The title molecule, featuring an intramolecular O—H⋯O hydrogen bond, is non-planar as seen in the dihedral angle between the pyridyl rings of 7.45 (7)°. In the crystal, supramolecular chains are formed via π(pyridin-2-yl)–π(pyridin-3-yl) interactions.
Keywords: crystal structure; propane-1,3-dione; hydrogen bond; Hirshfeld surface
Abstract
The title compound, C13H10N2O2 [also called 1-(pyridin-2-yl)-3-(pyridin-3-yl)propane-1,3-dione], features an almost planar (r.m.s. deviation = 0.0095 Å) central C3O2 core consolidated by an intramolecular hydroxy-O—H⋯O(carbonyl) hydrogen bond. Twists are evident in the molecule, as seen in the dihedral angles between the central core and the 2- and pyridin-3-yl rings of 8.91 (7) and 15.88 (6)°, respectively. The conformation about the C=C bond [1.3931 (17) Å] is Z, and the N atoms lie to the same side of the molecule. In the molecular packing, supramolecular chains along the a axis are mediated by π(pyridin-2-yl)–π(pyridin-3-yl) interactions [inter-centroid distance = 3.7662 (9) Å]. The observation that chains pack with no directional interactions between them is consistent with the calculated electrostatic potential, which indicates that repulsive interactions dominate.
Chemical context
The β-diketonates of virtually all metals are known (Lamprey, 1960 ▸) because of the stability of the resulting six-membered metallocycle formed from bidentate coordination through the two oxygen atoms and the ability of the ligand to be accommodated within the common octahedral, tetrahedral and square-pyramidal coordination geometries. There has been an interest over the last few years to introduce extra donor functionality such as nitrile and pyridyl to this class of ligand to generate heterometallic complexes and novel coordination networks. Dipyridyl β-diketonates, for example, have been used to synthesize mixed-metal–organic frameworks. Burrows and co-workers employed di(pyridin-4-yl)propane-1,3-dione to prepare the corresponding AlIII and GaIII octahedral building blocks for network structures linked by AgI ions (Burrows et al., 2010 ▸). Carlucci and co-workers used the same ligand to make FeIII metalloligands that were again joined by coordination to AgI ions. The type of the resulting two- or three-dimensional coordination polymer depended on the nature of the counter-ion to silver (Carlucci et al., 2011 ▸). By comparison, the di(pyridin-2-yl)propane-1,3-dione ligand, which also has extra donor functionality available for coordination, is sterically hindered to allow network formation. Tan and co-workers prepared the CdII and CuII complexes from this ligand and did indeed observe chelation through the 2,2′-nitrogen atoms (Tan et al., 2012 ▸). However, they did not observe solid-state network formation from bridging oxygen-atom, μ2-Cl or μ3-Cl donors in the CdII complexes; the CuII complex was a tetranuclear oligomer linked via bridging water and acetate counter-ions (Tan et al., 2012 ▸). Less work has been performed with the unsymmetrical pyridyl β-diketonates. Zhang and co-workers have made the FeIII salt of 3-(pyridin-4-yl)-2,4-pentanedione as well as the mixed-MOF with AgNO3 in a two-dimensional honeycomb structure while at higher AgI concentrations, a one-dimensional ladder motif was formed (Zhang et al., 2008 ▸). This ligand and the symmetrical 4,4′- and 3,3′- variants have been treated with hydrazine to give the corresponding pyrazoles that were used to prepare strongly photoluminescent CuI coordination polymers (Zhan et al., 2011 ▸).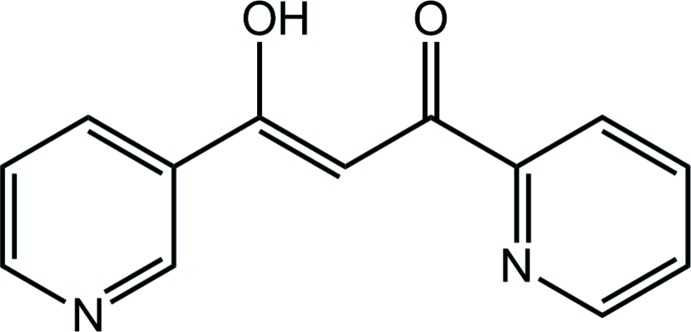
All of the mentioned dipyridyl ligands can be conveniently prepared by the Claisen condensation of an acetylpyridine with a pyridine carboxylic ester. The title compound, (I), has not previously been reported, but was prepared in this way from 2-acetylpyridine and ethyl nicotinate, and crystals suitable for X-ray crystallography were obtained by recrystallization from a mixture of dichloromethane and hexane. Herein, the crystal structure analysis of (I) is described along with a detailed investigation of the molecular packing by a Hirshfeld surface analysis.
