Abstract
Nonpathogenic Escherichia coli strain Nissle 1917 (O6:K5:H1) is used as a probiotic agent in medicine, mainly for the treatment of various gastroenterological diseases. To gain insight on the genetic level into its properties of colonization and commensalism, this strain's genome structure has been analyzed by three approaches: (i) sequence context screening of tRNA genes as a potential indication of chromosomal integration of horizontally acquired DNA, (ii) sequence analysis of 280 kb of genomic islands (GEIs) coding for important fitness factors, and (iii) comparison of Nissle 1917 genome content with that of other E. coli strains by DNA-DNA hybridization. PCR-based screening of 324 nonpathogenic and pathogenic E. coli isolates of different origins revealed that some chromosomal regions are frequently detectable in nonpathogenic E. coli and also among extraintestinal and intestinal pathogenic strains. Many known fitness factor determinants of strain Nissle 1917 are localized on four GEIs which have been partially sequenced and analyzed. Comparison of these data with the available knowledge of the genome structure of E. coli K-12 strain MG1655 and of uropathogenic E. coli O6 strains CFT073 and 536 revealed structural similarities on the genomic level, especially between the E. coli O6 strains. The lack of defined virulence factors (i.e., alpha-hemolysin, P-fimbrial adhesins, and the semirough lipopolysaccharide phenotype) combined with the expression of fitness factors such as microcins, different iron uptake systems, adhesins, and proteases, which may support its survival and successful colonization of the human gut, most likely contributes to the probiotic character of E. coli strain Nissle 1917.
In recent years, it became evident that mobile and accessory genetic elements, such as bacteriophages, plasmids, and genomic islands (GEIs), play an important role in bacterial evolution and may contribute significantly to bacterial genome content. Bacterial genomes consist of two gene pools: the core gene pool and the flexible gene pool. Whereas the core gene pool is conserved and required for basic cellular functions, the flexible gene pool is variable, sometimes strain specific, and important for adaptation to special growth conditions, e.g., colonization of new ecological niches, symbiosis, host-cell interaction, and pathogenicity. Many DNA regions which belong to the flexible gene pool represent or are derived from mobile genetic elements, and these regions are involved in adaptation to the above-mentioned growth conditions (24, 25).
The species Escherichia coli comprises nonpathogenic (commensal) as well as pathogenic strains. Since commensal E. coli strains generally represent normal and ecologically important inhabitants of the human and animal intestinal tracts (38) which may develop into pathogenic variants and vice versa, it is important to study the evolutionary aspects of the development of traits of colonization and commensalism in particular strains. Both nonpathogenic and pathogenic E. coli strains are able to colonize the gut and are well adapted to the conditions found in the large intestine. Nonpathogenic and pathogenic E. coli strains differ in the presence of genetic information that may contribute to specific virulence traits or to successful survival and fitness of the bacteria in the host (26, 28). Most of the determinants coding for such factors are thought to be acquired by horizontal gene transfer and are often clustered on the chromosome on GEIs (23, 25).
E. coli serogroup O6 is very heterogeneous, including nonpathogenic and pathogenic variants (7). The O6 serotype is commonly detected among intestinal commensal as well as diarrheagenic E. coli isolates (4, 27, 34), but it is also widely distributed among uropathogenic E. coli (UPEC) strains (44). In the case of uropathogenic O6 strains, the gut may serve as a reservoir for recurrent urinary tract infections (2).
E. coli strain Nissle 1917 (O6:K5:H1) is a typical example of a nonpathogenic, commensal E. coli isolate and forms the basis of the probiotic preparation Mutaflor, which is used for treatment of various intestinal disorders and is known to be a successful colonizer of the human gut (36, 37, 48; J. Schulze and U. Sonnenborn, Letter, Infection 23:184-188, 1999). This strain lacks P- and S-fimbrial adhesin determinants, but it expresses type 1 and F1C fimbriae (7). It exhibits a semirough lipopolysaccharide (LPS) phenotype and serum sensitivity and does not produce known protein toxins (7, 20). Additionally, strain Nissle 1917 expresses two microcins (46), strongly produces curli and cellulose in a temperature-independent manner (U. Dobrindt and L. Grozdanov, unpublished data), and possesses a surprisingly high number of iron uptake systems (enterobactin, yersiniabactin, aerobactin, salmochelin, ferric dicitrate transport system, and the chu heme transport locus).
In this study, we report on the structure of four GEIs carrying several fitness-conferring determinants of strain Nissle 1917. In addition, we provide insights into this strain's genome organization, thus elucidating the genetic background of its ability to efficiently colonize the intestinal tract. Comparison of our results with the available data on the genome organization of two UPEC O6 strains (strains 536 and CFT073) reveals detailed genetic differences responsible for the absence of virulence traits in E. coli Nissle 1917.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
E. coli strain Nissle 1917 (Mutaflor, DSM 6601, serotype O6:K5:H1) was kindly supplied by Ardeypharm GmbH, Herdecke, Germany. The UPEC strains 536 (O6:K15:H31) and CFT073 (O6:K2?:H1) have been isolated from patients with acute pyelonephritis and urosepsis, respectively (3, 39). E. coli strains MG1655, EN99, and AAEC189, also used in this study, have been described before (5, 6, 11). A collection of 324 E. coli strains, consisting of 70 UPEC O6 strains (58, 59), 29 E. coli isolates from the Escherichia coli Reference Collection (ECOR) (10), extraintestinal pathogenic E. coli (ExPEC) and intestinal pathogenic E. coli (IPEC) strains belonging to the strain collection of the Institute for Molecular Biology of Infectious Diseases (IMIB) (19), and 135 fecal isolates obtained from healthy volunteers (40), was screened for the presence of strain Nissle 1917-specific DNA regions by PCR. The cosmids and plasmids used in this study are listed in Table 1. The E. coli strains were routinely grown in Luria-Bertani medium (49) with or without 1.5% Bacto Agar (Difco Laboratories, Detroit, Mich.). Where appropriate, ampicillin was added to the growth medium at a concentration of 50 μg/ml.
TABLE 1.
Plasmids used in this study
| Designation | Description | Reference |
|---|---|---|
| pUC19 | Aprorif1 lacZ | 57 |
| SuperCos 1 | Cosmid vector, Apr | Stratagene |
| pCos3Y20 | Cosmid clone, insert contains the fimNissle 1917 gene cluster, Apr | This study |
| pCos9YB4 | Cosmid clone, insert contains the iuc gene cluster and sat of strain Nissle 1917, Apr | This study |
| pCos2YE4 | Cosmid clone, insert overlaps with pCos9YB4 and contains ihaNissle 1917, Apr | This study |
| pCos2RF2 | Cosmid clone, insert overlaps with pCos2YE4 and contains the K5-specific kps gene cluster of strain Nissle 1917, Apr | This study |
| pCos3YE4 | Cosmid clone, insert contains the mch-mcm, foc and iro gene clusters of strain Nissle 1917, Apr | This study |
| pCos2RA4 | Cosmid clone, insert overlaps with pCos3YE4, Apr | This study |
| pCos1YA7 | Cosmid clone, insert contains argW of strain Nissle 1917, Apr | This study |
| pPSD1917 | 7-kb EcoRI/HindIII fragment from pCos9YB4 containing satNissle 1917 subcloned in EcoRI/HindIII-digested pUC19, Apr | This study |
DNA technology.
Isolation of DNA and recombinant DNA techniques were performed as described previously (49). Restriction enzymes were obtained from Amersham-Pharmacia Biotech (Freiburg, Germany) and used as recommended by the supplier. DNA primers were purchased from Sigma-ARK (Steinheim, Germany). Construction of genomic library of strain Nissle 1917 was done by using a Gigapack III Gold packaging extract kit (Stratagene, Heidelberg, Germany), following the instructions of the manufacturer. High-molecular-weight DNA required for genome size analysis and comparison of whole-genome restriction patterns by pulsed-field gel electrophoresis (PFGE) was isolated and digested as described before (45, 58). The high-molecular-weight DNA was separated in 0.8% (wt/vol) agarose gels. The gels were subjected to electrophoresis for 21 to 24 h with pulsed periods of 0.5 to 50 s.
