The crystal structure of a second polymorph of sodium dihydrogen citrate has been solved and refined using laboratory X-ray powder diffraction data, and optimized using density functional techniques. The powder pattern of a commercial sample did not match that corresponding to the known crystal structure (NAHCIT).
Keywords: crystal structure, powder diffraction, density functional theory, citrate, sodium, polymorphs
Abstract
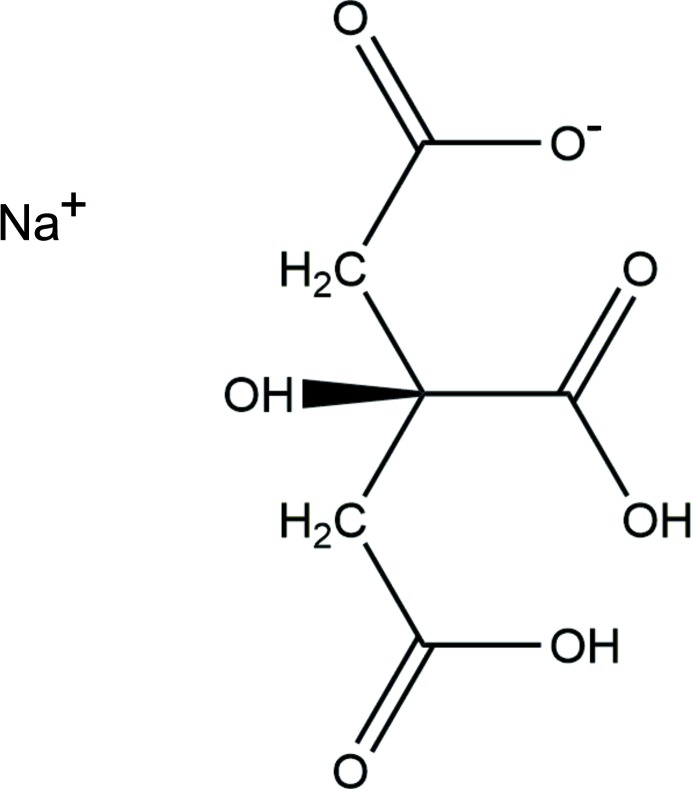
The crystal structure of a second polymorph of sodium dihydrogen citrate, Na+·H2C6H5O7 −, has been solved and refined using laboratory X-ray powder diffraction data, and optimized using density functional techniques. The powder pattern of the commercial sample used in this study did not match that corresponding to the known crystal structure [Glusker et al. (1965). Acta Cryst. 19, 561–572; refcode NAHCIT]. In this polymorph, the [NaO7] coordination polyhedra form edge-sharing chains propagating along the a axis, while in NAHCIT the octahedral [NaO6] groups form edge-sharing pairs bridged by two hydroxy groups. The most notable difference is that in this polymorph one of the terminal carboxyl groups is deprotonated, while in NAHCIT the central carboxylate group is deprotonated, as is more typical.
Chemical context
In the course of a systematic study of the crystal structures of Group 1 (alkali metal) citrate salts to better understand the anion’s conformational flexibility, deprotonation mode, coordination tendencies, and hydrogen bonding, we have determined several new crystal structures. Most of the new structures were solved using powder diffraction data (laboratory and/or synchrotron), but single crystals were used where available. The general trends and conclusions about the 16 new compounds and 12 previously characterized structures are being reported separately (Rammohan & Kaduk, 2016a
▸). Three of the new structures – NaKHC6H5O7, NaK2C6H5O7, and Na3C6H5O7 – have been published recently (Rammohan & Kaduk, 2016b
▸,c
▸,d
▸).
