Abstract
Bordetella pertussis, the causative agent of whooping cough, has remained endemic in The Netherlands despite extensive nationwide vaccination since 1953. In the 1990s, several epidemic periods have resulted in many cases of pertussis. We have proposed that strain variation has played a major role in the upsurges of this disease in The Netherlands. Therefore, molecular characterization of strains is important in identifying the causes of pertussis epidemiology. For this reason, we have developed a multiple-locus variable-number tandem repeat analysis (MLVA) typing system for B. pertussis. By combining the MLVA profile with the allelic profile based on multiple-antigen sequence typing, we were able to further differentiate strains. The relationships between the various genotypes were visualized by constructing a minimum spanning tree. MLVA of Dutch strains of B. pertussis revealed that the genotypes of the strains isolated in the prevaccination period were diverse and clearly distinct from the strains isolated in the 1990s. Furthermore, there was a decrease in diversity in the strains from the late 1990s, with a remarkable clonal expansion that coincided with the epidemic periods. Using this genotyping, we have been able to show that B. pertussis is much more dynamic than expected.
In many developed countries, whooping cough is an endemic disease with regular epidemic outbreaks. In the 1990s, a significant increase in pertussis incidence was observed in several countries with a high level of vaccination coverage (6, 14, 24). This is remarkable because in these countries nationwide vaccination of children against pertussis had been introduced 50 years ago, and this has resulted in high vaccination coverage. Although the greatest morbidity is observed in children, pertussis is now also considered an important infectious disease of adults (14, 29). Studies in a number of countries have revealed that the circulating Bordetella pertussis population changed significantly after the introduction of the vaccine (3, 13, 22, 23, 25). Significant antigenic divergence between vaccine strains and circulating strains has been observed in vaccinated populations (15, 24). We have provided evidence that the mismatch between vaccine strains and circulating strains has played a role in the reemergence of pertussis in The Netherlands (6, 17, 24, 25, 32, 36, 37). However, in other countries increased awareness, improved diagnosis, and waning immunity have been suggested to be the cause of the resurgence of pertussis (5).
The homogeneity of B. pertussis has hampered the development of molecular typing methods to monitor changes in the pathogen population. Several approaches have been used to study changes in the B. pertussis populations. IS1002 r estriction fragment length polymorphism (RFLP) DNA fingerprinting has revealed temporal changes in the genotypes in the Dutch B. pertussis population, which were associated with the emergence of novel pertussis toxin and pertactin types (35, 37). Pulsed-field gel electrophoresis has also been used to demonstrate genomic polymorphism and temporal trends in B. pertussis populations (2, 30). However, both methods are laborious, and the results are difficult to compare between laboratories. Recently, multilocus sequence typing (MLST) was introduced as a new approach for studying the molecular epidemiology of bacterial pathogens (7-9, 20). An important advantage of MLST, in which sequences of housekeeping genes are used, is that it is a portable technique, making it well suited for interlaboratory comparisons and for studying long-term and global epidemiology. Due to the homogeneity of the B. pertussis species, very little polymorphism could be found in B. pertussis housekeeping genes (36). However, the latter study also showed that analysis of genes encoding surface proteins yielded more allelic variation, which could be used for typing.
In the study presented here, we introduce molecular typing of B. pertussis using variation in direct repeat regions in the B. pertussis genome. This method, known as multiple-locus variable-number tandem repeat analysis (MLVA) does not require culturing and can be applied directly to nasal or pharyngeal swabs. Variable-number tandem repeat (VNTR) analysis revealed considerable heterogeneity of the B. pertussis genome and clonal expansion during epidemic periods.
MATERIALS AND METHODS
Strains and swabs.
For this study, we included 13 B. pertussis strains collected in the prevaccination period from 1949 until 1952 and 185 B. pertussis strains from collected in the years 1992 to 2000. All strains were collected in The Netherlands. Approximately 20 strains collected between July and December of each year were used. In addition, 11 Bordetella parapertussis strains originating from various countries, a Bordetella bronchiseptica strain from The Netherlands, and a Bordetella holmesii isolate from France were used for MLVA. A table with details on the origin of the strains is available from us.
Strains were grown on Bordet-Gengou agar supplemented with 1% glycerol and 4% sheep blood at 35°C for 3 to 4 days. For some strains, purified DNA was prepared as described before (37), and this DNA was used in the PCRs. In the majority of the cases, Bordetella cells were scraped off the plates, suspended in water, and heated for 5 min at 99°C to kill and lyse the cells. The lysate was centrifuged for 2 min at 13,000 × g, and the supernatant was used in the PCRs.
Twenty Bordetella nasal and pharyngeal swabs that were originally positive by IS481 PCR (33) were included in the study. These swabs, kindly provided by A. van der Zee, were collected from patients from various regions in The Netherlands in 2002. Material from the swabs was suspended in 200 μl of saline and stored at 4°C until used in the PCRs.
In silico tandem repeat searches.
At the time of this study, the whole genome of B. pertussis was sequenced and available from the Sanger Institute web site (http://www.sanger.ac.uk/Projects/B_pertussis/). The genome sequence of B. pertussis was screened for the presence of tandem repeat sequences using the Tandem Repeats Finder program, version 2.02 (1).
MLVA.
Each VNTR locus was amplified using a 5′ 6-carboxyfluorescein-labeled forward primer and an unlabeled reverse primer (Table 1). Optimized VNTR PCRs were performed in 20-μl volumes in Applied Biosystems (Foster City, Calif.) 9700 PCR machines. Either 2 μl of 1:400-diluted heat-treated B. pertussis lysate, 1 μl of 10-ng/μl purified genomic DNA, or 1 μl of suspension from a swab was added to a mixture containing 10 pmol of each primer, 10 μl of HotStar Taq master mix (QIAGEN, Hilden, Germany), and 4 μl of betain (catalogue no. W4502; Sigma-Aldrich Chemie, Zwijndrecht, The Netherlands). The PCR program used for lysates or purified DNA was 15 min at 95°C, followed by 28 cycles of amplification that each consisted of 20 s at 95°C, 30 s at 67°C, and 1 min at 72°C, with a final step of 30 min at 68°C to ensure complete terminal transferase activity of the Taq DNA polymerase. For PCR analysis of material from swabs, a touchdown PCR protocol with an extended number of cycles was used. The PCR program started with 15 min at 95°C, followed by a 10-cycle touchdown protocol that consisted of 20 s at 94°C, 30 s at 70 to 60°C, and 30 s at 72°C. At the touchdown temperature, PCR was completed with 40 cycles, each consisting of 20 s at 94°C, 30 s at 60°C, and 2 min at 72°C, with a final step of 30 min at 68°C.
TABLE 1.