PCR.
A description of the primers used in this study is available as supplementary material (http://www.uni-wuerzburg.de/infektionsbiologie/imi-start.htm). The Taq DNA polymerase, used for the detection of defined DNA regions and genes in different E. coli strains, was purchased from QIAGEN (Hilden, Germany). When proofreading activity was required, or when long-distance PCRs were carried out, the Expand Long Template PCR system (Roche Molecular Biochemicals, Mannheim, Germany) was used. Grouping of the ECOR strain collection into the main phylogenetic lineages was done by a triplex PCR described before (16). The tRNA screening approach was performed as described before (42), using genomic DNA of completely sequenced E. coli strains MG1655 (K-12), CFT073 (O6:K2?:H1), and EDL 933 (O157:H7) as controls. All monocistronic tRNA-encoding operons, the most promoter-distal tRNA-encoding gene of polycistronic tRNA operons, and the tmRNA-encoding gene ssrA were included into this screening. For this purpose, primer pairs which are specific for the open reading frame (ORF) immediately up- or downstream of these tRNA-encoding genes in the genome of E. coli strain MG1655 were generated, enabling amplification of the tRNA-encoding gene together with its flanking sequences.
Southern hybridization.
Where necessary, the presence of certain genes on the identified cosmid clones was demonstrated by Southern hybridization. After digestion of genomic DNA of strain Nissle 1917 by the use of appropriate restriction enzymes and agarose gel electrophoresis, the DNA was transferred to Biodyne B nylon membranes (PALL, Rossdorf, Germany). The probes were obtained by PCR. Hybridization and detection were carried out using an enhanced chemiluminescence labeling and signal detection system (Amersham-Pharmacia Biotech) according to the manufacturer's recommendations.
DNA sequence analysis and sequence annotation.
Small insert libraries (2 to 2.5 kb) were generated by mechanical DNA shearing of the identified cosmid clones (41). After end repair with T4 polymerase, the fragments were ligated into the prepared pTZ19R vector. Isolated plasmids were sequenced from both ends by dye terminator chemistry and analyzed on ABI337 sequencers (Applied Biosystems, Munich, Germany). The Phrap software in the Staden software package was used for assembling and editing the sequence data (52). For the left-hand region of GEI II of strain Nissle 1917 (GEI IINissle 1917; between pheV and shiA), primer walking, amplified with proofreading Taq DNA polymerase, was performed directly on a PCR product.
Homology searches as well as searches for conserved protein domains were performed with the BLASTN, BLASTX, and PSI- and PHI-BLAST programs of the National Center for Biotechnology Information (1) (http://www.ncbi.nlm.nih.gov/BLAST/). Putative ORFs were identified with Vector NTI (InforMax, Oxford, United Kingdom) and the National Center for Biotechnology Information ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html).
Genome comparison by DNA-DNA hybridization.
In order to assess the E. coli K-12-specific fraction of the strain Nissle 1917 genome, this strain's total genomic DNA was used to probe Panorama E. coli gene arrays (Sigma-Genosys, Cambridge, United Kingdom). In addition, the E. coli “pathoarray” for detection of various virulence- and fitness-related genes distributed in E. coli and Shigella strains was used. DNA labeling, array hybridization, and data analysis were performed as described previously (18).
Phenotypic characterization of gene expression.
F1C fimbria expression was detected by immunoagglutination with anti-FocA polyclonal serum. For this purpose, the cosmid pCos3YE4 was transformed into nonfimbriated E. coli strain AAEC189 (6). Immunoagglutination was performed by mixing 10 μl of overnight growth culture of the transformant with 10 μl of anti-FocA antibody dissolved in phosphate-buffered saline on microscope slides, followed by incubation on ice until aggregation of bacterial cells was clearly observed. E. coli strain Nissle 1917 was used as a positive control, and E. coli strain AAEC189 was used as a negative control.
Microcin production was assessed by the presence of clear zones of growth inhibition of indicator strain E. coli DH5α around the colonies of the tested strains after overnight incubation at 37°C on M9 agar plates. Expression of the siderophore aerobactin was assessed as described before (11) by the presence of growth zones of the iron-deficient indicator strain EN99 around colonies of the tested strains on medium supplied with dipyridyl.
For the preparation of proteins secreted by strain Nissle 1917, 10 ml of an overnight growth culture was centrifuged, and the supernatant was mixed with 1 g of trichloroacetic acid by vortexing it until the acid was completely dissolved. After overnight incubation at 4°C, precipitated proteins were collected by centrifugation and dissolved in 100 μl of 0.1 N NaOH. After being heated for 10 min at 90°C in loading buffer, the proteins were separated by polyacrylamide gel electrophoresis as described before (33). After electrophoresis, the gel was stained for 2 h in 1% (wt/vol) Coomassie blue solution, followed by incubation in destaining solution for 2 to 6 h.
For detection of serine protease activity, the EnzCheck protease assay kit E-6638 (Molecular Probes, Leiden, The Netherlands) was used, following the instructions of the manufacturer.
Nucleotide sequence accession numbers.
E. coli strain Nissle 1917-specific sequences of GEI INissle 1917 to GEI IIINissle 1917 determined in this study were submitted to the EMBL nucleotide sequence database and are available under accession numbers AJ586887, AJ586888, and AJ586889, respectively.
RESULTS
E. coli strain Nissle 1917 genome size determination and grouping into main phylogenetic E. coli lineages.
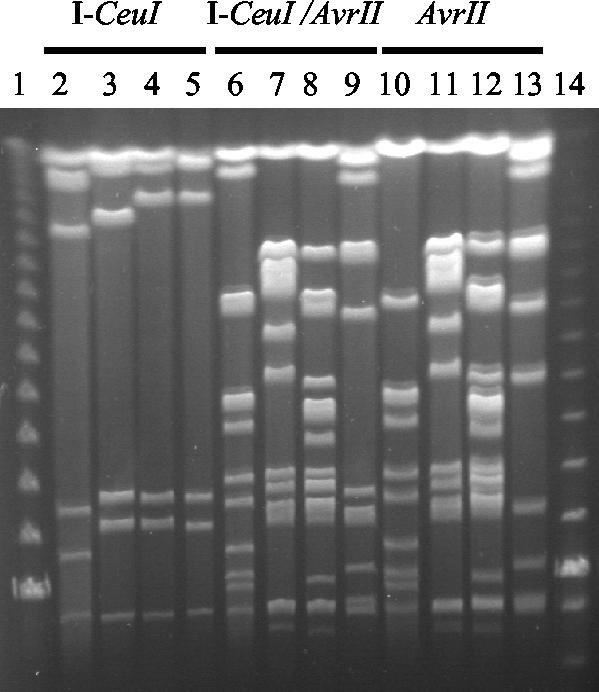
The genome size of nonpathogenic E. coli strain Nissle 1917 was assessed by I-CeuI restriction followed by PFGE (Fig. 1). The 5.1-Mb genome is approximately 0.4 Mb larger than that of K-12 strain MG1655 (4.7 Mb), about 0.1 Mb larger than that of UPEC strain 536 (5.0 Mb), and roughly 0.13 Mb smaller than that of UPEC strain CFT073 (5.23 Mb) (5, 18, 56). The affiliation with the major phylogenetic lineages of E. coli has been determined for these four strains by PCR (16). Whereas strain MG1655 belongs to ECOR group A, both UPEC strains as well as the probiotic strain Nissle 1917 are members of ECOR group B2 (data not shown). ECOR group B2 is known to comprise isolates with considerable amounts of horizontally acquired DNA, including pathogenicity islands (PAIs) (10).
FIG. 1.