Structural commentary
In (I), the assignment of carbonyl- versus hydroxy-O atoms is not readily confirmed by a great disparity in the C1—O1 [1.2871 (14) Å] and C3—O2 [1.3041 (14) Å] bond lengths. The assignment was based on an unrestrained refinement of the H1O atom which resulted in a O2—H1O bond length of 1.090 (18) Å. More certainty is associated with the assignment of the nitrogen atoms in the pyridyl rings. Thus, the short C5—N1 and C4—N1 [1.3325 (17) and 1.3484 (15) Å] and C10—N2 and C11—N2 [1.3371 (17) and 1.3397 (18) Å] bond lengths cf. the C—C bonds in the rings confirm their assignment. The central C3O2 residual in (I), Fig. 1 ▸, is essentially planar with the r.m.s. deviation of the five atoms being 0.0095 Å. The syn arrangement of the oxygen atoms enables the formation of an intramolecular hydroxy-O—H⋯O(carbonyl) hydrogen bond, Table 1 ▸. The dihedral angles formed between the central plane and the N1- and N2-pyridinyl rings are 8.91 (7) and 15.88 (6)°, respectively, indicating twists in the molecule. The dihedral angle between the pyridyl rings is 7.45 (7)°. The conformation about the C2=C3 [1.3931 (17) Å] is Z, and, to a first approximation, the N1 and N2 atoms lie to the same side of the molecule.
Figure 1.
The molecular structure of (I), showing the atom-labelling scheme and displacement ellipsoids at the 70% probability level.
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O2—H2O⋯O1 | 0.85 (2) | 1.65 (1) | 2.4673 (14) | 160 (2) |
Supramolecular features
The molecular packing in the crystal is dominated by π–π interactions formed between the N1- and N2-pyridinyl rings of translationally related molecules [Cg(N1-pyridinyl)⋯Cg(N2-pyridinyl) = 3.7662 (9) Å, angle of inclination = 7.45 (6)° for symmetry operation 1 + x, y, z]. The result is the formation of a linear supramolecular chain, Fig. 2 ▸ a. The chains pack with no directional interactions between them in accord with the distance criteria in PLATON (Spek, 2009 ▸), Fig. 2 ▸ b.
Figure 2.
Molecular packing in (I): (a) a view of the supramolecular chain along the a axis sustained by π–π interactions and (b) unit-cell contents shown in projection down the a axis. The π–π interactions are shown as purple dashed lines.
Hirshfeld surface analysis
The program Crystal Explorer 3.1 (Wolff et al., 2012 ▸) was used to generate Hirshfeld surfaces mapped over the electrostatic potential, d norm, shape-index and curvedness. The electrostatic potential was calculated with TONTO (Spackman et al., 2008 ▸; Jayatilaka et al., 2005 ▸), integrated in Crystal Explorer, using the experimental geometry as the input. The electrostatic potentials were mapped on the Hirshfeld surface using the STO-3G basis set at the Hartree–Fock level of theory over a range ±0.06 au. The contact distances d i and d e from the Hirshfeld surface to the nearest atom inside and outside, respectively, enables the analysis of the intermolecular interactions through the mapping of d norm. The combination of d e and d i in the form of a two-dimensional fingerprint plot (McKinnon et al., 2004 ▸) provides a summary of the intermolecular contacts in the crystal.
