Abstract
F+ strains of Escherichia coli infected with donor-specific bacteriophage such as M13 are sensitive to bile salts. We show here that this sensitivity has two components. The first derives from secretion of bacteriophage particles through the cell envelope, but the second can be attributed to expression of the F genes required for the formation of conjugative (F) pili. The latter component was manifested as reduced or no growth of an F+ strain in liquid medium containing bile salts at concentrations that had little or no effect on the isogenic F− strain or as a reduced plating efficiency of the F+ strain on solid media; at 2% bile salts, plating efficiency was reduced 104-fold. Strains with F or F-like R factors were consistently more sensitive to bile salts than isogenic, plasmid-free strains, but the quantitative effect of bile salts depended on both the plasmid and the strain. Sensitivity also depended on the bile salt, with conjugated bile salts (glycocholate and taurocholate) being less active than unconjugated bile salts (deoxycholate and cholate). F+ cells were also more sensitive to sodium dodecyl sulfate than otherwise isogenic F− cells, suggesting a selectivity for amphipathic anions. A mutation in any but one F tra gene required for the assembly of F pili, including the traA gene encoding F pilin, substantially restored bile salt resistance, suggesting that bile salt sensitivity requires an active system for F pilin secretion. The exception was traW. A traW mutant was 100-fold more sensitive to cholate than the tra+ strain but only marginally more sensitive to taurocholate or glycocholate. Bile salt sensitivity could not be attributed to a generalized change in the surface permeability of F+ cells, as judged by the effects of hydrophilic and hydrophobic antibiotics and by leakage of periplasmic β-lactamase into the medium.
Normally, coliform bacteria are resistant to noxious agents, such as bile salts, likely to be encountered at high levels in the mammalian digestive tract. Resistance is the result of the synergistic effects of the permeability barrier established by the outer membrane of gram-negative cells and broad-specificity efflux pumps that capture and expel toxic molecules that do penetrate the outer membrane (38-40). The number of genes and the amount of metabolic energy devoted to these defenses reflect the magnitude of the selective pressures exerted by exposure to these agents.
We report here that the type IV secretion system encoded by the F plasmid can render F+ cells sensitive to certain anionic detergents, including bile salts and sodium dodecyl sulfate (SDS). Type IV secretion systems of gram-negative bacteria are broadly distributed, versatile, macromolecular transporters (9, 10, 12, 29). One subclass of these systems mediates conjugal DNA transfer among bacterial cells or between bacterial and eukaryotic cells. This subclass is distinguished by a requirement for conjugative pili, which are extended surface filaments that mediate the initial cell-cell contact stages of DNA transfer (42).
The type IV secretion system encoded by F is the archetype of one class of conjugal DNA transfer systems (18, 20, 29). Of the 26 F-encoded Tra proteins, 19 localize or are predicted to localize to the cell envelope (inner membrane, outer membrane, or periplasm). Of these, 15 are required for the assembly of functional F pili or to regulate F pilus number and length distributions (18). A 16th gene, traC, is also required for F pilus assembly as a peripheral inner membrane protein (45). A working hypothesis is that these proteins assemble and function in concert at the cell surface to mediate the formation and function of F pili, and presumably DNA transfer as well (24, 46). Such assemblies are probably common to all type IV secretion systems (10, 12), and their composition, structure, and function(s) are of considerable interest.
Previous studies have hinted that the presence of F or F-like R factors sensitizes Escherichia coli to anionic detergents, such as bile salts or SDS, among other compounds (2, 53). In this report, we confirm and extend these observations. We show that F+ strains of E. coli are sensitive to bile salts at concentrations that do not affect isogenic F− strains. By genetic criteria, we show that sensitivity can be attributed to genes required for the elaboration of functional F pili. We propose that sensitivity occurs when F pilin secretion transiently opens the cell envelope to the surrounding medium, allowing entry of the anionic detergents. Elsewhere, we have identified the Tra proteins primarily responsible for this activity and have proposed additional features of their structure and function (P. M. Silverman et al., unpublished data).
MATERIALS AND METHODS
Bacterial strains and bacteriophage.
All strains used in this study are derivatives of E. coli K-12. JC3272 (1) and CC118 (34) have been described elsewhere. AE2086 is a spontaneous Nalr derivative, and M1174 is a His+ recombinant (5) of JC3272. A529 (tolA529), D21 (rfa+), and the deep rough mutant D21f2 (rfa-1 rfa-3) were acquired from the E. coli Genetic Stock Center at Yale University. F plasmids are derivatives of JCFL0 (1) or F′13-1 (35) (both F′ lac), or of pOX38, a tra+ F replicon lacking the transposable sequences of F itself (23); these plasmids are assumed to be isogenic at all tra loci. R100 and R100drd1 (4) were from our laboratory stocks, as was bacteriophage M13K07 (48). To construct strain AE2086/M13K07, an isolate of HfrH infected with the bacteriophage was grown to stationary phase. M13K07 phagemid DNA was isolated and used to transform AE2086, selecting for kanamycin-resistant colonies. These were tested for M13K07 production by plaque assay of supernatant fluid from overnight cultures on HfrH host bacteria.
