Abstract
Dental cementum (DC) covers the tooth root and has important functions in tooth attachment and position. DC can be lost to disease, and regeneration is currently unpredictable due to limited understanding of DC formation. This study used a model of experimentally-induced apposition (EIA) in mice to identify proteins associated with new DC formation. Mandibular first molars were induced to super-erupt for 6 and 21 days after extracting opposing maxillary molars. Decalcified and formalin-fixed paraffin-embedded mandible sections were prepared for laser capture microdissection. Microdissected protein extracts were analyzed by liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS), and the data submitted to repeated measure ANOVA test (RM-ANOVA, alpha=5%). A total of 519 proteins were identified, with 97 (18.6%) proteins found exclusively in EIA sites and 50 (9.6%) proteins exclusively expressed in control sites. Fifty six (10.7%) proteins were differentially regulated by RM-ANOVA (p<0.05), with 24 regulated by the exclusive effect of EIA (12 proteins) or the interaction between EIA and time (12 proteins), including serpin 1a, procollagen C-endopeptidase enhancer, tenascin X (TNX), and asporin (ASPN). In conclusion, proteomic analysis demonstrated significantly altered protein profile in DC under EIA, providing new insights on DC biology and potential candidates for tissue engineering applications.
Keywords: dental cementum, proteomic analysis, periodontal ligament, tissue apposition
Graphical abstract

1. Introduction
Dental cementum (DC) is a mineralized tissue that covers the tooth root surface and is part of the periodontal attachment complex. DC is present in human teeth in two primary types, acellular and cellular cementum. Acellular cementum anchors collagen fibers from the periodontal ligament (PDL), promoting attachment to the surrounding alveolar bone (AB). The apical cellular cementum adjusts the post-eruptive tooth position. DC is produced by cementoblasts, derived from the differentiation of ectomesenchymal cells from the dental follicle [1, 2]. During cementogenesis of the cellular type of DC, some cementoblasts are entrapped within the cementoid and become cementocytes, residents of the DC that dwell within lacunae and feature dendritic processes within a lacunocanalicular system, similar to osteocytes of the bone [3]. While it is now understood that osteocytes modulate bone homeostasis and remodeling, act as mechanical sensors, and play a role in endocrine regulation of mineral metabolism [4], it remains unclear whether cementocytes play an active role in either cementum formation or repair.
Like bone, the DC extracellular matrix (ECM) is primarily composed of collagens (predominantly type I and smaller amounts of types III, IV, V, XI, and XII), several non-collagenous proteins thought to regulate mineralization, such as bone sialoprotein (BSP) and osteopontin (OPN), and proteoglycans, including biglycan (BGN) and decorin (DCN) [1, 5]. Unlike bone, DC is avascular, non-innervated, and grows by apposition with no physiological role for remodeling or turnover. While it is known that DC regeneration is possible, current clinical strategies to regenerate periodontal tissues often lack a biologic basis for treatment and have unpredictable outcomes with limited regeneration, especially for DC [6]. Studies focusing on transgenic animals have identified novel regulators of cementogenesis [7–9]. Comparative proteomic analysis of human DC vs. alveolar bone (AB) identified differentially expressed proteins that may be associated with physiological differences between the two tissues [10].
In order to identify factors involved in neocementogenesis, or new cementum formation, that may be novel targets for cementum regeneration, we used a previously established model of mouse molar super-eruption [11, 12]. Experimentally-induced apposition (EIA) in mandibular molars of mice, by extraction of maxillary molars, was shown to generate new DC formation. By using laser capture microdissection (LCM) on EIA and control tissues, we harvested DC and performed proteomic profiling by liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS). The resulting analysis identified significantly differentially expressed proteins resulting from EIA, as well as evidence for cementocyte cell activity.
2. Materials and Methods
2.1. Animal model and histological processing
Animals were housed within the University of Campinas animal facility at the Piracicaba Dental School, and all experimental procedures were approved by the University Committee for Ethics in Animal Research (Protocol # 2160-1). Male Swiss Webster mice (35 days of age) were kept in plastic cages on a 12-hour light/dark cycle at 25 to 30° C, with rodent chow and tap water ad libitum. To promote experimentally induced apposition (EIA) of DC, we used a previously verified mouse model wherein extraction of maxillary molars removed mandibular molars from occlusion (i.e. they become unloaded), promoting increased appositional growth of cellular DC on the tooth apex [11, 12]. The experimental side (right or left) was chosen randomly, and first and second maxillary molars were extracted while the mouse was under anesthesia (Figure 1A). The contralateral side was used as the untreated control. Experimental groups were divided as follows: 1) Control at 6 days after surgery (control-6D) (n=7); 2) Control at 21 days after surgery (control-21D) (n=7); 3) EIA at 6 days after tooth extraction (EIA-6D) (n=7); and 4) EIA at 21 days after tooth extraction (EIA-21D) (n=7). Following euthanasia, mandibles were dissected, fixed in 10% Protocol® buffered formalin (Fisher Diagnostics, USA), and processed for histology according to a protocol previously described for laser capture microdissection (LCM) of decalcified and formalin-fixed paraffin-embedded (FFPE) samples [13]. Longitudinal 5 μm thick sections of the mandibular first molars were obtained and mounted onto PEN membrane glass slides (Applied Biosystems, USA). Sections were deparaffinized in two changes of xylene for 2 and 5 minutes respectively, air dried for 5 minutes, and then immediately microdissected.
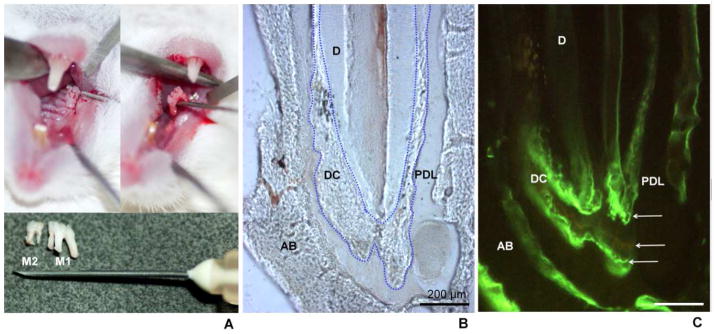
Figure 1. Experimentally-induced dental cementum apposition model in mouse.
(A) Extraction of first (M1) and second (M2) maxillary molars. The antagonist lower molars were kept out of occlusion, inducing super-eruption and apposition of dental cementum (DC) and alveolar bone (AB). (B) Light and (C) fluorescence microscopy images of a longitudinal section of the mesial root of the first mandibular molar. Blue dotted lines in B indicate the DC-dentin (D) border. The white arrows in panel C point to the fluorescent labels formed by the incorporation of fluorochrome markers during DC apposition. Calcein (administered at 24 hours and 17 days after surgery) is observed as two intense green lines, while tetracycline (administered 9 days after surgery) is observed as the faint yellow line between the tetracycline labels.
2.2. Fluorescence microscopy
To verify DC apposition in EIA experimental groups, animals were administered fluorochromes to label mineralizing surfaces. Five animals were given two intraperitoneal injections of calcein (20 mg/kg) at 24 hours and 17 days, and one injection of tetracycline (20 mg/kg) at 9 days after tooth extraction. At day 21 post-surgery, mandibles were harvested, fixed in 10% formalin solution for 48 hours, rinsed in water for 72 hours, and embedded in 0.5% methyl methacrylate. Longitudinal 500 μm-thick sections of mandibles were obtained using a diamond blade mounted on a low speed saw (South Bay Technology, USA) and prepared by manual grinding to a final thickness of 50–100 μm. Sections were mounted on glass slides and imaged by fluorescence microscopy (Leica DMLP, Germany) (Figure 1B, C).