Assessment of the genome size of E. coli strain Nissle 1917 by I-CeuI restriction, followed by PFGE. Genomic DNA of E. coli strains MG1655, Nissle 1917, CFT073, and 536 was digested with the indicated restriction enzyme(s). Restriction fragments were separated by PFGE. Lane 1, lambda ladder PFGE marker; lanes 2, 6, and 10, E. coli strain MG1655; lanes 3, 7, and 11, E. coli strain Nissle 1917; lanes 4, 8, and 12, E. coli strain CFT073; lanes 5, 9, and 13, E. coli strain 536; lane 14, low-range PFGE marker.
Detection of horizontally acquired DNA in E. coli strain Nissle 1917 genome by tRNA screening.
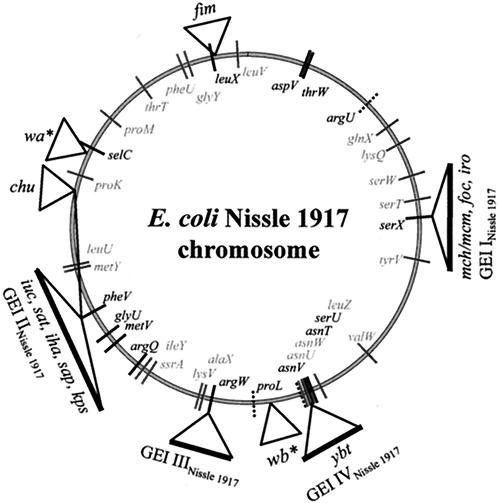
The genome size differences of the four strains included in this study may result from the presence or absence of horizontally acquired DNA regions. Since tRNA genes are common sites for integration of foreign DNA elements, including bacteriophages, plasmids, and GEIs (26), a PCR-based tRNA screening of the E. coli strain Nissle 1917 genome was performed in order to investigate sequence context alterations of tRNA genes due to insertion or deletion of chromosomal DNA regions compared to those of E. coli K-12 strain MG1655. Size differences in the PCR products obtained or the lack of a PCR product indicate genomic variations or even chromosomal integration of foreign DNA at the 3′ end of the respective tRNA gene which is absent in K-12 strain MG1655 (42). Such genes, which could not be amplified with K-12-specific primers, were then screened with one primer binding to the region immediately downstream of this tRNA-encoding gene in the genome of E. coli strain CFT073 (56), in combination with the primer that binds upstream of the respective tRNA-encoding gene in E. coli K-12 strain MG1655. The results from the tRNA screening are shown in Fig. 2. When K-12-specific primers were used, sequence context variations in 15 out of 37 tRNA loci tested were detected in strain Nissle 1917 compared to E. coli K-12 strain MG1655. The immediate downstream regions of 11 of these loci were identical in the nonpathogenic, probiotic strain Nissle 1917 and in UPEC strain CFT073. Among them are most of the tRNA-encoding genes, which represent already known integration sites of foreign DNA in other E. coli strains (asnT, argW, leuX, pheV, serX, and thrW). This finding may indicate the presence of identical or similar GEIs in both strains. However, similar bacteriophage integrase-encoding genes are frequently located downstream of a tRNA-encoding gene in different E. coli strains, although the DNA contents of the corresponding islands are not identical. The sequence context of tRNA-encoding genes in UPEC strain 536 also seems to be very similar to that of E. coli strain Nissle 1917 when E. coli K-12-specific primers were used. In both strains, the pheU gene could be amplified with K-12-specific primers, suggesting that the strains lack the pap-containing island (coding for P fimbriae) associated with this tRNA-encoding gene in strain CFT073.
FIG. 2.
Comprehensive genomic map of E. coli strain Nissle 1917 based on the chromosome of E. coli strain MG1655. GEIs INissle 1917 to IVNissle 1917 and smaller genomic islets coding for fitness factors have been indicated according to their chromosomal insertion site next to tRNA-encoding genes. The positions of the tRNA-encoding genes, which seem to be possible chromosomal insertion sites for horizontally transferred DNA, are indicated as well as those of chromosomal restriction sites of CeuI. Grey marks indicate tRNA genes with sequence contexts identical to that of K-12 strain MG1655. Black marks indicate tRNA genes with sequence contexts identical to that of UPEC O6 strain CFT073. Dotted marks indicate tRNA genes with an as-yet-unknown downstream region. fim, type 1 fimbrial determinant; mch/mcm, microcin M- and H47-encoding determinants; foc, F1C fimbrial determinant; iro, salmochelin-encoding determinant; ybt, yersiniabactin-encoding determinant; iuc, aerobactin-encoding determinant; sat, Sat protease-encoding determinant; iha, Iha adhesin-encoding determinant; sap, Sap-like autotransporter-encoding determinant; kps, capsule determinant; chu, Chu hemin uptake determinant; wa*/wb*, gene clusters required for LPS biosynthesis.
In summary, the larger genome size of nonpathogenic, probiotic E. coli strain Nissle 1917 compared to that of nonpathogenic E. coli K-12 strain MG1655 mirrors the presence of several horizontally acquired DNA regions which may also be, at least partially, present in other E. coli isolates (e.g., E. coli strains 536 and CFT073) and which may contribute to the phenotypic traits of strain Nissle 1917.
Analysis of the genome content of strain Nissle 1917 by DNA-DNA hybridization using DNA arrays.
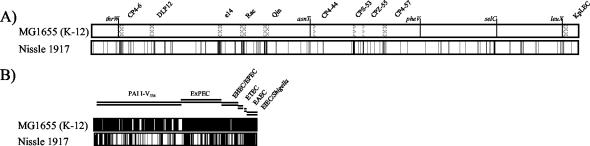
To further substantiate the analysis of the genome structure of strain Nissle 1917, the overall genome content was analyzed by DNA-DNA hybridization experiments. The fraction of the conserved E. coli core genome was assessed by hybridization with E. coli K-12 strain MG1655-specific DNA arrays. Hybridization with the E. coli pathoarray (18) allowed the rapid detection of virulence- and fitness-associated genes of pathogenic E. coli belonging to the flexible E. coli gene pool. The results of the DNA-DNA hybridization of genomic DNA isolated from strain Nissle 1917 with E. coli K-12 gene arrays demonstrated that 3,893 translatable ORFs present in the nonpathogenic reference strain (5) were detectable in strain Nissle 1917 (90.7% of all translatable ORFs of E. coli K-12 strain MG1655) (Table 2). Based on the functional classification of the GenProtEC database of the chromosomally encoded genes and proteins of E. coli K-12 (http://genprotec.mbl.edu), the majority of these missing ORFs can be functionally grouped as coding for hypothetical, not experimentally classified, or unknown gene products. A great diversity of ORFs which represent mobile and accessory genetic elements, e.g., insertion sequence (IS) elements, or which code for structural components of the cell in the K-12 genome were not detectable in strain Nissle 1917. Ten prophages found in E. coli strain MG1655 were not detected in strain Nissle 1917 (Fig. 3A). The chromosomal context of several tRNA-encoding genes (e.g., serX, argW, ileY, pheV, and leuX) was found to contain alterations in strain Nissle 1917 in comparison to the corresponding sequences in E. coli MG1655, implying the presence of horizontally acquired genetic information downstream of these tRNA loci.
TABLE 2.
Assessment of E. coli K-12 strain MG1655-specific genes detectable in strain Nissle 1917 core genome by E. coli Panorama DNA array hybridization
| ORF functional category | No. of ORFs detected in E. coli strain:
|
|
|---|---|---|
| MG1655 | Nissle 1917 | |
| Hypothetical, not experimentally classified, or unknown | 1,634 | 1,415 |
| Phage, transposon, or plasmid | 87 | 55 |
| Cell structure | 182 | 163 |
| Other | 2,387 | 2,260 |
| Total | 4,290 | 3,893 |
FIG. 3.
Assessment of the genome content of strain Nissle 1917 by DNA arrays. (A) Genome comparison of nonpathogenic E. coli strain Nissle 1917 and E. coli K-12 strain MG1655 by Panorama E. coli gene arrays. The individual chromosomes are displayed linearly and in equal length. Missing and/or undetectable ORFs are marked by vertical black lines in the individual chromosomes. The positions of the undetectable ORFs refer to the E. coli MG1655 chromosome. The positions of tRNA genes frequently used as chromosomal insertion sites of horizontally acquired DNA elements, those of 10 prophages of strain MG1655, and the chromosomal origin and terminus of replication are marked within the map of E. coli strain MG1655. (B) Detection of various genes of the flexible gene pool of E. coli and Shigella in nonpathogenic E. coli strain Nissle 1917 by the E. coli pathoarray. The genes are grouped by typical E. coli pathotypes. Missing and/or undetectable ORFs are marked by vertical black lines.
