Abstract
It is well known that the ykvU-ykvV operon is under the regulation of the σE-associated RNA polymerase (EσE). In our study, we observed that ykvV is transcribed together with the upstream ykvU gene by EσE in the mother cell and monocistronically under EσG control in the forespore. Interestingly, alternatively expressed ykvV in either the forespore or the mother cell increased the sporulation efficiency in the ykvV background. Studies show that the YkvV protein is a member of the thioredoxin superfamily and also contains a putative Sec-type secretion signal at the N terminus. We observed efficient sporulation in a mutant strain obtained by replacing the putative signal peptide of YkvV with the secretion signal sequence of SleB, indicating that the putative signal sequence is essential for spore formation. These results suggest that YkvV is capable of being transported by the putative Sec-type signal sequence into the space between the double membranes surrounding the forespore. The ability of ykvV expression in either compartment to complement is indeed intriguing and further introduces a new dimension to the genetics of B. subtilis spore formation. Furthermore, electron microscopic observation revealed a defective cortex in the ykvV disruptant. In addition, the expression levels of σK-directed genes significantly decreased despite normal σG activity in the ykvV mutant. However, immunoblotting with the anti-σK antibody showed that pro-σK was normally processed in the ykvV mutant, indicating that YkvV plays an important role in cortex formation, consistent with recent reports. We therefore propose that ykvV should be renamed spoIVH.
The gram-positive bacterium Bacillus subtilis forms dormant and environmentally resistant spores in response to nutrient deprivation (10). Early in sporulation, cells divide into two unequal compartments, a larger mother cell and a smaller forespore (24, 34). Just after septation, RNA polymerase sigma factors σF and σE govern gene expressions in the forespore and in the mother cell, respectively (14, 19). Later in sporulation, after the completion of the engulfment of the forespore by the mother cell, σG and σK become activated and replace σF and σE in the forespore and mother cell compartments, respectively (23, 25, 42). Prior to the completion of the engulfment process, an inactive precursor protein pro-σK, which contains an N-terminal extension of 20 amino acids (aa), is produced in the mother cell (6, 20, 26). The processing of pro-σK into an active form requires the expression of the signaling protein SpoIVB in the forespore under the control of σG (4, 46). A processing complex consisting of SpoIVFA, SpoIVFB, and BofA receives the signal via SpoIVB that engulfment is completed, and then pro-σK is processed into active σK in the mother cell (5, 27, 36). The engulfment process culminates in two bilayer membranes surrounding the forespore. A thick peptidoglycan layer is then deposited between the two membranes of the forespore to form the spore cortex that confers heat resistance on the spore (13). The coordinated functions of this cascade of sigma factors ensure distinct regulation of hundreds of sporulation-specific genes, including many whose functions are not yet known.
With the successful completion of the B. subtilis genome-wide sequence (21), the current focus of the B. subtilis functional genomics project is to identify the roles of all genes of unknown functions by gene disruption with insertional mutagenesis and pMUTIN vectors (31). Within the framework of this project, the sporulation-deficient spoIVH mutant was identified.
Recently, Eichenberger et al. (8) and Feucht et al. (11) reported that spoIVH is required for efficient sporulation and is transcribed from the consensus sequence of the σE-recognized promoter located upstream of ykvU. In this paper, we report that spoIVH is expressed in both compartments under the control of σE and σG. This is the first instance of nonspecific compartment expression of a sporulation gene during spore formation in B. subtilis.
MATERIALS AND METHODS
Measurement of sporulation frequencies.
Sporulation efficiency was measured by incubating B. subtilis cells in DSM (Difco sporulation medium) (39) at 37°C for 24 h. The number of spores per milliliter of culture (CFU) was determined as the number of heat-resistant (80°C for 10 min) colonies on tryptose blood agar base.
Plasmid and strain constructions.
Table 1 lists the bacterial strains and plasmids used in this study. B. subtilis was transformed and plasmids were constructed in Escherichia coli JM105 by standard methods (7, 37).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype and/or relevant characteristics | Source, reference, or constructiona |
|---|---|---|
| E. coli JM105 | supE endA sbcB15 hsdR4 rpsL thi Δ(lac-proAB)F′ [traD36 proAB+lacIqlacZΔM15] | 47 |
| B. subtilis | ||
| 168 | trpC2 | Laboratory stock |
| YKVVd | trpC2 spoIVH::pMUTIN2MCS | 31 |
| YKVUd | trpC2 ykvU::pMUTIN2MCS | 31 |
| TS001 | trpC2 spoIVH::pJMIVH | This study |
| TS002 | trpC2 ykvU::pJMVU | This study |
| SSPEd | trpC2 sspE::pMUTIN2MCS | 31 |
| RL13 | spβ::gerE-lacZ | R. Losick |
| REZ | trpC2 spβ::gerE-lacZ | RL13 → 168 |
| PE6 | amyE::spoIIE-gus::cat thr::pCW63(cotD-lacZ)::erm divIB::spc | R. Losick |
| TDZ | trpC2 thr::pCW63(cotD-lacZ)::erm | PE6 → 168 |
| TS003 | trpC2 spoIVH::pJMIVH sspE::pMUTIN2MCS | TS001 → SSPEd |
| TS004 | trpC2 spoIVH::pJMIVH spβ::gerE-lacZ | TS001 → REZ |
| TS005 | trpC2 spoIVH::pJMIVH thr::pCW63(cotD-lacZ)::erm | TS001 → TDZ |
| TS006 | trpC2 thrC::pTCE10(spoIVH) | This study |
| TS007 | trpC2 thrC::pTCE11(PspoIVH-spoIVH) | This study |
| TS008 | trpC2 thrC::pTCE12(PykvU-spoIVH) | This study |
| TS009 | trpC2 thrC::pTCE13(PsspE-spoIVH) | This study |
| TS010 | trpC2 spoIVH::pJMIVH thrC::pTCE10 | TS006 → TS001 |
| TS011 | trpC2 spoIVH::pJMIVH thrC::pTCE11 | TS007 → TS001 |
| TS012 | trpC2 spoIVH::pJMIVH thrC::pTCE12 | TS008 → TS001 |
| TS013 | trpC2 spoIVH::pJMIVH thrC::pTCE13 | TS009 → TS001 |
| TS014 | trpC2 thrC::pTCE14(PspoIVH-spoIVHΔsignal) | This study |
| TS015 | trpC2 thrC::pTCE15(PsleB-signalsleB-spoIVHΔsignal) | This study |
| TS016 | trpC2 spoIVH::pJMIVH thrC::pTCE14 | TS014 → TS001 |
| TS017 | trpC2 spoIVH::pJMIVH thrC::pTCE15 | TS015 → TS001 |
| TF97 | trpC2 spo0A::cat | K. Kobayashi |
| TF85 | trpC2 sigH::cat | K. Kobayashi |
| TF83 | trpC2 sigF::cat | K. Kobayashi |
| TF82 | trpC2 sigE::cat | K. Kobayashi |
| TF84 | trpC2 sigG::cat | K. Kobayashi |
| TF99 | trpC2 spoIIIC::cat | K. Kobayashi |
| Plasmids | ||
| pMUTIN2MCS | Vector carrying bla erm | 31 |
| pJM114 | Vector carrying bla kan | 33 |
| pHY300PLK | Vector carrying bla tet | 18 |
| pCBB31 | Vector carrying bla cat | 38 |
| pTCC0 | pCBB31 carrying a thrC 5′ region | This study |
| pTCC1 | Integration vector at thrC carrying bla and cat | This study |
| pUC18 | Vector carrying bla | 43 |
| pUC19 | Vector carrying bla | 47 |
| pE194 | Vector carrying erm | 16 |
| pTC1 | Vector carrying bla | This study |
| pTC2 | Vector carrying bla erm | This study |
| pTC3 | Vector carrying bla erm | This study |
| pTCE1 | Integration vector at thrC carrying bla and erm | This study |
| pJMVU | pJM114 carrying an internal region of ykvU | This study |
| pJMIVH | pJM114 carrying an internal region of spoIVH | This study |
| pTCE10 | pTCE1 carrying spoIVH | This study |
| pTCE11 | pTCE1 carrying PspoIVH-spoIVH | This study |
| pTCE12 | pTCE1 carrying PykvU-spoIVH | This study |
| pTCE13 | pTCE1 carrying PsspE-spoIVH | This study |
| pTCE14 | pTCE1 carrying PspoIVH-spoIVHΔsignal | This study |
| pTCE15 | pTCE1 carrying PsleB-signalsleB-spoIVH+79Δsignal | This study |
Arrows indicate transformation from the donor DNA to the recipient strain.