Primers used in MAST, MLVA, cloning, and DNA sequencing
| Primer name | Sequence | Coordinatea | Purpose(s) |
|---|---|---|---|
| prn-AF | GCCAATGTCACGGTCCAA | 1098595 | MAST |
| prn-AR | GCAAGGTGATCGACAGGG | 1099163 | MAST |
| ptx-PF | AATCGTCCTGCTCAACCGCC | 3987811 | MAST |
| ptxPR | GGTATACGGTGGCGGGAGGA | 3988365 | MAST |
| TcfAF | TTCTTGCGCGTCGTGTCTTC | 1264378 | MAST |
| TcfAR3 | GCGGTTGCGGACCTTCAT | 1265355 | MAST |
| BP-VNTR1-DF | 5′ FAM-CCTGGCGGCGGGAGACGTGGTGGTG | 2194507 | MLVA |
| BP-VNTR1-DR | AAAATTGCGGCATGTGGGCTGACTCTGA | 2194862 | MLVA |
| VNTR-1SEQF | GGCCGAGGTGCCCAATCCCGACAACATCC | 2194285 | Sequencing |
| VNTR-1SEQR | CGGAAGCGCCCGGGTAGGTCGCGGTGAT | 2195009 | Sequencing |
| BP-VNTR2-BF | 5′ FAM-CGCGCCGCCTACGACCGCTATGG | 2647550 | MLVA, sequencing |
| BP-VNTR2-BR | CCCGCGCCGAAGATCTCGCCAAAGATAT | 2647412 | MLVA, sequencing |
| BP-VNTR3-BF | 5′ FAM-GCCTCGGCGAAATTGCTGAAC | 2591464 | MLVA/Sequencing |
| BP-VNTR3-BR | GCGGGCGAGGAAACGCCCGAGACC | 2591350 | MLVA/Sequencing |
| BP-VNTR3-SQF | CACGCTGTTCGTGGTGCTG | 2591089 | Cloning, sequencing |
| BP-VNTR3-SQR | CAGTTCCTGGCGGATCATCAC | 2591768 | Cloning, sequencing |
| BP-VNTR4-CF | 5′ FAM-CGTGCCCTGCGCCTGGACCTG | 185211 | MLVA |
| BP-VNTR4-BR | GCCGCTGCTCGACGCCAGGGACAA | 185000 | MLVA |
| VNTR-4SEQF | GCTGGCCGGGCTGCGGCTGCCAATCC | 185381 | Sequencing |
| VNTR-4SEQR | CACGCCGCCGCCCGACAGCGACGAG | 184676 | Sequencing |
| BP-VNTR5-BF | 5′ FAM-GAAGCCGGCCCACCCGAGCTCCAGGCTCTT | 1005290 | MLVA, sequencing |
| BP-VNTR5-BR | TGCCGGGTTTCGGCATCTCGATGGGATACG | 1005177 | MLVA, sequencing |
| BPP-VNTR5-F | 5′ FAM-TGCCGGGCTTCGGCATCTCGATGGGGTATG | 1767062 | MLVA |
| BP-VNTR6-EF | 5′ FAM-CCAACGGCGGTCTGCTGGGTGGTC | 2099525 | MLVA |
| BP-VNTR6-FR | CGCCGCCCGCTGCGCCGCTACC | 2099315 | MLVA |
| VNTR-6SEQF | GGCCTGCTGGCCGGCGTGACCGGTAACCT | 2099566 | Sequencing |
| VNTR-6SEQR | GGGGGTCGCCCGAGGAGACTCGTTGGTCTT | 2099177 | Sequencing |
After PCR, samples were diluted 1:125, and 1 μl of the diluted samples was mixed with 10 μl of Hi-Di formamide (Applied Biosystems) containing 0.05 μl of MapMarker Rox 400 Low (Eurogentec, Seraing, Belgium). After heat denaturation for 5 min at 95°C and rapid cooling on ice, the fragments were separated on an ABI 3700 DNA sequencer using the standard GeneScan module with filter set D.
The GeneScan data were imported into the Bionumerics 3.0 software package (Applied Maths, Sint-Martens-Latem, Belgium), and the fragment size and the number of repeats were determined with a custom-made script. Each isolate was defined by a string of six integers (the MLVA profile), corresponding to the number of repeats found at the six different VNTR loci in the order VNTR1, VNTR3a, VNTR3b, VNTR4, VNTR5, and VNTR6. Each unique MLVA profile was assigned an MLVA type.
To confirm the accuracy of the sizing as determined by capillary electrophoresis and the translation of the fragment sizes into repeat numbers, the VNTR PCR fragments of five clinical B. pertussis isolates, the B5 vaccine strain, and the Tohama strain were sequenced. The sequence results showed that the sizing was not accurate and that a slight correction was required to calculate the correct number of repeats. The inaccurate sizing was probably the result of the secondary structure of the PCR product.
MAST.
For multiple antigen sequence typing (MAST), parts of the genes coding for pertactin (prn), the tracheal colonization factor (tcfA), and the pertussis toxin promoter (ptxP) were sequenced, essentially as described previously (36). The primer sequences used for PCR and sequencing are displayed in Table 1. PCRs were performed with HotStar Taq master mix (QIAGEN) with 10 pmol of each primer added to the mixture. After a 15-min incubation at 95°C, prn was amplified in 30 cycles, each cycle consisting of 45 s at 94°C, 45 s at 55°C, and 60 s at 72°C. For the amplification of ptxP and tcfA, touchdown PCRs were used. For the tcfA PCR, the temperature range was 72 to 62°C; for the ptxP gene, the range was 70 to 60°C. For each strain, every unique sequence of the prn, ptxP, and tcfA loci received a distinct allele number. The compositions of these loci were expressed in an allelic profile designated prn-ptxP-tcfA, e.g., 2-1-2.
Data analysis.
The MLVA profiles and MAST profiles were clustered with the Bionumerics software (beta version 3.5) by using a categorical coefficient and a graphing method called minimum spanning tree. The categorical coefficient results in an integer score (cost) depending on the number of VNTR loci that differ between two profiles. If two profiles differ in a single locus, the cost will be 1; if two loci differ, the cost will be 2, etc. In graph theory, minimum spanning trees are well known and are standard tools. In general, a weighted graph consists of a set of vertices or nodes (V) [V = (V1…Vn)] and a set of weighted edges (E) [E = (E1…Em)] connecting these vertices. A minimum spanning tree consists of all vertices and a subset (S) of the edges, so that all vertices are connected and the sum of the weights of the edges in subset S is minimized. In other words, each node is connected to another node at the lowest cost possible. Examples of algorithms that construct such a tree are Kruskal's algorithm or Prim's algorithm (26). Similar to maximum-parsimony phylogenetic tree reconstruction methods, a justification for this model can be found in the fact that identical alleles found in different genotypes are likely to be caused by common ancestry rather than convergent evolution. For most data sets, many equivalent minimum spanning tree solutions exist. This freedom was used to further refine the algorithm, with additional criteria being used to differentiate between the minimum spanning tree solutions. The criteria used for this purpose were taken from the BURST method (http://www.mlst.net/) and adapted for use in the minimum spanning trees. Later addition allows the use of priority rules to first link types that have the highest number of single- and/or double-locus variants, the highest number of entries, and the most frequent states. This method also enables the creation of complexes by setting the maximum-neighbor distance as a rule to include a particular type in a complex. The connections between types differing in more than a single locus are less likely to represent an evolutionary relationship but indicate the connection to the most similar type.