From the Hirshfeld surface mapped over electrostatic potential, Fig. 3 ▸, the negative potentials around the oxygen atoms of the hydroxy and carbonyl groups as well as about the nitrogen atoms of pyridyl rings prevent their participation in intermolecular interactions in the crystal of (I) due to the electrostatic repulsion that would eventuate. The presence of a short intermolecular C⋯C contact between the C5 and C10 atoms [C5⋯C10 = 3.313 (2) Å; symmetry code: −1 + x, y, z], which fall within the π–π contacts between pyridyl rings (Fig. 2 ▸ a), is viewed as bright-red spots near these atoms on the Hirshfeld surface mapped over d norm, Fig. 4 ▸.
Figure 3.
A view of the Hirshfeld surface mapped over electrostatic potential for (I). The red and blue regions represent negative and positive electrostatic potentials, respectively.
Figure 4.
Two views of the Hirshfeld surface mapped over d norm for (I): the bright-red spots at (a) C5 and (b) C10 indicate their involvement in short intermolecular C⋯C contacts.
The overall 2D fingerprint plot, Fig. 5 ▸ a, and those delineated into H⋯H, C⋯C, O⋯H/H⋯O, C⋯H/H⋯C and N⋯H/H⋯N contacts are illustrated in Fig. 5 ▸ b–f, respectively; their relative contributions to the surface are quantified in Table 2 ▸. The interatomic H⋯H contacts (McKinnon et al., 2007 ▸) appear as the scattered points over the greater part of the plot shown in Fig. 5 ▸ b, with a single peak at (d e, d i) less than the van der Waals separation corresponding to a short H13⋯H13 contact of 2.33 Å (symmetry code: 1 − x, −y, −z). The short interatomic C5⋯C10 contact and π–π stacking interactions appear as an arrow-like distribution of points with the tip at d e + d i ∼ 3.3 Å (Fig. 5 ▸ c). The presence of π–π stacking interactions between the pyridyl rings is also apparent from the appearance of red and blue triangle pairs on the Hirshfeld surface mapped with shape-index property identified with arrows in the image of Fig. 6 ▸, and in the flat region on the Hirshfeld surface mapped over curvedness in Fig. 7 ▸.
Figure 5.
The two-dimensional fingerprint plots for (I): (a) all interactions, and delineated into (b) H⋯H, (c) C⋯C, (d) O⋯H/H⋯O, (e) C⋯H/H⋯C and (f) N⋯H/H⋯N interactions.
Table 2. Percentage contribution of the different intermolecular interactions to the Hirshfeld surface of (I).
| Contact | % |
|---|---|
| H⋯H | 36.2 |
| O⋯H/H⋯O | 13.2 |
| C⋯H/H⋯C | 22.9 |
| N⋯H/H⋯N | 14.6 |
| C⋯C | 6.1 |
| C⋯O/O⋯C | 2.9 |
| C⋯N/N⋯C | 2.8 |
| O⋯O | 0.9 |
| N⋯N | 0.4 |
Figure 6.
A view of the Hirshfeld surface mapped with shape-index property for (I). The red and blue triangles identified with arrows indicate π–π stacking interactions.
Figure 7.
A view of the Hirshfeld surface mapped over curvedness for (I). The flat regions highlight the involvement of rings in the π–π stacking interactions.
The two-dimensional fingerprint plots delineated into O⋯H/H⋯O, C⋯H/H⋯C and N⋯H/H⋯N interactions exhibit their usual characteristic features in their respective plots; Fig. 4 ▸ d–f. However, the points are distributed at (d e, d i) distances greater than their respective van der Waals separations. This is consistent with the repulsion between the atoms having electrostatic negative potential dominating the molecular packing, hence the lack of specific intermolecular interactions between supramolecular chains.