Integration plasmids pJMVU and pJMIVH were constructed as follows. PCR-amplified products were generated. The 237-bp internal segment of ykvU was generated with primers ykvU-F (5′-CCGGAATTCTATGATTTTGGCGCGGG-3′, the EcoRI site is underlined) and ykvU-R (5′-CGCGGATCCGGAATAAACGGAAGCGC-3′, the BamHI site is underlined), and the 168-bp internal segment of spoIVH was generated with primers IVH-F (5′-CCGGAATTCCTGCTGTTCCCGCTGTT-3′, the EcoRI site is underlined) and IVH-R (5′-CGCGGATCCCACTGTCGGATGGATGG-3′, the BamHI site is underlined). These products were trimmed with the respective restriction enzymes and then ligated with pJM114 (33) digested with EcoRI/BamHI. The resulting plasmids, pJMVU and pJMIVH, were used to transform competent cells of B. subtilis 168 to generate strains TS002 and TS001, respectively.
To obtain the vector pTCE1 to enable the introduction of DNA fragments into the thrC region by double crossover, a 993-bp 5′ region and a 990-bp 3′ region of the thrC gene were amplified with primer pairs thrC-UF (5′-CGGGGTACCTTGAAGCCAGTGTTGCC-3′, the KpnI site is underlined) and thrC-UR (5′-CGCGGATCCTGTAAAGTTAGCGCCGG-3′, the BamHI site is underlined) and thrC-DF (5′-AAAACTGCAGGAAATCACCGATTGCCC-3′, the PstI site is underlined) and thrC-DR (5′-CCCAAGCTTGTCCGCTTCAGACAGCT-3′, the HindIII site is underlined), respectively. The plasmid that was used, pCBB31 (38), harbors a Cmr cassette flanked by unique KpnI/BamHI and PstI/HindIII sites. The 993-bp PCR product was trimmed with the enzymes KpnI and BamHI and then ligated with pCBB31 digested with KpnI/BamHI, resulting in the plasmid pTCC0. The 990-bp product was cut with PstI and HindIII and then ligated with pTCC0 digested with PstI/HindIII, resulting in the plasmid pTCC1. Next, the multiple cloning sequence region of pHY300PLK (18) was digested with EcoRI and HindIII, and the small 39-bp fragment was ligated into pUC19 (47) to obtain plasmid pTC1. A 1,197-bp HpaII and BanIII fragment of the erm gene from pE194 (16) was cloned into the AccI site of pTC1 to generate plasmid pTC2. To obtain plasmid pTC3, a 1,218-bp BamHI and XbaI fragment of the erm gene of pTC2 was cloned into the BamHI and XbaI site of pUC18 (43). A 1,213-bp BamHI and BglII fragment of the erm gene of pTC3 was cloned into the BamHI and BglII site of pTCC1, resulting in a BamHI- and BglII-digestible plasmid named pTCE1.
To obtain the plasmids pTCE10 and pTCE11, which harbor the SpoIVH coding region with and without promoter, respectively, DNA segments of positions −115 to +515 and −28 to +515 relative to the spoIVH start codon were amplified from strain 168 chromosomal DNA with primers IVH-115X (5′-TGCTCTAGAGCAAAGCATTGAAGGTA-3′, the XbaI site is underlined) or IVH-28X (5′-TGCTCTAGACTAATTGAAAAGCATGA-3′, the XbaI site is underlined) each against primer IVH-RB (5′-CGCGGATCCAGAGTCTATGCTCTCAG-3′, the BamHI site is underlined), respectively. These PCR products were trimmed with the respective restriction enzymes and then ligated with XbaI/BglII-digested pTCE1. The resulting plasmids, pTCE10 and pTCE11, were linearized with ScaI and then used for the introduction of spoIVH (without promoter) and PspoIVH-spoIVH into the thrC locus of B. subtilis strain 168 through a double-crossover event. Erythromycin-resistant transformants were selected to obtain strains TS007 and TS006 with and without spoIVH promoter, respectively.
For plasmids pTCE12 and pTCE13, which harbor the SpoIVH coding region plus the ykvU (σE) or sspE (σG) promoter region, respectively, the DNA segment containing positions −28 to +515 of the spoIVH region was amplified with IVH-28H (5′-CCCAAGCTTCTAATTGAAAAGCATGA-3′, the HindIII site is underlined) and IVH-RB, and then the promoter regions of ykvU (positions −113 to −13 from the initiation codon of the ykvU gene) and sspE (positions −58 to −12 from the initiation codon of the sspE gene) were amplified with primers ykvU-113 (5′-TGCTCTAGAATTTGTCTCAGCTGTGC-3′, the XbaI site is underlined) and ykvU-13 (5′-CCCAAGCTTTGTCTCTTGTACTACCA-3′, the HindIII site is underlined) and primers sspE-58 (5′-TGCTCTAGAAAAAGAGGAATAGCTAT-3′, the XbaI site is underlined) and sspE-12 (5′-CCCAAGCTTCCACGGTCATTAGAATG-3′, the HindIII site is underlined), respectively. These PCR products were trimmed with the respective restriction enzymes and then ligated with XbaI/BglII-digested pTCE1.
For plasmid pTCE14 with the putative PspoIVH promoter plus the signal sequence-less SpoIVH coding region, the DNA segment from −115 to +3 of spoIVH and the DNA segment from +79 to +515 relative to the spoIVH start codon were amplified with the primer pairs IVH-115X and IVH-3H (5′-CCCAAGCTTCATGGAATCTTCCTTTC-3′, the HindIII site is underlined) and IVH-sig-H (5′-CCCAAGCTTGAGGAAAAACAGCCTGC-3′, the HindIII site is underlined) and IVH-RB, respectively. These PCR products were trimmed with the respective restriction enzymes and then ligated with pTCE1 digested with XbaI/BglII. Plasmid pTCE15, harboring the PsleB promoter to the sleB signal sequence coding region plus the signal sequence-less SpoIVH coding region, was constructed by amplifying the DNA segment from −79 to +87 of sleB and the DNA segment from +79 to +515 with primer pairs sleB-FX (5′-TGCTCTAGAAAGGAAAGAGTGTCTAA-3′, the XbaI site is underlined) and sleB-RH (5′-CCCAAGCTTGGCAGAGATCGTTTCAG-3′, the HindIII site is underlined) and IVH-sig-H and IVH-RB, respectively. These PCR products were trimmed with each restriction enzyme and then ligated with pTCE1 digested with XbaI/BglII to obtain the plasmid pTCE15.