2.3. Laser capture microdissection (LCM)
LCM to capture DC samples was performed using a combination of infrared (IR) and ultraviolet (UV) laser cutting on an ArcturusXT Microdissection Instrument (Applied Biosystems, USA). LCM settings were adjusted to capture the projected area of tissue under a microscope (10×): UV cutting speed of 100 mm/s; IR laser power of 70–90 mW, duration 20 ms, spot size of 30 μm. For each sample, 8–10 unstained histological sections were microdissected, and the total captured area of each sample was calculated (average of 8.75 μm2, ± 1.73) using the ArcturusXT 3.1.0.0 software (firmware 2.1 XT). The microdissected area was used to normalize the amount of tissue/protein for LC-MS/MS analysis. The cap containing the microdissected tissues was adapted to a 0.5 mL microcentrifuge tube and stored at −80° C.
2.4. Preparation of peptide samples
Caps containing microdissected tissues were thawed on ice and then incubated with 30 μl of 8M urea for 30 minutes at room temperature for protein denaturation. Extracted proteins were reduced by incubation with 0.5 M dithiothreitol at 56° C for 25 minutes, alkylated by incubation with 0.5 M iodoacetamide at room temperature for 30 minutes, and further incubated for 15 minutes in 0.5 M dithiothreitol at room temperature. Samples were then diluted into 50 mM ammonium bicarbonate to a final concentration of 1.6 M urea, and 0.1 M calcium chloride was added to the mixture of proteins. Samples were digested by incubation with 2 μg of sequencing-grade modified trypsin (Promega, Madison, WI, USA) overnight at 37° C. All incubation steps above were performed with the cap coupled to a microcentrifuge tube placed in an inverted position to allow reagents to contact the tissues. After digestion, the caps were discarded and the peptide mixtures were equilibrated to pH 2.0 by addition of 0.1% formic acid. The resulting tryptic peptide samples were lyophilized and then reconstituted with 20 μl of 0.1% formic acid, centrifuged at 10,000 rpm for 5 minutes, and stored at −20° C for analysis.
2.5. Liquid chromatography and tandem mass spectrometry (LC-MS/MS)
For protein analysis, 4.5 μl of the resulting mixture of peptides from each microdissected sample were loaded on a mass spectrometer LTQ Velos Orbitrap (Thermo Fisher Scientific, Waltham, MA) connected to a nanoflow LC (NLC-MS / DM) for EASY-NLC system (Proxeon Biosystems, West Palm Beach, FL, USA) through a Proxeon nanoelectrospray ion source. Peptides were separated by 2%–90% acetonitrile gradient in 0.1% formic acid using a PicoFrit analytical column (20 cm x ID75 μm, 5 μm particle size, New Objective, Woburn, MA), with a flow rate of 300 nL/min for 45 minutes. The nanoelectrospray voltage was set to 1.7 kV with the source temperature at 275° C. All the equipment instrumental methods were set up in the data-dependent acquisition mode. Full scans of MS spectra (m/z 300–1600) were acquired by the Orbitrap analyzer after accumulation to a target value of 1e6. The resolution in the Orbitrap was set to the r = 60,000, and the 5 most intense peptide ions with charge states ≥ 2 were sequentially isolated to a target value of 5,000 and fragmented in the linear ion trap by low-energy CID (normalized collision energy of 35%). The signal threshold to trigger an MS/MS event was set to 500 counts. Dynamic exclusion was enabled with an exclusion size list of 500, exclusion duration of 60 seconds, and repeat count of 1. An activation q = 0.25 and time of 10 ms were used.
2.6. Proteomic data analysis
The MS/MS spectra (msf) were generated from the raw data files using Proteome Discover version 1.3 (Thermo Fisher Scientific) with Sequest (Thermo Finnigan, San Jose, CA, USA; version 1.3.0.339) search engine and searched against ipi.MOUSE.v3.86.fasta (version unknown, 58.667 entries) assuming a non-specific enzymatic digestion with carbamidomethylation in cysteine residues (+57.021Da) as a fixed modification, oxidation of methionine (+15.995 Da) as a variable modification, a precursor tolerance of 10 ppm, and 1.0 Da for fragment ions. For protein quantification, the data files were analyzed in Scaffold Q+ (version Scaffold_3.6.5, Proteome Software, Inc., Portland, OR, USA). The quantitative value (normalized spectral counts) was obtained with the protein thresholds set at a minimum 90% probability containing at least one peptide with thresholds established at a minimum 60% probability and XCorr cutoffs +1 > 1.8, +2 >2.2, +3 >2.5 and +4 >3.5 to have less than 1% FDR. Only peptides with a minimum of five amino acid residues, which showed significant threshold (p < 0.05) in the Sequest-based score system, were considered as a product of peptide cleavage. The peptide was considered unique when it differed by at least 1 amino acid residue; covalently modified peptides, including N- or C-terminal elongation (i.e. missed cleavages) were counted as unique, and different charge states of the same peptide and modifications were not counted as unique. For label-free quantification of endogenous peptides, the spectral count and the number of unique peptides were assessed. Resulting spectrum count values were used to analyze the distribution of identified proteins throughout the samples and experimental groups. Proteins were considered as an identified protein when detected in at least one of the seven individual samples per group.
2.7. Statistical analysis
Quantitative values (spectrum count values) were statistically analyzed to identify differentially expressed proteins between the experimental groups. The generalized linear mixed model of analysis of variance (ANOVA test) of repeated measures data (RM-ANOVA) was applied using statistical software (SAS release 9.3, 2012; SAS Institute, Inc., Cary, NC, USA). Averages of significant effects were compared by Tukey’s test, followed by t-test when differences were not detected. Statistical tests considered a significance level of 5% (p <0.05).
Gene Ontology (GO) enrichment analysis of identified proteins for each experimental group was also performed by using the Functional Classification tool on PANTHER Classification System data analysis platform (PANTHER version 10.0, released 2015-05-15, http://pantherdb.org/) [14, 15]. To determine over- or under-represented GO-Slim categories, the classifications of each protein list were compared to the Mus musculus reference database using the binomial test, followed by Bonferroni correction, on the Statistical Overrepresentation Test [16]. All statistical tests considered a significance level of 5% (p < 0.05) and only significant enriched GO terms are included in the results.
2.8. Immunohistochemistry (IHC)
Selected proteins identified in DC by proteomic analysis were confirmed by IHC using normal 2-month-old mouse mandibles with EIA and control samples at 21 days post-surgery. IHC was performed on paraffin sections using an avidin-biotinylated peroxidase enzyme complex (ABC) based kit (Vector Labs, Burlingame, CA, USA) with 3-amino-9-ethylcarbazole (AEC) chromogenic substrate (Vector Labs), as described previously [17]. Primary antibodies included: Goat polyclonal IgG anti-asporin (ASPN; Abcam Inc., Cambridge, MA, USA) [18]; Rabbit polyclonal IgG anti-biglycan (BGN; LF-159, provided by Dr. Larry Fisher, NIDCR/NIH, Bethesda, MD, USA) [10, 19]; Rabbit polyclonal IgG anti-bone sialoprotein (BSP; Provided by Dr. Renny Franceschi, University of Michigan, Ann Arbor, MI)[20]; Rabbit polyclonal IgG anti-collagen type I alpha 1 (COL1A1; LF-68, provided by Dr. Larry Fisher)[19]; Rabbit polyclonal IgG anti-collagen type XI alpha 1 (COL11A1; Abcam Inc.); Rabbit polyclonal IgG anti-collagen type XII (COL12; KR-33, provided by Dr. Manual Koch, University of Cologne, Germany)[21]; Rabbit polyclonal IgG anti-decorin (DCN; LF-114, provided by Dr. Larry Fisher)[10, 19]; Rabbit polyclonal IgG anti-fibromodulin (FMOD; LF-150, provided by Dr. Larry Fisher)[10]; Rabbit polyclonal IgG anti-osteopontin (OPN; LF-175, provided by Dr. Larry Fisher) [17]; Rabbit polyclonal IgG anti-periostin (POSTN; Abcam Inc.)[10]; and Rabbit polyclonal anti-osteonectin/secreted protein acidic and rich in cysteine (SPARC; BON-I, provided by Dr. Larry Fisher). Immunostaining of COL11A1 employed a trypsin digest antigen retrival step (Thermo Fisher Scientific, Waltham, MA, USA).