Database survey
A survey of the Cambridge Structural Database (Groom et al., 2016 ▸) revealed that there are two closely related pyridyl-substituted propane-1,3-dione structures in the crystallographic literature. These are the mono-pyridyl derivatives 3-hydroxy-1-phenyl-3-(pyridin-3-yl)prop-2-en-1-one (II) and 3-hydroxy-1-phenyl-3-(pyridin-4-yl)prop-2-en-1-one (III), both published by Dudek et al. (2011 ▸). Each structure features a very similar central core with the intramolecular O—H⋯O hydrogen bond. In each of (II) and (III), the pyridyl ring is connected to the carbon atom bearing the hydroxy group. As seen from the overlay diagram (Fig. 8 ▸) and as quantified in Table 3 ▸, the three structures (I)–(III) have very similar conformations.
Figure 8.
Overlay diagram of molecules of (I) (red image), (II) (green) and (III) (blue). The molecules have been overlapped so that the central five-membered rings are coincident.
Table 3. Dihedral angle (°) data for (I)–(III).
| Structure | C3O2/n-pyridyl | C3O2/pyridin-2-yl or phenyl | ring/ring | CSD refcodea | Reference |
|---|---|---|---|---|---|
| (I) | n = 3; 15.88 (6) | 8.91 (7) | 7.45 (7) | – | This work |
| (II) | n = 3; 2.23 (9) | 4.20 (8) | 4.38 (9) | XIOXID | Dudek et al. (2011 ▸) |
| (III)b | n = 4; 8.10 (5) | 11.41 (5) | 3.88 (5) | BEDREJ | Dudek et al. (2011 ▸) |
Notes: (a) Groom et al. (2016 ▸); (b) isolated as a 1:1 co-crystal with benzoic acid.
Synthesis and crystallization
2-Acetylpyridine (3.0562 g, 25.2 mmol) was added to a suspension of NaH (60% dispersion in mineral oil, 2.0058 g, 50.0 mmol) in anhydrous THF (10 ml) at room temperature with stirring. Ethyl nicotinate (7.5675g, 50.1 mmol) in anhydrous THF (10ml) was added dropwise to the mixture over 3 min. The yellow mixture was refluxed under a nitrogen atmosphere for 1.3 h and then quenched with ice–water (50 ml). Glacial acetic acid was added to adjust the pH to 6–7. The resulting yellow precipitate was collected by filtration, washed with cold water and dried under vacuum. Recrystallization from dichloromethane–hexane (1:1 v/v) solution afforded colourless crystals. Yield: 4.03 g (70.7%). M.p: 377–378 K. IR (KBr pellet) νmax/cm−1: 3121 (m), 3053 (m), 2922 (m), 2853 (m), 1611 (s), 1595 (s), 1539 (m), 1458 (m), 1418 (m), 1221 (m), 1188 (m), 1146 (m), 1115 (m), 1067 (m), 1018 (m), 989 (m), 926 (m), 775 (s), 739 (m), 679 (s), 611 (m). Analysis calculated for C13H10N2O2: C, 69.03; H, 4.42; N, 12.19. Found: C, 68.73; H, 4.54; N, 12.16. MS: m/z 226. 1H NMR (400 MHz, d 6–DMSO) δ 9.22 (1H, s), 8.82 (2H, m), 8.44 (1H, d, J = 7.9 Hz), 8.17 (1H, d, J = 7.8 Hz), 8.09 (1H, m), 7.70 (1H, m), 7.63 (2H, m).
Refinement details
Crystal data, data collection and structure refinement details are summarized in Table 4 ▸. Carbon-bound H atoms were placed in their calculated positions (C—H = 0.95 Å) and were included in the refinement in the riding-model approximation, with U iso(H) set to 1.2U eq(C). The hydroxy-H atom was located in a difference map and refined with O—H = 0.82±0.01Å, and with U iso(H) set to 1.5U eq(O).