Plasmids pTCE12, pTCE13, pTCE14, and pTCE15 were used for the introduction of PykvU-spoIVH, PsspE-spoIVH, PspoIVH-spoIVH Δsignal, and PsleB-signalsleB-spoIVH Δsignal into the thrC locus of B. subtilis strain 168 through a double-crossover event by selecting for erythromycin-resistant transformants to generate strains TS008, TS009, TS014, and TS015, respectively. Proper constructions were verified by PCR and DNA sequencing.
Electron microscopy.
B. subtilis cells that were grown in casein growth medium at 37°C and induced to sporulate by the resuspension method for 6 h were collected by centrifugation. Transmission electron micrographs were taken at UltraStructure Research Laboratories (Kanagawa, Japan). Samples were prefixed in 2% (wt/vol) glutaraldehyde in 0.1 M phosphate buffer (pH 7.4), fixed with 1% osmic acid, and successively stained with 2% uranyl acetate. Epoxy Spurr resin (Okenshoji Co., Ltd) was used for embedding the cells. Sections (800 Å) of the cells were prepared with an LKB Co. U5 ultramicrotome (Amersham Pharmacia) and examined with a JEOL Co. JEM 100S electron microscope.
DPA quantification.
The dipicolinic acid (DPA) content in sporulating cells was determined. At hourly intervals until 12 h after the end of log-phase growth (T12) and then subsequently T24, suspended cells and the culture medium were harvested by centrifugation (13,000 × g, 2 min) from 1.5 ml of culture. The pellet was resuspended in 1 ml of sterile distilled water, boiled for 20 min, cooled for 15 min on ice, and then separated by centrifugation at 9,000 × g for 2 min. The supernatant (600 μl) was reacted with 200 μl of 50 mM sodium acetate (25 ml, pH 4.6, adjusted with acetic acid) containing 25 mg of l-cysteine, 0.31 g of FeSO4 · 7H2O, and 80 mg of (NH4)2SO4. The DPA content was determined as the optical density at 440 nm (1).
β-Galactosidase assay.
Activities of β-galactosidase were determined as described by Miller (28) with o-nitrophenyl-β-d-galactopyranoside as the substrate. Enzyme-specific activity is expressed as nanomoles of substrate (o-nitrophenyl-β-d-galactopyranoside) hydrolyzed per milligram per minute.
Immunoblot analysis.
To detect pro-σK and σK by Western immunoblotting, B. subtilis cells were grown in casein growth medium at 37°C and induced to sporulate by the resuspension method (41). Protein samples were extracted from cultures taken at different time points. Samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and analyzed by Western immunoblotting with polyclonal anti-σK serum (12).
Northern hybridization.
Samples of cultures in DSM at 37°C were drawn at intervals, and total RNA was extracted from harvested cells as described previously (17). Total RNA (5 μg) resolved by electrophoresis was blotted onto a positively charged nylon membrane (Hybond N+; Amersham Pharmacia) and hybridized by using digoxigenin-labeled RNA probes (10 ng) according to the manufacturer's instructions (Roche). The following oligonucleotide primers were used to amplify the specific templates for probe generation: ykvU, ykvU-F2 (5′-AATGCCTTGAATTTGTCGTC-3′) and ykvU-T7R (5′-TAATACGACTCACTATAGGGCGAGAAATCAGCACAGCCATGTC-3′); spoIVH, IVH-F1 (5′-ATGTTGACGAAGCGCTTGC-3′) and IVH-T7R (5′-TAATACGACTCACTATAGGGCGACAATCGGAAACGTCAGCTTG-3′).
Primer extension.
Cells were grown in DSM at 37°C and withdrawn at T−1, T3, and T4. Total RNA was extracted from harvested cells as described previously (17). One hundred micrograms of total RNA and 1 pmol of the infrared-dye (IRD)-labeled oligonucleotide primer IVH-EX (5′-GCACCATGACGTCCAAAAATGGAG-3′), which is complementary to the nucleotide sequence of the spoIVH gene and the 3′ end of the primer located 172 nucleotides (nt) downstream from the initiation codon, were mixed and heated at 80°C for 15 min. Samples were incubated at 25°C for 10 min, and then reverse transcription reactions were carried out with 400 U of SuperScript III reverse transcriptase (Invitrogen) at 55°C for 60 min. Reactions were inactivated at 70°C for 15 min and treated with RNaseH. Ethanol-precipitated products were run on 5% polyacrylamide-6 M urea gels with a sequencing ladder. The DNA fragments amplified by PCR with IVH-115 and IVH-RB were sequenced with IVH-EX to generate a sequence ladder. IRD was detected with a LI-COR DNA sequencer model 4200 (Aloka).
Compartmental localization of β-galactosidase activity.
Cells in 200 μl of cultures were sedimented by centrifugation and resuspended into 200 μl of 50 μM fluorescein di-β-d-galactopyranoside (FDG) substrate reagent (Marker Gene Technologies, Inc.). Samples were incubated for 2 min at 37°C, and then FDG loading was terminated by the addition of 900 μl of ice-cold phosphate-buffered saline buffer (100 ml, pH 7.6, containing 0.8 g of NaCl, 0.02 g of KCl, 0.29 g of Na2HPO4 · 12H2O, 0.02 g of KH2PO4, and 0.28 g of HEPES). Cells were placed on ice until examination by fluorescence microscopy (22).
Fluorescence microscopy.
B. subtilis cells incubated in hydrolyzed casein growth medium at 37°C were induced to sporulate by the resuspension method of Sterlini and Mandelstam (41), as specified by Nicholson and Setlow (30) and Partridge et al. (32). The resuspension medium was supplemented with FM4-64 (final concentration, 0.5 μg/ml; Molecular Probes) for staining of the cell membranes. Samples mounted on glass slides coated with 0.1% poly-l-lysine (Sigma) were observed with an Olympus BX50 microscope with a 100× UplanApo objective. Images were captured by using a SenSys charge-coupled device camera (Photometrics). FM4-64 and FDG were visualized by using a wide interference green filter set (Olympus) or a fluorescein isothiocyanate filter set (Olympus) and processed by using Metamorph, version 4.5, software (Universal Image) and Adobe Photoshop, version 4.0.1J.
RESULTS
Disruption of spoIVH (ykvV) blocks stage IV of sporulation.
