 0.09801 (7) | 0.0188 (3) | |
| C12 | 0.84562 (17) | −0.12686 (16) | 0.07252 (7) | 0.0217 (3) | |
| H12 | 0.9215 | −0.1060 | 0.0458 | 0.026* | |
| C13 | 0.81037 (18) | −0.26753 (17) | 0.08526 (8) | 0.0254 (3) | |
| H13 | 0.8620 | −0.3414 | 0.0672 | 0.030* | |
| C14 | 0.70126 (18) | −0.29942 (17) | 0.12382 (7) | 0.0244 (3) | |
| H14 | 0.6775 | −0.3949 | 0.1329 | 0.029* | |
| C15 | 0.62638 (18) | −0.19130 (17) | 0.14925 (7) | 0.0243 (3) | |
| H15 | 0.5502 | −0.2127 | 0.1758 | 0.029* | |
| C16 | 0.66181 (17) | −0.05090 (16) | 0.13627 (7) | 0.0214 (3) | |
| H16 | 0.6092 | 0.0225 | 0.1540 | 0.026* | |
| C17 | 0.70037 (16) | 0.15610 (15) | −0.04944 (7) | 0.0180 (3) | |
| C18 | 0.77075 (17) | 0.24493 (16) | −0.08755 (7) | 0.0212 (3) | |
| H18 | 0.7800 | 0.3471 | −0.0764 | 0.025* | |
| C19 | 0.82656 (17) | 0.18773 (16) | −0.14060 (7) | 0.0214 (3) | |
| H19 | 0.8735 | 0.2487 | −0.1671 | 0.026* | |
| C20 | 0.81369 (16) | 0.03855 (16) | −0.15535 (7) | 0.0190 (3) | |
| C21 | 0.74181 (17) | −0.05225 (16) | −0.11828 (7) | 0.0207 (3) | |
| H21 | 0.7316 | −0.1544 | −0.1297 | 0.025* | |
| C22 | 0.68649 (17) | 0.00643 (16) | −0.06567 (7) | 0.0204 (3) | |
| H22 | 0.6379 | −0.0547 | −0.0396 | 0.025* | |
| C23 | 0.9661 (2) | 0.06002 (19) | −0.23777 (7) | 0.0297 (4) | |
| H23A | 1.0146 | −0.0009 | −0.2682 | 0.045* | |
| H23B | 1.0409 | 0.1233 | −0.2079 | 0.045* | |
| H23C | 0.9083 | 0.1192 | −0.2599 | 0.045* | |
| N1 | 0.52777 (14) | 0.29681 (13) | 0.00553 (6) | 0.0173 (2) | |
| H1 | 0.471 (2) | 0.3091 (19) | −0.0271 (9) | 0.021* | |
| N2 | 0.98391 (15) | 0.36924 (15) | 0.11821 (7) | 0.0272 (3) | |
| O1 | 1.01892 (15) | 0.37644 (15) | 0.06681 (6) | 0.0391 (3) | |
| O2 | 1.00019 (16) | 0.47097 (14) | 0.15869 (7) | 0.0418 (3) | |
| O3 | 0.87109 (12) | −0.02940 (12) | −0.20477 (5) | 0.0229 (2) | |
| C24 | 1.07078 (17) | 0.27498 (15) | 0.60018 (7) | 0.0186 (3) | |
| C25 | 1.19481 (18) | 0.23268 (16) | 0.62786 (7) | 0.0222 (3) | |
| H25 | 1.2510 | 0.1700 | 0.6046 | 0.027* | |
| C26 | 1.23137 (18) | 0.28475 (17) | 0.68904 (7) | 0.0245 (3) | |
| H26 | 1.3142 | 0.2580 | 0.7102 | 0.029* | |
| C27 | 1.14771 (19) | 0.37763 (17) | 0.72123 (7) | 0.0252 (3) | |
| H27 | 1.1756 | 0.4136 | 0.7643 | 0.030* | |
| C28 | 1.02574 (18) | 0.42001 (17) | 0.69332 (7) | 0.0224 (3) | |
| H28 | 0.9715 | 0.4842 | 0.7169 | 0.027* | |
| C29 | 0.98380 (16) | 0.36824 (15) | 0.63114 (7) | 0.0182 (3) | |
| C30 | 0.86833 (17) | 0.38853 (16) | 0.58743 (7) | 0.0184 (3) | |
| C31 | 0.88891 (16) | 0.30774 (15) | 0.53304 (6) | 0.0178 (3) | |
| C32 | 0.73865 (18) | 0.46891 (17) | 0.59473 (7) | 0.0222 (3) | |
| H32 | 0.6627 | 0.4216 | 0.5600 | 0.027* | |
| C33 | 0.6680 (2) | 0.45620 (18) | 0.65431 (8) | 0.0279 (4) | |
| H33A | 0.5817 | 0.5077 | 0.6551 | 0.033* | |
| H33B | 0.7389 | 0.5043 | 0.6901 | 0.033* | |
| C34 | 0.76883 (17) | 0.62946 (16) | 0.58830 (7) | 0.0203 (3) | |