Structural commentary
The asymmetric unit of the title compound is shown in Fig. 1 ▸. The root-mean-square deviation of the non-hydrogen atoms in the Rietveld-refined and DFT-optimized structures is 0.148 Å. The maximum deviation is 0.318 Å, at the sodium ion. The good agreement between the two structures (Fig. 2 ▸) is strong evidence that the experimental structure is correct (van de Streek & Neumann, 2014 ▸). This discussion uses the DFT-optimized structure. All of the bond lengths, bond angles, and most torsion angles fall within the normal ranges indicated by a Mercury Mogul geometry check (Macrae et al., 2008 ▸). Only the C2—C3—C4—C5 torsion angle is flagged as unusual. It lies in the tail of a minority gauche population of similar torsion angles. The citrate anion occurs in the gauche,trans-conformation, which is one of the two low-energy conformations of an isolated citrate ion. The central carboxylate group and the hydroxy group occur in the normal planar arrangement. The citrate chelates to one Na19 ion through the central carboxyl O9 atom and the hydroxy group O13, and to a second Na19 ion through the terminal carboxyl atom O12 and the hydroxy group O13. The Na+ ion is seven-coordinate (pentagonal–bipyramidal), and has a bond-valence sum of 1.12.
Figure 1.
The asymmetric unit, showing the atom numbering. The atoms are represented by 50% probability spheroids.
Figure 2.
Comparison of the refined and optimized structures of sodium dihydrogen citrate. The refined structure is in red, and the DFT-optimized structure is in blue.
Supramolecular features
In this polymorph, the [NaO7] coordination polyhedra (Fig. 3 ▸) form edge-sharing chains propagating along the a axis, while in NAHCIT (Glusker et al., 1965 ▸), the octahedral [NaO6] units form edge-sharing pairs bridged by two hydroxy groups.
Figure 3.
Crystal structure of NaH2C6H5O7, viewed down the a axis.
The conformations of the citrate ions in the two structures are similar. The root-mean-square displacement of the non-hydrogen atoms is 0.11 Å. The conformations of the hydroxy groups differ, reflecting differences in coordination and hydrogen bonding. The most notable difference is that in this polymorph, one of the terminal carboxyl groups is deprotonated, while in NAHCIT the central carboxylate group is deprotonated, as is more typical.
In this form, the hydrogen bonds occur in layers in the ab plane, while in NAHCIT the hydrogen bonds form double-ladder chains along the c axis. The hydrogen bonds in this form contribute about 4.3 kcal mol−1 more to the lattice energy than those in NAHCIT, and seem to include a C—H⋯O hydrogen bond (Table 1 ▸). Comparison of the DFT energies of the two polymorphs shows that this polymorph is 3.24 kcal mol−1 higher in energy than NAHCIT. Presumably it was crystallized at a higher temperature than NAHCIT, which was crystallized at 343 K.
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O7—H20⋯O11 | 1.01 | 1.61 | 2.627 | 176 |
| O10—H21⋯O12 | 1.04 | 1.46 | 2.498 | 175 |
| O13—H16⋯O8 | 0.97 | 2.50 | 3.033 | 114 |
| C2—H15⋯O8 | 1.09 | 2.50 | 3.166 | 119 |
Database survey
Details of the comprehensive literature search for citrate structures are presented in Rammohan & Kaduk (2016a ▸). The crystal structure of sodium dihydrogen citrate is reported in Glusker et al. (1965 ▸), and the powder pattern calculated from this structure is PDF entry 02-063-5032. The observed powder pattern matched PDF entry 00-016-1182 (de Wolff et al., 1966 ▸) A reduced cell search of the cell of the observed polymorph in the Cambridge Structural Database (Groom et al., 2016 ▸) (increasing the default tolerance from 1.5 to 2.0%) yielded 223 hits, but limiting the chemistry to C, H, Na, and O only resulted in no hits. The powder pattern is now contained in the the Powder Diffraction File (ICDD, 2015 ▸) as entry 00-063-1340.
Synthesis and crystallization
The sample was purchased from Sigma–Aldrich (lot #BCBC0142). Before measuring the powder pattern, a portion of the sample was ground in a Spex 8000 mixer/mill and blended with a NIST SRM 640b silicon internal standard.
Refinement details
The powder pattern was indexed using DICVOL06 (Louër & Boultif, 2007 ▸). The background and Kα2 peaks were removed using Jade (MDI, 2012 ▸), and Powder4 (Dragoe, 2001 ▸) was used to convert the data into an XYE file. The 10–52.22° portion of the pattern was processed in DASH 3.2 (David et al., 2006 ▸), which suggested P212121 as the most probable space group. Citrate and Na fragments were used to solve the structure in this space group using DASH.