3. Results and Discussion
3.1. Proteomic profile of DC under EIA
Experimentally-induced apposition (EIA) of dental cementum (DC) was produced by extraction of opposing maxillary molars, removing mandibular molars from occlusion and putting them into super-eruption (Figure 1A). DC apposition was confirmed by fluorochrome labeling and fluorescence microscopy (Figure 1B, C). The DC proteome was analyzed by liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) after 6 and 21 days under EIA. Proteins were identified by Sequest against a mouse IPI database (ipi.MOUSE.v3.86.fasta) and validated using the Scaffold Q+. Peptide mixtures from seven samples/group were individually analyzed and averaged for a statistical analysis. In total, 519 proteins were identified and their distribution is displayed in the Venn diagram in Figure 2. Among the 519 proteins, 246 (47.4%) were commonly present in all experimental groups and 97 proteins (18.7%) were only detected in the EIA groups, being 35 (6.7%) and 41 (7.9%) proteins found exclusively at 6 and 21 days, respectively. In addition, 50 proteins (9.6%) were exclusively detected in the control groups, with 31 (5.9%) and 15 (2.9%) proteins found exclusively at 6 and 21 days, respectively. RM-ANOVA data analysis showed that a total of 56 proteins were differentially expressed in DC under the effects of EIA treatment and/or time of experiment (Table 1). All identified and quantified proteins, as well as statistical results, are presented in Supplemental Material S1 and S2, respectively.
Figure 2. Proteomic profile of dental cementum.
Area-proportional Venn diagram showing the distribution of the total 519 proteins identified in dental cementum (DC) under experimentally-induced apposition for 6 and 21 days (EIA groups), as well as respective control groups at the same time points. Proteins were considered as an identified protein when detected in, at least, one of the seven individual samples. The number of exclusive proteins to each group and shared proteins in every intersection of the diagram are shown.
Table 1.
List of differentially expressed proteins for the experimentally-induced apposition (EIA) and control groups. Average of spectra counts of seven samples of each group and p-values (repeated measures ANOVA test) are shown.
| Accession Number | Description | Spectrum counts | ANOVA test | |||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| EIA-6D | Ctrl-6D | EIA-21D | Ctrl-21D | p-Value Time | p-Value Treatment | p-Value Interaction | ||
| P97427 | Dihydropyrimidinase-related protein 3 | 0.00 | 0.64 | 0.68 | 0.10 | 0.752 | 0.893 | 0.025 |
| P17742 | Peptidyl-prolyl cis-trans isomerase | 0.36 | 0.97 | 0.98 | 0.12 | 0.667 | 0.632 | 0.026 |
| P02089 | Hemoglobin subunit beta-2 | 8.67 | 3.54 | 4.12 | 3.33 | 0.017 | 0.006 | 0.024 |
| Q9QUN8 | Beta-globin | 14.62 | 6.73 | 6.62 | 4.42 | 0.004 | 0.004 | 0.047 |
| Q60770 | Isoform 2 of Syntaxin-binding protein 3 | 1.14 | 0.00 | 0.00 | 0.00 | 0.016 | 0.016 | 0.016 |
| Q07079 | Insulin-like growth factor-binding protein 5 | 0.00 | 0.82 | 0.74 | 0.35 | 0.581 | 0.394 | 0.043 |
| P48036 | Annexin A5 | 3.76 | 6.98 | 5.38 | 4.96 | 0.778 | 0.088 | 0.038 |
| P08121 | Collagen alpha-1(III) chain | 0.38 | 1.25 | 1.23 | 1.06 | 0.169 | 0.148 | 0.048 |
| P35441 | Uncharacterized protein | 4.18 | 4.93 | 6.65 | 3.32 | 0.609 | 0.159 | 0.044 |
| Q58ES8 | major urinary protein 10 | 0.60 | 0.31 | 3.52 | 0.71 | 0.015 | 0.020 | 0.043 |
| P09411 | Phosphoglycerate kinase 1 | 0.00 | 0.00 | 0.59 | 0.00 | 0.043 | 0.043 | 0.043 |
| Q61245 | Isoform Short of Collagen alpha-1(XI) chain | 0.00 | 0.35 | 1.67 | 0.09 | 0.009 | 0.016 | 0.002 |
| P07214 | SPARC | 1.24 | 1.53 | 4.18 | 1.62 | 0.024 | 0.066 | 0.030 |
| Q00623 | Apolipoprotein A-I | 0.00 | 0.40 | 1.31 | 0.11 | 0.152 | 0.247 | 0.043 |
| P07310 | Creatine kinase M-type | 3.91 | 1.76 | 1.26 | 2.52 | 0.213 | 0.536 | 0.046 |
| P50608 | Fibromodulin | 5.80 | 3.23 | 1.51 | 4.23 | 0.097 | 0.934 | 0.020 |
| P97298 | Pigment epithelium-derived factor | 4.02 | 5.59 | 6.90 | 4.86 | 0.187 | 0.760 | 0.047 |
| P51885 | Lumican | 2.49 | 4.10 | 4.34 | 3.04 | 0.487 | 0.778 | 0.034 |
| P19221 | Prothrombin (Fragment) | 4.58 | 6.80 | 10.34 | 7.14 | 0.033 | 0.675 | 0.050 |
| P10923 | Osteopontin | 2.33 | 4.66 | 6.82 | 5.58 | 0.004 | 0.404 | 0.026 |
| P07724 | Serum albumin | 15.25 | 22.17 | 25.31 | 18.63 | 0.113 | 0.948 | 0.008 |
| Q80Z71 | Tenascin-N | 0.35 | 5.84 | 1.21 | 2.22 | 0.098 | 0.004 | 0.019 |
| hemoglobin subunit beta-1-like isoform 5 | 12.61 | 6.78 | 5.97 | 4.32 | 0.002 | 0.006 | 0.061 | |
| Q00896 | Alpha-1-antitrypsin 1-3 | 1.14 | 0.62 | 2.78 | 0.70 | 0.046 | 0.009 | 0.063 |
| Q5FW85 | Extracellular matrix protein 2 | 0.00 | 0.16 | 1.04 | 2.08 | 0.001 | 0.037 | 0.095 |
| Q9QXS1 | plectin isoform 1 | 0.85 | 2.12 | 0.22 | 0.88 | 0.049 | 0.044 | 0.451 |
| Q8K440 | Isoform 1 of ATP-binding cassette sub-family A member 8-B | 3.34 | 0.42 | 1.64 | 1.21 | 0.520 | 0.048 | 0.114 |
| E9Q616 | AHNAK nucleoprotein isoform 1 | 1.12 | 0.31 | 1.20 | 0.58 | 0.536 | 0.037 | 0.721 |
| Uncharacterized protein | 0.72 | 0.28 | 0.83 | 0.15 | 0.964 | 0.024 | 0.535 | |
| Q6XAS3 | Uncharacterized protein | 0.76 | 0.38 | 0.97 | 0.29 | 0.746 | 0.029 | 0.461 |
| P07758 | Serpina1a protein | 1.14 | 0.62 | 2.51 | 0.70 | 0.090 | 0.017 | 0.122 |
| Q61398 | Procollagen C-endopeptidase enhancer 1 | 2.52 | 1.61 | 3.70 | 1.38 | 0.460 | 0.037 | 0.291 |
| P62270 | 40S ribosomal protein S18 | 0.89 | 0.00 | 0.75 | 0.38 | 0.502 | 0.008 | 0.158 |
| P09405 | Nucleolin | 0.17 | 0.00 | 0.82 | 0.00 | 0.100 | 0.026 | 0.100 |
| Q3V5L4 | Tenascin-X | 0.00 | 0.31 | 0.00 | 1.10 | 0.207 | 0.045 | 0.207 |