| C35 | 0.89706 (19) | 0.72001 (17) | 0.61460 (7) | 0.0241 (3) | |
| H35 | 0.9719 | 0.6818 | 0.6360 | 0.029* | |
| C36 | 0.9164 (2) | 0.86699 (18) | 0.60972 (7) | 0.0272 (4) | |
| H36 | 1.0045 | 0.9280 | 0.6278 | 0.033* | |
| C37 | 0.8086 (2) | 0.92481 (18) | 0.57886 (8) | 0.0296 (4) | |
| H37 | 0.8214 | 1.0254 | 0.5767 | 0.035* | |
| C38 | 0.6826 (2) | 0.83509 (19) | 0.55131 (8) | 0.0297 (4) | |
| H38 | 0.6091 | 0.8733 | 0.5291 | 0.036* | |
| C39 | 0.66297 (18) | 0.68832 (18) | 0.55610 (7) | 0.0252 (3) | |
| H39 | 0.5757 | 0.6273 | 0.5370 | 0.030* | |
| C40 | 0.80143 (17) | 0.28679 (16) | 0.47391 (7) | 0.0186 (3) | |
| C41 | 0.78144 (17) | 0.40508 (16) | 0.44448 (7) | 0.0204 (3) | |
| H41 | 0.8257 | 0.4998 | 0.4637 | 0.024* | |
| C42 | 0.69952 (17) | 0.38631 (17) | 0.38876 (7) | 0.0216 (3) | |
| H42 | 0.6880 | 0.4669 | 0.3684 | 0.026* | |
| C43 | 0.63290 (17) | 0.24818 (17) | 0.36192 (7) | 0.0213 (3) | |
| C44 | 0.65272 (18) | 0.12873 (17) | 0.39006 (7) | 0.0231 (3) | |
| H44 | 0.6082 | 0.0341 | 0.3709 | 0.028* | |
| C45 | 0.73698 (18) | 0.14931 (16) | 0.44560 (7) | 0.0219 (3) | |
| H45 | 0.7516 | 0.0683 | 0.4651 | 0.026* | |
| C46 | 0.4670 (2) | 0.1044 (2) | 0.28258 (8) | 0.0346 (4) | |
| H46A | 0.4059 | 0.1135 | 0.2451 | 0.052* | |
| H46B | 0.4035 | 0.0696 | 0.3138 | 0.052* | |
| H46C | 0.5345 | 0.0355 | 0.2712 | 0.052* | |
| N3 | 1.01008 (14) | 0.23898 (13) | 0.54058 (6) | 0.0186 (3) | |
| H3 | 1.050 (2) | 0.193 (2) | 0.5094 (9) | 0.022* | |
| N4 | 0.61859 (17) | 0.30122 (17) | 0.66247 (8) | 0.0351 (4) | |
| O4 | 0.55989 (17) | 0.21888 (16) | 0.61780 (8) | 0.0474 (4) | |
| O5 | 0.6370 (2) | 0.26938 (18) | 0.71306 (8) | 0.0575 (4) | |
| O6 | 0.54974 (13) | 0.24217 (13) | 0.30834 (5) | 0.0278 (3) |
(II) 2-(4-Methoxyphenyl)-3-(2-nitro-1-phenylethyl)-1H-indole . Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0182 (7) | 0.0134 (6) | 0.0185 (6) | −0.0002 (5) | −0.0014 (5) | 0.0033 (5) |
| C2 | 0.0184 (7) | 0.0157 (7) | 0.0230 (7) | 0.0027 (6) | −0.0002 (6) | 0.0054 (5) |
| C3 | 0.0219 (8) | 0.0159 (7) | 0.0263 (7) | 0.0025 (6) | 0.0044 (6) | 0.0034 (6) |
| C4 | 0.0270 (8) | 0.0195 (7) | 0.0189 (7) | 0.0001 (6) | 0.0012 (6) | 0.0029 (5) |
| C5 | 0.0226 (8) | 0.0170 (7) | 0.0197 (7) | 0.0002 (6) | −0.0027 (6) | 0.0056 (5) |
| C6 | 0.0180 (7) | 0.0117 (6) | 0.0209 (7) | −0.0004 (5) | −0.0022 (5) | 0.0045 (5) |
| C7 | 0.0164 (7) | 0.0136 (6) | 0.0231 (7) | 0.0004 (5) | −0.0009 (5) | 0.0053 (5) |
| C8 | 0.0157 (7) | 0.0131 (6) | 0.0229 (7) | 0.0013 (5) | 0.0004 (5) | 0.0041 (5) |
| C9 | 0.0194 (8) | 0.0192 (7) | 0.0276 (8) | 0.0037 (6) | −0.0007 (6) | 0.0078 (6) |
| C10 | 0.0229 (8) | 0.0216 (8) | 0.0295 (8) | 0.0031 (6) | −0.0026 (6) | 0.0082 (6) |
| C11 | 0.0180 (7) | 0.0173 (7) | 0.0208 (7) | 0.0020 (6) | −0.0053 (5) | 0.0059 (5) |
| C12 | 0.0199 (8) | 0.0226 (7) | 0.0228 (7) | 0.0048 (6) | −0.0024 (6) | 0.0043 (6) |