To construct the traB::cat allele used in these studies, the entire traB gene was amplified from strain RD17/pOX38 (tra+) using primers CCTGCCAGCCAAGCTTACTGGCAGG (tra nucleotides [nt] 4175 to 4197) and CATACCGGGCGGGATCCCTGGCACGCC (tra nt 5708 to 5686) (tra nucleotide numbering is from reference 20). HindIII and BamHI sites were introduced into the respective primers. Amplified DNA was digested with both enzymes and cloned into pUC19 also digested with both enzymes. Most of traB was removed from pUCtraB by digestion with NaeI and SphI. Ends were blunted by digestion with mung bean nuclease. The 875-bp cat fragment was isolated from pBR325 (7), digested with TaqI, and blunt ended with Klenow fragment. After ligation, Camr Ampr transformants of AE2086 were isolated and the structure of pUCtraB::cat plasmids was confirmed by restriction digestion and DNA sequence analyses. To cross the traB::cat allele onto F, AE2086/pUCtraB::cat (optical density at 600 nm [OD600] = 0.5) and an equal volume of JC3272/JCFL0 at the same density were incubated together for 60 min at 37°C. One OD unit of AE2248 (thr-34::Tn10) was then added, and incubation continued for an additional 60 min. Tetr Camr Amps Lac+ transconjugants were isolated and tested for complementation by a traB+ plasmid. DNA sequence analysis confirmed the cat insertion in traB, Western blotting failed to detect material cross-reacting with anti-TraB antibodies (24), and donor functions were substantially restored by transformation with a traB+ plasmid.
Bacteria were routinely grown in Luria-Bertani (LB) medium supplemented with antibiotics where appropriate. Cultures were incubated at 37°C in 5 ml of medium using a New Brunswick TC-6 roller drum set at maximum speed. Solid media were prepared with 1.5% agar.
Assays for bile salt and SDS sensitivity.
Difco bile salts no. 3 (Becton-Dickinson and Co., Sparks, Md.) and individual bile salts (Sigma Chemical Co., St. Louis, Mo.) were prepared in water as 10 or 25% solutions (wt/vol) and filter sterilized. SDS (10%, wt/vol) was prepared the same way. Appropriate dilutions (2.5 ml) were distributed in test tubes with an equal volume of 2× LB medium. Overnight cultures were diluted to about 104 cells/ml, and 0.1 ml was added to each tube. Cultures were incubated for 18 h before the OD600 was read. In some figures, data were normalized to the OD of the culture incubated without bile salts or SDS.
To compare plating efficiencies, overnight cultures were diluted and 0.1-ml aliquots of the appropriate dilutions were spread on agar plates containing bile salts no. 3 at the levels indicated for individual experiments.
Antibiotic sensitivity.
Overnight cultures were used to prepare agar overlays, as for a bacteriophage assay. Antibiotic disks (BD-BBL, obtained from Fisher Scientific Corp.) were placed on the surface, and the plates were incubated for 18 h at 37°C.
β-Lactamase leakage.
Leakage was estimated by cross-protection and serial dilution assays (6). Cross-protection was determined by transforming appropriate strains with pUC18 DNA and selecting for ampicillin-resistant (100 μg of ampicillin/ml) transformants. Areas around transformant colonies were inspected for satellite colonies of sensitive cells. Relative β-lactamase activity in culture supernatants was estimated on lawns of ampicillin-sensitive bacteria (JC3272). Disks containing 10 μg of ampicillin were deposited on agar overlays of JC3272 bacteria. Immediately thereafter, dilutions of culture supernatants (30 μl) were deposited on the disks. After overnight incubation at 37°C, the plates were inspected to determine the highest dilution that gave complete resistance. The transition from resistance to sensitivity occurred over a twofold dilution of a given sample.
RESULTS
Effect of bile salts no. 3 on M13-infected cells.
We initially sought a genetic selection for F+ cells that are unable to elaborate functional F pili. Since F pili are required for infection by filamentous DNA bacteriophage, such as M13 and f1, and since it is well-established that f1 infection sensitizes cells to bile salts (6, 44), among other agents, we reasoned that, in a mixture of infected and uninfected cells, only the latter would form colonies on media containing bile salts. We were in fact able to achieve selectivity of more than 3 orders of magnitude in reconstruction experiments with F− cells and HfrH infected with M13K07 plated on 1% bile salts no. 3 (data not shown).
In the course of these experiments, we noticed that as the bile salt concentration was increased, the colony size of uninfected HfrH became smaller. We did not observe the same effect of bile salts no. 3 on the F− strain. These observations suggested that F itself might sensitize cells to bile salts independently of filamentous DNA bacteriophage infection. Yoshida et al. (53) earlier reported that cells with an R factor were more sensitive to cholate than R− cells.
Differential effect of bile salts no. 3 on F+ and F− strains of E. coli.
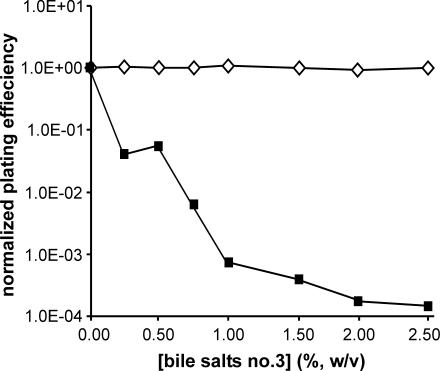
We first tested F− strain JC3272 and the same strain carrying JCFL0, an F′ lac plasmid. Both the strain and the plasmid were used in the earliest genetic studies of F tra genes (1, 26, 52) and in many other studies since. We observed that, whereas the plating efficiency and colony size of JC3272 were unaltered as a function of bile salt concentration up to at least 2.5%, the plating efficiency of the same strain with JCFL0 diminished drastically with increasing bile salt levels (Fig. 1). At 2% bile salts no. 3, the plating efficiency was reduced 4 orders of magnitude, comparable to that of M13-infected HfrH.
FIG. 1.
Plating efficiencies of E. coli F− strain JC3272 (⋄) and the F′ lac+ tra+ strain JC3272/JCFL0 (▪) as a function of bile salts no. 3 concentration.