Table 4. Experimental details.
| Crystal data | |
| Chemical formula | C13H10N2O2 |
| M r | 226.23 |
| Crystal system, space group | Orthorhombic, P b c a |
| Temperature (K) | 273 |
| a, b, c (Å) | 7.2124 (9), 14.1782 (19), 20.794 (3) |
| V (Å3) | 2126.4 (5) |
| Z | 8 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.10 |
| Crystal size (mm) | 0.20 × 0.20 × 0.19 |
| Data collection | |
| Diffractometer | Bruker D8-Quest CCD |
| Absorption correction | Multi-scan (SADABS; Sheldrick, 1996 ▸) |
| T min, T max | 0.981, 0.982 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 46632, 2647, 2227 |
| R int | 0.057 |
| (sin θ/λ)max (Å−1) | 0.671 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.042, 0.121, 1.05 |
| No. of reflections | 2647 |
| No. of parameters | 157 |
| No. of restraints | 1 |
| Δρmax, Δρmin (e Å−3) | 0.33, −0.24 |
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S205698901600832X/hb7587sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S205698901600832X/hb7587Isup2.hkl
Supporting information file. DOI: 10.1107/S205698901600832X/hb7587Isup3.cml
CCDC reference: 1481225
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
We acknowledge the financial support from the Brunei Research Council (BRC) Science and Technology grant (S&T17).
supplementary crystallographic information
Crystal data
| C13H10N2O2 | Dx = 1.413 Mg m−3 |
| Mr = 226.23 | Mo Kα radiation, λ = 0.71073 Å |
| Orthorhombic, Pbca | Cell parameters from 9904 reflections |
| a = 7.2124 (9) Å | θ = 3.0–28.3° |
| b = 14.1782 (19) Å | µ = 0.10 mm−1 |
| c = 20.794 (3) Å | T = 273 K |
| V = 2126.4 (5) Å3 | Block, colourless |
| Z = 8 | 0.20 × 0.20 × 0.19 mm |
| F(000) = 944 |
Data collection
| Bruker D8-Quest CCD diffractometer | 2647 independent reflections |
| Radiation source: sealed tube | 2227 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.057 |
| Detector resolution: 8.366 pixels mm-1 | θmax = 28.5°, θmin = 3.0° |
| φ and ω scans | h = −8→9 |
| Absorption correction: multi-scan (SADABS; Sheldrick, 1996) | k = −18→18 |
| Tmin = 0.981, Tmax = 0.982 | l = −27→27 |
| 46632 measured reflections |
Refinement
| Refinement on F2 | 1 restraint |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.042 | w = 1/[σ2(Fo2) + (0.0588P)2 + 0.9349P] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.121 | (Δ/σ)max < 0.001 |
| S = 1.05 | Δρmax = 0.33 e Å−3 |
| 2647 reflections | Δρmin = −0.24 e Å−3 |
| 157 parameters |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | −0.09862 (13) | 0.14710 (7) | 0.02692 (4) | 0.0286 (2) | |
| O2 | 0.19541 (13) | 0.05816 (7) | 0.02776 (4) | 0.0276 (2) | |
| H2O | 0.1025 (19) | 0.0929 (11) | 0.0188 (8) | 0.041* | |
| N1 | −0.33075 (15) | 0.12603 (7) | 0.17723 (5) | 0.0246 (2) | |