The powder pattern (Fig. 4 ▸) was indexed using Jade 9.5 (MDI, 2012 ▸). Pseudo-Voigt profile coefficients were as parameterized in Thompson et al. (1987 ▸) with profile coefficients for Simpson’s rule integration of the Pseudo-Voigt function according to Howard (1982 ▸). The asymmetry correction of Finger et al. (1994 ▸) was applied and microstrain broadening by Stephens (1999 ▸).
Figure 4.
Rietveld plot for the refinement of NaH2C6H5O7. The vertical scale is not the raw counts but the counts multiplied by the least-squares weights. This plot emphasizes the fit of the weaker peaks. The red crosses represent the observed data points, and the green line is the calculated pattern. The magenta curve is the difference pattern, plotted at the same scale as the other patterns. The lower row of black tick marks indicates the reflection positions for the major phase and the upper row of red tick marks is for the silicon internal standard.
The structure was refined by the Rietveld method using GSAS/EXPGUI (Larson & Von Dreele, 2004 ▸: Toby, 2001 ▸). All C—C and C—O bond lengths were restrained, as were all bond angles. The hydrogen atoms were included at fixed positions, which were recalculated during the course of the refinement using Materials Studio (Dassault Systemes, 2014 ▸). The U iso values of the atoms in the central and outer portions of the citrate were constrained to be equal, and the U iso values of the hydrogen atoms were constrained to be 1.3× those of the atoms to which they are attached.
The Bravais–Friedel–Donnay–Harker (Bravais, 1866 ▸; Friedel, 1907 ▸; Donnay & Harker, 1937 ▸) morphology suggests that we might expect a blocky morphology for this phase. A 4th-order spherical harmonic texture model was included in the refinement. The texture index was 1.374, indicating that preferred orientation was significant for this rotated-flat-plate specimen.
DFT calculations
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. After the Rietveld refinement, a density functional geometry optimization (fixed experimental unit cell) was carried out using CRYSTAL09 (Dovesi et al., 2005 ▸). The basis sets for the C, H, and O atoms were those of Gatti et al. (1994 ▸), and the basis set for Na was that of Dovesi et al. (1991 ▸). The calculation used 8 k-points and the B3LYP functional, and took about 60 h on a 2.4 GHz PC. The U iso from the Rietveld were assigned to the optimized fractional coordinates.
Table 2. Experimental details.
| Phase 1 | Phase 2 | |
|---|---|---|
| Crystal data | ||
| Chemical formula | Na+·C6H7O7 − | Si |
| M r | 214.10 | 28.09 |
| Crystal system, space group | Orthorhombic, P212121 | Cubic, F
d
 m
m
|
| Temperature (K) | 300 | 300 |
| a, b, c (Å) | 7.4527 (3), 7.7032 (3), 13.4551 (4) | 5.43105, 5.43105, 5.43105 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| V (Å3) | 772.45 (5) | 160.20 |
| Z | 4 | 8 |
| Radiation type | Kα1, Kα2, λ = 1.540629, 1.544451 Å | Kα1, Kα2, λ = 1.540629, 1.544451 Å |
| Specimen shape, size (mm) | Flat sheet, 25 × 25 | Flat sheet, 25 × 25 |
| Data collection | ||
| Diffractometer | Bruker D2 Phaser | Bruker D2 Phaser |
| Specimen mounting | Bruker PMMA holder | Bruker PMMA holder |
| Data collection mode | Reflection | Reflection |
| Scan method | Step | Step |
| 2θ values (°) | 2θmin = 5.042 2θmax = 100.048 2θstep = 0.020 | 2θmin = 5.042 2θmax = 100.048 2θstep = 0.020 |
| Refinement | ||
| R factors and goodness of fit | R p = 0.063, R wp = 0.084, R exp = 0.024, R(F 2) = 0.0780, χ2 = 12.180 | R p = 0.063, R wp = 0.084, R exp = 0.024, R(F 2) = 0.0780, χ2 = 12.180 |
| No. of parameters | 76 | 76 |
| No. of restraints | 29 | 29 |
The same symmetry and lattice parameters were used for the DFT calculation. Computer programs: DIFFRAC.Measurement (Bruker, 2009 ▸), Powder4 (Dragoe, 2001 ▸), DASH (David et al., 2006 ▸), GSAS (Larson & Von Dreele, 2004 ▸), EXPGUI (Toby, 2001 ▸), DIAMOND (Crystal Impact, 2015 ▸) and publCIF (Westrip, 2010 ▸).