For calculation of the genetic diversity of the Shannon-Weiner diversity index (H), the equation H = − ∑ PilnPi was used, where Pi is the frequency of the ith type divided by the number of strains with this particular type (21).
Cloning of PCR fragments.
Cloning of PCR fragments obtained from B. pertussis was performed using the TOPO TA cloning kit (Invitrogen, Groningen, The Netherlands).
DNA sequencing.
For DNA sequencing reactions, fluorescence-labeled dideoxynucleotide technology was used (Applied Biosystems). PCR products were purified using Qiaquick PCR purification kits (QIAGEN). Sequence reaction mixtures were analyzed on an ABI 3700 automated DNA sequencer. The collected sequences were assembled and edited using Kodon 1.0 software (Applied Maths).
RESULTS
Identification of VNTR loci.
With the Tandem Repeats Finder software, 13 sequences that contained tandem repeats in the B. pertussis genome were initially identified. Primer sequences were designed for all 13 loci and tested on a panel of 12 B. pertussis strains with divergent IS1002 RFLP patterns to assess the level of polymorphism of these loci. Analysis showed that some of the loci were not polymorphic and that others were not present in each strain. This finding resulted in the initial selection of six VNTR primer sets that were deemed to be suitable for typing purposes (Table 2). All VNTR loci were located in open reading frames. Three of the six VNTR loci (VNTR3, VNTR5, and VNTR6) were located in B. pertussis pseudogenes encoding a putative membrane protein, a sulfate transport system permease protein, and a putative membrane protein, respectively. In contrast, in B. bronchiseptica and B. parapertussis these genes are intact. The remaining three VNTRs (VNTR1, VNTR2, and VNTR4) were located in apparently intact genes for a putative efflux inner membrane protein, the chaperone protein DnaJ, and a putative exported protein, respectively. Thus, with the exception of VNTR2, all VNTRs were located in genes coding for membrane-associated or -exported proteins.
TABLE 2.
Characteristics of VNTR loci in B. pertussis, B. bronchiseptica, and B. parapertussis
| VNTR | Size (bp) | Repeat sequence |
B. pertussisNC_02929
|
B. bronchisepticaNC_02927
|
B. parapertussisNC_02928
|
Putative function of open reading frame involved | |||
|---|---|---|---|---|---|---|---|---|---|
| Genome coordinate | No. of repeats | Genome coordinate | No. of repeats | Genome coordinate | No. of repeats | ||||
| VNTR1 | 15 | GAACCCGCCAAGCAG | 2194649 | 9 | 3578750 | 1 | 1883884 | 2 | Efflux system inner membrane |
| VNTR2 | 12 | CCGCCCATGCCG | 2647463 | 5 | 4182223 | 2 | 3768291 | 3 | Chaperone protein DnaJ |
| VNTR3 | 5 | CTGGC | 2591401a | 7 | 4130277 | 2 | 3715072 | 2 | Membrane protein |
| VNTR4 | 12 | CAAGGACAAGGG | 185045 | 9 | 890522 | 7 | 806607 | 4 | Exported protein |
| VNTR5 | 6 | TGGTGC | 1005228a | 7 | 3696052 | 2 | 1767113 | 3 | Sulfate transport system permease |
| VNTR6 | 9 | CGAGCCGCC | 2099406a | 11 | 1922442 | 2 | 2531131 | 5 | Membrane protein |
Pseudogene.
Characteristics of VNTRs in B. pertussis strains.
The 198 strains collected from Dutch whooping cough patients were subjected to MLVA. This revealed that the repeat size of the VNTRs ranged from 5 to 15 bp and that the number of repeats varied between 1 and 13 repeats per VNTR locus (Table 3). The sequences of the repeats in the different VNTR loci were diverse, and the high G+C content of the B. pertussis genome was reflected in the composition of the repeats. The average number of repeats per VNTR showed only modest variation. In 86% (170 of 198) of the strains, VNTR1 carried eight repeats, approximately 90% of the strains carried seven repeats in VNTR3 (179 of 198) and VNTR4 (184 of 198), and 90% (179 of 198) had six repeats in VNTR5. For VNTR6, 40% (80 of 198) of the strains carried seven repeats, and in another 48% (95 of 198), nine repeats were found. During this analysis, VNTR2 hardly displayed polymorphism and had a Shannon-Weiner diversity index of only 0.20. In fact, of the strains tested, only one had two repeats in the VNTR2 locus, one strain had five repeats, and all other strains carried three repeats in VNTR2. For this reason, VNTR2 was omitted from further cluster analyses. The Shannon-Weiner diversity index for VNTR1, VNTR3, VNTR4, and VNTR5 ranged from 0.52 to 0.69, whereas VNTR6 had a considerably higher diversity index of 1.15.
TABLE 3.
Variation of VNTRs in B. pertussis strains obtained in The Netherlands
| VNTR | No. of repeats | No. of variants | Shanon-Weiner index |
|---|---|---|---|
| VNTR1 | 2-10 | 7 | 0.61 |
| VNTR2 | 2-5 | 3 | 0.20 |
| VNTR3a | 1-12 | 10 | 0.69 |
| VNTR3b | 7-9 | 4 | 0.35 |
| VNTR4 | 1-9 | 8 | 0.52 |
| VNTR5 | 3-13 | 7 | 0.47 |
| VNTR6 | 3-12 | 8 | 1.15 |
Stability of the MLVA pattern.