| N2 | 0.43366 (17) | −0.09680 (9) | 0.21246 (6) | 0.0335 (3) | |
| C1 | −0.13288 (17) | 0.11505 (8) | 0.08360 (6) | 0.0217 (3) | |
| C2 | −0.01053 (16) | 0.05315 (8) | 0.11536 (5) | 0.0214 (3) | |
| H2 | −0.0402 | 0.0290 | 0.1557 | 0.026* | |
| C3 | 0.15550 (17) | 0.02859 (8) | 0.08551 (5) | 0.0211 (2) | |
| C4 | −0.30743 (16) | 0.14778 (8) | 0.11462 (5) | 0.0203 (2) | |
| C5 | −0.48525 (19) | 0.15721 (9) | 0.20520 (6) | 0.0286 (3) | |
| H5 | −0.5044 | 0.1425 | 0.2483 | 0.034* | |
| C6 | −0.61954 (19) | 0.21046 (9) | 0.17384 (7) | 0.0294 (3) | |
| H6 | −0.7244 | 0.2313 | 0.1957 | 0.035* | |
| C7 | −0.59415 (18) | 0.23185 (9) | 0.10951 (7) | 0.0284 (3) | |
| H7 | −0.6819 | 0.2669 | 0.0871 | 0.034* | |
| C8 | −0.43511 (18) | 0.19984 (8) | 0.07917 (6) | 0.0245 (3) | |
| H8 | −0.4141 | 0.2129 | 0.0360 | 0.029* | |
| C9 | 0.29989 (16) | −0.03006 (8) | 0.11646 (5) | 0.0206 (2) | |
| C10 | 0.30159 (18) | −0.04710 (9) | 0.18265 (6) | 0.0269 (3) | |
| H10 | 0.2055 | −0.0225 | 0.2073 | 0.032* | |
| C11 | 0.57090 (19) | −0.13133 (10) | 0.17598 (7) | 0.0315 (3) | |
| H11 | 0.6644 | −0.1656 | 0.1960 | 0.038* | |
| C12 | 0.58095 (19) | −0.11875 (9) | 0.10999 (7) | 0.0308 (3) | |
| H12 | 0.6782 | −0.1445 | 0.0865 | 0.037* | |
| C13 | 0.44368 (18) | −0.06720 (9) | 0.07980 (6) | 0.0257 (3) | |
| H13 | 0.4474 | −0.0575 | 0.0356 | 0.031* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0318 (5) | 0.0325 (5) | 0.0214 (4) | 0.0031 (4) | 0.0021 (3) | 0.0078 (3) |
| O2 | 0.0272 (5) | 0.0349 (5) | 0.0208 (4) | 0.0036 (4) | 0.0036 (3) | 0.0050 (3) |
| N1 | 0.0235 (5) | 0.0280 (5) | 0.0224 (5) | −0.0003 (4) | −0.0005 (4) | 0.0020 (4) |
| N2 | 0.0316 (6) | 0.0394 (6) | 0.0294 (6) | 0.0077 (5) | −0.0005 (5) | 0.0056 (5) |
| C1 | 0.0229 (6) | 0.0215 (5) | 0.0207 (5) | −0.0035 (4) | −0.0010 (4) | −0.0003 (4) |
| C2 | 0.0222 (6) | 0.0232 (6) | 0.0188 (5) | −0.0003 (4) | −0.0006 (4) | 0.0020 (4) |
| C3 | 0.0226 (6) | 0.0211 (5) | 0.0195 (5) | −0.0033 (4) | −0.0009 (4) | −0.0010 (4) |
| C4 | 0.0225 (6) | 0.0173 (5) | 0.0211 (5) | −0.0022 (4) | −0.0023 (4) | −0.0002 (4) |
| C5 | 0.0282 (6) | 0.0333 (7) | 0.0243 (6) | 0.0001 (5) | 0.0023 (5) | 0.0003 (5) |
| C6 | 0.0250 (6) | 0.0277 (6) | 0.0355 (7) | 0.0030 (5) | 0.0034 (5) | −0.0046 (5) |
| C7 | 0.0275 (6) | 0.0220 (6) | 0.0357 (7) | 0.0049 (5) | −0.0053 (5) | 0.0007 (5) |
| C8 | 0.0286 (6) | 0.0209 (5) | 0.0240 (6) | 0.0006 (5) | −0.0034 (5) | 0.0013 (4) |
| C9 | 0.0202 (5) | 0.0191 (5) | 0.0224 (5) | −0.0030 (4) | −0.0001 (4) | −0.0010 (4) |
| C10 | 0.0247 (6) | 0.0320 (6) | 0.0239 (6) | 0.0044 (5) | 0.0020 (5) | 0.0022 (5) |
| C11 | 0.0263 (7) | 0.0309 (7) | 0.0373 (7) | 0.0055 (5) | −0.0021 (5) | 0.0043 (5) |
| C12 | 0.0276 (6) | 0.0279 (6) | 0.0369 (7) | 0.0066 (5) | 0.0059 (5) | −0.0008 (5) |
