Abstract
Bacterial persisters are phenotypic variants that exhibit an impressive ability to tolerate antibiotics. Persisters are hypothesized to cause relapse infections, and therefore, understanding their physiology may lead to novel therapeutics to treat recalcitrant infections. However, persisters have yet to be isolated due to their low abundance, transient nature, and similarity to the more highly abundant viable but non-culturable cells (VBNCs), resulting in limited knowledge of their phenotypic state. This technical hurdle has been addressed through the use of fluorescence activated cell sorting (FACS) and quantification of persister levels in the resulting sorted fractions. These assays provide persister phenotype distributions, which can be compared to the phenotype distributions of the entire population, and can also be used to examine persister heterogeneity. Here we describe two detailed protocols for analysis of persister physiology with FACS. One protocol assays the metabolic state of persisters using a fluorescent metabolic stain, whereas the other assays the growth state of persisters with use of a fluorescent protein.
Keywords: persister, antibiotic, fluorescence activated cell sorting (FACS), phenotypic heterogeneity, viable but non-culturable cell (VBNC), Redox Sensor Green (RSG)
1. Introduction
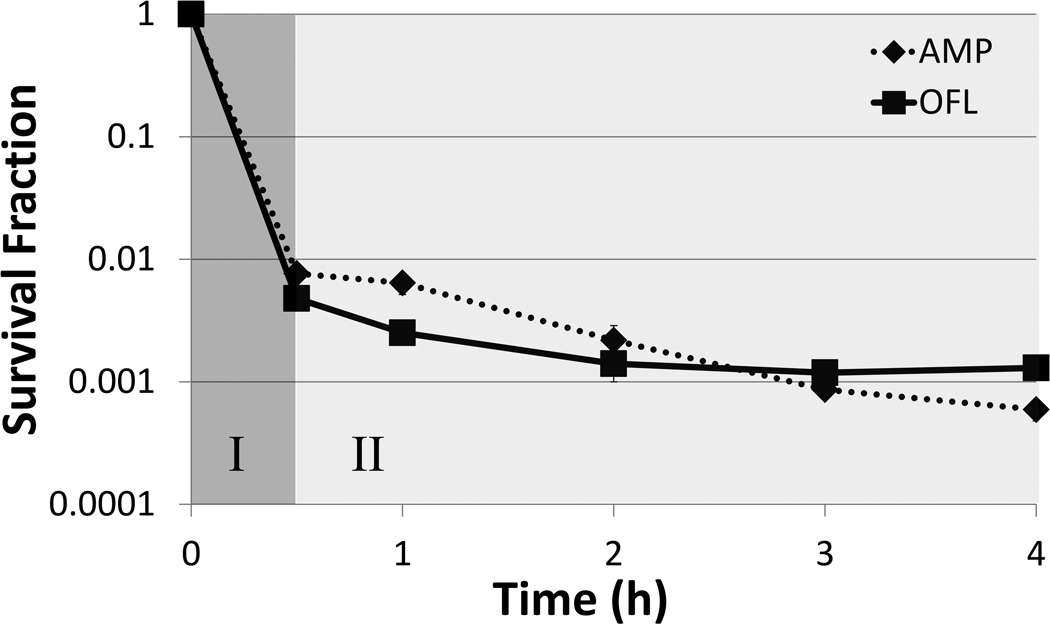
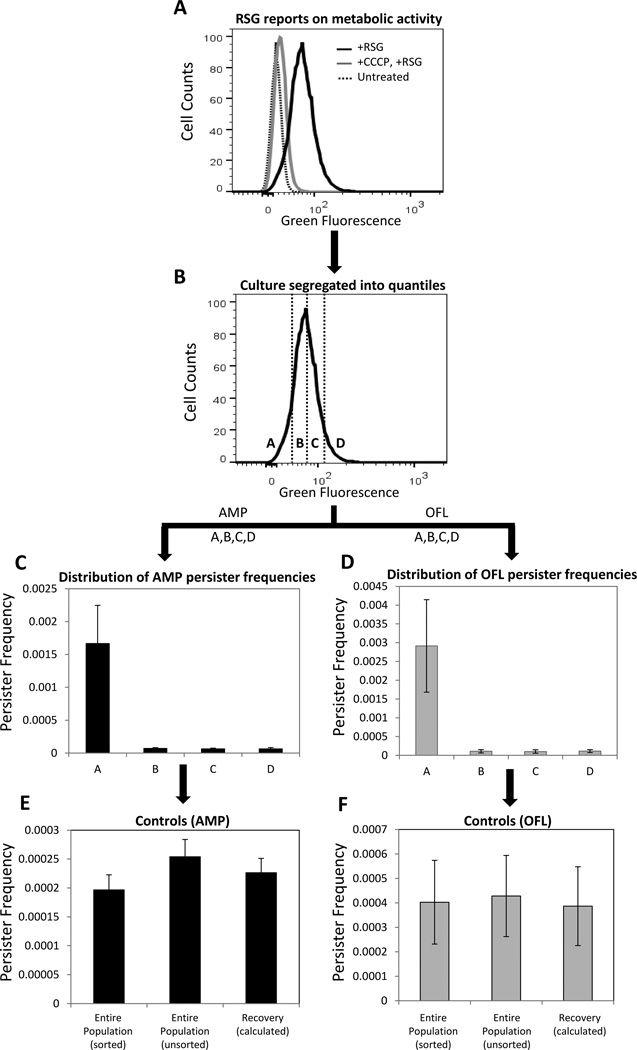
Bacterial cultures treated with high concentrations of bactericidal antibiotics often exhibit two regimes of killing (Figure 1). The first regime is characterized by a rapid killing rate, depicting the death of normal cells, whereas the second regime, characterized by a much slower or non-existent killing rate, indicates the presence of a separate subpopulation of cells (1, 2). When these “persisters” are subsequently cultured, they give rise to a population with antibiotic sensitivity identical to that of the original culture, demonstrating that they are not antibiotic-resistant mutants. Rather, persisters are phenotypic variants that tolerate extraordinary levels of antibiotics due to their physiological state at the time of treatment (3, 4). Persisters have been hypothesized to underlie the propensity of biofilm infections to relapse, and strategies to eliminate them have the potential to impact over half of infections treated in hospitals (5, 6). To facilitate the identification of such potential therapeutics, a greater understanding of persister physiology is desirable. However, persisters are rare, generally 1 in 102–106, transient, and by all measures to date, extremely similar to another more highly abundant subpopulation in bacterial cultures known as viable but non-culturable cells (VBNCs) (7–10). Both persisters and VBNCs exclude propidium iodide (PI) (which stains dead cells), harbor