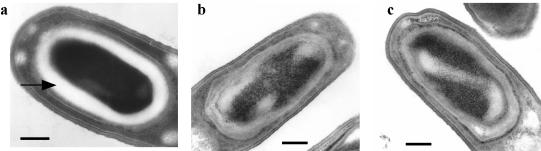
The ykvV (named spoIVH) pMUTIN2MCS insertional mutant YKVVd was identified as a heat-sensitive spore phenotype during a screening of disruptants of genes of unknown function in B. subtilis. Consistent with a recent report (8), the spoIVH mutant TS001 sporulated in DSM at a low frequency of about 0.0001 (Table 2). The forespores were mostly phase dark (data not shown). We further examined the structure of the forespores by electron microscopy. After engulfment is completed, the cortex layer is synthesized at the inner space between double membranes surrounding the forespore in B. subtilis. The wild-type cortex was obvious as a white layer at T6 (Fig. 1a). Although we observed more than 20 cells, the spoIVH mutant did not form a visible cortex layer (Fig. 1b and c), suggesting that the absence of SpoIVH may have some effects on spore cortex formation.
TABLE 2.
Defective sporulation in spoIVH disruptanta
| Strain | Description | No. (CFU/ml) of:
|
Frequencyb | |
|---|---|---|---|---|
| Viable cells | Spores | |||
| 168 | Wild type | 4.7 × 108 | 3.4 × 108 | 0.72 |
| TS001 | spoIVH::pJMIVH | 2.7 × 108 | 1.3 × 104 | 4.8 × 10−5 |
| TS002 | ykvU::pJMVU | 4.8 × 108 | 2.5 × 108 | 0.52 |
Cells were grown in DSM.
Frequency is the ratio of the number of spores to that of viable cells for each strain.
FIG. 1.
Transmission electron micrographs of typical sporulating cells. Wild-type 168 (a) and spoIVH mutant TS001 (b and c) were allowed to sporulate for 6 h after the initiation of sporulation. The arrow indicates the wild-type cortex layer. Bar, 0.2 μm.
DPA accumulation significantly decreased in mutant spores.
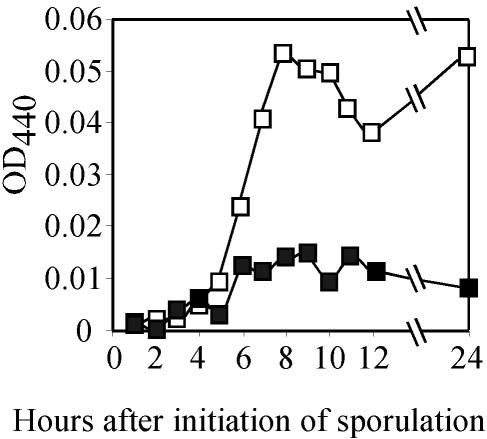
Several previous studies indicate that the accumulation of DPA is impaired in mutants with a defective cortex (1, 2). We therefore proceeded to examine the cortex integrity of the spoIVH mutant by quantifying the DPA content in pellets of centrifuged culture (Fig. 2). We observed significantly less DPA accumulation in the spoIVH mutant than in wild-type cells. Together, these results confirm defective cortex formation in spoIVH mutant spores. Based on these late-stage characteristics, we classified ykvV as a stage IV sporulation gene and renamed it spoIVH.
FIG. 2.
Quantification of DPA. Cultures of 168 (□) and TS001 (▪) were induced to sporulate in resuspension medium. Aliquots were withdrawn at the indicated time points. Time zero (0) corresponds to the initiation of sporulation. The optical density at 440 nm (OD440) directly reflects the DPA concentration.
Effect of spoIVH mutation on the sigma cascade.
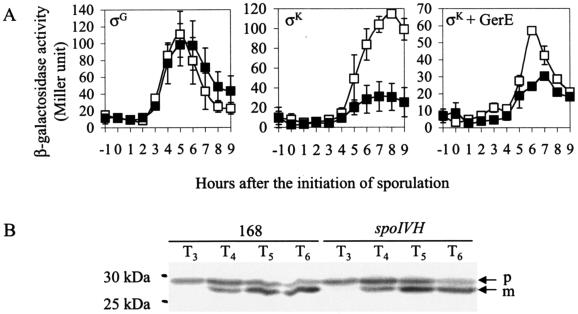
To determine the effect of a spoIVH mutation on the activity of the sporulation-specific RNA polymerase sigma cascade comprising σF, σE, σG, and σK, we compared the expressions of various lacZ fusions in the presence or absence of an intact spoIVH gene. Although σF, σE, and σG activities were normal (Fig. 3A and data not shown), the expression of σK-directed gerE-lacZ and σK-directed GerE-dependent cotD-lacZ was inhibited (Fig. 3A). These results suggest that spoIVH mutation affects the sigma cascade after the activation of forespore-specific σG and partially shuts down mother cell-specific σK activation late in sporulation. The expression of low σK-directed genes and normal σF-, σE-, and σG-directed genes in the spoIVH mutant suggests that the spoIVH gene product is necessary for efficient pro-σK processing.
FIG. 3.
Effects of spoIVH mutation on sigma cascade. Strains carrying lacZ fusions along with intact (□) or disrupted (▪) spoIVH were induced to sporulate, and β-galactosidase activities were assayed. σG, σG-directed sspE-lacZ expression (□, SSPEd; ▪, TS003). σK, σK-directed gerE-lacZ expression (□, REZ; ▪, TS004). σK+GerE, σK-directed and GerE-dependent cotD-lacZ expression (□, TDZ; ▪, TS005). Averages of the results from three or two independent experiments are shown. Error bars represent standard deviations. (B) Western blots of pro-σK processing in strain 168 and the spoIVH mutant (TS001). Cells were induced into sporulation and collected at the indicated time points. Whole-cell extracts were Western blotted with antibody that recognizes σK. p and m indicate pro-σK and mature σK, respectively.
To examine this hypothesis, we Western blotted the spoIVH mutant with anti-σK antibody. Figure 3B shows that both the spoIVH mutant and the wild-type strain contained the mature form of σK from T4 to T6. Densitometry analysis showed that the levels of both pro-σK and σK observed in the wild type and the spoIVH mutant at every examined time point were not significantly different (data not shown). These results indicate that pro-σK processing is not impaired by spoIVH inactivation, suggesting that decreased activity of σK may result from an indirect effect of the spoIVH mutation, most probably caused by an abnormal condition in the mother cell resulting from impaired sporulation.
Transcriptional analysis of spoIVH.