The genome of strain Nissle 1917 was also screened for the presence of DNA sequences which belong to the flexible gene pool of UPEC strain 536, as well as for typical fitness-associated genes and genes specific for ExPEC and IPEC strains, as well as Shigella, with the previously described E. coli pathoarray (18) (Table 3). A considerable amount of sequences specific for PAIs I536 to V536 and pathogenicity islets as well as genes specific for ExPEC or homologous sequences thereof were detectable in this strain (55% of the probes specific for ORFs of PAIs I536 to V536 and 37% of the probes for other ExPEC-specific genes spotted with the E. coli pathoarray). Generally, the hybridization signals confirmed the results obtained from the characterization of GEIs of strain Nissle 1917. The presence of several determinants encoding adhesins (type 1 and F1C fimbriae, Iha, curli, AIDA-I/Sap-like), proteases (Sat and Tsh), microcins, and multiple-gene clusters coding for proteins involved in iron acquisition (yersiniabactin, aerobactin, salmochelin, and Chu hemin receptor) was confirmed. The gene cluster required for capsule biosynthesis and many putative ORFs located on PAIs of strains CFT073 and 536 were detected as well. About 18% of the probes for IPEC-specific genes showed a clear hybridization signal (Fig. 3B; Table 3). Most of these probes are complementary to fimbrial determinants (indicating a possible cross-reaction of probes designed for the detection of ExPEC fimbria-encoding determinants) or to putative ORFs present on PAIs of the enterohemorrhagic E. coli (EHEC) O157:H7 strain EDL 933 (47). However, the detection of these genes or homologues thereof does not show whether they are intact and functional. They may have premature stop codons, insertions, or deletions, thus rendering them nonfunctional, which may also explain why strain Nissle 1917 is nonpathogenic. Importantly, known protein toxin-encoding determinants of pathogenic E. coli have not been detected in strain Nissle 1917 by this approach.
TABLE 3.
Assessment of the flexible gene pool of E. coli strain Nissle 1917 by DNA-DNA hybridization using the E. coli pathoarray
| Gene specificity | No. of genes detectable by:
|
|
|---|---|---|
| E. coli pathoarray | Screening E. coli strain Nissle 1917 | |
| PAI I536-PAI V536 | 212 | 116 |
| ExPEC | 100 | 37 |
| IPEC | 95 | 17 |
| Total | 407 | 170 |
These results demonstrate that such factors as adhesins, iron uptake systems, and proteases do not necessarily have to be considered virulence-associated factors but can also contribute to the fitness and adaptability of nonpathogenic bacteria. Although many DNA regions which belong to the flexible gene pool of pathogenic E. coli or at least homologues thereof can be detected in the genome of nonpathogenic strain Nissle 1917, those coding for important virulence factors of UPEC strains are absent (protein toxin- or P-fimbria-encoding genes).
Characterization of GEIs of E. coli strain Nissle 1917.
According to the results of the tRNA screening and DNA-DNA hybridization experiments, several horizontally acquired DNA regions may be chromosomally inserted downstream of tRNA-encoding genes in the E. coli Nissle 1917 genome. To verify this assumption and to subclone parts of GEIs contributing to this strain's probiotic character, a cosmid genomic library of strain Nissle 1917 was screened for clones containing tRNA genes of interest and their downstream sequence contexts. The genetic structure of the GEI IVNissle 1917 equivalent has been determined for other E. coli and Yersinia isolates and was confirmed for strain Nissle 1917 by PCR screening and sample sequencing with primers described previously (13, 32).
Characterization of GEI INissle 1917.
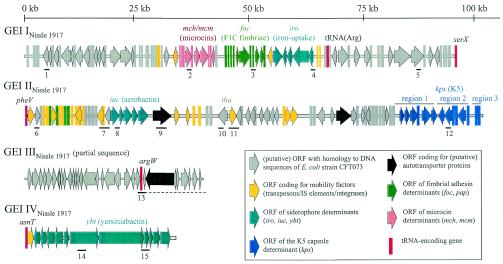
A 97,174-bp genomic region of E. coli strain Nissle 1917 covering the serX tRNA locus and its downstream-located sequences was characterized. For this purpose, two overlapping cosmid clones (pCos3YE4 and pCos2RA4) and four overlapping PCR products were identified or generated and sequenced. This DNA region covers the mch and mcm microcin determinants of strain Nissle 1917 (46), as well as the foc and iro gene clusters coding for F1C fimbriae and the salmochelin iron uptake system, respectively (Fig. 4). The genetic organization and DNA content of this chromosomal fragment are almost identical (99% homology on the nucleotide level) to those of a putative horizontally acquired chromosomal region of E. coli strain CFT073 (accession number AE014075) inserted at serX (56). Consequently, this region represents a GEI of E. coli strain Nissle 1917 (GEI INissle 1917). Thus, the structure and DNA content of this GEI, which carries several determinants coding for fitness-conferring traits (microcins, adhesion, and iron uptake), are highly similar in both strains. In UPEC strain 536, however, PAI III536 (accession number AF302690) shows a different overall genetic organization and chromosomal insertion site (thrW), although this island also carries the iro- and another foc-related S adhesin-encoding determinant sfa(I) as well as remnants of the mcm operon (19).
FIG. 4.
Genetic structure of E. coli Nissle 1917-specific GEIs (GEI INissle 1917 to GEI IVNissle 1917). Important GEI regions or fitness-conferring determinants are highlighted with different colors or patterns. The argW downstream region, identified as a part of GEI IIINissle 1917, is underlined. The localization of tRNA-encoding genes is indicated as well as that of the DNA regions included in the PCR-screening approach for the detection of E. coli strain Nissle 1917-specific sequences.
Characterization of GEI IINissle 1917.
The assembly of overlapping inserts of three cosmids (pCos9YB4, pCos2YE4, and pCos2RF2) resulted in a 103,135-bp DNA region which was termed GEI IINissle 1917 (Fig. 4). This GEI carries the iuc, sat, and iha genes which encode the aerobactin siderophore system, the serine protease Sat (autotransporter), and the putative adherence-conferring protein Iha, respectively, as well as the determinant required for K5 capsule biosynthesis. Additionally, GEI IINissle 1917 contains many putative ORFs with as-yet-unknown functions, a considerable number of transposon- and IS element-related features, and fragments of integrase-encoding genes (Fig. 4). The G+C content of GEI IINissle 1917 (46.9%) differs markedly from that of the E. coli K-12-specific chromosomal backbone (50.8%). The organization and DNA content of this GEI partially resemble those of the pheV-located island of E. coli CFT073.
Two autotransporter proteins are encoded on GEI IINissle 1917: one protein homologous to antigen 43, which may be involved in autoaggregation and biofilm formation (17), and the Sat serine protease. Since the mutational inactivation of sat in E. coli strain CFT073 did not affect its virulence capacity (21, 22), the encoded protease cannot be considered a virulence factor, and the importance of the Sat protein for infection or colonization of the host in vivo remains to be elucidated.