Supplementary Material
Crystal structure: contains datablock(s) RAMM012A_publ, ramm012a_DFT, RAMM012A_overall, RAMM012A_phase_1, RAMM012A_phase_2, RAMM012A_p_01. DOI: 10.1107/S2056989016008343/hb7585sup1.cif
Additional supporting information: crystallographic information; 3D view; checkCIF report
supplementary crystallographic information
Crystal data
| C6H7NaO7 | c = 13.4551 Å |
| Mr = 214.10 | V = 772.45 Å3 |
| Orthorhombic, P212121 | Z = 4 |
| a = 7.4527 Å | None; DFT calculation radiation |
| b = 7.7032 Å | T = 300 K |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.86930 | 0.18689 | 0.07976 | 0.02580* | |
| C2 | 0.76864 | 0.31082 | 0.14718 | 0.01910* | |
| C3 | 0.80887 | 0.50489 | 0.13526 | 0.01910* | |
| C4 | 0.72870 | 0.60356 | 0.22535 | 0.01910* | |
| C5 | 0.54276 | 0.54256 | 0.25725 | 0.02580* | |
| C6 | 1.01121 | 0.54559 | 0.13579 | 0.02580* | |
| O7 | 0.79690 | 0.02935 | 0.07081 | 0.02580* | |
| O8 | 1.01065 | 0.22287 | 0.03877 | 0.02580* | |
| O9 | 1.07983 | 0.64886 | 0.07729 | 0.02580* | |
| O10 | 1.09698 | 0.46975 | 0.20891 | 0.02580* | |
| O11 | 0.51997 | 0.49361 | 0.34617 | 0.02580* | |
| O12 | 0.42047 | 0.54898 | 0.19138 | 0.02580* | |
| O13 | 0.73153 | 0.57264 | 0.04623 | 0.02580* | |
| H14 | 0.79934 | 0.27249 | 0.22351 | 0.02500* | |
| H15 | 0.62504 | 0.29365 | 0.13548 | 0.02500* | |
| H16 | 0.75707 | 0.49090 | −0.00712 | 0.02500* | |
| H17 | 0.71973 | 0.74075 | 0.20546 | 0.02500* | |
| H18 | 0.81890 | 0.59174 | 0.28862 | 0.03350* | |
| Na19 | 0.91363 | −0.18930 | −0.03840 | 0.03460* | |
| H20 | 0.67516 | 0.02054 | 0.10415 | 0.03900* | |
| H21 | 0.23334 | 0.49718 | 0.20429 | 0.03900* |
Bond lengths (Å)
| C1—C2 | 1.516 | C4—H17 | 1.092 |
| C1—O7 | 1.334 | C4—H18 | 1.089 |
| C1—O8 | 1.221 | C5—O11 | 1.266 |
| C2—C3 | 1.533 | C5—O12 | 1.272 |
| C2—H14 | 1.093 | C6—O9 | 1.230 |
| C2—H15 | 1.090 | C6—O10 | 1.311 |
| C3—C4 | 1.550 | O7—H20 | 1.014 |
| C3—C6 | 1.540 | O10—H21i | 1.040 |
| C3—O13 | 1.428 | O13—H16 | 0.974 |
| C4—C5 | 1.525 | H21—O10ii | 1.040 |
Symmetry codes: (i) x+1, y, z; (ii) x−1, y, z.
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O7—H20···O11 | 1.01 | 1.61 | 2.627 | 176 |
| O10—H21···O12 | 1.04 | 1.46 | 2.498 | 175 |
| O13—H16···O8 | 0.97 | 2.50 | 3.033 | 114 |
| C2—H15···O8 | 1.09 | 2.50 | 3.166 | 119 |
References
- Bravais, A. (1866). In Etudes Cristallographiques. Paris: Gauthier Villars.