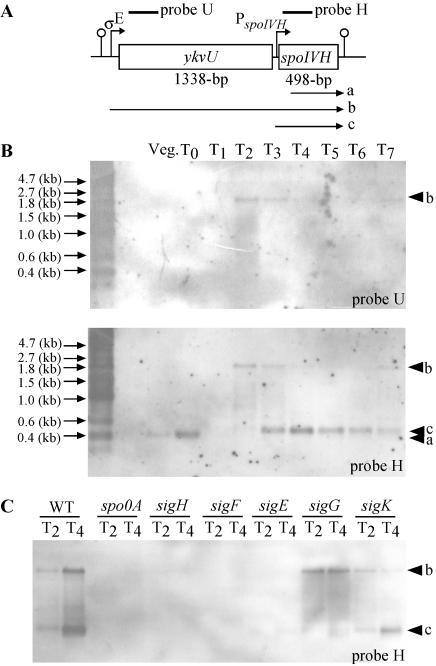
In a recent report, Eichenberger et al. (8) and Feucht et al. (11) indicated that ykvU, located upstream of spoIVH (Fig. 4A), and spoIVH were cotranscribed by EσE by microarray analysis. In addition, Eichenberger et al. (8) identified a transcriptional start site of the ykvU-spoIVH operon and found a promoter very similar to the consensus sequence recognized by EσE, 5′-Ata-16 bp-cATAcanT-3′ (15), in the −27 to −56 segment of ykvU (8). However, plasmid integration in ykvU, whose mutation is expected to prevent the expression of spoIVH, did not abolish the sporulation efficiency (Table 2). These findings indicate that there may be a promoter located just upstream of spoIVH. To investigate the transcription of the ykvU and spoIVH genes, we examined RNA synthesis by Northern blotting of RNA extracted from growing and sporulating cells (Fig. 4B). The ykvU gene probe detected a band at approximately 2.0 kb (band b, T2 and T3), corresponding to the predicted length of a transcript initiated at the ykvU promoter and terminating at the putative terminator located downstream of the spoIVH coding region. The spoIVH gene-specific probe detected bands of about 2.0 kb (T2 to T3), 0.5 kb (T3 to T7), and 0.4 kb (vegetative cells and T0). The largest band corresponded to band b, which was detected with the ykvU probe. A smaller band (band c) corresponded to the predicted length of a transcript initiated upstream of spoIVH and terminating in a stem and loop structure at the end of the ykvU-spoIVH transcriptional unit. The smallest band (band a), detected at the vegetative stage and at T0, seems slightly shorter than band c and the spoIVH gene (498 bp). Probably, the band a signal is not specific for spoIVH mRNA. However, even if spoIVH is expressed in the vegetative phase and is functional, it may be one of several similar proteins including paralogous genes, so its inactivation had little or no effect on growth.
FIG. 4.
Transcriptional analysis of spoIVH region. (A) Arrangement of spoIVH and upstream gene ykvU. Bars indicate the positions of sequences corresponding to the Northern blotting probe. Loops with lines indicate putative terminators. Arrows with lines indicate (putative) promoters. Arrows under the physical map indicate observed mRNA. (B) Northern blots of whole RNA extracted from wild-type 168 cells. Arrows indicate positions of molecular size markers. Cells were grown on DSM and harvested at the indicated time points. (C) Northern blot of whole RNA extracted from 168 with disruptions of the indicated genes. WT, 168; spo0A, TF97; sigH, TF85; sigF, TF83; sigE, TF82; sigG, TF84; sigK, TF99; arrowheads, mRNA signals; Veg., vegetative cells.
To determine the dependence of spoIVH expression, we analyzed transcripts from the two promoters that function in the sporulation phase at T2 and T4 in spo0A (Spo0A), spo0H (σH), spoIIAC (σF), spoIIGB (σE), spoIIIG (σG), and spoIIIC (σK) mutant backgrounds (Fig. 4C). A band (band b) was detected in the wild type and the spoIIIG and spoIIIC mutants but not in the spo0A, spo0H, spoIIAC, and spoIIGB mutants. The transcript (band c) was detected only in the wild type and in the spoIIIC mutant (Fig. 4C). These results indicate that spoIVH is transcribed together with ykvU from T2 to T3 by EσE from the promoter located upstream of ykvU, consistent with previous reports (8, 11), and transcribed monocistronically from T3 under EσG control from promoter PspoIVH, indicated in Fig. 4A.
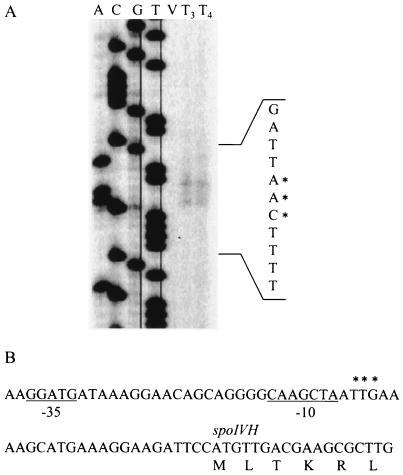
To determine the 5′ end of spoIVH mRNA transcribed by EσG, we carried out the primer extension analysis with total RNA extracted from strain 168 (Fig. 5A). Although the transcription start site was not detected at the vegetative phase, it was located 23 to 25 nt upstream of the initiation codon of spoIVH at T3 and T4. Consistent with our results, we found a region highly similar to the consensus sequence recognized by σG, 5′-gnATA/G-18 bp-cAtnnTA-3′ (15), in the spoIVH segment from −27 to −56 from its initiation codon (Fig. 5B). An extension reaction was primed from 172 nt downstream of the initiation codon of spoIVH with IRD-labeled primer IVH-EX. However, a vegetative-phase-specific extension product was not detected (data not shown), suggesting that band a, detected as shown in Fig. 4B, may not be specific for spoIVH mRNA.
FIG. 5.
Mapping of the transcription start sites of spoIVH by primer extension analysis. (A) Total RNA was prepared from wild-type 168 of exponentially growing cells (lane V) or at T3 (lane T3) or T4 (lane T4). (B) Nucleotide sequence of the upstream region of spoIVH. The regions with similar consensus sequences recognized by σG are underlined. The positions of primer-extended products are indicated with asterisks.
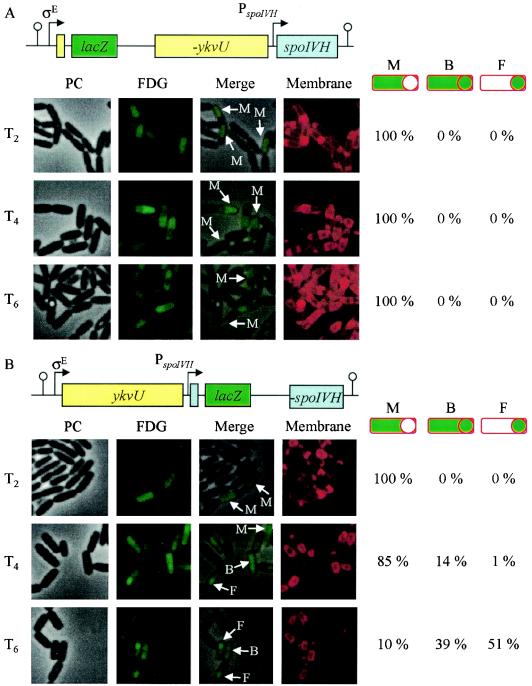
To confirm the compartment localization of spoIVH expression, we used fluorescence microscopy and the β-galactosidase sensitive substrate FDG, which can show compartment expression of the lacZ gene (22). Fluorescence from FDG in spoIID-lacZ (σE) and sspE-lacZ (σG) strains was detected in the mother cell and in the forespore compartment, respectively (data not shown). No fluorescence from FDG was detected under our conditions in the wild-type strain (data not shown). In the ykvU and spoIVH insertional mutants with the pMUTIN2MCS vector, the lacZ gene was integrated into each of the genes, meaning β-galactosidase is expressed from the promoter for ykvU and spoIVH. Figure 6 shows that FDG fluorescence was detected in the mother cell at T2 in both strains. At T4, in the strain carrying ykvU-lacZ (YKVUd), fluorescence was still detectable in the mother cells (Fig. 6A) and in both compartments of the ykvU-spoIVH-lacZ (YKVVd) strain (Fig. 6B). Furthermore, the signal became more intense in the forespore than in the mother cell at T6 in the ykvU-spoIVH-lacZ strain. The percentage of compartment expression in cells that showed FDG fluorescence is shown to the right of the pictures (Fig. 6). At least 377 cells were counted in each sample. In the ykvU-lacZ strain, although the number of cells showing FDG green fluorescence decreased from T2 to T6 (35, 22, and 17% of cells at T2, T4, and T6, respectively), FDG fluorescence was observed only in the mother cell compartment at every observed time point. In contrast, ykvU-spoIVH-lacZ expression was detected only in the mother cell at T2; however, it was observed in both compartments in 14 and 39% of cells at T4 and T6, respectively. In the ykvU-spoIVH-lacZ strain, FDG fluorescence was observed only in the forespore in 51% of cells at T6. These results suggest that ykvU and spoIVH are expressed only in the mother cell early in sporulation; however, spoIVH alone is expressed in the forespore during late sporulation.