metabolic activity, and do not grow (though exceptions for persisters exist (10, 11)). Indeed, the only known differentiating characteristic between VBNCs and persisters is that persisters can divide and produce colonies after antibiotic treatment on standard media, whereas VBNCs cannot (though some non-standard media can revive some VBNCS (12)). Unfortunately, those resulting colonies are no longer persisters, because the cells had exited the persistent state and initiated cell division once again. These technical challenges necessitate that persisters be studied while in their transient, antibiotic-tolerant state, surrounded by other, more highly abundant cell-types, such as VBNCs. The difficulties posed by VBNCs for the interrogation of persister physiology have only recently been recognized (7–10), and this revelation suggests that two previous methods for “isolating” persisters (13, 14) actually provided only persister-enriched samples that contained many more other cell-types. A recent attempt at isolating persisters was published by Canas-Duarte and colleagues, where Escherichia coli were treated with lysis solutions and biphasic killing was observed (15). Unfortunately, the authors did not test the surviving bacteria for antibiotic tolerance, which is the defining characteristic of persistence. Further, the VBNC levels of the resulting cell suspensions were not quantified, which is of particular concern, since a previous lysis-based technique (13) was found to yield far more VBNCs than persisters (9).Without these controls it is not possible to ascertain whether the method of Canas-Duarte and colleagues was able to segregate persisters from other cell types. Therefore, at present, isolation of persisters has yet to be realized, and biomarkers able to distinguish persisters from VBNCs have yet to be found. In the absence of techniques to separate persisters from other cell-types, fluorescence activated cell sorting (FACS) has become the gold standard technique to examine persister physiology (8–10, 16, 17). In essence, bacterial populations are segregated into groups (quantiles) based on a quantitative characteristic (e.g., expression of a fluorescent protein), and although the existence of persisters within those distributions are unknown at the time of sorting, persistence assays can be performed on the resulting quantiles to quantify the abundance of persisters (Figure 2). In this manner, a persister phenotype distribution is obtained, which can differ quite significantly from that of the total bacterial population (10). Beyond providing one of the only means to quantify persister physiology to date, FACS approaches quantify persister heterogeneity, which has become a topic of increasing interest due to the challenges it poses for eradicating chronic, relapsing infections (18).