- Bruker (2009). DIFFRAC. Measurement. Bruker AXS Inc., Madison, Wisconsin, USA.
- Crystal Impact (2015). DIAMOND. Crystal Impact GbR, Bonn, Germany. http://www.crystalimpact.com/diamond.
- Dassault Systemes (2014). Materials Studio. BIOVIA, San Diego, CA, USA.
- David, W. I. F., Shankland, K., van de Streek, J., Pidcock, E., Motherwell, W. D. S. & Cole, J. C. (2006). J. Appl. Cryst. 39, 910–915.
- Donnay, J. D. H. & Harker, D. (1937). Am. Mineral. 22, 446–467.
- Dovesi, R., Orlando, R., Civalleri, B., Roetti, C., Saunders, V. R. & Zicovich-Wilson, C. M. (2005). Z. Kristallogr. 220, 571–573.
- Dovesi, R., Roetti, C., Freyria-Fava, C., Prencipe, M. & Saunders, V. R. (1991). Chem. Phys. 156, 11–19.
- Dragoe, N. (2001). J. Appl. Cryst. 34, 535.
- Finger, L. W., Cox, D. E. & Jephcoat, A. P. (1994). J. Appl. Cryst. 27, 892–900.
- Friedel, G. (1907). Bull. Soc. Fr. Mineral. 30, 326–455.
- Gatti, C., Saunders, V. R. & Roetti, C. (1994). J. Chem. Phys. 101, 10686–10696.
- Glusker, J. P., Van Der Helm, D., Love, W. E., Dornberg, M., Minkin, J. A., Johnson, C. K. & Patterson, A. L. (1965). Acta Cryst. 19, 561–572.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Howard, C. J. (1982). J. Appl. Cryst. 15, 615–620.
- ICDD (2015). PDF-4+ 2015 and PDF-4 Organics 2016 (Databases), edited by S. Kabekkodu. International Centre for Diffraction Data, Newtown Square, PA, USA.
- Larson, A. C. & Von Dreele, R. B. (2004). General Structure Analysis System (GSAS). Report LAUR, 86–784 Los Alamos National Laboratory, New Mexico, USA.
- Louër, D. & Boultif, A. (2007). Z. Kristallogr. Suppl. 2007, 191–196.
- Macrae, C. F., Bruno, I. J., Chisholm, J. A., Edgington, P. R., McCabe, P., Pidcock, E., Rodriguez-Monge, L., Taylor, R., van de Streek, J. & Wood, P. A. (2008). J. Appl. Cryst. 41, 466–470.
- MDI (2012). JADE. Materials Data Inc., Livermore, CA, USA.
- Rammohan, A. & Kaduk, J. A. (2016a). Acta Cryst. B. Submitted.
- Rammohan, A. & Kaduk, J. A. (2016b). Acta Cryst. E72, 170–173. [DOI] [PMC free article] [PubMed]
- Rammohan, A. & Kaduk, J. A. (2016c). Acta Cryst. E72, 403–406. [DOI] [PMC free article] [PubMed]
- Rammohan, A. & Kaduk, J. A. (2016d). Acta Cryst. E72, 793–796. [DOI] [PMC free article] [PubMed]
- Stephens, P. W. (1999). J. Appl. Cryst. 32, 281–289.
- Streek, J. van de & Neumann, M. A. (2014). Acta Cryst. B70, 1020–1032. [DOI] [PMC free article] [PubMed]
- Thompson, P., Cox, D. E. & Hastings, J. B. (1987). J. Appl. Cryst. 20, 79–83.
- Toby, B. H. (2001). J. Appl. Cryst. 34, 210–213.
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
- Wolff, P. de (1966). ICDD Grant-in-Aid, PFD entry 00-016-1182.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) RAMM012A_publ, ramm012a_DFT, RAMM012A_overall, RAMM012A_phase_1, RAMM012A_phase_2, RAMM012A_p_01. DOI: 10.1107/S2056989016008343/hb7585sup1.cif
Additional supporting information: crystallographic information; 3D view; checkCIF report