FIG. 6.
Compartmental expression of spoIVH. Strains YKVUd (A) and YKVVd (B) were induced to sporulate, and after the indicated time, samples were prepared for fluorescence microscopy and stained with FDG. PC, phase contrast; FDG, FDG fluorescent signal; Merge, merge of PC and FDG; Membrane, FM4-64-stained membrane. Arrows indicate typical cells. M, F, and B indicate the cells in which the FDG signal was observed in the mother cell, forespore, and both compartments, respectively. Cells were induced to sporulation by the resuspension method and observed at the indicated time points. Ratios of compartmental expression are shown to the right of the pictures. At least 377 cells were counted at the indicated time points. This number did not include cells that showed no fluorescence.
Alternative expression of spoIVH.
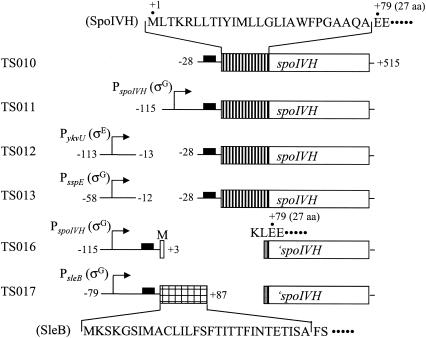
To determine the essentiality of spoIVH expression in the forespore and mother cell for efficient sporulation, we constructed strains in which spoIVH is transcribed in the forespore only or in the mother cell only (Fig. 7). Although strain TS010, which has intact spoIVH without promoter, did not support wild-type levels of sporulation (frequency of 0.0017), the expression of spoIVH from the putative σG promoter located just upstream of spoIVH was completely sufficient for proper spore formation (0.81 in TS011), as shown in Table 3. This is consistent with the observation that the ykvU insertional mutant sporulated normally. In the event of a polar effect, the transcription of spoIVH from ykvU may be blocked in the ykvU mutant and the expression of spoIVH would be effected only from its own promoter. Interestingly, the sporulation deficiency of the spoIVH mutant strain was compensated completely (0.94) or partially (0.13) by the introduction of the σG (PsspE, TS013)- and σE (PykvU, TS012)-directed spoIVH genes, respectively (Table 3). These results suggest that spoIVH is capable of contributing to spore formation from both sporulating cell compartments. It is, however, quite puzzling how spoIVH functions from both compartments.
FIG. 7.
Schematic representation of strains used in experiments analyzed in Tables 3 and 4. The putative Sec-type signal sequence in SpoIVH is hatched. The Sec-type signal sequence of SleB is checked. Black boxes indicate putative ribosome binding sites. Arrows with lines indicate promoters of genes and are shown to the left. spoIVH at the native locus was disrupted in all strains. The amino acid sequences of signal peptides and flanking regions are indicated. The peptide of KL was encoded by the HindIII restriction site (gray boxes) in TS016 and TS017. The positions of DNA fragments relative to the start codons of the original genes are indicated at the ends of the fragments.
TABLE 3.
Alternative expression of spoIVH in mother cell or foresporea
| Strain | Original spoIVH | Promoter for spoIVH in thrC | No. (CFU/ml) of:
|
Frequencyb | |
|---|---|---|---|---|---|
| Viable cells | Spores | ||||
| 168 | Intact | 2.8 × 108 | 2.0 × 108 | 0.71 | |
| TS001 | Disrupted | 2.0 × 108 | 1.3 × 104 | 6.5 × 10−5 | |
| TS010 | Disrupted | No promoter | 1.1 × 108 | 1.9 × 105 | 1.7 × 10−3 |
| TS011 | Disrupted | spoIVH | 2.1 × 108 | 1.7 × 108 | 0.81 |
| TS012 | Disrupted | ykvU (σE) | 1.2 × 108 | 1.5 × 107 | 0.13 |
| TS013 | Disrupted | sspE (σG) | 1.8 × 108 | 1.7 × 108 | 0.94 |
Cells were grown in DSM.
Frequency is the ratio of the number of spores to that of viable cells for each strain.
We could not detect SpoIVH localization by using the SpoIVH-GFP strain, probably due to the low level of spoIVH expression. However, a peptide in the N-terminal region (1 to 26 aa) of SpoIVH closely resembles the Sec-type signals, which are involved in one of the major pathways for translocation in B. subtilis (45). Furthermore, the C-terminal region of the SpoIVH signal sequence contains a consensus amino acid sequence A-X-A, which serves as an SPase I cleavage site (44). The sleB gene, which encodes a putative spore-cortex-lytic enzyme, is translocated across the forespore inner membrane by a secretion Sec-type signal peptide and is deposited in the cortex layer synthesized between the forespore inner and outer membranes (29). To investigate the importance of the signal peptide for SpoIVH function, the region encoding the putative signal peptide (1 to 26 aa) of the SpoIVH protein was removed (TS016) or substituted (TS017) with a fragment (−79 to +87) containing the σG-recognized promoter region of sleB to the region encoding the signal peptide (1 to 29 aa) of the SleB protein (Fig. 7). As shown in Table 4, in the mutant strain without the SpoIVH signal sequence (TS016), as well as in the spoIVH single mutant (TS001), sporulation was inhibited, indicating that the signal sequence of SpoIVH is indispensable for SpoIVH to function efficiently. In contrast, in the other spoIVH mutant strain (TS017) in which the SleB signal domain replaced the signal sequence of the SpoIVH protein, the sporulation efficiency compared to that of the wild-type strain was not significantly different, indicating that the SpoIVH signal domain is functionally similar to that of the SleB protein. These results suggest that SpoIVH could act in the inner space between the double membranes where the cortex is formed.
TABLE 4.
Sporulation by fused protein of SleB signal sequence and SpoIVHa
| Strain | Description | No. (CFU/ml) of:
|
Frequencyb | |
|---|---|---|---|---|
| Viable cells | Spores | |||
| 168 | Wild type | 2.8 × 108 | 2.0 × 108 | 0.71 |
| TS001 | spoIVH::pJMIVH | 2.0 × 108 | 1.3 × 104 | 6.5 × 10−5 |
| TS016 | spoIVHΔsignalc | 1.0 × 108 | 1.5 × 104 | 1.5 × 10−4 |
| TS017 | PsleB-signalsleB- spoIVHΔsignalc | 2.9 × 108 | 1.5 × 108 | 0.52 |
Cells were grown in DSM.