| C13 | 0.0255 (9) | 0.0196 (7) | 0.0296 (8) | 0.0062 (6) | −0.0080 (6) | 0.0006 (6) |
| C14 | 0.0272 (9) | 0.0171 (7) | 0.0270 (8) | −0.0014 (6) | −0.0116 (6) | 0.0074 (6) |
| C15 | 0.0236 (8) | 0.0250 (8) | 0.0233 (7) | −0.0018 (6) | −0.0039 (6) | 0.0084 (6) |
| C16 | 0.0205 (8) | 0.0202 (7) | 0.0234 (7) | 0.0048 (6) | −0.0029 (6) | 0.0036 (6) |
| C17 | 0.0159 (7) | 0.0179 (7) | 0.0200 (7) | 0.0030 (6) | −0.0021 (5) | 0.0037 (5) |
| C18 | 0.0227 (8) | 0.0162 (7) | 0.0260 (7) | 0.0057 (6) | 0.0007 (6) | 0.0053 (6) |
| C19 | 0.0217 (8) | 0.0198 (7) | 0.0244 (7) | 0.0038 (6) | 0.0022 (6) | 0.0083 (6) |
| C20 | 0.0156 (7) | 0.0234 (7) | 0.0179 (7) | 0.0043 (6) | −0.0022 (5) | 0.0028 (5) |
| C21 | 0.0205 (8) | 0.0157 (7) | 0.0244 (7) | 0.0007 (6) | −0.0011 (6) | 0.0012 (5) |
| C22 | 0.0198 (8) | 0.0185 (7) | 0.0223 (7) | 0.0000 (6) | 0.0003 (6) | 0.0045 (5) |
| C23 | 0.0347 (10) | 0.0337 (9) | 0.0195 (7) | −0.0004 (7) | 0.0050 (7) | 0.0048 (6) |
| N1 | 0.0175 (6) | 0.0176 (6) | 0.0174 (6) | 0.0048 (5) | −0.0016 (5) | 0.0041 (4) |
| N2 | 0.0180 (7) | 0.0245 (7) | 0.0394 (8) | 0.0016 (5) | −0.0014 (6) | 0.0094 (6) |
| O1 | 0.0357 (8) | 0.0405 (8) | 0.0374 (7) | −0.0061 (6) | 0.0004 (6) | 0.0074 (6) |
| O2 | 0.0470 (9) | 0.0270 (7) | 0.0482 (8) | 0.0013 (6) | 0.0062 (7) | −0.0006 (6) |
| O3 | 0.0264 (6) | 0.0230 (5) | 0.0185 (5) | 0.0029 (5) | 0.0020 (4) | 0.0021 (4) |
| C24 | 0.0212 (8) | 0.0158 (7) | 0.0178 (6) | −0.0010 (6) | −0.0010 (5) | 0.0048 (5) |
| C25 | 0.0234 (8) | 0.0185 (7) | 0.0247 (7) | 0.0028 (6) | −0.0012 (6) | 0.0055 (6) |
| C26 | 0.0231 (8) | 0.0250 (8) | 0.0252 (8) | 0.0001 (6) | −0.0065 (6) | 0.0105 (6) |
| C27 | 0.0299 (9) | 0.0271 (8) | 0.0168 (7) | −0.0023 (7) | −0.0030 (6) | 0.0062 (6) |
| C28 | 0.0258 (8) | 0.0230 (7) | 0.0173 (7) | 0.0004 (6) | 0.0010 (6) | 0.0038 (5) |
| C29 | 0.0195 (7) | 0.0179 (7) | 0.0172 (6) | 0.0004 (6) | 0.0002 (5) | 0.0056 (5) |
| C30 | 0.0189 (7) | 0.0183 (7) | 0.0179 (6) | 0.0007 (6) | 0.0005 (5) | 0.0048 (5) |
| C31 | 0.0191 (7) | 0.0163 (7) | 0.0180 (7) | 0.0001 (6) | −0.0011 (5) | 0.0057 (5) |
| C32 | 0.0220 (8) | 0.0213 (7) | 0.0237 (7) | 0.0026 (6) | 0.0020 (6) | 0.0055 (6) |
| C33 | 0.0297 (9) | 0.0270 (8) | 0.0284 (8) | 0.0050 (7) | 0.0059 (7) | 0.0073 (6) |
| C34 | 0.0227 (8) | 0.0210 (7) | 0.0184 (7) | 0.0051 (6) | 0.0055 (6) | 0.0042 (5) |
| C35 | 0.0278 (9) | 0.0262 (8) | 0.0183 (7) | 0.0036 (7) | 0.0014 (6) | 0.0040 (6) |
| C36 | 0.0349 (10) | 0.0245 (8) | 0.0197 (7) | −0.0024 (7) | 0.0072 (6) | 0.0002 (6) |
| C37 | 0.0421 (11) | 0.0208 (8) | 0.0288 (8) | 0.0062 (7) | 0.0147 (7) | 0.0077 (6) |
| C38 | 0.0317 (9) | 0.0304 (9) | 0.0329 (9) | 0.0118 (7) | 0.0099 (7) | 0.0149 (7) |
| C39 | 0.0235 (8) | 0.0276 (8) | 0.0262 (8) | 0.0049 (7) | 0.0043 (6) | 0.0079 (6) |
| C40 | 0.0181 (7) | 0.0210 (7) | 0.0164 (6) | 0.0013 (6) | −0.0002 (5) | 0.0049 (5) |