Figure 1. Biphasic killing of E. coli treated with antibiotics.
Survival fraction of exponential phase E. coli treated with 200µg/mL ampicillin (AMP) or 5µg/mL ofloxacin (OFL) as measured by CFU. Initial phase of killing (I: dark gray) corresponds to death of normal cells, whereas the second phase of killing (II: light gray) represents colonies derived from persisters.
Figure 2. FACS method to study persister metabolism.
(A) Exponential phase cells (E. coli MG1655::ΔcyoA) were stained with RSG. RSG produces a stable green fluorescent signal when reduced by bacterial reductases. Staining was diminished when cells were pre-treated with CCCP, which depletes proton motive force. (B) RSG stained cells were sorted from the indicated regions (gates) in order to quantify the persister distribution within the quantiles. Gates A, B, C and D compromise 10, 40, 40, and 10%, respectively, of the entire population. (C–D) Persister frequencies were quantified after 5h antibiotic treatment of FACS sorted cells from regions A, B, C, and D. The frequency is the ratio of persisters to initial number of FACS sorted cells. (E–F) Persister frequencies in control samples were similarly quantified after 5h antibiotic treatment. “Entire population (sorted)” corresponds to samples that were sorted without gating, “Entire population (unsorted)” corresponds to samples that did not enter the sorter, and “Recovery (calculated)” is the frequency of persisters one would expect from the total population, as calculated from the persister frequencies measured from the segregated quantiles (A, B, C, D). We note that these three quantities should be indistinguishable from one another. Genetic deletion for MG1655::ΔcyoA strain was transduced from the Keio collection using the standard P1 phage method (33) and the mutation was confirmed with PCR.
Here we describe FACS procedures to assay both the metabolic and cell division states of exponential phase E. coli persisters (10). These cellular qualities were chosen as model characteristics because they involve the use of both a fluorescent stain and protein, and therefore can serve as templates for the interrogation of cellular properties that can be fluorescently labeled by either means.
2. Materials
2.1 Bacterial strains
The methods described here have been used to examine metabolic activity and cell division in persisters of E. coli MG1655 (10). To monitor cell division, the methods described here make use of MO001, which is an MG1655 strain with a chromosomally integrated lacIq promoter in place of the lacI promoter, and a chromosomally integrated T5p-mCherry in place of lacZYA (10).
2.2 Chemicals
Redox sensor green (RSG) (Life Technologies, Invitrogen, Grand Island, NY)
Isopropyl β-D-1-thiogalactopyranoside (IPTG) (Gold Biotechnology, St. Louis, MO)
Carbonyl cyanide m-chlorophenyl hydrazine (CCCP) (Life Technologies, Invitrogen, Grand Island, NY)
Potassium cyanide (KCN)
Luria-Bertani (LB) medium (tryptone, yeast extract, NaCl)
Agar
Phosphate buffered saline (PBS)
Antibiotics: Ampicillin (AMP), Ofloxacin (OFL), and Chloramphenicol (CAM)
-
Fluoresbrite Plain YG 1 micron microspheres (Polysciences, Inc., Warrington, PA) or equivalent.
Unless otherwise stated, all chemicals were purchased from Fisher Scientific or Sigma-Aldrich.
2.3 Media
LB medium was used for planktonic growth. LB medium was prepared by dissolving 10g tryptone, 5g yeast extract, and 10g NaCl in 1L deionized (dI) water and autoclaving for 30 mins at 121°C.
2x-concentrated LB medium was used after FACS. The medium was prepared by dissolving 20g tryptone, 10g yeast extract, and 10g NaCl in 1L dI water and autoclaving for 30 mins at 121°C. Only 1× NaCl was included in this medium, as the 2x-concentrated LB is mixed with PBS, which contains NaCl.
LB agar plates were used for enumeration of colony forming units (CFUs). 15g pure agar powder was added to 1L LB medium as described above. After autoclaving for 30 mins at 121°C and allowing media to cool to 50–60°C, approximately 30mL LB agar was poured into each square petri dish.