| Q61781 | Keratin. type I cytoskeletal 14 | 2.52 | 4.67 | 1.64 | 2.97 | 0.111 | 0.045 | 0.575 |
| Q99MQ4 | Asporin | 2.80 | 5.36 | 4.08 | 6.38 | 0.201 | 0.023 | 0.871 |
| P62242 | 40S ribosomal protein S8 | 0.17 | 1.23 | 0.54 | 0.67 | 0.708 | 0.038 | 0.082 |
| Q91VB8 | Putative uncharacterized protein | 7.42 | 8.85 | 3.13 | 3.22 | 0.008 | 0.574 | 0.619 |
| P62908 | 40S ribosomal protein S3 | 1.30 | 1.35 | 0.30 | 0.30 | 0.018 | 0.951 | 0.940 |
| Q9R0T7 | trypsin 4 | 10.69 | 7.67 | 6.87 | 4.68 | 0.020 | 0.053 | 0.712 |
| O88207 | Collagen alpha-1(V) chain | 0.00 | 0.26 | 1.72 | 0.95 | 0.002 | 0.330 | 0.075 |
| Uncharacterized protein | 0.00 | 0.15 | 0.98 | 0.77 | 0.004 | 0.862 | 0.333 | |
| Q921I1 | Serotransferrin | 3.31 | 5.73 | 8.16 | 7.49 | 0.008 | 0.342 | 0.116 |
| Uncharacterized protein | 3.15 | 5.73 | 7.42 | 7.04 | 0.009 | 0.187 | 0.092 | |
| P16294 | Coagulation factor IX | 0.35 | 0.00 | 1.07 | 0.86 | 0.012 | 0.247 | 0.761 |
| Q8R138 | Transmembrane protein 119 | 0.00 | 0.00 | 0.81 | 0.32 | 0.016 | 0.204 | 0.204 |
| P33587 | Vitamin K-dependent protein C | 0.00 | 0.00 | 0.65 | 0.38 | 0.018 | 0.429 | 0.429 |
| P20152 | Vimentin | 3.29 | 3.86 | 5.20 | 5.24 | 0.030 | 0.617 | 0.664 |
| P29699 | Alpha-2-HS-glycoprotein | 12.03 | 13.91 | 17.83 | 18.43 | 0.031 | 0.525 | 0.739 |
| P07759 | Serine protease inhibitor A3K | 0.00 | 1.06 | 2.17 | 1.22 | 0.031 | 0.891 | 0.052 |
| Q9QZ47 | Troponin I. fast skeletal muscle | 0.00 | 0.00 | 0.29 | 0.50 | 0.032 | 0.473 | 0.473 |
| Q640N1 | Isoform 1 of Adipocyte enhancer-binding protein 1 | 4.62 | 6.73 | 10.57 | 7.28 | 0.033 | 0.634 | 0.062 |
| Q8R054 | Uncharacterized protein | 0.00 | 0.00 | 0.40 | 0.66 | 0.033 | 0.524 | 0.524 |
| P21550 | Beta-enolase | 0.00 | 0.00 | 0.83 | 1.01 | 0.039 | 0.810 | 0.810 |
| P12023 | Isoform APP770 of Amyloid beta A4 protein (Fragment) | 1.53 | 1.26 | 3.19 | 2.24 | 0.043 | 0.284 | 0.539 |
3.2. Identification of DC protein markers
To validate our approach of LC-MS/MS analysis of decalcified and microdissected formalin-fixed paraffin-embedded (FFPE) samples, we first examined the proteomic profile (in both control and EIA groups) to confirm previously identified DC markers. Our analysis identified the presence of a number of DC-associated proteins, and immunohistochemistry (IHC) was used to further validate localization of selected markers in the DC ECM (Figure 3). Collagens were among the most abundant of the identified proteins in DC, including COL1A1 and COL12A1 (Figure 3A, B), together with COL6A3 and COL5A2, which were found in almost all samples analyzed. COL6A1, COL3A1, and COL11A1 were also identified, but in less abundance. These collagens are consistent with the previously reported composition of DC [5, 22].
Figure 3. Immunohistochemistry for identification of dental cementum markers.
Immunohistochemistry (IHC) was used to localize (by red or brown-red reaction color) selected protein markers identified by proteomic analysis of dental cementum (DC). All photomicrographs were taken at original magnification of 100×. Blue dotted lines in all panels indicate the DC-dentin (DE) border. (A) Collagen type I alpha 1 (COL1A1) localizes to DC, DE, and alveolar bone (AB). (B) Collagen type XII (COL12) localizes to periodontal ligament (PDL) and DC-PDL interface. (C) Bone sialoprotein (BSP) stains DC and AB. (D) Osteopontin (OPN) is found in DC and AB. (E) Dentin matrix protein 1 (DMP1) is found in AB and DC, around osteocytes and cementocytes, respectively, as well as in tubules of DE. (F) Tissue nonspecific alkaline phosphatase (TNAP) is rich in the PDL and at the borders of DC-PDL and AB-PDL. (G) Decorin (DCN) localizes to PDL and DC-PDL interface. (H) Biglycan (BGN) is found in DC and PDL. (I) Fibromodulin (FMOD) is observed in PDL, and in DC around cementocytes lacunae. (J) Asporin (ASPN) stains the PDL and DC-PDL interface. (K) Periostin (POSTN) is richly present in the PDL and associated with embedded collagen fibers in DC and AB. (L) A negative control (NEG) section that lacked primary antibody is included for comparison to IHC-stained sections.
Several classes of non-collagenous ECM proteins were identified in DC. Key markers associated with DC formation included secreted phosphoproteins, bone sialoprotein (BSP), osteopontin (OPN), and dentin matrix protein 1 (DMP1), and mineralization-regulating enzyme tissue-nonspecific alkaline phosphatase (TNAP) (Figure 3C–F) [9, 17, 20, 23]. Several proteoglycans were identified, including decorin (DCN), biglycan (BGN), fibromodulin (FMOD), asporin (ASPN), lumican (LUM), and osteomodulin/osteoadherin (OSAD), and these potential mineralization regulators have been identified in DC (Figure 3G–J) and the periodontium [10, 18, 24–26]. We also identified periostin (POSTN) (Figure 3K), vimentin (VIM), tenascin (TNN), and vitronectin (VTN), which have been identified in DC and/or associated embedded PDL tissues [5, 10, 27–29].
Importantly, these data demonstrate that the approach taken for sample preparation and microdissection resulted in high quality protein extracts, allowing for the detection of abundant components of DC ECM including collagens, non-collagenous proteins, and proteoglycans, as well factors present in lower abundance, such as growth factors EGF and IGF. This approach was also capable of detecting proteins that strongly interact with hydroxyapatite, such as BSP and OPN [30], where decalcification could potentially result in loss of protein and an inability for their detection, as we have noted in previous proteomic analysis using an alternate tissue extraction and grinding approach [10].