| C41 | 0.0214 (8) | 0.0192 (7) | 0.0195 (7) | −0.0007 (6) | −0.0001 (6) | 0.0044 (5) |
| C42 | 0.0225 (8) | 0.0228 (7) | 0.0204 (7) | 0.0017 (6) | −0.0011 (6) | 0.0092 (6) |
| C43 | 0.0199 (8) | 0.0271 (8) | 0.0162 (6) | 0.0008 (6) | −0.0024 (5) | 0.0056 (6) |
| C44 | 0.0261 (8) | 0.0209 (7) | 0.0204 (7) | 0.0001 (6) | −0.0026 (6) | 0.0027 (6) |
| C45 | 0.0258 (8) | 0.0200 (7) | 0.0197 (7) | 0.0024 (6) | −0.0023 (6) | 0.0059 (6) |
| C46 | 0.0381 (11) | 0.0361 (10) | 0.0236 (8) | −0.0102 (8) | −0.0110 (7) | 0.0062 (7) |
| N3 | 0.0206 (7) | 0.0196 (6) | 0.0152 (6) | 0.0030 (5) | −0.0014 (5) | 0.0028 (5) |
| N4 | 0.0287 (8) | 0.0320 (8) | 0.0469 (9) | 0.0046 (7) | 0.0123 (7) | 0.0104 (7) |
| O4 | 0.0411 (8) | 0.0394 (8) | 0.0608 (10) | −0.0023 (7) | 0.0144 (7) | 0.0094 (7) |
| O5 | 0.0674 (11) | 0.0575 (10) | 0.0579 (10) | 0.0112 (8) | 0.0167 (8) | 0.0370 (8) |
| O6 | 0.0310 (7) | 0.0303 (6) | 0.0194 (5) | −0.0034 (5) | −0.0103 (5) | 0.0078 (4) |
(II) 2-(4-Methoxyphenyl)-3-(2-nitro-1-phenylethyl)-1H-indole . Geometric parameters (Å, º)
| C1—N1 | 1.3377 (18) | C24—N3 | 1.3478 (18) |
| C1—C2 | 1.387 (2) | C24—C25 | 1.394 (2) |
| C1—C6 | 1.410 (2) | C24—C29 | 1.400 (2) |
| C2—C3 | 1.343 (2) | C25—C26 | 1.345 (2) |
| C2—H2A | 0.9500 | C25—H25 | 0.9500 |
| C3—C4 | 1.396 (2) | C26—C27 | 1.388 (2) |
| C3—H3A | 0.9500 | C26—H26 | 0.9500 |
| C4—C5 | 1.376 (2) | C27—C28 | 1.381 (2) |
| C4—H4 | 0.9500 | C27—H27 | 0.9500 |
| C5—C6 | 1.366 (2) | C28—C29 | 1.372 (2) |
| C5—H5 | 0.9500 | C28—H28 | 0.9500 |
| C6—C7 | 1.429 (2) | C29—C30 | 1.432 (2) |
| C7—C8 | 1.341 (2) | C30—C31 | 1.343 (2) |
| C7—C9 | 1.510 (2) | C30—C32 | 1.509 (2) |
| C8—N1 | 1.3820 (19) | C31—N3 | 1.3830 (19) |
| C8—C17 | 1.440 (2) | C31—C40 | 1.441 (2) |
| C9—C10 | 1.494 (2) | C32—C33 | 1.490 (2) |
| C9—C11 | 1.530 (2) | C32—C34 | 1.531 (2) |
| C9—H9 | 1.0000 | C32—H32 | 1.0000 |
| C10—N2 | 1.512 (2) | C33—N4 | 1.509 (2) |
| C10—H10A | 0.9900 | C33—H33A | 0.9900 |
| C10—H10B | 0.9900 | C33—H33B | 0.9900 |
| C11—C16 | 1.381 (2) | C34—C35 | 1.389 (2) |
| C11—C12 | 1.382 (2) | C34—C39 | 1.391 (2) |
| C12—C13 | 1.394 (2) | C35—C36 | 1.396 (2) |
| C12—H12 | 0.9500 | C35—H35 | 0.9500 |
| C13—C14 | 1.371 (2) | C36—C37 | 1.383 (3) |
| C13—H13 | 0.9500 | C36—H36 | 0.9500 |
| C14—C15 | 1.380 (2) | C37—C38 | 1.379 (3) |
| C14—H14 | 0.9500 | C37—H37 | 0.9500 |
| C15—C16 | 1.394 (2) | C38—C39 | 1.393 (2) |
| C15—H15 | 0.9500 | C38—H38 | 0.9500 |
| C16—H16 | 0.9500 | C39—H39 | 0.9500 |
| C17—C18 | 1.384 (2) | C40—C45 | 1.383 (2) |
| C17—C22 | 1.391 (2) | C40—C41 | 1.395 (2) |
| C18—C19 | 1.356 (2) | C41—C42 | 1.357 (2) |
| C18—H18 | 0.9500 | C41—H41 | 0.9500 |
| C19—C20 | 1.385 (2) | C42—C43 | 1.385 (2) |
| C19—H19 | 0.9500 | C42—H42 | 0.9500 |
| C20—O3 | 1.3398 (17) | C43—O6 | 1.3389 (17) |
| C20—C21 | 1.386 (2) | C43—C44 | 1.389 (2) |