The effects of bile salts no. 3 in liquid culture were similar to those on solid media. In these experiments, we inoculated 5-ml cultures containing different concentrations of bile salts with about 1,000 cells. Culture ODs were measured after 18 h of incubation at 37°C. JC3272 grew to about the same density at bile salt concentrations of up to 2%, whereas JC3272/JCFL0 failed to grow at bile salt concentrations of ≥0.5% (Fig. 2). Note, however, that the sensitivity of this assay is only about 100-fold, whereas plating efficiency can detect much greater differences (Fig. 1).
FIG. 2.
M13 infection and F independently sensitize E. coli JC3272 to bile salts no. 3 in liquid culture. ⋄, JC3272; ○, AE2086/M13K07; □, JC3272/JCFL0.
In order to prove that F+ cells and cells infected with donor-specific, filamentous DNA bacteriophage are each independently sensitized to bile salts, we transformed the F− strain AE2086, a nalidixic acid-resistant derivative of JC3272, with phagemid DNA isolated from HfrH/M13K07, selecting for kanamycin resistance. AE2086/M13K07 yielded small colonies on LB plates, and bacteriophage could be detected by plaque assay of cell-free medium after overnight growth of transformants. These cells, like JC3272/JCFL0, were sensitive to bile salts no. 3 by the liquid growth assay (Fig. 2). Hence, production of filamentous DNA bacteriophage or the presence of JCFL0 independently sensitizes E. coli to bile salts no. 3. Interestingly, the effects were not multiplicative; HfrH/M13K07 was no more sensitive than JC3272/JCFL0 or AE2086/M13K07.
The differential effects of bile salts no. 3 on strains with and without F or F-like R factors depended upon both the strain and the plasmid. We first tested F− and JCFL0 derivatives of the strain CC118 (34) by the liquid growth assay. CC118/JCFL0 responded to increasing bile salt concentrations much like JC3272/JCFL0 (data not shown). However, CC118 itself grew poorly at bile salt concentrations of >0.75%. Consequently, the differential effects of bile salts no. 3 were not as dramatic as with JC3272. We note that some E. coli K-12 strains have accumulated mutations in efflux pumps that would alter their resistance to agents such as bile salts (43). This might also be true of CC118, though we have not explored this possibility further.
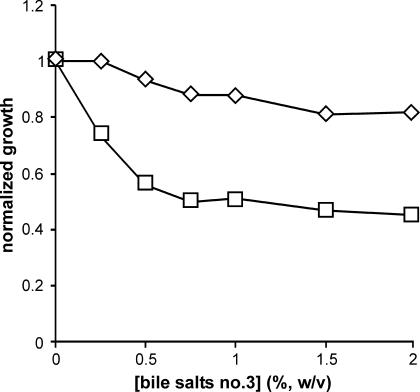
We also tested strain AE2086 with the F-like R factor R100 and its derepressed derivative, R100drd1 (4). The R100 derivative was as resistant to bile salts no. 3 as AE2086 itself over the range of 0 to 2%. The R100drd1 derivative was more sensitive, but only to about 50% growth inhibition at 0.75% bile salts (Fig. 3). At higher levels, the effect of bile salts no. 3 on the R100drd1 strain paralleled that on the R100 strain. These data suggest population heterogeneity, perhaps owing to loss of R100drd1 in the absence of antibiotics to select for plasmid retention during incubation in bile salts no. 3. Yoshida et al. (53) reported that Hfr derivatives of several R+ strains were more sensitive to cholate than the same strains with the R factors as autonomous plasmids, also suggesting that bile salts strongly selected for cells that had lost the R factor.
FIG. 3.
Effect of bile salts no. 3 on strain AE2086 with R100 or the derepressed mutant R100drd1 in liquid culture. ⋄, AE2086/R100; □, AE2086/R100drd1.
Bile salt sensitivity can be attributed to an active type IV secretion system.
The data on R100 and R100drd1 strains imply that expression of the R100 tra genes is responsible for bile salt sensitivity, since such expression is derepressed by the drd1 mutation. To confirm that the F tra genes are responsible for bile salt sensitivity of F+ strains and to identify which, if not all, are important, we tested several tra mutants for plating efficiency on LB agar containing 1.1% bile salts no. 3. We focused on the tra genes required for F+ cells to synthesize normal numbers of extended F pili, since those genes encode surface proteins (18). Mutations were ranked according to the bile salt resistance of mutant strains (Table 1).
TABLE 1.
Effect of tra mutations on sensitivity to bile salt mix
| tra mutant allele | Mutation (reference) | Normalized plating efficiencya |
|---|---|---|
| F− | 1.1 | |
| JCFL0 (F′ lac) | tra+ (1) | <6 × 10−3 |
| F′ traB100 | Insertion (this study) | 0.98 |
| F′ traL311 | UGA (cited in ref. 35) | 0.98 |
| F′ traV100 | Insertion (24) | 0.85 |
| F′ traA1 | UAG (52) | 0.83 |
| F′ traK105 | Frameshift (52) | 0.58 |
| pOX38 traU347 | Insertion (36) | 0.57 |
| F′ traH80 | UAG (52) | 0.47 |
| F′ traG106 | Frameshift (52) | 0.40 |
| pOX38 trbI463 | Insertion (33) | 0.34 |
| F′ traF13 | UAG (52) | 0.23 |
| F′ traE18 | UAG (52) | 0.17 |
| F′ traC5 | UAG (52) | 0.14 |
| F′ traW546 | UAG (35) | <4 × 10−3 |
[CFU on LB agar containing 1.1% (wt/vol) bile salt mix]/(CFU on LB agar).
Mutations in any of four genes, traB, traL, traV, or traA, restored resistance essentially to that of JC3272 (Table 1). Mutations in any of eight other tra genes increased resistance to within an order of magnitude of the F− strain, but the plating efficiencies of the mutants were still reduced at least twofold (traK, traU, and traH) and as much as sevenfold (traC). Since the tra mutations we tested are not necessarily all null alleles, modest differences in resistance (<10-fold) may not be significant. The traH, traF, and especially traE mutants formed obviously smaller colonies on bile salt plates, suggesting that these mutants remained somewhat more sensitive than their plating efficiencies indicated. Of all the mutants we tested, only the traW mutant was clearly as (or more) sensitive to bile salts no. 3 as the tra+ strain.