Stability of the MLVA profile during repeated subculturing of B. pertussis strains is essential for its suitability for molecular typing. To determine the stability of the MLVA patterns, three Tohama strains originating from laboratories in The Netherlands, France, and the United States were analyzed by MLVA. Furthermore, three batches of one of the vaccine strains used in The Netherlands originating from 1956, 1975, and 1987 and isolates from three different patients from a pertussis outbreak in a Dutch monastery were also subjected to MLVA. The strains from the monastery were assumed to be epidemiologically related, as they were isolated during the same time period and location from individuals suffering from pertussis. Also, these strains appeared to have identical IS1002 RFLP patterns (35). The MLVA analysis revealed identical MLVA profiles for the vaccine strains subcultured at different times and for the three strains from the monastery outbreak. This indicated that the composition of the VNTR loci is relatively stable and does not change after prolonged storage or subculture in a laboratory. Also, the homogeneity of the epidemiologically linked strains from the monastery outbreak suggests that the VNTR loci remained stable through multiple transmissions and hosts. However, we have not tested the stability and heterogeneity within the bacterial population after extensive subculturing of individual colonies of B. pertussis strains.
Duplication of the VNTR3 locus.
With 15 Dutch strains, the VNTR3 PCR yielded two fragments differing in size by 5 nucleotides (one repeat unit), and with 1 strain, the VNTR3 PCR yielded two fragments differing in size by 10 nucleotides (two repeat units). To determine whether this was caused by a PCR artifact or by duplication of the VNTR3 region, a new primer set was used to amplify a larger segment encompassing the VNTR3 region. PCR was performed on five B. pertussis strains that yielded double bands in the VNTR3 PCR. On agarose gels, only a single band could be distinguished. However, the resolution of the agarose gel was insufficient to separate larger fragments differing only by 5 bp. The 700-bp fragment was sequenced, and this revealed a mixture of sequences that probably differed by one or two repeat units in length. The PCR fragments from the five strains were cloned, and the inserts of six clones of each cloning experiment were analyzed by DNA sequencing. Sequence analysis revealed the presence of two types of sequences that differed in size by one or two repeat units, and this was in full agreement with results obtained in the MLVA analysis. This result showed that a segment of DNA containing the VNTR3 locus had been duplicated in some B. pertussis strains. The exact size of the duplication could not be deduced from these experiments, but based on our sequencing results it should amount to more than 700 bp.
As it was impossible to determine which of the two bands had to be used for the MLVA, we split VNTR3 into VNTR3a and VNTR3b and considered them separate loci. Comparison of the minimum spanning tree constructed with MLVA-MAST data using only the VNTR3a locus showed only minor differences from that constructed with both the VNTR3a and VNTR3b loci.
VNTRs in other Bordetella species.
To determine if the B. pertussis VNTRs were unique for this species, we subjected DNA from B. bronchiseptica, B. parapertussis, and B. holmesii to the VNTR PCRs. The B. bronchiseptica strain yielded fragments in all VNTR PCRs, resulting in a unique VNTR profile, and the composition of the VNTR loci was identical to that found in the genome sequence of B. bronchiseptica (GenBank accession number NC_02927). Similarly, the nine B. parapertussis strains all yielded fragments with five of the six VNTR PCRs. However, the PCR on the VNTR6 locus was successful only after the use of a newly designed reverse primer that fitted the slightly different sequences flanking the VNTR in B. parapertussis. The profiles of the 11 B. parapertussis strains displayed only limited variation. Nine of the 11 B. parapertussis strains were MLVA type 1, and the 2 other strains each had unique MLVA types (MLVA types 2 and 3) differing in only in the VNTR5 and VNTR6 loci. Analysis of the composition of the VNTR loci in the B. parapertussis genome sequence (GenBank accession number NC_02928) revealed that this strain was also MLVA type 1. When a B. holmesii strain was used in the VNTR analysis, only PCRs on VNTR2 and VNTR3 yielded fragments corroborating the divergent character of this Bordetella species. Sequence analysis of the VNTR PCR products of B. bronchiseptica, B. parapertussis, and B. holmesii strains confirmed the presence of repeats virtually identical to those of B. pertussis.
Studying the molecular epidemiology of B. pertussis with MLVA.
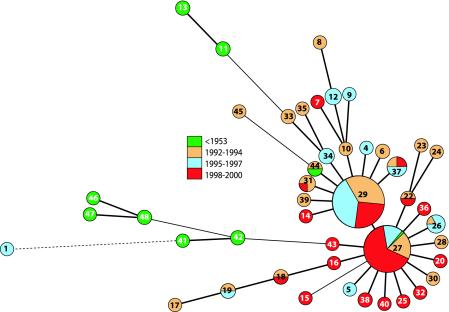
To determine the utility of the MLVA for molecular typing of B. pertussis, a collection of 198 well-defined Dutch B. pertussis isolates were subjected to this typing technique. Thirteen of these strains were collected in the period prior to the introduction of the pertussis vaccine in 1953, and 185 strains were collected from 1992 to 2000, before, during, and after the two recent pertussis epidemics in The Netherlands in 1996 and 1999. Typing resulted in 45 different B. pertussis MLVA profiles (Table 4). A regular categorical clustering showed that the majority of the recent strains isolated from 1998 to 2000 clustered together (data not shown). A second large cluster was composed of strains isolated mainly from 1992 to 1997. The strains isolated in the prevaccination era formed a distinct cluster. Although different clusters could be distinguished, the dendrogram was difficult to interpret because of its size and complexity. For this reason, we used a different approach and constructed a minimum spanning tree. Figure 1 shows the minimum spanning tree based on categorical clustering of the MLVA profiles of the 198 Dutch B. pertussis isolates. Consistent with IS1002 typing (35), the strains isolated during the prevaccination era were clearly distinct from the strains isolated in the 1990s. Furthermore, the tree showed that there were two main MLVA profiles (MLVA types 27 and 29), each representing about 30% (58 and 66 strains, respectively) of all B. pertussis isolates used in the study. The other MLVA profiles radiated from these two dominant ancestral profiles. The majority of MLVA type 27 strains were isolated after 1997 (38 of 58), and there were no strains isolated in 1995 and 1996 with this MLVA type. Although the majority of the strains with MLVA type 29 were isolated before 1999 (58 of 66), this type could be found in all periods studied. The Shannon-Weiner diversity index was calculated for the Dutch strains, stratified by year of isolation (Fig. 2). There was a sharp decrease in the diversity of MLVA types after the 1996 epidemic, with the lowest diversity occurring in 1998, after which diversity increased again. The decrease in genetic diversity suggests a clonal expansion, particularly during the latest epidemic period. There was a single B. pertussis strain that carried an MLVA profile that closely resembled that of the B. parapertussis strains. The speciation of this strain was rechecked to exclude a possible mistake in strain identification. The strain indeed was a B. pertussis strain with an unusual MLVA profile but with B. pertussis-specific MAST characteristics.
TABLE 4.