| C13 | 0.0284 (6) | 0.0245 (6) | 0.0243 (6) | 0.0005 (5) | 0.0036 (5) | −0.0012 (5) |
Geometric parameters (Å, º)
| O1—C1 | 1.2871 (14) | C5—H5 | 0.9300 |
| O2—C3 | 1.3041 (14) | C6—C7 | 1.3839 (19) |
| O2—H2O | 0.853 (9) | C6—H6 | 0.9300 |
| N1—C5 | 1.3325 (17) | C7—C8 | 1.3855 (18) |
| N1—C4 | 1.3484 (15) | C7—H7 | 0.9300 |
| N2—C10 | 1.3371 (17) | C8—H8 | 0.9300 |
| N2—C11 | 1.3397 (18) | C9—C13 | 1.3907 (17) |
| C1—C2 | 1.4090 (16) | C9—C10 | 1.3975 (16) |
| C1—C4 | 1.4888 (17) | C10—H10 | 0.9300 |
| C2—C3 | 1.3931 (17) | C11—C12 | 1.386 (2) |
| C2—H2 | 0.9300 | C11—H11 | 0.9300 |
| C3—C9 | 1.4800 (16) | C12—C13 | 1.3815 (18) |
| C4—C8 | 1.3915 (16) | C12—H12 | 0.9300 |
| C5—C6 | 1.3905 (19) | C13—H13 | 0.9300 |
| C3—O2—H2O | 102.4 (12) | C6—C7—C8 | 118.55 (12) |
| C5—N1—C4 | 116.73 (11) | C6—C7—H7 | 120.7 |
| C10—N2—C11 | 117.16 (12) | C8—C7—H7 | 120.7 |
| O1—C1—C2 | 121.93 (11) | C7—C8—C4 | 118.71 (11) |
| O1—C1—C4 | 116.69 (10) | C7—C8—H8 | 120.6 |
| C2—C1—C4 | 121.37 (10) | C4—C8—H8 | 120.6 |
| C3—C2—C1 | 119.02 (11) | C13—C9—C10 | 117.90 (11) |
| C3—C2—H2 | 120.5 | C13—C9—C3 | 119.94 (11) |
| C1—C2—H2 | 120.5 | C10—C9—C3 | 122.12 (11) |
| O2—C3—C2 | 121.32 (11) | N2—C10—C9 | 123.63 (12) |
| O2—C3—C9 | 115.20 (10) | N2—C10—H10 | 118.2 |
| C2—C3—C9 | 123.47 (10) | C9—C10—H10 | 118.2 |
| N1—C4—C8 | 123.38 (11) | N2—C11—C12 | 123.53 (12) |
| N1—C4—C1 | 116.91 (10) | N2—C11—H11 | 118.2 |
| C8—C4—C1 | 119.69 (11) | C12—C11—H11 | 118.2 |
| N1—C5—C6 | 123.92 (12) | C13—C12—C11 | 118.73 (12) |
| N1—C5—H5 | 118.0 | C13—C12—H12 | 120.6 |
| C6—C5—H5 | 118.0 | C11—C12—H12 | 120.6 |
| C7—C6—C5 | 118.69 (12) | C12—C13—C9 | 119.05 (12) |
| C7—C6—H6 | 120.7 | C12—C13—H13 | 120.5 |
| C5—C6—H6 | 120.7 | C9—C13—H13 | 120.5 |
| O1—C1—C2—C3 | 2.34 (18) | N1—C4—C8—C7 | 0.49 (18) |
| C4—C1—C2—C3 | −176.70 (10) | C1—C4—C8—C7 | −178.19 (11) |
| C1—C2—C3—O2 | −3.26 (17) | O2—C3—C9—C13 | −14.19 (16) |
| C1—C2—C3—C9 | 175.33 (10) | C2—C3—C9—C13 | 167.14 (11) |
| C5—N1—C4—C8 | −0.14 (17) | O2—C3—C9—C10 | 163.35 (11) |
| C5—N1—C4—C1 | 178.58 (11) | C2—C3—C9—C10 | −15.33 (18) |
| O1—C1—C4—N1 | −170.37 (10) | C11—N2—C10—C9 | 0.2 (2) |
| C2—C1—C4—N1 | 8.72 (17) | C13—C9—C10—N2 | 0.14 (19) |
| O1—C1—C4—C8 | 8.40 (16) | C3—C9—C10—N2 | −177.44 (12) |
| C2—C1—C4—C8 | −172.51 (10) | C10—N2—C11—C12 | −0.6 (2) |
| C4—N1—C5—C6 | −0.55 (19) | N2—C11—C12—C13 | 0.6 (2) |
| N1—C5—C6—C7 | 0.9 (2) | C11—C12—C13—C9 | −0.27 (19) |
| C5—C6—C7—C8 | −0.48 (19) | C10—C9—C13—C12 | −0.09 (18) |
| C6—C7—C8—C4 | −0.16 (18) | C3—C9—C13—C12 | 177.55 (11) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O2—H2O···O1 | 0.85 (2) | 1.65 (1) | 2.4673 (14) | 160 (2) |
References
- Brandenburg, K. (2006). DIAMOND. Crystal Impact GbR, Bonn, Germany.