Assay by growth in liquid (data not shown) largely confirmed the data obtained based on plating efficiency, except the traE mutant was more sensitive in liquid than would be expected from its plating efficiency. In any case, the sensitivity of F+ strains to bile salts no. 3 can be attributed to the expression of the F tra genes required for F pilus formation or function, including traA, the F pilin structural gene.
To determine if tra genes directly involved in the later stages of DNA transfer are also required for bile salt sensitivity, we tested the effect of plasmid pTG801. This plasmid contains all the tra genes required for F pilus formation but lacks nearly all the tra genes required for the later stages of DNA transfer (21). Consequently, pTG801 cells are Tra− but elaborate F pili visible by electron microscopy and are sensitive to bacteriophage that adsorb to F pili (21). In strain M1174, pTG801 caused a dramatic increase in bile salt sensitivity compared with M1174 itself. The saturation OD of M1174 grown in LB medium containing 2% bile salts no. 3 was 85% of that when the cells were grown without bile salts (OD600 = 1.34 and 1.63, respectively), similar to results with the JC3272 parent strain. In contrast, M1174 containing pTG801 grew to a saturation OD of 0.71 in the absence of bile salts but failed to grow (OD600 < 0.01) in the presence of 0.25% (or higher) bile salts no. 3. We conclude that the bile salt sensitivity of tra+ strains does not require donor activities specifically related to DNA transfer, all of which cells with pTG801 lack (21), but instead reflects the function of the F-encoded type IV secretion system specifically as it relates to the F pilus assembly pathway.
Bile salt specificity.
The preceding experiments all utilized a commercial bile salt mixture (see Materials and Methods). We therefore separately tested the unconjugated bile salts cholate and deoxycholate and the conjugated salts glycocholate and taurocholate. In addition to JC3272 and JC3272/JCFL0, we tested E. coli K-12 strains A529 (tolA529) and D21f2 (rfa-1 rfa-3). tolA is part of the tolQRA cluster at 17 min on the E. coli genetic map (50). A mutation in any of the tolQRA/tolB/pal genes of E. coli leads to profound functional defects in the outer membrane, manifested by hypersensitivity to bile salts, among other agents, and leakage of periplasmic proteins into the medium (38, 50). In addition, the TolA protein acts as a secondary receptor for filamentous DNA bacteriophages that bind initially to the tips of F pili, are drawn to the cell surface as the filaments retract, and there interact with the TolA protein via the bacteriophage pIII protein before infection can be completed (13, 16, 17, 27, 30). Conceivably, F-encoded Tra proteins might also interact with TolA, even in the absence of bacteriophage, in such a way as to render cells partial TolA− phenocopies. If so, any bile salt selectivity should be similar for the tolA mutant and JCFL0 cells.
Strain D21f2 is a deep rough mutant whose lipopolysaccharide (LPS) lacks heptose and other LPS oligosaccharides that require heptose for their addition (40). Deep rough mutants are sensitive to various hydrophobic compounds, including bile salts, to which E. coli is normally resistant (40).
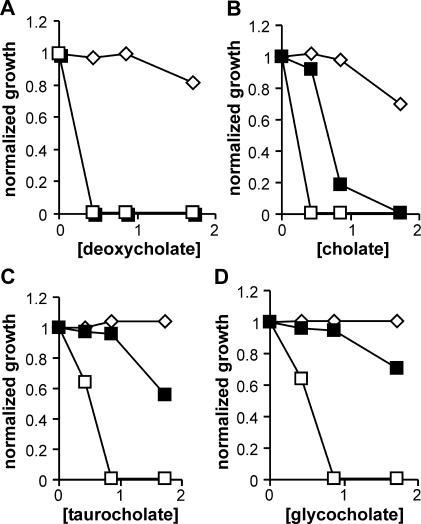
Both JC3272/JCFL0 and A529 cells failed to grow in liquid medium at 0.4% deoxycholate (Fig. 4A). However, with the other three bile salts, the sensitivities of the two strains diverged. With cholate and especially with taurocholate and glycocholate, JC3272/JCFL0 cells were more resistant than A529, whereas there was little difference in taurocholate and glycocholate sensitivities between the F− and F+ strains except at the highest bile salt concentration used, 1.6% (Fig. 4B to D). These data argue against the hypothesis that F+ strains are simply TolA− phenocopies.
FIG. 4.
Effects of different bile salts on F−, F+, and tolA mutant cells in liquid culture. ⋄, JC3272 (F−); ▪, JC3272/JCFL0; □, A529 (tolA529). (For deoxycholate sensitivity, the data points for the JC3272/JCFL0 and A529 strains overlap.).
Bile salt selectivity was even more pronounced with the traW546 mutant, the only tra mutant we tested that remained sensitive to the bile salt mixture (Table 1). The traW546 mutant was slightly more sensitive than the tra+ strain to the conjugated bile salts taurocholine and glycocholine, but at 0.4%, a concentration that had minimal effects on the F− and tra+ strains (Fig. 4B), cholate completely inhibited the growth of the traW546 mutant (Table 2).
TABLE 2.