Frequency is the ratio of the number of spores to that of viable cells for each strain.
Native spoIVH was disrupted in strains TS016 and TS017.
DISCUSSION
Our work reveals a remarkable aspect of spoIVH as a gene that is expressed not only in the mother cell by EσE but also in the forespore under the control of EσG. The strain with σE-directed spoIVH has a somewhat reduced sporulation efficiency relative to that of the strain with σG-directed spoIVH, suggesting a greater functional significance of the forespore-specific expression of spoIVH than that of the mother cell. However, its ability to produce a number of viable spores in either strain with the σE- and σG-directed spoIVH gene was 2 to 3 orders of magnitude greater than that produced by the strain with spoIVH lacking its promoter (Table 3). This result demonstrates that both the SpoIVH proteins produced in the forespore and the mother cell have a role in sporulation. We also showed that the SpoIVH protein possesses an irremovable N-terminal signal sequence, composed of 26 aa, but that the SpoIVH protein with a substitution of the SleB signal domain in place of its signal sequence was functional. We therefore conclude that the mature SpoIVH is primarily localized in the inner space between the double membranes where the cortex is formed.
How spoIVH acquired such a dual control system, however, is quite intriguing. However, ykvU is one of the paralogous genes of spoVB, with a BLAST score of 180, which is expressed by EσE (35). It appears that ykvU may have moved to the present position from the spoVB region through transposition. Presumably, spoIVH may have acquired this transcription system under the dual control of σE and σG from the spoVB gene through evolution. spoIVH is also known to belong to the AhpC/thiol-specific antioxidant protein family (http://bacillus.genome.ad.jp/) and is paralogous to trxA (thioredoxin) and resA (thiol-disulfide oxidoreductase), with BLAST scores of 40 and 89, respectively. Proteins of this family participate in reduction and are widely conserved (3). SpoIVH may act as a thiol-specific antioxidant or thiol/disulfide bond interchange protein during sporulation. Schiott and Hederstadt (40) have reported that the CcdA protein, which is required for c-type cytochrome synthesis, is also required for the late stage of sporulation in B. subtilis. Erlendsson and Hederstedt (9) also speculated that CcdA is related to the role of SpoIVH. If the SpoIVH protein has a disulfide bond isomerase activity that modifies the tertiary structure of some protein(s) required for cortex formation, it may be possible that SpoIVH plays a significant role in the thiol-disulfide exchange between cysteine residues of proteins in the inner double membrane space. Among products of the many identified cortex formation genes, SpoVB, SpoVD, SpoVE, and YabQ have plural numbers of cysteine residues and membrane spanning domains, suggesting that they may be targets for SpoIVH.
In addition, it is possible that spoIVH is important for maintaining the redox state of some protein(s) in the space between the mother cell and the forespore. Presumably, the redox states of many proteins in the space differ relative to the mother cell and the forespore. If the SpoIVH protein has an antioxidant enzymatic activity, it may be altered to compensate for the reduction in the space between the inner and outer forespore membranes; thus, the activities of various proteins involved in spore cortex formation may be impaired in the spoIVH mutant. Since there is little detail on cortex formation in the space, further investigation is required to understand the activity of SpoIVH and reveal the target of SpoIVH during sporulation.
Acknowledgments
We thank the Japanese and European Consortia for Functional Analysis of the B. subtilis Genome for providing the pMUTIN strains. We especially thank Richard Losick for providing B. subtilis strains, Masaya Fujita for providing the σK antibody, Hideaki Nanamiya, Sawako Yoshida, and Fujio Kawamura for assistance with the primer extension analysis, and Samuel Amiteye for critically reading the manuscript.
This study was supported by a grant-in-aid for scientific research on the priority area Genome Biology from the Ministry of Education, Science, Sports, and Culture of Japan.
REFERENCES
- 1.Beall, B., and C. P. Moran, Jr. 1994. Cloning and characterization of spoVR, a gene from Bacillus subtilis involved in spore cortex formation. J. Bacteriol. 176:2003-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Catalano, F. A., J. Meador-Parton, D. L. Popham, and A. Driks. 2001. Amino acids in the Bacillus subtilis morphogenetic protein SpoIVA with roles in spore coat and cortex formation. J. Bacteriol. 183:1645-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chae, H. Z., K. Robinson, L. B. Poole, G. Church, G. Storz, and S. G. Rhee. 1994. Cloning and sequencing of thiol-specific antioxidant from mammalian brain: alkyl hydroperoxide reductase and thiol-specific antioxidant define a large family of antioxidant enzymes. Proc. Natl. Acad. Sci. USA 91:7017-7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cutting, S., A. Driks, R. Schmidt, B. Kunkel, and R. Losick. 1991. Forespore-specific transcription of a gene in the signal transduction pathway that governs pro-σK processing in Bacillus subtilis. Genes Dev. 5:456-466. [DOI] [PubMed] [Google Scholar]
- 5.Cutting, S., S. Roels, and R. Losick. 1991. Sporulation operon spoIVF and the characterization of mutations that uncouple mother-cell from forespore gene expression in Bacillus subtilis. J. Mol. Biol. 221:1237-1256. [DOI] [PubMed] [Google Scholar]
- 6.Cutting, S., V. Oke, A. Drinks, R. Losick, S. Lu, and L. Kroos. 1990. A forespore checkpoint for mother cell gene expression during development in Bacillus subtilis. Cell 62:239-250. [DOI] [PubMed] [Google Scholar]
- 7.Dubnou, D., and R. Davidoff-Abelson. 1971. Fate of transforming DNA following uptake by competent Bacillus subtilis. Formation and properties of the donor-recipient complex. J. Mol. Biol. 56:209-221. [DOI] [PubMed] [Google Scholar]
- 8.Eichenberger, P., S. T. Jensen, E. M. Conlon, C. V. Ooij, J. Silvaggi, J.-E. Gonzalez-Pastor, M. Fujita, S. Ben-Yehuda, P. Stragier, J. S. Liu, and R. Losick. 2003. The σE regulon and the identification of additional sporulation genes in Bacillus subtilis. J. Mol. Biol. 327:945-972. [DOI] [PubMed] [Google Scholar]
- 9.Erlendsson, L. S., and L. Hederstedt. 2002. Mutations in the thiol-disulfide oxidoreductases BdbC and BdbD can suppress cytochrome c deficiency of CcdA-defective Bacillus subtilis cells. J. Bacteriol. 184:1423-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Errington, J. 1993. Bacillus subtilis sporulation: regulation of gene expression and control of morphogenesis. Microbiol. Rev. 57:1-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feucht, A., L. Evans, and J. Errington. 2003. Identification of sporulation genes by genome-wide analysis of the σE regulon of Bacillus subtilis. Microbiology 149:3023-3034. [DOI] [PubMed] [Google Scholar]
- 12.Fujita, M. 2000. Temporal and selective association of multiple sigma factors with RNA polymerase during sporulation in Bacillus subtilis. Genes Cells 5:79-88. [DOI] [PubMed] [Google Scholar]
- 13.Gerhardt, P., and R. E. Marquis. 1989. Spore thermoresistance mechanisms, p. 43-63. In I. Smith, R. A. Slepecky, and P. Setlow (ed.), Regulation of prokaryotic development. American Society for Microbiology, Washington, D.C.