| C21—C22 | 1.351 (2) | C44—C45 | 1.363 (2) |
| C21—H21 | 0.9500 | C44—H44 | 0.9500 |
| C22—H22 | 0.9500 | C45—H45 | 0.9500 |
| C23—O3 | 1.4240 (19) | C46—O6 | 1.425 (2) |
| C23—H23A | 0.9800 | C46—H46A | 0.9800 |
| C23—H23B | 0.9800 | C46—H46B | 0.9800 |
| C23—H23C | 0.9800 | C46—H46C | 0.9800 |
| N1—H1 | 0.886 (19) | N3—H3 | 0.875 (18) |
| N2—O1 | 1.1841 (19) | N4—O5 | 1.186 (2) |
| N2—O2 | 1.1856 (19) | N4—O4 | 1.193 (2) |
| N1—C1—C2 | 128.86 (14) | N3—C24—C25 | 129.20 (14) |
| N1—C1—C6 | 106.46 (13) | N3—C24—C29 | 105.99 (13) |
| C2—C1—C6 | 124.68 (13) | C25—C24—C29 | 124.81 (14) |
| C3—C2—C1 | 116.96 (14) | C26—C25—C24 | 116.94 (15) |
| C3—C2—H2A | 121.5 | C26—C25—H25 | 121.5 |
| C1—C2—H2A | 121.5 | C24—C25—H25 | 121.5 |
| C2—C3—C4 | 119.63 (14) | C25—C26—C27 | 119.82 (15) |
| C2—C3—H3A | 120.2 | C25—C26—H26 | 120.1 |
| C4—C3—H3A | 120.2 | C27—C26—H26 | 120.1 |
| C5—C4—C3 | 123.24 (14) | C28—C27—C26 | 122.88 (14) |
| C5—C4—H4 | 118.4 | C28—C27—H27 | 118.6 |
| C3—C4—H4 | 118.4 | C26—C27—H27 | 118.6 |
| C6—C5—C4 | 118.74 (14) | C29—C28—C27 | 119.17 (15) |
| C6—C5—H5 | 120.6 | C29—C28—H28 | 120.4 |
| C4—C5—H5 | 120.6 | C27—C28—H28 | 120.4 |
| C5—C6—C1 | 116.72 (14) | C28—C29—C24 | 116.38 (14) |
| C5—C6—C7 | 134.97 (14) | C28—C29—C30 | 134.77 (14) |
| C1—C6—C7 | 108.31 (12) | C24—C29—C30 | 108.84 (12) |
| C8—C7—C6 | 105.06 (13) | C31—C30—C29 | 105.15 (13) |
| C8—C7—C9 | 122.53 (13) | C31—C30—C32 | 122.19 (14) |
| C6—C7—C9 | 132.32 (13) | C29—C30—C32 | 132.51 (13) |
| C7—C8—N1 | 110.65 (13) | C30—C31—N3 | 110.11 (13) |
| C7—C8—C17 | 127.68 (14) | C30—C31—C40 | 128.18 (14) |
| N1—C8—C17 | 121.66 (13) | N3—C31—C40 | 121.70 (13) |
| C10—C9—C7 | 112.94 (13) | C33—C32—C30 | 113.41 (13) |
| C10—C9—C11 | 109.64 (12) | C33—C32—C34 | 108.23 (12) |
| C7—C9—C11 | 114.72 (12) | C30—C32—C34 | 115.66 (13) |
| C10—C9—H9 | 106.3 | C33—C32—H32 | 106.3 |
| C7—C9—H9 | 106.3 | C30—C32—H32 | 106.3 |
| C11—C9—H9 | 106.3 | C34—C32—H32 | 106.3 |
| C9—C10—N2 | 112.11 (13) | C32—C33—N4 | 113.00 (14) |
| C9—C10—H10A | 109.2 | C32—C33—H33A | 109.0 |
| N2—C10—H10A | 109.2 | N4—C33—H33A | 109.0 |
| C9—C10—H10B | 109.2 | C32—C33—H33B | 109.0 |
| N2—C10—H10B | 109.2 | N4—C33—H33B | 109.0 |
| H10A—C10—H10B | 107.9 | H33A—C33—H33B | 107.8 |
| C16—C11—C12 | 118.10 (14) | C35—C34—C39 | 118.41 (14) |
| C16—C11—C9 | 122.57 (13) | C35—C34—C32 | 122.49 (14) |
| C12—C11—C9 | 119.33 (14) | C39—C34—C32 | 119.08 (14) |
| C11—C12—C13 | 121.32 (15) | C34—C35—C36 | 120.23 (16) |
| C11—C12—H12 | 119.3 | C34—C35—H35 | 119.9 |
| C13—C12—H12 | 119.3 | C36—C35—H35 | 119.9 |
| C14—C13—C12 | 120.07 (15) | C37—C36—C35 | 120.76 (16) |
| C14—C13—H13 | 120.0 | C37—C36—H36 | 119.6 |
| C12—C13—H13 | 120.0 | C35—C36—H36 | 119.6 |
| C13—C14—C15 | 119.29 (14) | C38—C37—C36 | 119.40 (15) |
| C13—C14—H14 | 120.4 | C38—C37—H37 | 120.3 |
| C15—C14—H14 | 120.4 | C36—C37—H37 | 120.3 |