3.3. DC protein composition is significantly regulated by EIA
Differential analysis was performed in order to identify significant changes in quantitative values (spectrum counts) of proteins located in EIA vs. control DC at 6 and 21 days. Among the differentially expressed proteins, 24 were regulated by the exclusive effect of EIA (12 proteins), or the interaction between EIA and time (12 proteins). Most of the differentially expressed proteins (70%) were up-regulated after EIA. The direction of regulation of these proteins is shown in Figure 4A. Proteins regulated only by EIA (up- or down-regulated at both 6 and 21 days; Figure 4B), included three secreted factors: serpin 1a (fold-change = 1.84/3.60, 6D/21D), procollagen C-endopeptidase enhancer 1 (fold-change = 1.57/2.67, 6D/21D), tenascin X (TNX) (fold-change = −0.31/−1.1, 6D/21D), and asporin (ASPN) (fold-change = −1.9/−1.6, 6D/21D).
Figure 4. Dental cementum protein expression is significantly regulated by experimentally-induced apposition.
(A) Venn diagram showing the distribution of 24 differentially expressed proteins under the effect of EIA alone, or including the interaction with time. Identities and fold-changes for these 24 statistically significant differentially expressed proteins are described in bar graphs in panels B–D. (B) Bar graph showing the fold-regulation for 12 proteins under the exclusive effect of EIA, at 6 and 21 days. (C) Bar graph showing the fold-regulation of 6 proteins at 6 days. (D) Bar graph showing the fold-regulation of 6 proteins at 21 days.
Serpin 1a is a secreted serine protease inhibitor with a wide range of substrates including elastase, a potent enzyme released during inflammatory processes [31]. Mutations in the SERPINA1 gene have been linked to cirrhosis and emphysema [32]. Although serpin 1a protein has generally not been reported in mineralized tissues, cells of the osteoblastic niche were reported to express high levels of Serpina1 mRNA [33]. While it is presently unclear what role serpin 1a protein may have in the context of EIA, it is intriguing to consider how the inflammation modulatory functions and regulatory role in maintenance of hematopoietic progenitor cell populations may come into play during cementogenesis.
TNX and ASPN were both down-regulated in DC by EIA at both 6 and 21 days. TNX is an ECM glycoprotein expressed in muscle and loose connective tissue in association with collagen type I, and mutations in the TNXB gene are associated with the connective tissue disorder, Ehlers-Danlos syndrome [34–36]. Although the role of TNX has been described in several tissues, little is known about the role of TNX in the skeleton or dentition [37].
Expression of ASPN, a class II small leucine rich proteoglycan (SLRP), has been reported in dentin, alveolar bone, DC, and PDL [10, 18, 38, 39]. ASPN is reported to bind collagen and regulate bio-mineralization, as well as affecting fibroblast growth factor (FGF) signaling and inflammatory response [40–43]. Evidence that ASPN competes with DCN for collagen type I binding [41], and specifically modulates mineralization in PDL, is intriguing in the context of new DC formation, and the significant reduction of ASPN in EIA groups may create a permissive environment for DC mineralization.
While the above factors were consistently regulated up or down at both time points, other factors were significantly regulated at only 6 days (Figure 4C) or 21 days (Figure 4D). Data analysis showed that the expression of collagenous proteins, with roles in the fibrillar structure of DC-ECM, was significantly affected by EIA. Collagen type III alpha 1 (COL3A1), present in soft connective tissues in association with the fibrillary structure of collagen type I, was down-regulated by EIA at day 6 (fold-change = −3.3), while collagen type XI alpha 1 (COL11A1) was up-regulated at day 21 (fold-change = 19.5). COL11A1 is associated with collagen type II fibrils in cartilage [44], but pro-α2 transcripts have been described in odontoblasts [45], indicating it is not necessarily cartilage-specific.
Osteonectin (secreted protein acidic and rich in cysteine; SPARC) and thrombospondin 1 (THBS1), two multifunctional glycoproteins, were up-regulated (fold-change = 2.6 and 2.0, respectively) at 21 days. SPARC functions as an adhesion protein, regulates cell-matrix interactions, and is implicated in tissue remodeling and repair [46, 47]. SPARC is part of the secretory calcium-binding phosphoprotein (SCPP) family that includes several mineral regulatory ECM proteins such as BSP, OPN, and DMP1 [48]. Importantly, SPARC was previously shown to be up-regulated in the periodontium of unloaded super-erupting mouse teeth [12], and we confirm that finding here.
THBS1 mediates cell-cell and cell-matrix interactions via its multiple binding partners and receptors, and is implicated in regulation of angiogenesis [49–51]. THBS1 is expressed in osteoblasts [52, 53] and odontoblasts [54], especially during early stages of differentiation and mineralization [54, 55]. Interestingly, THBS1 was up-regulated in periodontia under orthodontic treatment [56], indicating a potential function in remodeling periodontal tissues.
We confirmed localization of selected ECM markers in EIA and control tissues at 21 days, including ASPN, COL11A1, and SPARC (Figure 5A–F). While IHC is not quantitative and small fold-changes would not be expected to be apparent in a relatively small volume of new DC, the representative images serve to confirm localization of identified proteins in DC, and DC-associated PDL and bone. Overall, EIA was associated with significant changes in ECM proteins including structural and non-structural elements, and factors involved with cell signaling through cell-cell and cell-matrix interactions. Proteins previously associated with DC and the periodontium, ASPN and SPARC, are identified as factors of interest in neocementogenesis, while SERPINA1, TNX, COL11A1, and THBS1 warrant consideration as factors of interest in periodontal homeostasis.
Figure 5. Immunohistochemistry confirmation of proteins in control and experimentally-induced apposition tissues.

Immunohistochemistry (IHC) was used to localize (by red or brown-red reaction color) selected protein markers identified by proteomic analysis of dental cementum (DC) in control and experimentally-induced apposition (EIA) groups 21 days after surgery. All photomicrographs were taken at original magnification of 100×. Blue dotted lines in all panels indicate the DC-dentin (DE) border. (A, B) Asporin (ASPN) localizes to alveolar bone (AB), periodontal ligament (PDL) and DC in control and EIA tissues. (C, D) Collagen type XI alpha 1 (COL11A1) exhibits immunostaining in AB, PDL, DE, and DC in control and EIA tissues. (E, F) Secreted protein, acidic cysteine-rich (SPARC) immunostains widely in AB, PDL, DE, and DC in control and EIA groups. (G, H) Osteopontin (OPN) localizes to reversal lines in AB, as well as DC matrix in control and EIA tissues. (I, J) Fibromodulin (FMOD) is observed immunostaining AB, PDL, DE-pulp, and DC in control and EIA tissues.
3.4. Temporal changes in the DC proteomic profile
The factors discussed in the preceding section were significantly up- or down-regulated by EIA. Expression of a number of proteins was modulated over time, regardless of treatment group. Figure 6 illustrates quantitative values (spectrum counts) of selected differentially expressed proteins at 6 and 21 days. The full list can be accessed in Supplemental Material T2. Most of the proteins regulated over time exhibited increased expression at day 21 (76%), including collagen type V alpha 1 chain (COL5A1), vimentin (VIM), and alpha-2-HS-glycoprotein (AHSG). Some proteins were regulated exclusively in the EIA group over time, including significant increases (p < 0.05) in pigment epithelium-derived factor (PEDF), a potent inhibitor of angiogenesis, mineralization regulator OPN, and the proteoglycan lumican (LUM). Another SLRP, fibromodulin (FMOD), was decreased at 21 versus 6 days in the EIA group. IHC was used to confirm localization of selected temporal markers in EIA and control tissues at 21 days, including OPN and FMOD (Figure 5G–J). These findings suggest complex and dynamic processes associated with new DC formation and point to temporal roles for ECM proteins in this process. Interestingly, differences in proteomic profiles were noted between control group at 6 and 21 days. At 35 days of age (initiation of the experiment), mouse molars have only been in occlusion for approximately a week [57], therefore these changes in control mice may reflect physiological modeling/remodeling in the periodontium.
Figure 6. Temporal changes in the dental cementum proteomic profile.