Composition and frequency of MLVA types in Bordetella strains
| Species | MLVA type | Frequency | No. of repeats at VNTR locus:
|
|||||
|---|---|---|---|---|---|---|---|---|
| VNTR1 | VNTR3a | VNTR3b | VNTR4 | VNTR5 | VNTR6 | |||
| B. pertussis | 1 | 1 | 2 | 2 | 0 | 4 | 3 | 5 |
| 4 | 1 | 4 | 7 | 0 | 7 | 6 | 9 | |
| 5 | 1 | 6 | 7 | 0 | 7 | 6 | 7 | |
| 6 | 2 | 6 | 7 | 0 | 7 | 6 | 9 | |
| 7 | 1 | 7 | 7 | 0 | 5 | 6 | 9 | |
| 8 | 1 | 7 | 7 | 0 | 6 | 6 | 3 | |
| 9 | 4 | 7 | 7 | 0 | 7 | 6 | 3 | |
| 10 | 1 | 7 | 7 | 0 | 7 | 6 | 9 | |
| 11 | 2 | 7 | 7 | 0 | 8 | 5 | 8 | |
| 12 | 1 | 7 | 7 | 0 | 8 | 6 | 9 | |
| 13 | 2 | 7 | 7 | 0 | 9 | 5 | 8 | |
| 14 | 1 | 8 | 3 | 0 | 7 | 6 | 9 | |
| 15 | 1 | 8 | 5 | 0 | 7 | 7 | 7 | |
| 16 | 1 | 8 | 6 | 0 | 7 | 6 | 7 | |
| 17 | 1 | 8 | 6 | 7 | 6 | 6 | 9 | |
| 18 | 2 | 8 | 6 | 7 | 7 | 6 | 7 | |
| 19 | 2 | 8 | 6 | 7 | 7 | 6 | 9 | |
| 20 | 1 | 8 | 7 | 0 | 1 | 6 | 7 | |
| 22 | 2 | 8 | 7 | 0 | 6 | 6 | 7 | |
| 23 | 1 | 8 | 7 | 0 | 6 | 6 | 10 | |
| 25 | 1 | 8 | 7 | 0 | 7 | 5 | 7 | |
| 26 | 6 | 8 | 7 | 0 | 7 | 6 | 6 | |
| 27 | 58 | 8 | 7 | 0 | 7 | 6 | 7 | |
| 28 | 1 | 8 | 7 | 0 | 7 | 6 | 8 | |
| 29 | 66 | 8 | 7 | 0 | 7 | 6 | 9 | |
| 30 | 1 | 8 | 7 | 0 | 7 | 6 | 10 | |
| 31 | 4 | 8 | 7 | 0 | 7 | 7 | 9 | |
| 32 | 1 | 8 | 7 | 0 | 8 | 6 | 7 | |
| 33 | 2 | 8 | 7 | 0 | 8 | 6 | 8 | |
| 34 | 3 | 8 | 7 | 0 | 8 | 6 | 9 | |
| 35 | 1 | 8 | 7 | 0 | 8 | 7 | 9 | |
| 36 | 2 | 8 | 7 | 8 | 7 | 6 | 7 | |
| 37 | 8 | 8 | 7 | 8 | 7 | 6 | 9 | |
| 38 | 1 | 8 | 7 | 9 | 7 | 6 | 7 | |
| 39 | 1 | 8 | 8 | 0 | 7 | 6 | 9 | |
| 40 | 1 | 8 | 12 | 0 | 7 | 6 | 7 | |
| 41 | 1 | 9 | 5 | 0 | 4 | 12 | 7 | |
| 42 | 2 | 9 | 5 | 0 | 7 | 12 | 7 | |
| 43 | 1 | 9 | 7 | 0 | 7 | 6 | 7 | |
| 44 | 2 | 9 | 7 | 0 | 7 | 6 | 9 | |
| 45 | 1 | 9 | 8 | 0 | 7 | 6 | 6 | |
| 46 | 1 | 10 | 5 | 0 | 6 | 13 | 7 | |
| 47 | 1 | 10 | 5 | 0 | 7 | 11 | 7 | |
| 48 | 2 | 10 | 5 | 0 | 7 | 13 | 7 | |
| B. parapertussis | 1 | 9 | 2 | 2 | 0 | 4 | 3 | 5 |
| 2 | 1 | 2 | 2 | 0 | 4 | 6 | 5 | |
| 3 | 1 | 2 | 2 | 0 | 5 | 3 | 5 | |
| B. bronchiseptica | 86 | 1 | 1 | 2 | 0 | 7 | 2 | 2 |
FIG. 1.
Minimum spanning tree of MLVA of Dutch B. pertussis strains. A categorical coefficient and the BURST priority rule of the highest number of single-locus changes were used for the clustering. Each circle in the tree represents a different MLVA type, and the type number is indicated by the number in the circle. Heavy short lines connecting two MLVA types denote types differing by a single MLVA locus, thin longer lines connect double-locus variants, and dotted lines indicate the most likely connection between two types differing by more than two MLVA loci. The connections between types differing in more than a single locus are less likely to represent an evolutionary relationship but indicate the connection to the most similar type. The size of the circle indicates the number of strains with this particular MLVA type; large circles indicate predominant types. The colors indicate the period of time in which the strains included were isolated. If strains isolated from different time periods carry identical MLVA types, pie charts are used to indicate distribution. The predominant MLVA types are types 27 and 29.
FIG. 2.
Relationship between genetic diversity of the B. pertussis population and the number of pertussis notifications. (A and B) Shannon-Weiner diversity index (H) of the MLVA (A) and MAST (B) profiles of Dutch B. pertussis strains isolated in the 1990s; (C) pertussis notifications.
Trends in MAST and MLVA frequencies.