| C14—C15—C16 | 120.43 (15) | C37—C38—C39 | 119.95 (16) |
| C14—C15—H15 | 119.8 | C37—C38—H38 | 120.0 |
| C16—C15—H15 | 119.8 | C39—C38—H38 | 120.0 |
| C11—C16—C15 | 120.79 (14) | C34—C39—C38 | 121.22 (16) |
| C11—C16—H16 | 119.6 | C34—C39—H39 | 119.4 |
| C15—C16—H16 | 119.6 | C38—C39—H39 | 119.4 |
| C18—C17—C22 | 119.76 (13) | C45—C40—C41 | 119.30 (14) |
| C18—C17—C8 | 120.44 (13) | C45—C40—C31 | 120.40 (13) |
| C22—C17—C8 | 119.80 (13) | C41—C40—C31 | 120.31 (13) |
| C19—C18—C17 | 120.71 (14) | C42—C41—C40 | 120.70 (14) |
| C19—C18—H18 | 119.6 | C42—C41—H41 | 119.7 |
| C17—C18—H18 | 119.6 | C40—C41—H41 | 119.7 |
| C18—C19—C20 | 118.60 (14) | C41—C42—C43 | 119.13 (14) |
| C18—C19—H19 | 120.7 | C41—C42—H42 | 120.4 |
| C20—C19—H19 | 120.7 | C43—C42—H42 | 120.4 |
| O3—C20—C19 | 123.68 (14) | O6—C43—C42 | 114.42 (13) |
| O3—C20—C21 | 114.75 (13) | O6—C43—C44 | 124.49 (14) |
| C19—C20—C21 | 121.55 (13) | C42—C43—C44 | 121.09 (14) |
| C22—C21—C20 | 119.10 (14) | C45—C44—C43 | 118.99 (14) |
| C22—C21—H21 | 120.4 | C45—C44—H44 | 120.5 |
| C20—C21—H21 | 120.4 | C43—C44—H44 | 120.5 |
| C21—C22—C17 | 120.25 (14) | C44—C45—C40 | 120.77 (14) |
| C21—C22—H22 | 119.9 | C44—C45—H45 | 119.6 |
| C17—C22—H22 | 119.9 | C40—C45—H45 | 119.6 |
| O3—C23—H23A | 109.5 | O6—C46—H46A | 109.5 |
| O3—C23—H23B | 109.5 | O6—C46—H46B | 109.5 |
| H23A—C23—H23B | 109.5 | H46A—C46—H46B | 109.5 |
| O3—C23—H23C | 109.5 | O6—C46—H46C | 109.5 |
| H23A—C23—H23C | 109.5 | H46A—C46—H46C | 109.5 |
| H23B—C23—H23C | 109.5 | H46B—C46—H46C | 109.5 |
| C1—N1—C8 | 109.50 (12) | C24—N3—C31 | 109.90 (12) |
| C1—N1—H1 | 120.6 (11) | C24—N3—H3 | 125.5 (12) |
| C8—N1—H1 | 129.3 (11) | C31—N3—H3 | 123.7 (12) |
| O1—N2—O2 | 122.88 (15) | O5—N4—O4 | 123.94 (18) |
| O1—N2—C10 | 119.27 (14) | O5—N4—C33 | 118.26 (17) |
| O2—N2—C10 | 117.80 (15) | O4—N4—C33 | 117.78 (16) |
| C20—O3—C23 | 116.20 (12) | C43—O6—C46 | 116.24 (13) |
| N1—C1—C2—C3 | −178.82 (14) | N3—C24—C25—C26 | −179.75 (15) |
| C6—C1—C2—C3 | 2.2 (2) | C29—C24—C25—C26 | 0.8 (2) |
| C1—C2—C3—C4 | −1.0 (2) | C24—C25—C26—C27 | −0.8 (2) |
| C2—C3—C4—C5 | −0.5 (2) | C25—C26—C27—C28 | 0.3 (2) |
| C3—C4—C5—C6 | 1.0 (2) | C26—C27—C28—C29 | 0.3 (2) |
| C4—C5—C6—C1 | 0.1 (2) | C27—C28—C29—C24 | −0.4 (2) |
| C4—C5—C6—C7 | −179.51 (15) | C27—C28—C29—C30 | −179.21 (16) |
| N1—C1—C6—C5 | 179.10 (13) | N3—C24—C29—C28 | −179.75 (13) |
| C2—C1—C6—C5 | −1.8 (2) | C25—C24—C29—C28 | −0.2 (2) |
| N1—C1—C6—C7 | −1.20 (16) | N3—C24—C29—C30 | −0.62 (16) |
| C2—C1—C6—C7 | 177.94 (14) | C25—C24—C29—C30 | 178.93 (14) |
| C5—C6—C7—C8 | −179.37 (16) | C28—C29—C30—C31 | 179.30 (17) |
| C1—C6—C7—C8 | 1.01 (16) | C24—C29—C30—C31 | 0.40 (16) |
| C5—C6—C7—C9 | −2.8 (3) | C28—C29—C30—C32 | −5.2 (3) |
| C1—C6—C7—C9 | 177.56 (14) | C24—C29—C30—C32 | 175.88 (15) |
| C6—C7—C8—N1 | −0.45 (16) | C29—C30—C31—N3 | −0.02 (16) |