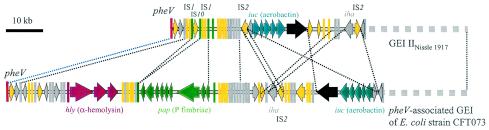
Many GEIs have the capacity to be completely or partially deleted from the chromosome due to homologous or site-specific recombination mediated by bacteriophage integrases (8, 55). In GEI IINissle 1917, two regions which may be deleted by site-specific recombination between flanking repeats were identified (Fig. 4). One of them is about 30 kb in size; contains the iuc, sat, and iha genes; and is flanked by inverted repeats formed by two IS2 elements oriented in opposite directions. Interestingly, the corresponding region of the pheV-associated island of UPEC strain CFT073 contains only one IS2 element and displays a slightly different order of the genes than GEI IINissle 1917 (Fig. 5). Another region of GEI IINissle 1917 also exhibits a transposon-like structure of about 4 kb which is flanked by two IS10 elements in opposite orientation and consists of putative ORFs with homology to transposase-encoding genes and remnants of the pap gene cluster (Fig. 4). This transposon-like structure is absent in UPEC O6 strain CFT073 but is replaced by a 30-kb region containing intact hly and pap determinants coding for the toxin alpha-hemolysin and the P-fimbrial adhesin, which are important UPEC virulence factors (Fig. 5). The pheV-associated GEI of UPEC O6 strain 536 shows a completely different genetic structure although it also contains the pix operon, which is similar to the pap determinant, as well as the K15 capsule-encoding determinant (unpublished data).
FIG. 5.
Comparison of the genetic organization of the left-hand end of GEI IINissle 1917 and the pheV-associated PAI of E. coli strain CFT073, demonstrating the loss of the alpha-hemolysin-encoding determinant (hly) and large parts of the P-fimbrial operon (pap) in strain Nissle 1917. Homologous regions between the two islands are highlighted by identical colors. The color code is identical to that shown on Fig. 4.
Characterization of GEI IIINissle 1917 and GEI IVNissle 1917.
Sequence analysis of the insert of cosmid clone pCos1YA7 demonstrated that the tRNA-encoding gene argW serves as a chromosomal insertion site of horizontally acquired DNA in the genome of strain Nissle 1917 (Fig. 4), which represents a part of GEI IIINissle 1917. The overall homology of this 37.9-kb Nissle 1917-specific genome fragment (47.9% G+C content) to E. coli CFT073-specific sequences is high (98%). The region downstream of argW is homologous to that of the argW-associated island of E. coli strain CFT073, which contains two putative autotransporter protein-encoding ORFs (56). The genetic organization of the argW upstream region differs between both strains. The argW sequence context in the genome of strain UPEC 536 differs from those of strains Nissle 1917 and CFT073 (Grozdanov and Dobrindt, unpublished).
The presence and genetic organization of the DNA region which encodes the siderophore system yersiniabactin (15) was verified in strain Nissle 1917 by sample sequencing of PCR products obtained with primers described before (13, 32). According to the results obtained, we conclude that by analogy to the high-pathogenicity island of Yersinia pseudotuberculosis, the asnT-associated GEI IVNissle 1917 is about 30.2 kb in size and has a G+C content of 57%. The left and right junctions of GEI IVNissle 1917 have already been sequenced, and flanking repeat structures have not been reported (51). This island is present in the same chromosomal insertion site in UPEC O6 strains CFT073 and 536 (19, 56), many isolates of nonhuman pathogenic Salmonella enterica subspecies III and VI, and other commensal E. coli isolates (18, 43).
Analysis of the distribution of E. coli strain Nissle 1917-specific sequences among various E. coli strains from different sources.
In order to study the distribution of E. coli strain Nissle 1917-specific sequences among 324 nonpathogenic and pathogenic E. coli isolates as well as to find out whether an identical repertoire of genetic information can be detected in other E. coli strains, PCRs were designed for the amplification of selected regions of GEI INissle 1917 to GEI IVNissle 1917. These regions include (putative) fitness factor determinants, IS elements, and the border regions of GEI INissle 1917 and GEI IINissle 1917 (Fig. 4). The complete list of the PCR screening results is available as supplementary material on the homepage of the IMIB research group “Enterobacteria” (http://www.uni-wuerzburg.de/infektionsbiologie/imi-start.htm). A large group of ExPEC and IPEC strains, as well as nonpathogenic E. coli strains, has been screened, including (i) ExPEC and IPEC strains from the IMIB collection obtained from various sources, including the ECOR collection, (ii) 70 UPEC isolates of serotype O6 (56, 57), and (iii) 135 nonpathogenic fecal E. coli strains isolated from healthy volunteers. The results obtained (Table 3 and 4) demonstrate that strain Nissle 1917-specific sequences were widely distributed among ExPEC strains, but less frequently present in IPEC strains and the nonpathogenic fecal isolates which are phylogenetically unrelated to E. coli Nissle 1917 relative to the O6:K5 strains. Interestingly, many regions of the GEI INissle 1917 and GEI II Nissle 1917 were highly specific for O6:K5 strains (data available as supplementary material). All of the strain Nissle 1917-specific sequences included into the PCR screening approach were detectable in two of the investigated O6:K5 isolates (strains RZ442 and RZ525), thus suggesting a common clonal origin of these strains. However, further comparison of their genome content using PCR primer pairs for amplification of the two cryptic plasmids of strain Nissle 1917, pMUT1 and pMUT2 (9), showed that both strains contained only pMUT1. In addition, the E. coli strains RZ442 and RZ525 differ phenotypically from strain Nissle 1917. These results imply that the flexible gene pool of E. coli strain Nissle 1917, although lacking typical virulence determinants of pathogenic E. coli strains, more closely resembles that of ExPEC O6:K5 strains. Nevertheless, these DNA regions are, to a certain extent, also detectable in nonpathogenic isolates.
TABLE 4.
Distribution of the investigated E. coli Nissle 1917-specific sequences among nonpathogenic E. coli, ExPEC, and IPEC isolates
| E. coli isolate (n) and/or sourceb | % of E. coli strain Nissle 1917-specific sequences by PCR analysis in regiona (GEI):
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 (I) | 2 (I) | 3 (I) | 4 (I) | 5 (I) | 6 (II) | 7 (II) | 8 (II) | 9 (II) | 10 (II) | 11 (II) | 12 (II) | 13 (III) | 14 (IV) | 15 (IV) | |
| O6:K5 UTI (34) | 100.0 | 97.1 | 94.1 | 100.0 | 100.0 | 100.0 | 23.5 | 97.1 | 97.1 | 100.0 | 67.6 | 100.0 | 76.5 | 100.0 | 100.0 |
| UTI Non-O6:K5 (88) | 55.7 | 21.6 | 28.4 | 71.6 | 69.3 | 54.5 | 6.8 | 30.7 | 14.8 | 29.5 | 8.0 | 13.6 | 5.7 | 96.6 | 77.3 |
| NBM or sepsis (28) | 14.3 | 3.6 | 7.1 | 67.9 | 21.4 | 10.7 | 3.6 | 46.4 | 7.1 | 14.3 | 3.6 | 3.6 | 0.0 | 100.0 | 75.0 |
| IPEC (17) | 29.4 | 0.0 | 5.9 | 17.6 | 5.9 | 29.4 | 0.0 | 23.5 | 0.0 | 41.2 | 5.9 | 0.0 | 0.0 | 70.6 | 29.4 |
| Nonpathogenic (157) | 27.4 | 2.5 | 10.2 | 43.9 | 24.2 | 59.2 | 5.1 | 49.7 | 10.8 | 24.8 | 4.5 | 1.9 | 0.6 | 58.0 | 18.5 |
| Total (324) | 41.7 | 17.6 | 23.5 | 58.0 | 43.5 | 56.5 | 7.1 | 47.8 | 20.1 | 34.0 | 12.0 | 15.4 | 9.9 | 76.5 | 49.1 |
The chromosomal localization of the amplified DNA regions is indicated in Fig. 4.
NBM, newborn meningitis; UTI, urinary tract infection.
DISCUSSION
In this study, the genome content and organization of E. coli strain Nissle 1917 were analyzed to elucidate the genetic basis of its probiotic nature as well as mechanisms underlying its evolution. The nucleotide sequence information of four GEIs carrying fitness factor-encoding determinants was combined with different whole-genome approaches, i.e., a PCR-based tRNA screening and DNA-DNA hybridizations using whole-genome macroarrays. This integrated picture of the strain Nissle 1917 genome was compared with the available information on the genome organization and content of nonpathogenic E. coli K-12 strain MG1655 and two UPEC O6 strains. Thus, it should be possible to define genomic differences responsible for the nonpathogenic, pathogenic, or probiotic nature of these E. coli strains and to speculate on their evolution (Fig. 1).