Bile salt selectivity of the traW546 mutant
| Bile salt (concn) | OD600 (% of control)
|
||
|---|---|---|---|
| F− | F′ lac tra+ | F′lac traW546 | |
| None | 1.43 | 1.32 | 1.00 |
| Glycocholate (0.4%) | 1.41 (99) | 1.29 (98) | 0.66 (66) |
| Taurocholate (0.4%) | 1.43 (100) | 1.26 (95) | 0.79 (79) |
| Cholate (0.4%) | 1.43 (100) | 1.32 (100) | <0.01 (<0.01) |
The sensitivity pattern of the deep rough mutant D21f2 also differed from those of JC3272/JCFL0 and the traW mutant. All three strains were more sensitive to cholate than to either of the conjugated bile salts (Fig. 4 and 5 and Table 2). However, whereas glycocholate and taurocholate had equivalent but relatively minor effects on growth of the F+ strains, especially at levels of <1% (Fig. 4 and Table 2), glycocholate was significantly more inhibitory to the growth of D21f2 than taurocholate (Fig. 5). This agrees with the expectation that biological membranes should be essentially impermeable to taurocholate (pKa = 1.4), which would be fully charged at neutral pH, more permeable to glycocholate (pKa = 4.4), and most permeable to cholate (pKa = 6.4) (39). The apparent equivalence of taurocholate and glycocholate indicates that factors other than membrane permeability dominate the bile salt sensitivity pattern of F+ strains.
FIG. 5.
Effects of different bile salts on the deep rough strain D21f2 in liquid culture. ⋄, cholate; ▵, glycocholate; □, taurocholate. Under the same conditions, D21, the rfa+ parent strain of D21f2, was inhibited 13, 28, and 42% at 1.6% taurocholate, glycocholate, and cholate, respectively.
Sensitivity of F+ strains to SDS and antibiotics.
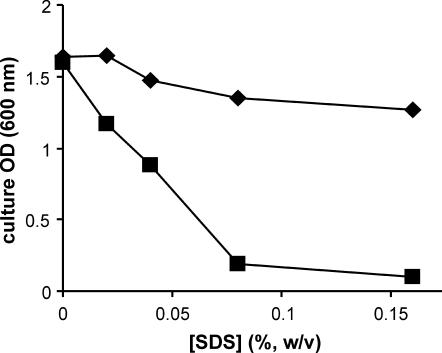
It has been reported that SDS in the range of 0.03 to 0.1% selectively lyses wild-type F+ cells relative to F− cells or F+ cells unable to produce F pili (2). The effect of SDS on the growth of JC3272 and JC3272/JCFL0 confirmed this observation (Fig. 6). The effects of SDS on strain A529 were more complicated. While growth appeared to be inhibited at the same SDS levels and to the same degree as in JC3272/JCFL0, A529 cultures incubated in 0.04 to 0.16% SDS contained large amounts of viscous cell debris; we did not observe any debris in JC3272 or JC3272/JCFL0 cultures incubated in SDS over this range of concentrations. We infer that A529 in exponential growth is not sensitive to SDS at the levels we tested, but that these cells became sensitive as they entered stationary phase and lysed, leaving the debris we observed.
FIG. 6.
F′ lac sensitizes cells to the anionic detergent SDS. ♦, JC3272 (F−); ▪, JC3272/JCFL0.
By disk assay, isogenic F+ and F− strains exhibited no significant differences in sensitivity to ampicillin (10 μg), kanamycin (30 μg), tetracycline (30 μg), nalidixic acid (30 μg), or novobiocin (30 μg). Both strains were resistant to bacitracin (10 IU) and rifampin (5 μg). In contrast, the deep rough mutant D21f2, but not its parent strain D21, was rifampin sensitive.
We also compared JC3272/JCFL0 and A529 for leakage of periplasmic enzymes by cross-protection and by a semiquantitative β-lactamase assay (6). The two strains and JC3272 itself were transformed with pUC19 and plated on LB containing ampicillin (100 μg/ml). Under these conditions, leakage of periplasmic β-lactamase, encoded by pUC19, hydrolyzes ampicillin in the vicinity of the transformant, allowing nearby sensitive cells to grow. The result is the formation of satellite colonies. Such colonies clearly surrounded A529 transformants but neither JC3272 nor JC3272/JCFL0 transformants (Fig. 7). We also estimated the relative amounts of β-lactamase in cell-free supernatants by the serial dilution assay described in Materials and Methods. The amounts of enzyme accumulated in cultures of JC3272 and JC3272/JCFL0 were indistinguishable from each other and at least fourfold less than the amount in cultures of A529.
FIG. 7.
Leakage of β-lactamase by cross-protection. Cells were transformed with pUC19, and ampicillin-resistant transformants were selected as described in Materials and Methods. (A) JC3272; (B) JC3272/JCFL0; (C) A529 (tolA529). Note the ampicillin-sensitive satellite colonies around the A529 transformants but neither the JC3272 nor the JC3272/JCFL0 transformants.
DISCUSSION
The outer membrane of gram-negative bacteria is an effective barrier against noxious agents likely to be encountered by these cells (38-40). The barrier properties of the outer membrane are supplemented by broad-specificity efflux pumps that remove molecules, including bile salts, that do penetrate (54, 56). Not surprisingly, many genes are devoted to these defenses. It is therefore of some interest that expression of the type IV secretion systems encoded by F and F-like R factors sensitizes host cells to bile salts (54), SDS (2), and perhaps to other compounds. This sensitivity might account at least in part for the elaborate genetic regulatory network characteristic of strains with F-like R factors (55). These networks allow for epidemic spread of the R factors by conjugation, but at the same time maintain minimal steady-state levels of tra gene expression (8, 14, 51). F, like R100drd1, is a derepressed mutant, and cells with either plasmid were more sensitive to a bile salt mixture than those with the repressed R100.