2.4 Persister assay materials
For persister assays, 5µg/mL OFL (19) or 200µg/mL AMP (7) were used. To generate a 5mg/mL OFL stock solution, the solution was titrated with sodium hydroxide (1M, dissolved in sterile dI H2O) to fully dissolve the OFL, and then filter-sterilized and stored at 4°C. The day of the experiment, a working solution of 500µg/mL OFL was generated by diluting the stock solution in sterile dI water. Sterile 20mg/mL AMP solution in dI water was prepared freshly on each experimental day.
PBS was used to wash the cells in order to remove the chemicals and antibiotics and to serially dilute the cells for plating.
All materials were purchased from Fisher Scientific or Sigma Aldrich. Other necessary materials include test tubes (glass and/or polypropylene), 96-well round-bottom plates, microcentrifuge tubes (Eppendorf, 1.5mL), syringes, 0.22µm filter units, and 5mL polystyrene round-bottom tubes and BD Falcon 35µm cell strainer capped tubes.
3. Methods
3.1 Fluorophore selection
Here we describe the use of two fluorophores to study persister physiology: RSG, which is a metabolic stain, and mCherry, a fluorescent protein (FP). RSG is a fluorogenic redox indicator that yields green fluorescence when reduced by bacterial reductases. Unlike tetrazolium salts, such as 5-cyano-2,3-ditolyl tetrazolium chloride (CTC), that are reduced to an insoluble, fluorescent formazan product, RSG is nontoxic and does not suppress cellular metabolism (10, 20–22). FPs can be used to monitor numerous cellular properties including promoter activity (transcriptional fusion), protein abundance (translational fusion), and cell division (FP dilution due to growth) (8, 10, 14, 16, 17). Here we describe the use of mCherry as a cell division reporter, where its expression is controlled by a synthetic, chromosomally-integrated expression system that is induced by IPTG (10). Cell division is monitored by fully inducing the expression system and then transferring cells to an inducer-free environment. mCherry is stable in E. coli during the time course of experiments, and therefore, a reduction in fluorescence would be accomplished by dilution through cell division.
In general, fluorophores used to study persister physiology should have negligible impact on culturability or persister levels. Below are template protocols to determine whether a fluorescent stain or protein can be used in persistence studies. We note that persisters are enumerated by measuring CFUs within the second regime of a biphasic kill curve (4, 19). To illustrate, Figure 1 depicts biphasic kill curves of E. coli cultures treated with either OFL or AMP. The first, rapid-killing regime depicts normal cell dying (I), whereas the second phase of killing demonstrates the presence of persisters (II). Therefore, to measure persisters it is important to establish that antibiotic treatments yield biphasic killing, otherwise the CFUs counted may not reflect the abundance of persisters in a population. It is also important to note that the killing rate of persisters need not be zero, it must only deviate significantly from that of normal cells (first regime of biphasic kill curve).
3.1.1 Determination of the impact of a fluorescent stain on culturability and persistence
Prepare an overnight (O/N) culture by inoculating cells from a 25% glycerol, −80°C stock into 2mL media in a test tube, and then incubate the culture at 37°C with shaking (250rpm). We generally use 16 or 24h O/Ns.
Dilute O/N cultures in fresh media to desired optical density (OD600), we suggest ≤0.01, and grow until desired OD600 is reached (we prefer OD600 ~0.1).
Place a 1mL aliquot of the exponential phase culture into a test tube (Note 1).
Remove 10µL of the sample and dilute into 90µL PBS. Plate 10-fold dilutions of this untreated sample onto LB agar plates (Note 2). CFUs from this sample will enumerate the number of cells in the culture before RSG treatment. For ease of serial dilutions, we recommend using a 96-well round-bottom plate, with each well containing 90µL PBS.
Add 1µL of 1mM RSG into 1mL of cell culture in 5 mL polystyrene tubes and incubate in the dark at room temperature for approximately 30 mins. For an unstained cell culture control, incubate 1mL of diluted culture in the dark at room temperature for 30 mins without staining.
Remove 10µL of the samples from both stained and unstained cultures, dilute into 90µL PBS, and plate onto LB agar. CFUs from these samples will enumerate the number of cells in the cultures after RSG treatment. This determines the impact of RSG staining on culturability.