At least four genomic islands were identified and partially characterized, which are absent in K-12 strain MG1655 and contain most of the determinants coding for known fitness factors of strain Nissle 1917 (7). Additional determinants which may contribute to fitness and which were previously unknown in this strain's genome were identified within these islands (sat, iha, and iro) as well as many putative ORFs coding for hypothetical proteins of unknown function. The overall genome structure and genetic organization of many GEIs of E. coli strain Nissle 1917 more closely resemble those of UPEC strain CFT073 than those of UPEC strain 536. It is tempting to speculate that the first two strains have a common clonal origin. This is also supported by genome restriction patterns analyzed by PFGE (data available as supplementary material). Although the overall genetic structure of the investigated GEIs is very similar, important differences exist which are responsible for the nonpathogenic nature of strain Nissle 1917, whereas strain CFT073 is a uropathogenic isolate.
The serX-associated GEI INissle 1917 is almost identical to a GEI of E. coli strain CFT073 chromosomally inserted at the same tRNA-encoding gene (56). GEI INissle 1917 and PAI III536 of strain 536 also exhibit some common structural features (19). Nevertheless, they differ significantly in the presence of functional determinants coding for fitness factors as the microcin determinants are truncated in the latter strain. In addition, PAI III536 is integrated at thrW, whereas the tRNA screening revealed the same sequence context of thrW in strain Nissle 1917 as in the E. coli CFT073 genome. Microcin expression, mediated by GEI INissle 1917, is probably involved in the antagonistic action of E. coli strain Nissle 1917, which contributes to the successful competition with other bacteria during intestinal colonization and is therefore believed to be important for this strain's probiotic effect (46).
GEI IINissle 1917 represents an island responsible for expression of several important fitness-conferring traits. Many putative ORFs coding for hypothetical proteins with unknown functions are located on GEI IINissle 1917, together with a surprisingly great number of ORFs with homology to mobile and accessory DNA elements, i.e., transposase-encoding genes and IS elements. The genetic organization and gene content of GEI IINissle 1917 closely resemble those of the pheV-associated PAI of E. coli strain CFT073 but also exhibit some important differences with respect to the structure and stability of that strain (Fig. 4 and 5). Whereas the K5 capsule-encoding determinant is present on GEI IINissle 1917, a presumed K2 capsule gene cluster is located on the corresponding island of strain CFT073 (56). K5 capsule expression is important for bacterial adhesion and colonization but does not contribute to serum resistance (14, 29). The expression of the aerobactin iron uptake system is important for the fitness of strain Nissle 1917, as the availability of free iron in the human body is limited. Thus, together with the expression of other systems involved in iron acquisition (ent, fec, ybt, iro, and chu) the aerobactin system contributes to competitiveness, successful survival, and colonization. From our point of view, the presence of six different determinants involved in iron uptake is noteworthy and seems to contribute to the great adaptability and fitness of E. coli strain Nissle 1917. The iha gene codes for a putative nonfimbrial adherence-conferring molecule initially identified as a putative siderophore receptor molecule present in the genomes of EHEC strains, as well as in the genome of UPEC strain CFT073 (47, 50, 54, 56). It has been demonstrated that the iha gene, when cloned into an E. coli K-12 strain, resulted in diffuse adherence to HeLa cells. However, adherence of an iha mutant of EHEC strain 86-24 was not significantly altered relative to the corresponding wild type. Though Iha is sufficient to confer adherence upon nonadherent E. coli, one has to be cautious about designating Iha a virulence factor, as its role during pathogenesis of EHEC and ExPEC is unclear (54). The serine protease-encoding gene sat is located between the iuc gene cluster and iha. Sat secretion and its protease activity were demonstrated (data not shown), but the importance of Sat for gut colonization and fitness of strain Nissle 1917 remains to be elucidated.
The iuc-sat-iha region on GEI IINissle 1917 is flanked by two IS2 elements with opposite orientations. In comparison, the corresponding region of the pheV-associated island of strain CFT073 contains fewer genes per ORF organized in a different order and is not flanked by IS2 copies. In contrast to GEI IINissle 1917, the pheV-associated island of strain CFT073 carries the complete hly and pap gene clusters coding for the important virulence factors alpha-hemolysin and P fimbria, respectively. These determinants, together with other putative hypothetical ORFs, represent a 30-kb region, which is presumably of crucial importance for this strain's virulence properties (12, 30, 31, 35). Interestingly, only a fragmented pap operon is present in a similar DNA context on GEI IINissle 1917 (Fig. 4), associated with a transposon-like element. It is tempting to speculate that during evolution of GEI IINissle 1917 from a pheV-associated island as present in strain CFT073, the intact pap gene cluster was disrupted and partially deleted due to insertion of IS10 elements and consecutive recombination events. These events were important for evolution of an ancestor of strain Nissle 1917 as they were responsible for inactivation of the P-fimbrial operon (and probably for the loss of the alpha-hemolysin-encoding determinant as well), reducing the hypothetical virulence capacity of the corresponding strain.
The organization of GEI IIINissle 1917 (as it is known so far) and of GEI IVNissle 1917 is similar to that of the islands inserted at argW and asnT in E. coli strain CFT073, whereas there is only a counterpart of GEI IVNissle 1917 in strain 536 (PAI IV536).
Since E. coli belongs to the normal human intestinal microflora (commensal as well as ExPEC variants) it is important to investigate the genetic mechanisms involved in the evolution of bacterial pathogens. It has been shown that a considerable fraction of genetic information of ExPEC, which has so far been considered as virulence associated, is also present in many commensal E. coli isolates. Thus, many of these features can be considered rather as contributing to fitness (e.g., iron uptake systems, bacteriocins, proteases, fimbriae, and other adhesins), thereby generally increasing adaptability, competitiveness, and the ability to efficiently colonize the human body, than as typical virulence factors directly involved in infection. This finding is supported by the results of this study, indicating that DNA regions of GEI INissle 1917 to GEI IVNissle 1917 can also be detected in nonpathogenic E. coli isolates. In addition, GEIs contain multiple copies of highly homologous functional and nonfunctional ORFs of mobile genetic elements which do not directly contribute to virulence (18). Whether a commensal E. coli will develop into a pathogen depends not only on the acquisition of fitness-conferring genetic information enabling successful colonization of the host, but also on the presence of functional genes directly contributing to pathogenesis.
From the available data on genome content and organization of E. coli O6 strains Nissle 1917, CFT073, and 536, it is tempting to speculate on the evolution of the nonpathogenic, probiotic character of Nissle 1917 and the uropathogenic character of the latter two isolates. According to the results of DNA-DNA hybridization experiments using E. coli K-12-specific DNA arrays (Fig. 3) and the complete genome sequence of strain CFT073, the overall K-12-specific genome content of strain Nissle 1917 does not differ significantly from that of other pathogenic and nonpathogenic E. coli strains, including UPEC strains CFT073 and 536 (18, 56). It seems that strain Nissle 1917 is characterized by a specific combination of traits, as it acquired a set of important determinants which enable successful survival and colonization of the human intestine. In addition, this strain does not express important UPEC virulence factors (i.e., alpha-hemolysin, P-fimbrial adhesins, serum resistance-conferring long-chain O6 LPS), and it exhibits some phenotypic features which are not typical for the two UPEC isolates included into this study: the presence of six different iron uptake systems, the temperature-independent expression of curli and cellulose (unpublished results), a FimB-independent type 1 fimbrial switch (53), the presence of two small plasmids (9), and a semirough LPS responsible for serum sensitivity (20). This specific combination of traits, which most likely represents the basis of its probiotic nature, might mirror the process of evolutionary withdrawal from the pathogenic E. coli serotype O6 lineages, indicating that horizontal gene transfer, subsequent loss of horizontally acquired genetic information, and point mutations have been involved in the evolution of strain Nissle 1917. There is no known E. coli strain that exhibits the same genotype and phenotype as strain Nissle 1917. Apart from that, the possibility cannot be excluded that other putative ORFs coding for as-yet-hypothetical proteins may be important for fitness or virulence of the investigated strains. The availability of the complete genome sequence of E. coli strain Nissle 1917 is essential for a better understanding of the processes involved in the evolution of E. coli O6 strains as well as for a deeper insight into the genetic basis of the probiotic nature of probiotic strain Nissle 1917.