- 14.Harry, E. J., K. Pogliano, and R. Losick. 1995. Use of immunofluorescence to visualize cell-specific gene expression during sporulation in Bacillus subtilis. J. Bacteriol. 177:3386-3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Helmann, J. D., and C. P. Moran, Jr. 2002. RNA polymerase and sigma factors, p. 289-312. In A. L. Sonenshein, J. H. Hock, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. American Society for Microbiology, Washington, D.C.
- 16.Horinouchi, S., and B. Weisblum. 1982. Nucleotide sequence and functional map of pE194, a plasmid that specifies inducible resistance to macrolide, lincosamide, and streptogramin type B antibiotics. J. Bacteriol. 150:804-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Igo, M. M., and R. Losick. 1989. Regulation of a promoter that is utilized by minor forms of RNA polymerase holoenzyme in Bacillus subtilis. J. Mol. Biol. 191:615-624. [DOI] [PubMed] [Google Scholar]
- 18.Ishiwa, H., and H. Shibahara. 1985. New shuttle vectors for Escherichia coli and Bacillus subtilis. II. Plasmid pHY300PLK, a multipurpose cloning vector with a polylinker, derived from pHY460. Jpn. J. Genet. 60:235-243. [Google Scholar]
- 19.Karow, M. L., P. Glaser, and P. J. Piggot. 1995. Identification of a gene, spoIIR, that links the activation of σE to the transcriptional activity of σF during sporulation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 92:2012-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kroos, L., B. Kunkel, and R. Losick. 1989. Switch protein alters specificity of RNA polymerase containing a compartment-specific sigma factor. Science 243:526-529. [DOI] [PubMed] [Google Scholar]
- 21.Kunst, F., N. Ogasawara, I. Moszer, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 22.Lewis, P. J., S. R. Partridge, and J. Errington. 1994. σ factors, asymmetry, and the determination of cell fate in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 91:3849-3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, Z., and P. J. Piggot. 2001. Development of a two-part transcription probe to determine the completeness of temporal and spatial compartmentalization of gene expression during bacterial development. Proc. Natl. Acad. Sci. USA 98:12538-12543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Losick, R., P. Youngman, and P. J. Piggot. 1986. Genetics of endospore formation in Bacillus subtilis. Annu. Rev. Genet. 20:625-669. [DOI] [PubMed] [Google Scholar]
- 25.Losick, R., and P. Stragier. 1992. Crisscross regulation of cell-type-specific gene expression during development in Bacillus subtilis. Nature 355:601-604. [DOI] [PubMed] [Google Scholar]
- 26.Lu, S., R. Halberg, and L. Kroos. 1990. Processing of the mother-cell σ factor, σK, may depend on events occurring in the forespore during Bacillus subtilis development. Proc. Natl. Acad. Sci. USA 87:9722-9726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu, S., S. Cutting, and L. Kroos. 1995. Sporulation protein SpoIVFB from Bacillus subtilis enhances processing of the sigma factor precursor Pro-σK in the absence of other sporulation gene products. J. Bacteriol. 177:1082-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller, J. H. 1972. Experiments in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 29.Moriyama, R., H. Fukuoka, S. Miyata, S. Kudoh, A. Hattori, S. Kozuka, Y. Yasuda, K. Tochikubo, and S. Makino. 1999. Expression of a germination-specific amidase, SleB, of bacilli in the forespore compartment of sporulating cells and its localization on the exterior side of the cortex in dormant spores. J. Bacteriol. 181:2373-2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicholson, W. L., and P. Setlow. 1990. Sporulation, germination and outgrowth, p. 391-450. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. Wiley, Chichester, United Kingdom.
- 31.Ogasawara, N. 2000. Systematic function analysis of Bacillus subtilis genes. Res. Microbiol. 151:129-134. [DOI] [PubMed] [Google Scholar]
- 32.Partridge, K. L., J. K. Grimsley, and J. Errington. 1993. The importance of morphological events and intercellular interactions in the regulation of respore-specific gene expression during sporulation in Bacillus subtilis. Mol. Microbiol. 8:945-955. [DOI] [PubMed] [Google Scholar]
- 33.Perego, M. 1993. Integrational vectors for genetic manipulation in Bacillus subtilis, p. 615-624. In A. L. Sonenshein, J. H. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria. American Society for Microbiology, Washington, D.C.
- 34.Piggot, P. J., and J. G. Coote. 1976. Genetic aspects of bacterial endospore formation. Bacteriol. Rev. 40:908-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Popham, D. L., and P. Stragier. 1991. Cloning, characterization, and expression of the spoVB gene of Bacillus subtilis. J. Bacteriol. 173:7942-7949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ricca, E., S. Cutting, and R. Losick. 1992. Characterization of bofA, a gene involved in intercompartmental regulation of pro-σK processing during sporulation in Bacillus subtilis. J. Bacteriol. 174:3177-3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 38.Sato, T., K. Harada, and Y. Kobayashi. 1996. Analysis of suppressor mutations of spoIVCA mutations: occurrence of DNA rearrangement in the absence of a site-specific DNA recombinase SpoIVCA in Bacillus subtilis. J. Bacteriol. 178:3380-3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schaeffer, P., J. Millet, and J. P. Aubert. 1965. Catabolic repression of bacterial sporulation. Proc. Natl. Acad. Sci. USA 54:704-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schiott, T., and L. Hederstadt. 2000. Efficient spore synthesis in Bacillus subtilis depends on the CcdA protein. J. Bacteriol. 182:2845-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sterlini, J. M., and J. Mandelstam. 1969. Commitment to sporulation on Bacillus subtilis and its relationship to the development of actinomycin resistance. Biochem. J. 113:29-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stragier, P., and R. Losick. 1996. Molecular genetics of sporulation in Bacillus subtilis. Annu. Rev. Genet. 30:297-341. [DOI] [PubMed] [Google Scholar]
- 43.Strom, M. S., and S. Lory. 1986. Cloning and expression of the pilin gene of Pseudomonas aeruginosa PAK in Escherichia coli. J. Bacteriol. 165:367-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tjalsma, H., A. G. Stöer, A. Driks, G. Venema, S. Bron, and J. M. van Dijl. 2000. Conserved serine and histidine residues are critical for activity of the ER-type signal peptidase SipW of Bacillus subtilis. J. Biol. Chem. 275:25102-25108. [DOI] [PubMed] [Google Scholar]
- 45.Van Dijl, J. M., A. Bolhuis, H. Tjalsma, J. D. H. Jongbloed, A. D. Jone, and S. Bron. 2002. Protein transport pathways in Bacillus subtilis: a genome-based road map, p. 337-355. In A. L. Sonenshein, J. H. Hock, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. American Society for Microbiology, Washington, D.C.
- 46.Wakeley, P. R., R. Dorazi, N. T. Hoa, J. R. Bowyer, and S. M. Cutting. 2000. Proteolysis of SpoIVB is a critical determinant in signaling of pro-σK processing in Bacillus subtilis. Mol. Microbiol. 36:1336-1348. [DOI] [PubMed] [Google Scholar]
- 47.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vector and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]