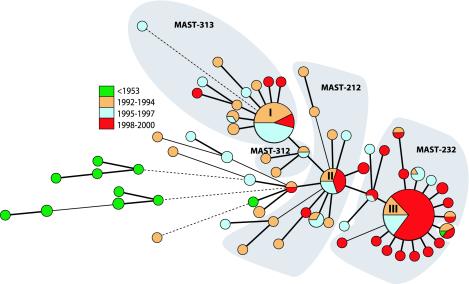
Previous work has revealed significant changes in the frequencies of prn, tcfA, and ptxP alleles ((25, 36); our unpublished results). To visualize this and compare these changes with changes in the MLVA profile, the alleles of the three virulence factors were combined into a MAST profile. Consistent with the MLVA data presented in the previous studies, the diversity of the MAST profiles seemed relatively stable until 1996, with a maximum fluctuation of 0.2 in diversity. However, after 1996 the diversity decreased considerably, from 1.4 in 1996 to 0.7 in 2000; in 2000, 80% of the strains were of MAST profile 2-3-2 (Fig. 2). In contrast, the diversity of the MLVA profiles appeared to decrease until 1998, after which diversity increased again, indicating that VNTRs evolve more rapidly than the virulence factors. The MAST and MLVA profiles were combined into large composite profiles, and a minimum spanning tree was constructed based on the combined profile (Fig. 3). The prevaccination strains were diverse and were positioned distantly from the other strains in the minimum spanning tree. There were three predominant MLVA-MAST profiles comprising more than half (102 of 198) of all strains included in the analysis. MAST-MLVA type I, comprising 35 strains, consisted mainly of strains isolated from 1992 to 1994 (43%) and 1995 to 1997 (46%). Only 11% of this type consisted of recent strains, isolated from 1998 to 2000. The second predominant MAST-MLVA type (type II), comprising 20 strains, was composed of virtually equal numbers of strains from each of the three periods of isolation. More than 47% (34 of 72) of the strains collected from 1998 to 2000 belonged to a single MAST-MLVA type, designated type III. In fact, more than 72% (34 of 37) of type III was made up of strains isolated from 1998 to 2000. In the minimum spanning tree, the transitions in MAST profiles (Fig. 3) are clearly visible. The first area, surrounding type I, indicates the combination MAST-313 (prn3-ptxP1-tcfA3). This group is connected to the small intermediate group MAST-312, in which there has been a change from tcfA3 to tcfA2. The area surrounding type II denotes the strains with MAST-212, in which prn3 has changed into prn2. In the last area, encompassing type III strains with MAST-232, ptxP1 has changed into ptxP3. This suggests a recent clonal expansion of B. pertussis in The Netherlands, leading to a predominance of strains with the MAST-232 and MLVA type 27 combination.
FIG. 3.
Minimum spanning tree of the combined MLVA-MAST profiles of Dutch B. pertussis strains. A categorical coefficient and the BURST priority rule of the highest number of single-locus changes were used for the clustering. Each circle in the tree represents a different MAST-MLVA type. The predominant combined types, I, II, and III, are indicated in the circles. The gray areas denote the combined types with particular MAST profiles. Further description of symbols and colors can be found in the legend to Fig. 1.
Direct MLVA of nasal and pharyngeal swabs.
One of the advantages of MLVA typing is that it is PCR based and does not require growth of bacteria and extraction and purification of DNA. Therefore, the method may also be used to type B. pertussis directly from clinical samples. To determine the feasibility of this approach, 20 swabs from patients with whooping cough were used. All swabs were retested in an IS481 PCR (34) to verify that the swabs still contained sufficient B. pertussis DNA for PCR detection. Of the 20 swabs, 17 yielded an IS481 PCR product upon retesting after storage, prior to performance of the VNTR PCRs. The three IS481 PCR-negative samples also did not yield any product in the VNTR PCRs, possibly because the Bordetella DNA had been degraded. Of the remaining 17 samples, 10 gave complete MLVA profiles, 4 samples generated incomplete profiles, and the remaining 3 samples did not yield any VNTR product. This result indicates that MLVA typing can also be performed directly on clinical samples, which may be helpful in molecular epidemiology studies.
DISCUSSION
In this study, we have shown that the MLVA is a valuable typing technique to characterize B. pertussis isolates. MLVA is a robust, simple, and portable method which can be used to create strain profiles that are easily electronically exchanged. MLVA has been successfully used to type several different bacterial species and proven to be an excellent method with high resolution, particularly useful for organisms with a low level of sequence diversity (4, 10-12, 16, 18, 19, 27, 28, 31). Although MLVA resulted in high-resolution typing of B. pertussis, the analysis was significantly enhanced after MLVA was combined with sequencing-based analysis of three B. pertussis virulence genes. The latter typing method, designated MAST, also yielded allelic profiles (36). The combined MAST-MLVA profiles were used to perform clustering analyses of the Dutch B. pertussis strains isolated before the introduction of the pertussis vaccine in 1953 and isolates obtained before and after the pertussis epidemics in the 1990s, a period in which neither vaccine formulation nor vaccine coverage have changed. The analysis showed that the profiles from strains predating the vaccination era were more diverse than those isolated in the 1990s and only distantly related to the recent strains. Furthermore, there was a strong decrease in diversity in the genotypes of the B. pertussis strains during and after the epidemics in the 1990s, suggesting that these epidemics were caused by a limited number of strains (clonal expansion). Interestingly, the decrease in MLVA diversity preceded that of MAST. Further, MLVA diversity increased again after 1998, while diversity based on MAST continued its downward trend. The different dynamics of MLVA and MAST diversity trends may be due to the fact that VNTRs evolve more rapidly than virulence genes. Consistent with this assumption, single-locus variants of the most recent predominant combined (MLVA-MAST) genotype were almost completely due to changes in the VNTR loci. Thus, VNTR analyses may be especially suitable to detect short-term changes in pathogen populations.
It is clear that changes in the virulence genes bear relevance for the biological processes involved in the increase of pertussis. MLVA markers may not reveal causal relationships but can be helpful to signal clonal expansions and thus visualize the spread of a subgroup of the B. pertussis population with increased fitness, e.g., because B. pertussis is able to escape host immunity. Previous studies showed that antibodies against Prn1, present in the current vaccine, are less efficient in protecting against pertussis in animals challenged with the other Prn variants (17). Variation of the prn gene is caused by variation in the number and composition of the repeats in this virulence gene. Hence, this is an example of a VNTR within a virulence gene in which variation results in antigenic change and possible vaccine escape. The functions of the VNTRs used for MLVA typing described in this study remain uncertain. However, it was noteworthy that, with the exception of VNTR3, the repeat units in the B. pertussis VNTRs were all 3-bp multiples. As virtually all Bordetella VNTRs are located in open reading frames encoding possibly surface-exposed proteins, this suggests that variation in the VNTRs may be related to antigenic variation. Our data suggest that some of these putative membrane protein genes do not vary in B. parapertussis and B. bronchiseptica. We did not observe variation in the number of VNTR3 repeats in the B. parapertussis strains tested. In addition, we showed that the number of VNTR3 repeats in these strains was the same in the B. bronchiseptica strain analyzed and in the strain containing the sequenced genome. These observations indicate that this gene may be essential in these two species and that variation in the number of VNTR3 repeats leads to lethal inactivation of this gene. In contrast, VNTR3 in B. pertussis can vary freely without affecting expression, as VNTR3 is located in an inactive gene in this species. The VNTR5 and VNTR6 loci are also located in inactive B. pertussis pseudogenes, genes which are intact in B. bronchiseptica and B. parapertussis. VNTR5 seems to vary in B. parapertussis and may be involved in antigenic variation. Yet, this locus also seems to vary in B. pertussis, where this gene is no longer active, and thus will not be involved in antigenic variation. VNTR6 is the locus with the highest degree of variation among the B. pertussis strains but appears to be stable in the B. parapertussis and B. bronchiseptica strains tested. VNTR1 and VNTR4 are located in apparently intact putative membrane-associated genes in B. pertussis. Variation in the number of repeats in these genes will result in altered amino acid composition and may represent antigenic variation. However, the Shannon-Weiner index of diversity of these VNTRs is similar to indices for VNTR3 and VNTR5, which reside in pseudogenes. VNTR2, located in the dnaJ gene, displayed the smallest amount of variation and was not used for MLVA for this reason. However, the number of repeats in this locus did vary, and the number of repeats in B. parapertussis differs from that in B. bronchiseptica, indicating that VNTR2 varies in these species as well. Apparently, variation in the dnaJ gene is limited, possibly due to the essential role of dnaJ in the bacterial cell. Overall, there is no clear indication that variation of the VNTRs is greater in loci located in pseudogenes than in loci in intact genes. This observation suggests that changes in the number of repeats do not cause antigenic variation as the result of immunological pressure, although the possibility of such a mechanism is not ruled out.