These findings support our view that E. coli strain Nissle 1917 exhibits a specific pattern of fitness factors but lacks prominent virulence factors which might contribute to its colonization efficiency and survival in the host body, therefore conferring the probiotic effect of this strain. In comparing the genome content of one nonpathogenic, probiotic E. coli O6 strain with that of two UPEC O6 isolates, it becomes clear that it is difficult to define true ExPEC virulence factors. If virulence correlates with the expression of virulence-related factors in pathogenic but not in closely related nonpathogenic variants, the presence of identical genes in pathogenic and nonpathogenic variants of one species indicates that some of their encoded factors (such as adhesins, iron uptake systems, or proteases) contribute to general adaptability, fitness, and competitiveness rather than to particular virulence traits. Consequently, it depends on the niche or growth conditions (a nonpathogenic milieu such as the gastrointestinal tract versus a pathogenic milieu such as the urinary tract) to show whether certain fitness factors can also promote virulence.
Acknowledgments
We are grateful to Ardeypharm GmbH and the Bayerische Forschungsstiftung for financial support. The Göttingen Genomics Laboratory received support from the Forschungsmittel des Landes Niedersachsen.
We thank B. Plaschke (Würzburg) for technical assistance, G. Blum-Oehler (Würzburg) for helpful advice, and Salam Khan (Würzburg) for providing a polyclonal anti-FocA serum.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Appelmelk, B. J., Y. Q. An, T. A. Hekker, L. G. Thijs, D. M. MacLaren, and J. de Graaf. 1994. Frequencies of lipopolysaccharide core types in Escherichia coli strains from bacteraemic patients. Microbiology 140:1119-1124. [DOI] [PubMed] [Google Scholar]
- 3.Berger, H., J. Hacker, A. Juarez, C. Hughes, and W. Goebel. 1982. Cloning of the chromosomal determinants encoding hemolysin production and mannose-resistant hemagglutination in Escherichia coli. J. Bacteriol. 152:1241-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bettelheim, K. A. 1997. Escherichia coli in the normal flora of humans and animals, p. 85-109. In M. Sussman (ed.), Escherichia coli—Mechanism of virulence. Cambridge University Press, Cambridge, England.
- 5.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 6.Blomfield, I. C., M. S. McClain, and B. I. Eisenstein. 1991. Type 1 fimbriae mutants of Escherichia coli K12: characterization of recognized afimbriate strains and construction of new fim deletion mutants. Mol. Microbiol. 5:1439-1445. [DOI] [PubMed] [Google Scholar]
- 7.Blum, G., R. Marre, and J. Hacker. 1995. Properties of Escherichia coli strains of serotype O6. Infection 23:234-236. [DOI] [PubMed] [Google Scholar]
- 8.Blum, G., M. Ott, A. Lischewski, A. Ritter, H. Imrich, H. Tschäpe, and J. Hacker. 1994. Excision of large DNA regions termed pathogenicity islands from tRNA-specific loci in the chromosome of an Escherichia coli wild-type pathogen. Infect. Immun. 62:606-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blum-Oehler, G., S. Oswald, K. Eiteljorge, U. Sonnenborn, J. Schulze, W. Kruis, and J. Hacker. 2003. Development of strain-specific PCR reactions for the detection of the probiotic Escherichia coli strain Nissle 1917 in fecal samples. Res. Microbiol. 154:59-66. [DOI] [PubMed] [Google Scholar]
- 10.Boyd, E. F., and D. L. Hartl. 1998. Chromosomal regions specific to pathogenic isolates of Escherichia coli have a phylogenetically clustered distribution. J. Bacteriol. 180:1159-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Braun, V., R. Gross, W. Koster, and L. Zimmermann. 1983. Plasmid and chromosomal mutants in the iron(III)-aerobactin transport system of Escherichia coli. Use of streptonigrin for selection. Mol. Gen. Genet. 192:131-139. [DOI] [PubMed] [Google Scholar]
- 12.Brauner, A., M. Katouli, and C. G. Ostenson. 1995. P-fimbriation and haemolysin production are the most important virulence factors in diabetic patients with Escherichia coli bacteraemia: a multivariate statistical analysis of seven bacterial virulence factors. J. Infect. 31:27-31. [DOI] [PubMed] [Google Scholar]
- 13.Buchrieser, C., C. Rusniok, L. Frangeul, E. Couve, A. Billault, F. Kunst, E. Carniel, and P. Glaser. 1999. The 102-kilobase pgm locus of Yersinia pestis: sequence analysis and comparison of selected regions among different Yersinia pestis and Yersinia pseudotuberculosis strains. Infect. Immun. 67:4851-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burns, S. M., and S. I. Hull. 1998. Comparison of loss of serum resistance by defined lipopolysaccharide mutants and an acapsular mutant of uropathogenic Escherichia coli O75:K5. Infect. Immun. 66:4244-4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carniel, E., I. Guilvout, and M. Prentice. 1996. Characterization of a large chromosomal “high-pathogenicity island” in biotype 1B Yersinia enterocolitica. J. Bacteriol. 178:6743-6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clermont, O., S. Bonacorsi, and E. Bingen. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Danese, P. N., L. A. Pratt, S. L. Dove, and R. Kolter. 2000. The outer membrane protein, antigen 43, mediates cell-to-cell interactions within Escherichia coli biofilms. Mol. Microbiol. 37:424-432. [DOI] [PubMed] [Google Scholar]
- 18.Dobrindt, U., F. Agerer, K. Michaelis, A. Janka, C. Buchrieser, M. Samuelson, C. Svanborg, G. Gottschalk, H. Karch, and J. Hacker. 2003. Analysis of genome plasticity in pathogenic and commensal Escherichia coli isolates by use of DNA arrays. J. Bacteriol. 185:1831-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dobrindt, U., G. Blum-Oehler, G. Nagy, G. Schneider, A. Johann, G. Gottschalk, and J. Hacker. 2002. Genetic structure and distribution of four pathogenicity islands (PAI I536 to PAI IV536) of uropathogenic Escherichia coli strain 536. Infect. Immun. 70:6365-6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grozdanov, L., U. Zähringer, G. Blum-Oehler, L. Brade, A. Henne, Y. A. Knirel, U. Schombel, J. Schulze, U. Sonnenborn, G. Gottschalk, J. Hacker, E. T. Rietschel, and U. Dobrindt. 2002. A single nucleotide exchange in the wzy gene is responsible for the semirough O6 lipopolysaccharide phenotype and serum sensitivity of Escherichia coli strain Nissle 1917. J. Bacteriol. 184:5912-5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guyer, D. M., I. R. Henderson, J. P. Nataro, and H. L. Mobley. 2000. Identification of sat, an autotransporter toxin produced by uropathogenic Escherichia coli. Mol. Microbiol. 38:53-66. [DOI] [PubMed] [Google Scholar]
- 22.Guyer, D. M., S. Radulovic, F. E. Jones, and H. L. Mobley. 2002. Sat, the secreted autotransporter toxin of uropathogenic Escherichia coli, is a vacuolating cytotoxin for bladder and kidney epithelial cells. Infect. Immun. 70:4539-4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hacker, J., and E. Carniel. 2001. Ecological fitness, genomic islands and bacterial pathogenicity. A Darwinian view of the evolution of microbes. EMBO Rep. 2:376-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hacker, J., U. Hentschel, and U. Dobrindt. 2003. Prokaryotic chromosomes and disease. Science 301:790-793. [DOI] [PubMed] [Google Scholar]
- 25.Hacker, J., and J. B. Kaper. 1999. The concept of pathogenicity islands, p. 1-11. In J. B. Kaper and J. Hacker (ed.), Pathogenicity islands and other mobile virulence elements. ASM Press, Washington D.C.