The bile salt sensitivity of F+ strains cannot be attributed to one or a few Tra proteins. Though cells overproducing bacteriophage f1 gene III protein are bile salt sensitive (6), F pilin is the only Tra protein sufficiently abundant to have comparable effects (41). In fact, the traA1 mutant, which contains no F pilin, was bile salt resistant. However, mutations in tra genes required for the assembly of F pili, or affecting F pilus number or length distributions, also reduced bile salt sensitivity. Several of these tra mutants were shown in previous studies to contain apparently normal membrane F pilin levels (37). Based on these data, we attribute the bile salt sensitivity of F+ strains to the functioning of the multicomponent type IV secretion system encoded by F. Moreover, the sensitivity of cells containing pTG801, which elaborate functional F pili but lack tra genes required for late stages of conjugal DNA transfer (21), indicates that sensitivity is related to F pilin secretion and not to DNA transfer itself.
Other macromolecular secretion systems may, under certain conditions, similarly alter the permeability of gram-negative bacteria. Daugelavicius et al. (15) reported that cells overexpressing plasmid RP4 genes roughly comparable to the F genes required for F pilus formation were more permeable than RP4− cells or cells with a complete RP4 conjugative transfer system expressed at normal levels. They attributed the permeability differences to active cell surface complexes of RP4 proteins, of which their cells contained about 80 each. F-encoded Tra proteins are also organized into surface complexes (24), though fewer than the corresponding RP4 proteins (Silverman et al., unpublished). More recently, Chen et al. (11) described a mutation in the Neisseria spp. pilQ gene that permeabilized mutant cells to several compounds, including heme, hydrophobic antibiotics, and nonionic detergents. PilQ is a member of the secretin superfamily required as an outer membrane complex for type IV pilus formation.
If, as we argue, the bile salt sensitivity of F+ strains requires active F pilin secretion, filament formation itself may not be essential. The traW546[Am] mutant, which was hypersensitive to cholate relative to a tra+ control, essentially lacked F pili visible by electron microscopy (33). While traW546 cells might still produce very short F pili (4), this in itself is not sufficient for bile salt sensitivity. Other mutants, such as those with the traH80 or traF13 alleles, may also produce very short filaments (4) but in the present study were nearly as resistant to bile salts as F− cells. Wild-type F pilin when overproduced (22) or certain mutant F pilins (32) do appear in the medium in forms other than filaments, suggesting that F pilin secretion might occur in the absence of filament formation.
Our data suggest a mechanism for F pilin secretion formally similar to the two-stage mechanisms proposed for bacterial type I secretion systems (47) and tripartite efflux pumps (28, 54, 56). Specifically, we propose that F pilus assembly occurs at Tra protein complexes that form a channel through the cell envelope. In our hypothesis, the channel would normally be closed to the exterior, opening transiently only in the presence of F pilin. This could occur during F pilus assembly from inner membrane F pilin or during F pilus disassembly (retraction). In either case, the open state would allow molecules of suitable size and chemistry to enter from the outside, thereby bypassing the otherwise intact outer membrane permeability barrier.
In this model, the TraW protein might mediate channel dynamics. In the absence of TraW, the channel would remain open longer than normal, accounting for the hypersensitivity of the traW mutant to cholate. Additionally, the failure to close at normal intervals (or at all) could limit the rate at which the channel complex delivered inner membrane F pilin for F pilus assembly, or it could favor F pilus retraction over assembly. Either effect would account for the drastically reduced length of F pili on traW mutant cells (4, 33).
This implies that the outer membrane permeability barrier of F+ strains should be largely intact. The similar effects of both hydrophobic and hydrophilic antibiotics on isogenic F+ and F− cells as well as measurements of β-lactamase leakage indicate that this is so. Moreover, the effects of different bile salts on F+ strains could be distinguished from their effects on D21f2, a deep rough LPS mutant whose sensitivity to lipophilic compounds is expected to reflect the ability of such compounds to pass through biological membranes (39, 40). We showed that D21f2 sensitivity was directly related to bile salt pKa and hence to the fraction of uncharged bile salt at neutral pH. This was not the case for F+ strains, which were similarly resistant to both glycocholate (pKa = 4.4) and taurocholate (pKa = 1.4). One possibility is that the larger sizes of the conjugated bile salts relative to cholate contribute to the differential effects of individual bile salts on F+ strains. Based on the crystal structures of the bile salts (3, 25, 31, 49), and not considering cations or bound water, a channel with a diameter of ∼6 to 8 Å could exclude taurocholate and glycocholate, or retard their passage, but allow entry of cholate. F pili themselves are hollow cylinders with a cationic lumen of ≅20 Å (19, 42) and are therefore unlikely to discriminate by size alone among the bile salts tested. Rather, bile salts and perhaps similar compounds may be useful as probes to investigate the structure and function of the type IV secretion apparatus encoded by F.
Acknowledgments
We thank Tim Mather, Oklahoma Medical Research Foundation, for assistance with bile salt structures.
This work was supported by National Science Foundation grant MCB-212365, the Oklahoma EPSCoR Infrastructure Improvement Award, and the Research Office of the University of Central Oklahoma. P.M.S. acknowledges support from the Marjorie Nichlos Chair in Medical Research.