| C9—C7—C8—N1 | −177.42 (12) | C32—C30—C31—N3 | −176.09 (13) |
| C6—C7—C8—C17 | 178.90 (14) | C29—C30—C31—C40 | 178.55 (14) |
| C9—C7—C8—C17 | 1.9 (2) | C32—C30—C31—C40 | 2.5 (2) |
| C8—C7—C9—C10 | 133.17 (15) | C31—C30—C32—C33 | 136.46 (16) |
| C6—C7—C9—C10 | −42.9 (2) | C29—C30—C32—C33 | −38.4 (2) |
| C8—C7—C9—C11 | −100.19 (17) | C31—C30—C32—C34 | −97.65 (17) |
| C6—C7—C9—C11 | 83.76 (19) | C29—C30—C32—C34 | 87.50 (19) |
| C7—C9—C10—N2 | −58.42 (17) | C30—C32—C33—N4 | −56.24 (19) |
| C11—C9—C10—N2 | 172.28 (13) | C34—C32—C33—N4 | 174.02 (14) |
| C10—C9—C11—C16 | 83.64 (18) | C33—C32—C34—C35 | 86.90 (18) |
| C7—C9—C11—C16 | −44.7 (2) | C30—C32—C34—C35 | −41.6 (2) |
| C10—C9—C11—C12 | −96.58 (17) | C33—C32—C34—C39 | −91.71 (17) |
| C7—C9—C11—C12 | 135.11 (15) | C30—C32—C34—C39 | 139.81 (14) |
| C16—C11—C12—C13 | −0.6 (2) | C39—C34—C35—C36 | 1.4 (2) |
| C9—C11—C12—C13 | 179.65 (14) | C32—C34—C35—C36 | −177.20 (13) |
| C11—C12—C13—C14 | −0.1 (2) | C34—C35—C36—C37 | 0.1 (2) |
| C12—C13—C14—C15 | 0.6 (2) | C35—C36—C37—C38 | −1.7 (2) |
| C13—C14—C15—C16 | −0.4 (2) | C36—C37—C38—C39 | 1.6 (2) |
| C12—C11—C16—C15 | 0.7 (2) | C35—C34—C39—C38 | −1.4 (2) |
| C9—C11—C16—C15 | −179.52 (14) | C32—C34—C39—C38 | 177.22 (14) |
| C14—C15—C16—C11 | −0.2 (2) | C37—C38—C39—C34 | −0.1 (2) |
| C7—C8—C17—C18 | −114.00 (18) | C30—C31—C40—C45 | −121.49 (18) |
| N1—C8—C17—C18 | 65.3 (2) | N3—C31—C40—C45 | 56.9 (2) |
| C7—C8—C17—C22 | 65.5 (2) | C30—C31—C40—C41 | 58.8 (2) |
| N1—C8—C17—C22 | −115.22 (16) | N3—C31—C40—C41 | −122.77 (16) |
| C22—C17—C18—C19 | −0.1 (2) | C45—C40—C41—C42 | 0.2 (2) |
| C8—C17—C18—C19 | 179.44 (14) | C31—C40—C41—C42 | 179.86 (14) |
| C17—C18—C19—C20 | −1.0 (2) | C40—C41—C42—C43 | 1.4 (2) |
| C18—C19—C20—O3 | −176.55 (14) | C41—C42—C43—O6 | 177.38 (14) |
| C18—C19—C20—C21 | 1.9 (2) | C41—C42—C43—C44 | −2.2 (2) |
| O3—C20—C21—C22 | 176.83 (14) | O6—C43—C44—C45 | −178.21 (15) |
| C19—C20—C21—C22 | −1.8 (2) | C42—C43—C44—C45 | 1.3 (2) |
| C20—C21—C22—C17 | 0.7 (2) | C43—C44—C45—C40 | 0.3 (2) |
| C18—C17—C22—C21 | 0.2 (2) | C41—C40—C45—C44 | −1.0 (2) |
| C8—C17—C22—C21 | −179.29 (14) | C31—C40—C45—C44 | 179.25 (15) |
| C2—C1—N1—C8 | −178.16 (14) | C25—C24—N3—C31 | −178.91 (14) |
| C6—C1—N1—C8 | 0.93 (16) | C29—C24—N3—C31 | 0.60 (16) |
| C7—C8—N1—C1 | −0.30 (17) | C30—C31—N3—C24 | −0.37 (17) |
| C17—C8—N1—C1 | −179.70 (13) | C40—C31—N3—C24 | −179.06 (13) |
| C9—C10—N2—O1 | −41.3 (2) | C32—C33—N4—O5 | 139.49 (17) |
| C9—C10—N2—O2 | 141.02 (15) | C32—C33—N4—O4 | −42.3 (2) |
| C19—C20—O3—C23 | 8.6 (2) | C42—C43—O6—C46 | −173.68 (15) |
| C21—C20—O3—C23 | −169.94 (13) | C44—C43—O6—C46 | 5.8 (2) |
(II) 2-(4-Methoxyphenyl)-3-(2-nitro-1-phenylethyl)-1H-indole . Hydrogen-bond geometry (Å, º)
Cg1, Cg2, Cg3, Cg6, Cg7 and Cg8 are the centroids of the N1/C1/C6–C8, C1–C6, C11–C16, N3/C24/C29–C31, C24–C29 and C34–C39 rings, respectively.