Bar graph showing the distribution of selected differentially expressed proteins over time in dental cementum (DC). (*) proteins regulated over time in the experimentally-induced apposition (EIA) groups.
3.5. Evidence of cementocyte activation by EIA
Based on the experimental strategy and precise sample microdissection, the only intracellular or cell membrane proteins anticipated to be present within DC samples would be attributed to cementocytes, the residents of the lacunocanalicular system within the bone-like cellular DC [3, 58]. While the morphology and ultrastructure of cementocytes have been previously reported, it is presently unknown whether the cells have in vivo functions related to cementum formation or repair. In addition to the largely secreted ECM proteins described above, our study aimed to identify whether evidence existed for cementocyte metabolic activity under EIA.
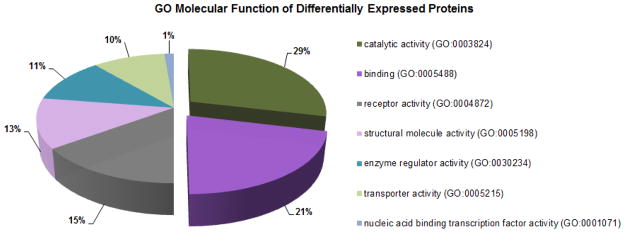
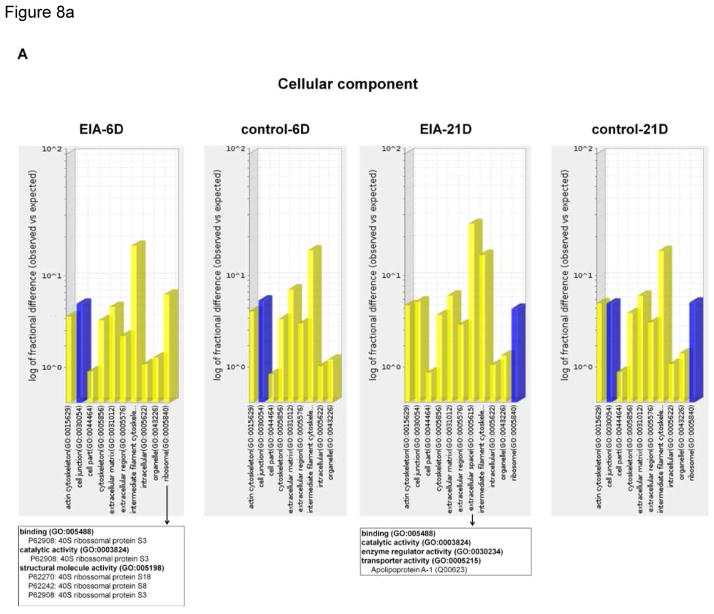
To identify whether sets of regulated genes indicated cellular activity, a bioinformatics gene ontology (GO) analysis was performed on differentially expressed proteins in the proteomic profiles of each experimental group, identifying significantly over- or under-represented GO terms. GO molecular function analysis of 56 significant differentially expressed proteins in DC under EIA showed that 50% of them were related to catalytic activity (29%) and binding (21%) functions, with receptor activity (15%) as the third most significant regulated function (Figure 7). The analysis of over-represented cellular components (Figure 8A) clearly identified the ribosome as an enriched location in the EIA-group. RPS18, RPS8, and RPS3, three proteins associated with protein synthesis via the 40S ribosomal subunit [59], were significantly regulated. Additional over-represented GO groups at 21 days (Figure 8B) included protein and receptor binding factors such as SPARC, ASPN, and AHSG, previously discussed in this article and having ECM functions, as well as regulating cell-cell and cell-matrix interactions.
Figure 7. Enriched gene ontology groups in dental cementum under experimentally induced apposition.
Enriched gene ontology (GO) terms of 56 differentially expressed proteins in dental cementum (DC) after experimentally-induced apposition (EIA) at 6 and 21 days, generated by the Functional Classification tool using GO Molecular function category in PANTHER Classification System. Only statistically significantly enriched GO terms are displayed, with the percentage of genes that hit compared against the total number of hits.
Figure 8. Gene ontology evidence of cementocyte activation by experimentally induced apposition.
Bar graphs indicating (A) GO-Slim cellular component and (B) molecular function classification of each protein group list were compared to the Mus musculus reference database using the binomial test, followed by Bonferroni correction, on the Statistical Overrepresentation Test in the PANTHER Classification System for gene ontology analysis. PANTHER-generated graphics display only significantly enriched terms. Blue and yellow bars represent significant terms with p-values <0.05 and <0.01, respectively. Arrows indicates child categories of molecular function of selected enriched terms with corresponding protein list involved with the GO term. Differentially regulated proteins for each term are listed in the boxes.
Overall, these data suggest activation of cementocyte metabolism by EIA, and provide some of the first evidence of cementocyte function in vivo. The concept that cementocytes may have an active role in neocementogenesis, DC apposition, and mineralization warrants additional studies of these cells and their role in periodontal development and homeostasis.
4. Conclusion
The novel approach of induction of experimentally-induced apposition (EIA) of dental cementum (DC), followed by liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) proteomic analysis, identified differentially expressed proteins associated with new DC formation. These included SerpinF1, tenascin-X (TNX), asporin (ASPN), collagen type XI alpha 1 (COL11A1), osteonectin (SPARC), thrombospondin 1 (THBS1), osteopontin (OPN), and fibromodulin (FMOD). These findings provide additional insights into periodontal biology, particularly the process of new DC formation, which remains poorly understood. The factors identified represent potential targets for DC regeneration in the clinic, and warrant further studies to better understand their functions in DC ECM and mineralization. Furthermore, our findings indicate that cementocytes are responsive to EIA, suggesting these cells may play an active role in controlling DC formation and regeneration.
Supplementary Material
Significance.
Dental cementum (DC) is a mineralized tissue that covers the tooth root surface and has important functions in tooth attachment and position. DC and other periodontal tissues can be lost to disease, and regeneration is currently unpredictable due to lack of understanding of DC formation. This study used a model of experimentally-induced apposition (EIA) in mice to promote new cementum formation, followed by laser capture microdissection (LCM) and liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) proteomic analysis. This approach identified proteins associated with new cementum formation that may be targets for promoting cementum regeneration.
Highlights.
Microdissection of FFPE tissues followed by LC-MS/MS was used successfully for proteomic analysis of dental cementum.
We identified 24 proteins in cementum regulated by experimentally-induced apposition.
We produced evidence for cementocyte activity during new cementum formation.
Acknowledgments
This work was supported by grant N° 2010/12486-7 (to EAS) from Agency São Paulo Research Foundation (FAPESP), and a seed grant from The Ohio State University College of Dentistry (to BLF). The authors thank Mariana P. F. Lazarim (State University of Campinas) for technical assistance with the histological sectioning.
Footnotes
Conflict of interest
The authors of manuscript entitled Global Proteome Profiling of Dental Cementum under Experimentally-Induced Apposition report no conflict of interest.