REFERENCES
- 1.Achtman, M., N. Willetts, and J. Clark. 1971. Beginning a genetic analysis of conjugational transfer determined by the F factor in Escherichia coli by isolation and characterization of transfer-deficient mutants. J. Bacteriol. 106:529-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adachi, H., M. Nakano, M. Inuzuka, and M. Tomoeda. 1972. Specific role of sex pili in the effective eliminatory action of sodium dodecyl sulfate on sex and drug resistance factors in Escherichia coli. J. Bacteriol. 109:1114-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adolph, H., P. Zwart, R. Meijers, I. Hubatsch, M. Kiefer, V. Lamzin, and E. Cedergren-Zeppezauer. 2000. Structural basis for substrate specificity differences of horse liver alcohol dehydrogenase isozymes. Biochemistry 39:12885-12897. [DOI] [PubMed] [Google Scholar]
- 4.Anthony, K., W. Klimke, J. Manchak, and L. Frost. 1999. Comparison of proteins involved in pilus synthesis and mating pair stabilization from the related plasmids F and R100-1: insights into the mechanism of conjugation. J. Bacteriol. 181:5149-5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beutin, L., and M. Achtman. 1979. Two Escherichia coli chromosomal cistrons, sfrA and sfrB, which are needed for expression of F factor tra functions. J. Bacteriol. 139:730-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boeke, J., P. Model, and N. Zinder. 1982. Effects of bacteriophage f1 gene III protein on the host cell membrane. Mol. Gen. Genet. 186:185-192. [DOI] [PubMed] [Google Scholar]
- 7.Bolivar, F. 1978. Construction and characterization of new cloning vehicles. III. Derivatives of plasmid pBR322 carrying unique EcoRI sites for selection of EcoRI generated recombinant DNA molecules. Gene 4:121-136. [DOI] [PubMed] [Google Scholar]
- 8.Broda, P. 1975. Transience of the donor state in an Escherichia coli K12 strain carrying a repressed R factor. Mol. Gen. Genet. 138:65-79. [DOI] [PubMed] [Google Scholar]
- 9.Cao, T., and M. Saier. 2001. Conjugal type IV macromolecular transfer systems of gram-negative bacteria: organismal distribution, structural constraints and evolutionary conclusions. Microbiology 147:3201-3214. [DOI] [PubMed] [Google Scholar]
- 10.Cascales, E., and P. Christie. 2003. The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 1:137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, C.-J., D. Tobiason, C. Thomas, W. Shafer, H. Seifert, and P. Sparling. 2004. A mutant form of the Neisseria gonorrhoea pilus secretin protein PilQ allows increased entry of heme and antimicrobial compounds. J. Bacteriol. 186:730-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christie, P., and J. Vogel. 2000. Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol. 8:354-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Click, E., and R. Webster. 1998. The TolQRA proteins are required for membrane insertion of the major capsid protein of the filamentous phage f1 during infection. J. Bacteriol. 180:1723-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cullum, J., J. Collins, and P. Broda. 1978. The spread of plasmids in model populations of Escherichia coli K12. Plasmid 1:545-556. [DOI] [PubMed] [Google Scholar]
- 15.Daugelavicius, R., J. Bamford, A. Grahn, E. Lanka, and D. Bamford. 1997. The IncP plasmid-encoded cell envelope-associated DNA transfer complex increases cell permeability. J. Bacteriol. 179:5195-5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng, L., and R. Perham. 2002. Delineating the site of interaction on the pIII protein of filamentous bacteriophage fd with the F-pilus of Escherichia coli. J. Mol. Biol. 319:603-614. [DOI] [PubMed] [Google Scholar]
- 17.Deng, L., P. Malik, and R. Perham. 1999. Interaction of the globular domains of pIII protein of filamentous bacteriophage fd with the F-pilus of Escherichia coli. Virology 253:271-277. [DOI] [PubMed] [Google Scholar]
- 18.Firth, N., K. Ippen-Ihler, and R. Skurray. 1996. Structure and function of the F factor and mechanism of conjugation, p. 2377-2401. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 19.Folkhard, W., K. Leanard, S. Malsey, D. Marvin, J. Dubochet, A. Engel, M. Achtman, and R. Helmuth. 1979. X-ray diffraction and electron microscope studies on the structure of F-pili. J. Mol. Biol. 130:145-160. [DOI] [PubMed] [Google Scholar]
- 20.Frost, L., K. Ippen-Ihler, and R. Skurray. 1994. Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol. Rev. 58:162-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grossman, T., and P. M. Silverman. 1989. Structure and function of conjugative pili: inducible synthesis of functional F pili by Escherichia coli K-12 containing a lac-tra operon fusion. J. Bacteriol. 171:650-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grossman, T., L. Frost, and P. Silverman. 1990. Structure and function of conjugative pili: monoclonal antibodies as probes for structural variants of F pili. J. Bacteriol. 172:1174-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guyer, M., R. Reed, J. Steitz, and K. Low. 1980. Identification of a sex-factor affinity site in E. coli as gamma delta. Cold Spring Harbor Symp. Quant. Biol. 45:135-140. [DOI] [PubMed] [Google Scholar]
- 24.Harris, R., V. Hombs, and P. Silverman. 2001. Evidence that F-plasmid proteins TraV, TraK, and TraB assemble into an envelope-spanning structure in Escherichia coli. Mol. Microbiol. 42:757-766. [DOI] [PubMed] [Google Scholar]
- 25.Hogan, A., S. Ealick, C. Bugg, and S. Barnes. 1984. Aggregation patterns of bile salts: crystal structure of calcium cholate chloride heptahydrate. J. Lipid Res. 25:791-798. [PubMed] [Google Scholar]
- 26.Ippen-Ihler, K., M. Achtman, and N. Willetts. 1972. Deletion map of the Escherichia coli K-12 sex factor F: the order of eleven transfer cistrons. J. Bacteriol. 110:857-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacobson, A. 1972. Role of F pili in the penetration of bacteriophage f1. J. Virol. 10:835-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson, J., and G. Church. 1999. Alignment and structure prediction of divergent protein families: periplasmic and outer membrane proteins of bacterial efflux pumps. J. Mol. Biol. 287:695-715. [DOI] [PubMed] [Google Scholar]