- 26.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 27.Hartley, C. L., C. S. Neumann, and M. H. Richmond. 1979. Adhesion of commensal bacteria to the large intestine wall in humans. Infect. Immun. 23:128-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hentschel, U., and J. Hacker. 2001. Pathogenicity islands: the tip of the iceberg. Microbes Infect. 3:545-548. [DOI] [PubMed] [Google Scholar]
- 29.Herias, M. V., T. Midtvedt, L. A. Hanson, and A. E. Wold. 1997. Escherichia coli K5 capsule expression enhances colonization of the large intestine in the gnotobiotic rat. Infect. Immun. 65:531-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hull, R. A., B. Nowicki, A. Kaul, R. Runyan, C. Svanborg, and S. I. Hull. 1994. Effect of pap copy number and receptor specificity on virulence of fimbriated Escherichia coli in a murine urinary tract colonization model. Microb. Pathog. 17:79-86. [DOI] [PubMed] [Google Scholar]
- 31.Johanson, I., R. Lindstedt, and C. Svanborg. 1992. Roles of the pap- and prs-encoded adhesins in Escherichia coli adherence to human uroepithelial cells. Infect. Immun. 60:3416-3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karch, H., S. Schubert, D. Zhang, W. Zhang, H. Schmidt, T. Ölschläger, and J. Hacker. 1999. A genomic island, termed high-pathogenicity island, is present in certain non-O157 Shiga toxin-producing Escherichia coli clonal lineages. Infect. Immun. 67:5994-6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 34.Lidin-Janson, G., B. Kaijser, K. Lincoln, S. Olling, and H. Wedel. 1978. The homogeneity of the faecal coliform flora of normal school-girls, characterized by serological and biochemical properties. Med. Microbiol. Immunol. (Berlin). 164:247-253. [DOI] [PubMed] [Google Scholar]
- 35.Linggood, M. A., and P. L. Ingram. 1982. The role of alpha haemolysin in the virulence of Escherichia coli for mice. J. Med. Microbiol. 15:23-30. [DOI] [PubMed] [Google Scholar]
- 36.Lodinova-Zadnikova, R., and U. Sonnenborn. 1997. Effect of preventive administration of a nonpathogenic Escherichia coli strain on the colonization of the intestine with microbial pathogens in newborn infants. Biol. Neonate 71:224-232. [DOI] [PubMed] [Google Scholar]
- 37.Lodinova-Zadnikova, R., H. Tlaskalova-Hogenova, and U. Sonnenborn. 1992. Local and serum antibody response in full-term and premature infants after artificial colonization of the intestine with the E. coli strain Nissle 1917 (Mutaflor). Pediatr. Allergy Immunol. 3:43-48. [Google Scholar]
- 38.Mason, T. G., and G. Richardson. 1981. Escherichia coli and the human gut: some ecological considerations. J. Appl. Bacteriol. 51:1-16. [DOI] [PubMed] [Google Scholar]
- 39.Mobley, H. L., D. M. Green, A. L. Trifillis, D. E. Johnson, G. R. Chippendale, C. V. Lockatell, B. D. Jones, and J. W. Warren. 1990. Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect. Immun. 58:1281-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mühldorfer, I., G. Blum, A. Donohue-Rolfe, H. Heier, T. Ölschläger, H. Tschäpe, U. Wallner, and J. Hacker. 1996. Characterization of Escherichia coli strains isolated from environmental water habitats and from stool samples of healthy volunteers. Res. Microbiol. 147:625-635. [DOI] [PubMed] [Google Scholar]
- 41.Oefner, P. J., S. P. Hunicke-Smith, L. Chiang, F. Dietrich, J. Mulligan, and R. W. Davis. 1996. Efficient random subcloning of DNA sheared in a recirculating point-sink flow system. Nucleic Acids Res. 24:3879-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ölschläger, T. A., U. Dobrindt, B. Janke, B. Middendorf, H. Karch, and J. Hacker. 2003. Analysis of pathogenicity islands of STEC. Methods Mol. Med. 73:99-112. [DOI] [PubMed] [Google Scholar]
- 43.Ölschläger, T. A., D. Zhang, S. Schubert, E. Carniel, W. Rabsch, H. Karch, and J. Hacker. 2003. The high-pathogenicity island is absent in human pathogens of Salmonella enterica subspecies I but present in isolates of subspecies III and VI. J. Bacteriol. 185:1107-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ørskov, I., F. Ørskov, B. Jann, and K. Jann. 1977. Serology, chemistry, and genetics of O and K antigens of Escherichia coli. Bacteriol. Rev. 41:667-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ott, M., L. Bender, G. Blum, M. Schmittroth, M. Achtman, H. Tschäpe, and J. Hacker. 1991. Virulence patterns and long-range genetic mapping of extraintestinal Escherichia coli K1, K5, and K100 isolates: use of pulsed-field gel electrophoresis. Infect. Immun. 59:2664-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patzer, S. I., M. R. Baquero, D. Bravo, F. Moreno, and K. Hantke. 2003. The colicin G, H and X determinants encode microcins M and H47, which might utilize the catecholate siderophore receptors FepA, Cir, Fiu and IroN. Microbiology 149:2557-2570. [DOI] [PubMed] [Google Scholar]
- 47.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 48.Rembacken, B. J., A. M. Snelling, P. M. Hawkey, D. M. Chalmers, and A. T. Axon. 1999. Non-pathogenic Escherichia coli versus mesalazine for the treatment of ulcerative colitis: a randomised trial. Lancet 354:635-639. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 50.Schmidt, H., W. L. Zhang, U. Hemmrich, S. Jelacic, W. Brunder, P. I. Tarr, U. Dobrindt, J. Hacker, and H. Karch. 2001. Identification and characterization of a novel genomic island integrated at selC in locus of enterocyte effacement-negative, Shiga toxin-producing Escherichia coli. Infect. Immun. 69:6863-6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schubert, S., A. Rakin, D. Fischer, J. Sorsa, and J. Heesemann. 1999. Characterization of the integration site of Yersinia high-pathogenicity island in Escherichia coli. FEMS Microbiol. Lett. 179:409-414. [DOI] [PubMed] [Google Scholar]
- 52.Staden, R., K. F. Beal, and J. K. Bonfield. 2000. The Staden package, 1998. Methods Mol. Biol. 132:115-130. [DOI] [PubMed] [Google Scholar]
- 53.Stentebjerg-Olesen, B., T. Chakraborty, and P. Klemm. 1999. Type 1 fimbriation and phase switching in a natural Escherichia coli fimB null strain, Nissle 1917. J. Bacteriol. 181:7470-7478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tarr, P. I., S. S. Bilge, J. C. Vary, Jr., S. Jelacic, R. L. Habeeb, T. R. Ward, M. R. Baylor, and T. E. Besser. 2000. Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect. Immun. 68:1400-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Turner, S. A., S. N. Luck, H. Sakellaris, K. Rajakumar, and B. Adler. 2001. Nested deletions of the SRL pathogenicity island of Shigella flexneri 2a. J. Bacteriol. 183:5535-5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Welch, R. A., V. Burland, G. Plunkett III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 99:17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 58.Zingler, G., G. Blum, U. Falkenhagen, I. Ørskov, F. Ørskov, J. Hacker, and M. Ott. 1993. Clonal differentiation of uropathogenic Escherichia coli isolates of serotype O6:K5 by fimbrial antigen typing and DNA long-range mapping techniques. Med. Microbiol. Immunol. (Berlin). 182:13-24. [DOI] [PubMed] [Google Scholar]
- 59.Zingler, G., M. Ott, G. Blum, U. Falkenhagen, G. Naumann, W. Sokolowska-Köhler, and J. Hacker. 1992. Clonal analysis of Escherichia coli serotype O6 strains from urinary tract infections. Microb. Pathog. 12:299-310. [DOI] [PubMed] [Google Scholar]