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···Cg3i | 0.886 (19) | 2.640 (19) | 3.3631 (15) | 139.6 (15) |
| N3—H3···Cg8ii | 0.875 (18) | 2.582 (19) | 3.3364 (15) | 144.9 (16) |
| C14—H14···Cg2iii | 0.95 | 2.58 | 3.4228 (18) | 149 |
| C21—H21···Cg2i | 0.95 | 2.69 | 3.4133 (17) | 134 |
| C22—H22···Cg1i | 0.95 | 2.68 | 3.4543 (17) | 138 |
| C23—H23B···Cg3iv | 0.98 | 2.79 | 3.6739 (18) | 150 |
| C37—H37···Cg6v | 0.95 | 2.86 | 3.7660 (18) | 160 |
| C41—H41···Cg6ii | 0.95 | 2.70 | 3.3793 (17) | 129 |
| C42—H42···Cg7ii | 0.95 | 2.67 | 3.3627 (17) | 130 |
Symmetry codes: (i) −x+1, −y, −z; (ii) −x+2, −y+1, −z+1; (iii) x, y−1, z; (iv) −x+2, −y, −z; (v) x, y+1, z.
References
- Burley, S. K. & Petsko, G. A. (1986). FEBS Lett. 203, 139–143. [DOI] [PubMed]
- Cordes, D. B., Hua, G., Slawin, A. M. Z. & Woollins, J. D. (2011). Acta Cryst. E67, o1606. [DOI] [PMC free article] [PubMed]
- Desiraju, G. R. & Steiner, T. (1999). In The Weak Hydrogen Bond in Structural Chemistry and Biology. Oxford University Press.
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst B72, 171–179. [DOI] [PMC free article] [PubMed]
- Kerr, J. R. (2013). PhD thesis, University of Aberdeen, Scotland.
- Kerr, J. R., Trembleau, L., Storey, J. M. D., Wardell, J. L. & Harrison, W. T. A. (2015). Acta Cryst. E71, 654–659. [DOI] [PMC free article] [PubMed]
- Kerr, J. R., Trembleau, L., Storey, J. M. D., Wardell, J. L. & Harrison, W. T. A. (2016). Acta Cryst. E72, 363–369. [DOI] [PMC free article] [PubMed]
- Krishna, R., Velmurugan, D., Babu, G. & Perumal, P. T. (1999). Acta Cryst. C55, 75–78.
- Lavanya, P., Ramaiah, S. & Anbarasu, A. (2014). Comput. Biol. Med. 46, 22–28. [DOI] [PubMed]
- Levitt, M. & Perutz, M. F. (1998). J. Mol. Biol. 201, 751–754. [DOI] [PubMed]
- Muñoz, M. A., Ferrero, R., Carmona, C. & Balón, M. (2004). Spectrochim. Acta Part A, 60, 193–200. [DOI] [PubMed]
- Rigaku (2012). CrystalClear. Rigaku Corporation, Tokyo, Japan.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
- Zhao, H., Jiang, L., Dong, H., Li, H., Hu, W. & Ong, B. (2009). ChemPhysChem, 10, 2345–2348. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, II, global. DOI: 10.1107/S2056989016006162/pk2578sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989016006162/pk2578Isup2.hkl
Structure factors: contains datablock(s) II. DOI: 10.1107/S2056989016006162/pk2578IIsup3.hkl
Supporting information file. DOI: 10.1107/S2056989016006162/pk2578Isup4.cml
Supporting information file. DOI: 10.1107/S2056989016006162/pk2578IIsup5.cml
Additional supporting information: crystallographic information; 3D view; checkCIF report