The authors report no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Foster BL, Somerman MJ. Cementum. In: McCauley LK, Somerman MJ, editors. Mineralized Tissues in Oral and Craniofacial Science: Biological Principles and Clinical Correlates. 1. Ames, IA: Wiley-Blackwell; 2012. pp. 169–92. [Google Scholar]
- 2.Foster BL, Nociti FH, Jr, Somerman MJ. Tooth Root Formation. In: Huang GTJ, Thesleff I, editors. Stem Cells, Craniofacial Development and Regeneration. 1. Wiley-Blackwell; 2013. pp. 153–77. [Google Scholar]
- 3.Zhao N, Nociti FH, Jr, Duan P, Prideaux M, Zhao H, Foster BL, et al. Isolation and Functional Analysis of an Immortalized Murine Cementocyte Cell Line, IDG-CM6. J Bone Mineral Res. 2015 doi: 10.1002/jbmr.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dallas SL, Prideaux M, Bonewald LF. The osteocyte: an endocrine cell ... and more. Endocr Rev. 2013;34:658–90. doi: 10.1210/er.2012-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosshardt D. Are cementoblasts a subpopulation of osteoblasts or a unique phenotype? J Dent Res. 2005;84:390–406. doi: 10.1177/154405910508400501. [DOI] [PubMed] [Google Scholar]
- 6.Bosshardt DD, Sculean A. Does periodontal tissue regeneration really work? Periodontol 2000. 2009;51:208–19. doi: 10.1111/j.1600-0757.2009.00317.x. [DOI] [PubMed] [Google Scholar]
- 7.Foster BL, Nagatomo KJ, Bamashmous SO, Tompkins KA, Fong H, Dunn D, et al. The progressive ankylosis protein regulates cementum apposition and extracellular matrix composition. Cells Tissues Organs. 2011;194:382–405. doi: 10.1159/000323457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foster BL, Ao M, Willoughby C, Soenjaya Y, Holm E, Lukashova L, et al. Mineralization defects in cementum and craniofacial bone from loss of bone sialoprotein. Bone. 2015;78:150–64. doi: 10.1016/j.bone.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zweifler LE, Patel MK, Nociti FH, Jr, Wimer HF, Millan JL, Somerman MJ, et al. Counter-regulatory phosphatases TNAP and NPP1 temporally regulate tooth root cementogenesis. Int J Oral Sci. 2015;7:27–41. doi: 10.1038/ijos.2014.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salmon CR, Tomazela DM, Ruiz KG, Foster BL, Paes Leme AF, Sallum EA, et al. Proteomic analysis of human dental cementum and alveolar bone. Journal of proteomics. 2013;91C:544–55. doi: 10.1016/j.jprot.2013.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walker CG, Dangaria S, Ito Y, Luan X, Diekwisch TG. Osteopontin is required for unloading-induced osteoclast recruitment and modulation of RANKL expression during tooth drift-associated bone remodeling, but not for super-eruption. Bone. 2010;47:1020–9. doi: 10.1016/j.bone.2010.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luan X, Ito Y, Holliday S, Walker C, Daniel J, Galang TM, et al. Extracellular matrix-mediated tissue remodeling following axial movement of teeth. J Histochem Cytochem. 2007;55:127–40. doi: 10.1369/jhc.6A7018.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salmon CR, Silverio KG, Giorgetti AP, Sallum EA, Casati MZ, Nociti FH., Jr Gene expression analysis in microdissected samples from decalcified tissues. Diagnostic molecular pathology : the American journal of surgical pathology, part B. 2012;21:120–6. doi: 10.1097/PDM.0b013e31823e9395. [DOI] [PubMed] [Google Scholar]
- 14.Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 2013;8:1551–66. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mi H, Muruganujan A, Thomas PD. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013;41:D377–86. doi: 10.1093/nar/gks1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas PD, Kejariwal A, Guo N, Mi H, Campbell MJ, Muruganujan A, et al. Applications for protein sequence-function evolution data: mRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Res. 2006;34:W645–50. doi: 10.1093/nar/gkl229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foster BL. Methods for studying tooth root cementum by light microscopy. Int J Oral Sci. 2012;4:119–28. doi: 10.1038/ijos.2012.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang L, Foster BL, Kram V, Nociti FH, Jr, Zerfas PM, Tran AB, et al. Fibromodulin and Biglycan Modulate Periodontium through TGFbeta/BMP Signaling. J Dent Res. 2014;93:780–7. doi: 10.1177/0022034514541126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fisher LW, Stubbs JT, 3rd, Young MF. Antisera and cDNA probes to human and certain animal model bone matrix noncollagenous proteins. Acta orthopaedica Scandinavica Supplementum. 1995;266:61–5. [PubMed] [Google Scholar]
- 20.Foster BL, Soenjaya Y, Nociti FH, Holm E, Zerfas PM, Wimer HF, et al. Deficiency in Acellular Cementum and Periodontal Attachment in Bsp Null Mice. J Dent Res. 2012 doi: 10.1177/0022034512469026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Veit G, Hansen U, Keene DR, Bruckner P, Chiquet-Ehrismann R, Chiquet M, et al. Collagen XII interacts with avian tenascin-X through its NC3 domain. J Biol Chem. 2006;281:27461–70. doi: 10.1074/jbc.M603147200. [DOI] [PubMed] [Google Scholar]
- 22.Foster BL, Popowics TE, Fong HK, Somerman MJ. Advances in defining regulators of cementum development and periodontal regeneration. Curr Top Dev Biol. 2007;78:47–126. doi: 10.1016/S0070-2153(06)78003-6. [DOI] [PubMed] [Google Scholar]
- 23.Foster BL, Nagatomo KJ, Nociti FH, Fong H, Dunn D, Tran AB, et al. Central role of pyrophosphate in acellular cementum formation. PLoS One. 2012;7:e38393. doi: 10.1371/journal.pone.0038393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saygin NE, Giannobile WV, Somerman MJ. Molecular and cell biology of cementum. Periodontol 2000. 2000;24:73–98. doi: 10.1034/j.1600-0757.2000.2240105.x. [DOI] [PubMed] [Google Scholar]
- 25.Ababneh K, Hall R, Embery G. The proteoglycans of human cementum: immunohistochemical localization in healthy, periodontally involved and ageing teeth. J Periodontal Res. 1999;34:87–96. doi: 10.1111/j.1600-0765.1999.tb02227.x. [DOI] [PubMed] [Google Scholar]
- 26.Couble ML, Bleicher F, Farges JC, Peyrol S, Lucchini M, Magloire H, et al. Immunodetection of osteoadherin in murine tooth extracellular matrices. Histochem Cell Biol. 2004;121:47–53. doi: 10.1007/s00418-003-0608-2. [DOI] [PubMed] [Google Scholar]
- 27.Horiuchi K, Amizuka N, Takeshita S, Takamatsu H, Katsuura M, Ozawa H, et al. Identification and characterization of a novel protein, periostin, with restricted expression to periosteum and periodontal ligament and increased expression by transforming growth factor beta. J Bone Miner Res. 1999;14:1239–49. doi: 10.1359/jbmr.1999.14.7.1239. [DOI] [PubMed] [Google Scholar]
- 28.Sculean A, Berakdar M, Windisch P, Remberger K, Donos N, Brecx M. Immunohistochemical investigation on the pattern of vimentin expression in regenerated and intact monkey and human periodontal ligament. Arch Oral Biol. 2003;48:77–86. doi: 10.1016/s0003-9969(02)00164-4. [DOI] [PubMed] [Google Scholar]
- 29.Zhang X, Schuppan D, Becker J, Reichart P, Gelderblom HR. Distribution of undulin, tenascin, and fibronectin in the human periodontal ligament and cementum: comparative immunoelectron microscopy with ultra-thin cryosections. J Histochem Cytochem. 1993;41:245–51. doi: 10.1177/41.2.7678270. [DOI] [PubMed] [Google Scholar]