Add 10µL of a freshly prepared stock solution of antibiotic at 100× the treatment concentration (Note 3) into both stained and unstained cell cultures. Be sure to add antibiotic directly to liquid, and gently shake tube several times so that the antibiotic is evenly dispersed and any cells that may be on the side of the tube are washed into the liquid sample.
Incubate the sample at 37°C with shaking at 250rpm.
At desired time points during the treatment, transfer the 1mL aliquot from one test tube to a microcentrifuge tube.
Centrifuge at 15,000rpm for 3 mins.
Remove 900µL of supernatant.
Add 900µL of PBS.
Repeat steps 10–12 until the antibiotic concentration is below the MIC. Do not add PBS to the last wash. Rather, resuspend the pellet in the remaining 100µL PBS (Note 4), resulting in a 10x-concentrated sample.
Plate serial dilutions of the 10x-concentrated sample on LB agar plates.
In order to increase the limit of detection, plate the remaining 80µL of the 10x-concentrated sample onto another LB agar plate.
Incubate plates at 37°C for 16h.
Count CFUs in both the treated and untreated samples (Note 5). Account for the 10× concentration of the treated samples and the dilution of the untreated sample. Biphasic kill curves are generated by plotting the CFU values in logarithmic scale with respect to duration of antibiotic treatment.
3.1.2. Determination of the impact of a FP cell division reporter on culturability and persistence
Perform the following protocol on both FP-expressing cells and cells not expressing FP (wild-type cells):
Prepare an O/N culture by inoculating cells from a 25% glycerol, −80°C stock into 2mL media in a test tube with an inducer (1mM IPTG), and then incubate the culture at 37°C with shaking (250rpm) for the desired O/N duration.
Remove the inducer by centrifuging 1mL of O/N culture for 3 mins at 15,000 rpm, and then removing the supernatant. Resuspend the cell pellet in 1mL of fresh media.
Dilute the resuspended cells in fresh media (OD600≤0.01) and culture at 37° with shaking (250rpm).
Take 1mL samples at desired time points from the exponential phase cultures.
Remove 10µL of the samples from both cultures, dilute into 90µL PBS, and plate on onto LB agar. CFUs from these samples will enumerate the number of cells in the cultures before antibiotic treatment. This also determines the impact of a FP on culturability.
Follow Steps 7–17 in Section 3.1.1 to determine the persister levels in cultures of both the FP expressing and wild-type strains.
An additional fluorophore characteristic to be mindful of is whether its fluorescence can exceed that of bacterial autofluorescence; fluorescence approaching that of autofluorescence would not reflect a physiological property. The fluorophore must also be compatible with the excitation and emission capabilities of the intended FACS system. Also, for two-color or higher-dimensional sorting experiments, the fluorophore should be chosen to minimize overlap of emission spectra, and appropriate single color control samples should be used to compensate for any fluorescence spillover caused by spectral overlap. For example, it is well known that fluorescein isothiocyanate (FITC) and R-phycoerythrin (PE) produce fluorescence that can be detected by photomultiplier tubes receiving emitted fluorescence through 525nm (green) and 575nm (orange) bandpass filters, respectively. Single stained samples can be used to determine the percentages of total FITC or PE fluorescence signals that spill over into the opposite detection channel and then can be appropriately subtracted out of subsequent double-stained samples.
3.2 Initial estimate of the number of cells needed to be sorted to quantify persister physiology
The fraction of persisters in a bacterial population can vary, often between 1 in 102–106, depending on the strain and environment of interest. To ensure that sufficient cells will be sorted to draw conclusions on the physiology of the persister subpopulation, persister levels under conditions identical to those that will be used on the sorted samples should be measured. Our experience has shown that >100 persisters in the entire sorted population are desirable and can yield statistical significance between sorted fractions. A general protocol to perform this experiment is outlined below.
3.2.1. Preliminary experiment to identify the number of cells needed to study persister physiology with sorting
Follow steps 1–17 in Section 3.1.1 to determine the number of persisters in both RSG stained and un-stained cultures (Note 6).
Follow steps 1–6 in Section 3.1.2 to determine the number of persisters in both wild type and FP-expressing strain cultures (Note 6).
Determine the persister fractions in the cultures by taking into account the CFUs at t=0h and t=5h during the antibiotic treatment (in our studies, 5h treatment is sufficient to reach the second killing regime within biphasic kill curves of exponential E. coli cultures).