In several B. pertussis strains, the VNTR3 locus and the flanking regions containing the inactive pseudogene were duplicated. The sequence analysis showed that the regions close to the VNTR locus were identical in both copies and that the only difference between the two copies was the number of repeats. This implies that duplication of this region took place after the putative gene became a pseudogene and that the number of VNTR3 repeats changed during or after duplication. This could also mean that more strains may carry a duplication of the VNTR3 region but that, in these cases, the number of repeats in the duplicated region is identical to that of the original genome segment. Such duplications would yield identical fragments in MLVA and therefore go unnoticed. None of the sequenced genomes of B. pertussis, B. parapertussis, and B. brochiseptica isolates contained a duplication of the VNTR3 region.
In this study, we have introduced the minimum spanning tree for analysis of the MLVA and MAST profiles. In biology, minimum spanning trees can easily be applied to multistate data such as MLVA, MAST, or MLST profiles. The vertices represent unique genotypes, and each pair of genotypes is connected by an edge, with a weight that is determined by the number of different alleles. A minimum spanning tree results in a topology that connects genotypes at the lowest possible cost, representing the minimum number of allele changes. In the trees, the predominant types can be considered the most likely ancestors, with their descendants radiating as variants of the ancestral type. The minimum spanning trees were easier to interpret than large dendrograms; in contrast to the BURST diagrams, they displayed possible relationships between the various clonal complexes.
MLVA has yielded a typing method with a high discriminatory power that can be used for high-throughput typing. In the study described here, we have used separate PCRs for each VNTR locus, but in the meantime we also successfully used multiplex PCRs to amplify three VNTR regions in a single reaction. The number of PCRs in a VNTR multiplex PCR is not limited by the number of compatible primer sets but rather by the number of different fluorescent labels that can be combined in a single channel on the automated DNA sequencer. Nevertheless, with the MLVA described in this report, two multiplex PCRs per sample would suffice. With an average run time of 3 h on the automated sequencer, 192 samples can be typed in a single day on a 96-channel automated sequencer. This may be particularly useful for the screening of a large number of samples, such as nasal or pharyngeal swabs. In the study presented here, we demonstrated that MLVA of such samples is feasible although it can be used only for molecular epidemiology. The method is not suited for diagnostic purposes, as too many samples will yield incomplete MLVA profiles and may require optimization to increase sensitivity before large-scale screening is started. The digital character of the MLVA will enable the creation of international databases to accurately determine geographical differences or similarities between B. pertussis strains isolated in various parts of the world. Particularly when combined with the MAST, this may reveal trends and possibly unexpected transmission routes of this respiratory pathogen.
REFERENCES
- 1.Benson, G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27:573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bisgard, K. M., C. D. Christie, S. F. Reising, G. N. Sanden, P. K. Cassiday, C. Gomersall, W. A. Wattigney, N. E. Roberts, and P. M. Strebel. 2001. Molecular epidemiology of Bordetella pertussis by pulsed-field gel electrophoresis profile: Cincinnati, 1989-1996. J. Infect. Dis. 183:1360-1367. [DOI] [PubMed] [Google Scholar]
- 3.Cassiday, P., G. Sanden, K. Heuvelman, F. Mooi, K. M. Bisgard, and T. Popovic. 2000. Polymorphism in Bordetella pertussis pertactin and pertussis toxin virulence factors in the United States, 1935-1999. J. Infect. Dis. 182:1402-1408. [DOI] [PubMed] [Google Scholar]
- 4.Coletta-Filho, H. D., M. A. Takita, A. A. de Souza, C. I. Aguilar-Vildoso, and M. A. Machado. 2001. Differentiation of strains of Xylella fastidiosa by a variable number of tandem repeat analysis. Appl. Environ. Microbiol. 67:4091-4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crowcroft, N. S., and J. Britto. 2002. Whooping cough—a continuing problem. Br. Med. J. 324:1537-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Melker, H. E., J. F. Schellekens, S. E. Neppelenbroek, F. R. Mooi, H. C. Rumke, and M. A. Conyn-van Spaendonck. 2000. Reemergence of pertussis in the highly vaccinated population of the Netherlands: observations on surveillance data. Emerg. Infect. Dis. 6:348-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dingle, K. E., F. M. Colles, D. R. A. Wareing, R. Ure, A. J. Fox, F. E. Bolton, H. J. Bootsma, R. J. L. Willems, R. Urwin, and M. C. J. Maiden. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enright, M. C., N. P. J. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049-3060. [DOI] [PubMed] [Google Scholar]
- 10.Farlow, J., D. Postic, K. L. Smith, Z. Jay, G. Baranton, and P. Keim. 2002. Strain typing of Borrelia burgdorferi, Borrelia afzelii, and Borrelia garinii by using multiple-locus variable-number tandem repeat analysis. J. Clin. Microbiol. 40:4612-4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farlow, J., K. L. Smith, J. Wong, M. Abrams, M. Lytle, and P. Keim. 2001. Francisella tularensis strain typing using multiple-locus, variable-number tandem repeat analysis. J. Clin. Microbiol. 39:3186-3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frothingham, R., and W. A. Meeker-O'Connell. 1998. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology 144:1189-1196. [DOI] [PubMed] [Google Scholar]
- 13.Fry, N. K., S. Neal, T. G. Harrison, E. Miller, R. Matthews, and R. C. George. 2001. Genotypic variation in the Bordetella pertussis virulence factors pertactin and pertussis toxin in historical and recent clinical isolates in the United Kingdom. Infect. Immun. 69:5520-5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guris, D., P. M. Strebel, B. Bardenheier, M. Brennan, R. Tachdjian, E. Finch, M. Wharton, and J. R. Livengood. 1999. Changing epidemiology of pertussis in the United States: increasing reported incidence among adolescents and adults, 1990-1996. Clin. Infect. Dis. 28:1230-1237. [DOI] [PubMed] [Google Scholar]