- 30.Goldberg HA, Warner KJ, Li MC, Hunter GK. Binding of bone sialoprotein, osteopontin and synthetic polypeptides to hydroxyapatite. Connect Tissue Res. 2001;42:25–37. doi: 10.3109/03008200109014246. [DOI] [PubMed] [Google Scholar]
- 31.Gooptu B, Lomas DA. Polymers and inflammation: disease mechanisms of the serpinopathies. J Exp Med. 2008;205:1529–34. doi: 10.1084/jem.20072080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lomas DA, Mahadeva R. Alpha1-antitrypsin polymerization and the serpinopathies: pathobiology and prospects for therapy. J Clin Invest. 2002;110:1585–90. doi: 10.1172/JCI16782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuiperij HB, van Pel M, de Rooij KE, Hoeben RC, Fibbe WE. Serpina1 (alpha1-AT) is synthesized in the osteoblastic stem cell niche. Exp Hematol. 2009;37:641–7. doi: 10.1016/j.exphem.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 34.Egging D, van Vlijmen-Willems I, van Tongeren T, Schalkwijk J, Peeters A. Wound healing in tenascin-X deficient mice suggests that tenascin-X is involved in matrix maturation rather than matrix deposition. Connect Tissue Res. 2007;48:93–8. doi: 10.1080/03008200601166160. [DOI] [PubMed] [Google Scholar]
- 35.Egging DF, van Vlijmen I, Starcher B, Gijsen Y, Zweers MC, Blankevoort L, et al. Dermal connective tissue development in mice: an essential role for tenascin-X. Cell Tissue Res. 2006;323:465–74. doi: 10.1007/s00441-005-0100-5. [DOI] [PubMed] [Google Scholar]
- 36.Mao JR, Taylor G, Dean WB, Wagner DR, Afzal V, Lotz JC, et al. Tenascin-X deficiency mimics Ehlers-Danlos syndrome in mice through alteration of collagen deposition. Nat Genet. 2002;30:421–5. doi: 10.1038/ng850. [DOI] [PubMed] [Google Scholar]
- 37.Chiovaro F, Chiquet-Ehrismann R, Chiquet M. Transcriptional regulation of tenascin genes. Cell adhesion & migration. 2015;9:34–47. doi: 10.1080/19336918.2015.1008333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park ES, Cho HS, Kwon TG, Jang SN, Lee SH, An CH, et al. Proteomics analysis of human dentin reveals distinct protein expression profiles. J Proteome Res. 2009;8:1338–46. doi: 10.1021/pr801065s. [DOI] [PubMed] [Google Scholar]
- 39.Leong NL, Hurng JM, Djomehri SI, Gansky SA, Ryder MI, Ho SP. Age-related adaptation of bone-PDL-tooth complex: Rattus-Norvegicus as a model system. PLoS One. 2012;7:e35980. doi: 10.1371/journal.pone.0035980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ikegawa S. Expression, regulation and function of asporin, a susceptibility gene in common bone and joint diseases. Curr Med Chem. 2008;15:724–8. doi: 10.2174/092986708783885237. [DOI] [PubMed] [Google Scholar]
- 41.Kalamajski S, Aspberg A, Lindblom K, Heinegard D, Oldberg A. Asporin competes with decorin for collagen binding, binds calcium and promotes osteoblast collagen mineralization. Biochem J. 2009;423:53–9. doi: 10.1042/BJ20090542. [DOI] [PubMed] [Google Scholar]
- 42.Yamaba S, Yamada S, Kajikawa T, Awata T, Sakashita H, Tsushima K, et al. PLAP-1/Asporin Regulates TLR2- and TLR4-induced Inflammatory Responses. J Dent Res. 2015;94:1706–14. doi: 10.1177/0022034515606859. [DOI] [PubMed] [Google Scholar]
- 43.Awata T, Yamada S, Tsushima K, Sakashita H, Yamaba S, Kajikawa T, et al. PLAP-1/Asporin Positively Regulates FGF-2 Activity. J Dent Res. 2015;94:1417–24. doi: 10.1177/0022034515598507. [DOI] [PubMed] [Google Scholar]
- 44.Eyre DR. Collagens and cartilage matrix homeostasis. Clin Orthop Relat Res. 2004:S118–22. doi: 10.1097/01.blo.0000144855.48640.b9. [DOI] [PubMed] [Google Scholar]
- 45.Hamada Y, Sumiyoshi H, Matsuo N, Yun-Feng W, Nakashima M, Yanagisawa S, et al. The pro-alpha2(XI) collagen gene is expressed in odontoblasts. Biochem Biophys Res Commun. 2010;392:166–70. doi: 10.1016/j.bbrc.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 46.Lane TF, Sage EH. The biology of SPARC, a protein that modulates cell-matrix interactions. FASEB J. 1994;8:163–73. [PubMed] [Google Scholar]
- 47.Brekken RA, Sage EH. SPARC, a matricellular protein: at the crossroads of cell-matrix communication. Matrix Biol. 2001;19:816–27. doi: 10.1016/s0945-053x(00)00133-5. [DOI] [PubMed] [Google Scholar]
- 48.Kawasaki K, Weiss KM. SCPP gene evolution and the dental mineralization continuum. J Dent Res. 2008;87:520–31. doi: 10.1177/154405910808700608. [DOI] [PubMed] [Google Scholar]
- 49.Chen H, Thomas MG, Hubbard BK, Losey HC, Walsh CT, Burkart MD. Deoxysugars in glycopeptide antibiotics: enzymatic synthesis of TDP-L-epivancosamine in chloroeremomycin biosynthesis. Proc Natl Acad Sci U S A. 2000;97:11942–7. doi: 10.1073/pnas.210395097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lopez-Dee ZP, Chittur SV, Patel B, Stanton R, Wakeley M, Lippert B, et al. Thrombospondin-1 type 1 repeats in a model of inflammatory bowel disease: transcript profile and therapeutic effects. PLoS One. 2012;7:e34590. doi: 10.1371/journal.pone.0034590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ren B, Yee KO, Lawler J, Khosravi-Far R. Regulation of tumor angiogenesis by thrombospondin-1. Biochim Biophys Acta. 2006;1765:178–88. doi: 10.1016/j.bbcan.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 52.Sherbina NV, Bornstein P. Modulation of thrombospondin gene expression during osteoblast differentiation in MC3T3-E1 cells. Bone. 1992;13:197–201. doi: 10.1016/8756-3282(92)90012-l. [DOI] [PubMed] [Google Scholar]
- 53.Carron JA, Bowler WB, Wagstaff SC, Gallagher JA. Expression of members of the thrombospondin family by human skeletal tissues and cultured cells. Biochem Biophys Res Commun. 1999;263:389–91. doi: 10.1006/bbrc.1999.1380. [DOI] [PubMed] [Google Scholar]
- 54.Ueno A, Yamashita K, Nagata T, Tsurumi C, Miwa Y, Kitamura S, et al. cDNA cloning of bovine thrombospondin 1 and its expression in odontoblasts and predentin. Biochim Biophys Acta. 1998;1382:17–22. doi: 10.1016/s0167-4838(97)00188-x. [DOI] [PubMed] [Google Scholar]
- 55.Liu J, Jin T, Chang S, Ritchie HH, Smith AJ, Clarkson BH. Matrix and TGF-beta-related gene expression during human dental pulp stem cell (DPSC) mineralization. In Vitro Cell Dev Biol Anim. 2007;43:120–8. doi: 10.1007/s11626-007-9022-8. [DOI] [PubMed] [Google Scholar]
- 56.Surlin P, Silosi I, Rauten AM, Cojocaru M, Foia L. Involvement of TSP1 and MMP9/NGAL in angiogenesis during orthodontic periodontal remodeling. TheScientificWorldJournal. 2014;2014:421029. doi: 10.1155/2014/421029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Popowics T, Boyd T, Hinderberger H. Eruptive and functional changes in periodontal ligament fibroblast orientation in CD44 wild-type vs. knockout mice. J Periodontal Res. 2014;49:355–62. doi: 10.1111/jre.12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao N, Foster BL, Bonewald LF. The cementocyte-An osteocyte relative? J Dent Res. 2015 doi: 10.1177/0022034516641898. In review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wool IG, Chan YL, Gluck A. Structure and evolution of mammalian ribosomal proteins. Biochem Cell Biol. 1995;73:933–47. doi: 10.1139/o95-101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.