- Bruker (2007). SMART and SAINT. Bruker AXS Inc., Madison, Wisconsin, USA.
- Burrows, A. D., Frost, C. G., Mahon, M. F., Raithby, P. R., Renouf, C. L., Richardson, C. & Stevenson, A. J. (2010). Chem. Commun. 46, 5067–5069. [DOI] [PubMed]
- Carlucci, L., Ciani, G., Proserpio, D. M. & Visconti, M. (2011). CrystEngComm, 13, 5891–5902.
- Dudek, M., Clegg, J. K., Glasson, C. R. K., Kelly, N., Gloe, K., Gloe, K., Kelling, A., Buschmann, H.-J., Jolliffe, K. A., Lindoy, L. F. & Meehan, G. V. (2011). Cryst. Growth Des. 11, 1697–1704.
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Gans, J. & Shalloway, D. (2001). J. Mol. Graphics Modell. 19, 557–559. [DOI] [PubMed]
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Jayatilaka, D., Grimwood, D. J., Lee, A., Lemay, A., Russel, A. J., Taylo, C., Wolff, S. K., Chenai, C. & Whitton, A. (2005). TONTO – A System for Computational Chemistry. Available at: http:// hirshfeldsurface. net/
- Lamprey, H. (1960). Ann. NY Acad. Sci. 88, 519–525. [DOI] [PubMed]
- McKinnon, J. J., Jayatilaka, D. & Spackman, M. A. (2007). Chem. Commun. pp. 3814–3816. [DOI] [PubMed]
- McKinnon, J. J., Spackman, M. A. & Mitchell, A. S. (2004). Acta Cryst. B60, 627–668. [DOI] [PubMed]
- Sheldrick, G. M. (1996). SADABS. University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
- Spackman, M. A., McKinnon, J. J. & Jayatilaka, D. (2008). CrystEngComm, 10, 377–388.
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Tan, J.-T., Zhao, W.-J., Chen, S.-P., Li, X., Lu, Y.-L., Feng, X. & Yang, X.-W. (2012). Chem. Pap. 66, 47–53.
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
- Wolff, S. K., Grimwood, D. J., McKinnon, J. J., Turner, M. J., Jayatilaka, D. & Spackman, M. A. (2012). Crystal Explorer. The University of Western Australia, Australia.
- Zhan, S.-Z., Li, M., Zhou, X.-P., Ni, J., Huang, X.-C. & Li, D. (2011). Inorg. Chem. 50, 8879–8892. [DOI] [PubMed]
- Zhang, Y., Chen, B., Fronczek, F. R. & Maverick, A. W. (2008). Inorg. Chem. 47, 4433–4435. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S205698901600832X/hb7587sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S205698901600832X/hb7587Isup2.hkl
Supporting information file. DOI: 10.1107/S205698901600832X/hb7587Isup3.cml
CCDC reference: 1481225
Additional supporting information: crystallographic information; 3D view; checkCIF report