- 15.He, Q., J. Makinen, G. Berbers, F. R. Mooi, M. K. Viljanen, H. Arvilommi, and J. Mertsola. 2003. Bordetella pertussis protein pertactin induces type-specific antibodies: one possible explanation for the emergence of antigenic variants? J. Infect. Dis. 187:1200-1205. [DOI] [PubMed] [Google Scholar]
- 16.Keim, P., A. M. Klevytska, L. B. Price, J. M. Schupp, G. Zinser, K. L. Smith, M. E. Hugh-Jones, R. Okinaka, K. K. Hill, and P. J. Jackson. 1999. Molecular diversity in Bacillus anthracis. J. Appl. Microbiol. 87:215-217. [DOI] [PubMed] [Google Scholar]
- 17.King, A. J., G. Berbers, H. F. van Oirschot, P. Hoogerhout, K. Knipping, and F. R. Mooi. 2001. Role of the polymorphic region 1 of the Bordetella pertussis protein pertactin in immunity. Microbiology 147:2885-2895. [DOI] [PubMed] [Google Scholar]
- 18.Klevytska, A. M., L. B. Price, J. M. Schupp, P. L. Worsham, J. Wong, and P. Keim. 2001. Identification and characterization of variable-number tandem repeats in the Yersinia pestis genome. J. Clin. Microbiol. 39:3179-3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, Y., M.-A. Lee, E.-E. Ooi, Y. Mavis, A.-L. Tan, and H.-H. Quek. 2003. Molecular typing of Salmonella enterica serovar Typhi isolates from various countries in Asia by a multiplex PCR assay on variable-number tandem repeats. J. Clin. Microbiol. 41:4388-4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maiden, M. C. J., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Margalef, R. 1958. Information theory in ecology. Gen. Syst. 3:36-71. [Google Scholar]
- 22.Mastrantonio, P., P. Spigaglia, H. van Oirschot, H. G. van der Heide, K. Heuvelman, P. Stefanelli, and F. R. Mooi. 1999. Antigenic variants in Bordetella pertussis strains isolated from vaccinated and unvaccinated children. Microbiology 145:2069-2075. [DOI] [PubMed] [Google Scholar]
- 23.Mooi, F. R., Q. He, H. van Oirschot, and J. Mertsola. 1999. Variation in the Bordetella pertussis virulence factors pertussis toxin and pertactin in vaccine strains and clinical isolates in Finland. Infect. Immun. 67:3133-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mooi, F. R., I. H. van Loo, and A. J. King. 2001. Adaptation of Bordetella pertussis to vaccination: a cause for its reemergence? Emerg. Infect. Dis. 7:526-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mooi, F. R., H. van Oirschot, K. Heuvelman, H. G. van der Heide, W. Gaastra, and R. J. L. Willems. 1998. Polymorphism in the Bordetella pertussis virulence factors P.69/pertactin and pertussis toxin in The Netherlands: temporal trends and evidence for vaccine-driven evolution. Infect. Immun. 66:670-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Rourke, J. 1998. Computational geometry in C, 2nd ed. Cambridge University Press, Cambridge, England.
- 27.Pourcel, C., Y. Vidgop, F. Ramisse, G. Vergnaud, and C. Tram. 2003. Characterization of a tandem repeat polymorphism in Legionella pneumophila and its use for genotyping. J. Clin. Microbiol. 41:1819-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sabat, A., J. Krzyszton-Russjan, W. Strzalka, R. Filipek, K. Kosowska, W. Hryniewicz, J. Travis, and J. Potempa. 2003. New method for typing Staphylococcus aureus strains: multiple-locus variable-number tandem repeat analysis of polymorphism and genetic relationships of clinical isolates. J. Clin. Microbiol. 41:1801-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Senzilet, L. D., S. A. Halperin, J. S. Spika, M. Alagaratnam, A. Morris, and B. Smith. 2001. Pertussis is a frequent cause of prolonged cough illness in adults and adolescents. Clin. Infect. Dis. 32:1691-1697. [DOI] [PubMed] [Google Scholar]
- 30.Syedabubakar, S. N., R. C. Matthews, N. W. Preston, D. Owen, and V. Hillier. 1995. Application of pulsed field gel electrophoresis to the 1993 epidemic of whooping cough in the United Kingdom. Epidemiol. Infect. 115:101-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Belkum, A., S. Scherer, W. van Leeuwen, D. Willemse, L. van Alphen, and H. Verbrugh. 1997. Variable number of tandem repeats in clinical strains of Haemophilus influenzae. Infect. Immun. 65:5017-5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Boven, M., H. E. de Melker, J. F. Schellekens, and M. Kretzschmar. 2000. Waning immunity and sub-clinical infection in an epidemic model: implications for pertussis in The Netherlands. Math. Biosci. 164:161-182. [DOI] [PubMed] [Google Scholar]
- 33.van der Zee, A., C. Agterberg, M. Peeters, J. Schellekens, and F. R. Mooi. 1993. Polymerase chain reaction assay for pertussis: simultaneous detection and discrimination of Bordetella pertussis and Bordetella parapertussis. J. Clin. Microbiol. 31:2134-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Zee, A., H. Groenendijk, M. Peeters, and F. R. Mooi. 1996. The differentiation of Bordetella parapertussis and Bordetella bronchiseptica from humans and animals as determined by DNA polymorphism mediated by two different insertion sequence elements suggests their phylogenetic relationship. Int. J. Syst. Bacteriol. 46:640-647. [DOI] [PubMed] [Google Scholar]
- 35.van der Zee, A., S. Vernooij, M. Peeters, J. van Embden, and F. R. Mooi. 1996. Dynamics of the population structure of Bordetella pertussis as measured by IS1002-associated RFLP: comparison of pre- and post-vaccination strains and global distribution. Microbiology 142:3479-3485. [DOI] [PubMed] [Google Scholar]
- 36.van Loo, I. H., K. J. Heuvelman, A. J. King, and F. R. Mooi. 2002. Multilocus sequence typing of Bordetella pertussis based on surface protein genes. J. Clin. Microbiol. 40:1994-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Loo, I. H., H. G. van der Heide, N. J. Nagelkerke, J. Verhoef, and F. R. Mooi. 1999. Temporal trends in the population structure of Bordetella pertussis during 1949-1996 in a highly vaccinated population. J. Infect. Dis. 179:915-923. [DOI] [PubMed] [Google Scholar]