- 29.Lawley, T., W. Klimke, M. Gubbins, and L. Frost. 2003. F factor conjugation is a true type IV secretion system. FEMS Microbiol. Lett. 224:1-15. [DOI] [PubMed] [Google Scholar]
- 30.Lubkowski, J., F. Hennecke, A. Pluckthun, and A. Wlodower. 1999. Filamentous phage infection: crystal structure of g3p in complex with its coreceptor, the C-terminal domain of TolA. Struct. Fold Des. 7:711-722. [DOI] [PubMed] [Google Scholar]
- 31.Lucke, C., F. Zhang, J. Hamilton, J. Sacchettin, and H. Ruterjans. 2000. Solution structure of ileal lipid binding protein in complex with glycocholate. Eur. J. Biochem. 267:2929-2938. [DOI] [PubMed] [Google Scholar]
- 32.Manchak, J., K. Anthony, and L. Frost. 2002. Mutational analysis of F-pilin reveals domains for pilus assembly, phage infection and DNA transfer. Mol. Microbiol. 43:195-205. [DOI] [PubMed] [Google Scholar]
- 33.Maneewannakul, S., K. Maneewannakul, and K. Ippen-Ihler. 1992. Characterization, localization, and sequence of F transfer region products: the pilus assembly gene product TraW and a new product, TrbI. J. Bacteriol. 174:5567-5574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manoil, C., and J. Beckwith. 1985. TnphoA: a transposon probe for protein export signals. Proc. Natl. Acad. Sci. USA 82:8129-8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miki, T., T. Horiuchi, and N. Willetts. 1978. Identification and characterization of four new tra cistrons on the E. coli K12 sex factor F. Plasmid 1:316-323. [DOI] [PubMed] [Google Scholar]
- 36.Moore, D., C. Hamilton, K. Maneewannakul, S. Maneewannakul, J. Wu, K. Ippen-Ihler, and D. Bradley. 1990. Characterization of the F-plasmid conjugation gene traU. J. Bacteriol. 172:4263-4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moore, D., B. Sowa, and K. Ippen-Ihler. 1981. The effect of tra mutations on the synthesis of the F-pilin membrane polypeptide. Mol. Gen. Genet. 184:260-264. [DOI] [PubMed] [Google Scholar]
- 38.Nikaido, H. 2003. Molecular basis of outer membrane permeability. Microbiol. Mol. Biol. Rev. 67:593-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nikaido, H. 1996. Outer membrane, p. 29-47. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 40.Nikaido, H., and M. Vaara. 1985. Molecular basis of outer membrane permeability. Microbiol. Rev. 49:1-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paiva, W., T. Grossman, and P. Silverman. 1992. Characterization of F-pilin as an inner membrane component of Escherichia coli K12. J. Biol. Chem. 267:26191-26197. [PubMed] [Google Scholar]
- 42.Paranchych, W., and L. Frost. 1988. The physiology and biochemistry of pili. Adv. Microb. Physiol. 29:53-114. [DOI] [PubMed] [Google Scholar]
- 43.Potrykus, J., S. Baranska, and G. Wegryzyn. 2002. Inactivation of the acrA gene is partially responsible for chloramphenicol sensitivity of Escherichia coli CM2555 strain expressing the chloramphenicol acetyltransferase gene. Microb. Drug Resist. 8:179-185. [DOI] [PubMed] [Google Scholar]
- 44.Roy, A., and S. Mitra. 1970. Increased fragility of Escherichia coli after infection with bacteriophage M13. J. Virol. 6:333-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schandel, K., M. Muller, and R. Webster. 1992. Localization of TraC, a protein involved in assembly of the F conjugative pilus. J. Bacteriol. 174:3800-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Silverman, P. 1997. Towards a structural biology of bacterial conjugation. Mol. Microbiol. 23:423-429. [DOI] [PubMed] [Google Scholar]
- 47.Thanabalu, T., E. Koronakis, C. Hughes, and V. Koronakis. 1998. Substrate-induced assembly of a contiguous channel for protein export from E. coli: reversible bridging of an inner-membrane translocase to an outer membrane exit pore. EMBO J. 17:6487-6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vieira, J., and J. Messing. 1987. Production of single-stranded plasmid DNA. Methods Enzymol. 153:3-11. [DOI] [PubMed] [Google Scholar]
- 49.Wang, X., C. Wang, J. Tang, F. Dyda, and X. Zhang. 1997. The crystal structure of bovine bile salt activated lipase: insights into the bile salt activation mechanism. Structure 5:1209-1218. [DOI] [PubMed] [Google Scholar]
- 50.Webster, R. 1991. The tol gene products and the import of macromolecules into Escherichia coli. Mol. Microbiol. 5:1005-1011. [DOI] [PubMed] [Google Scholar]
- 51.Willetts, N. 1974. The kinetics of inhibition of Flac transfer by R100 in E. coli. Mol. Gen. Genet. 129:123-130. [DOI] [PubMed] [Google Scholar]
- 52.Willetts, N., and M. Achtman. 1972. Genetic analysis of transfer by the Escherichia coli sex factor F, using P1 transductional complementation. J. Bacteriol. 110:843-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoshida, Y., N. Takamatsu, and M. Yoshikawa. 1978. Preferential inhibitory action of sodium cholate on an Escherichia coli strain carrying a plasmid in an integrated state. J. Bacteriol. 133:406-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu, E., J. Aires, and H. Nikaido. 2003. AcrB multidrug efflux pump of Escherichia coli: composite substrate-binding cavity of exceptional flexibility generates its extremely wide substrate specificity. J. Bacteriol. 185:5657-5664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zechner, E., F. de la Cruz, R. Eisenbrandt, A. Grahn, G. Koraimann, E. Lanka, G. Muth, W. Pansegrau, C. Thomas, B. Wilkins, and M. Zatyka. 2000. Conjugative DNA transfer processes, p. 87-174 In C. Thomas (ed.), The horizontal gene pool. Harwood Academic Publishers, Amsterdam, The Netherlands.
- 56.Zgurskaya, H., and H. Nikaido. 2000. Multidrug resistance mechanisms: drug efflux across two membranes. Mol. Microbiol. 37:219-225. [DOI] [PubMed] [Google Scholar]