3.3 Sample preparation for FACS
3.3.1. RSG staining
Prepare O/N and exponential phase cultures as indicated in Steps 1–2 in Section 3.1.1.
If necessary, dilute the cells at desired growth stage in filter-sterilized spent media from the same culture, i.e. media in which the cells have been previously grown, to obtain a cell density of approximately 107 cells/ml. The cell density should not exceed this value to prevent clogging of the cell sorter.
Add 1µL of 1mM RSG into 1mL of diluted cell cultures in 5mL polystyrene tubes and incubate in the dark at room temperature for approximately 30 mins before sorting. This sample can also be used as a positive control. Keep 1mL of unstained cell culture as a negative control for FACS analysis.
As controls, add 2µL of 5mM CCCP or 1µL of 1mM KCN into 1mL of diluted cell cultures for 5 mins prior to addition of RSG. The final concentrations of CCCP and KCN in the cultures should be 10µM and 1mM, respectively. KCN blocks respiration, and CCCP depletes proton motive force; therefore, pretreatment of cells with these inhibitors should reduce green fluorescence.
3.3.2. Cell division assay using FP
Prepare O/N and exponential phase cultures as indicated in Steps 1–3 in Section 3.1.2.
Take 1mL samples at desired time points from the exponential phase culture to sort population based on cell division. If necessary, dilute the cells in filter-sterilized spent media to obtain a cell density of approximately 107 cells/ml.
For a positive control, dilute fully induced O/N culture (see Step 1 in Section 3.1.2) in filter-sterilized spent media to reach a desired cell density for flow cytometric analysis (~107 cells/ml). This control is used to determine the gating for non-growing sub-populations.
For a negative control, incubate the cells without IPTG during the O/N growth, and inoculate in fresh media without inducer as described above.
To verify that the FP is not degraded during the time-frame of the experiment, dilute the washed O/N culture from Step 1 in fresh medium with 50µg/mL CAM (to inhibit protein synthesis) and culture at desired conditions, and then analyze 1mL samples with FACS.
3.4 FACS
We note that a basic working knowledge of flow cytometry and cell sorting is recommended prior to setting up and conducting FACS experiments (23–26). Prior to execution of the experiment, the internal tubing of the FACS instrument should be cleaned to ensure it is free from contaminating bacteria or particulate matter. Refer to manufacturer guidelines for proper system sterilization for your instrument (Note 7). Additionally, consideration must be given to the risk factor group and biosafety level designation of the organisms to be sorted. Cell sorting creates aerosols through droplet formation causing the potential risk for inhalation exposure, and the system can be under high pressure increasing the risk of splash exposure to liquids (27–29). Biosafety professionals should be consulted and proper precautions should be in place prior to conducting any FACS experiments.
When studying E. coli with FACS, care must be taken during system setup and alignment to ensure the proper differentiation between actual particles (E. coli) and electronic noise, since the size of E. coli approaches the limit of detection of many commercially available FACS systems. Electronic noise is seen as background signal present without cellular material running on the instrument (e.g., 0.22µm-filtered PBS) at a given set of conditions and can be affected by many factors. Photomultiplier tube (PMT) voltage and system threshold settings must be optimized to eliminate and/or minimize any signal contribution from electronic noise.
3.4.1. FACS method
Start up FACS system and give proper time for lasers to warm up and the stream to stabilize according to manufacturer’s recommendations (Note 8). A 488nm laser and ~530nm detection filter are required for RSG detection, whereas a 561nm laser and ~600nm detection filter are required for mCherry detection. Consult references or a flow cytometry specialist for proper setup choices for other dyes or FPs.
Align laser(s) and determine the proper droplet breakoff as per manufacturer’s recommendations for your system. Consult vendor or professional cytometrist as needed.
Set forward scatter (FSC), side scatter (SSC) and appropriate fluorescence parameters to log scale (Note 9).
Create FSC-A vs SSC-A and desired fluorescence parameter plots in acquisition software (Note 10).
Place a tube of clean, particle-free PBS (0.22µm-filtered) on the system as a sample and run.
Adjust FSC PMT voltage, SSC PMT voltage, and SSC threshold values while PBS is running to minimize electronic noise signal detected (Note 11).
Remove PBS tube and rinse the sample uptake line with clean dI water.
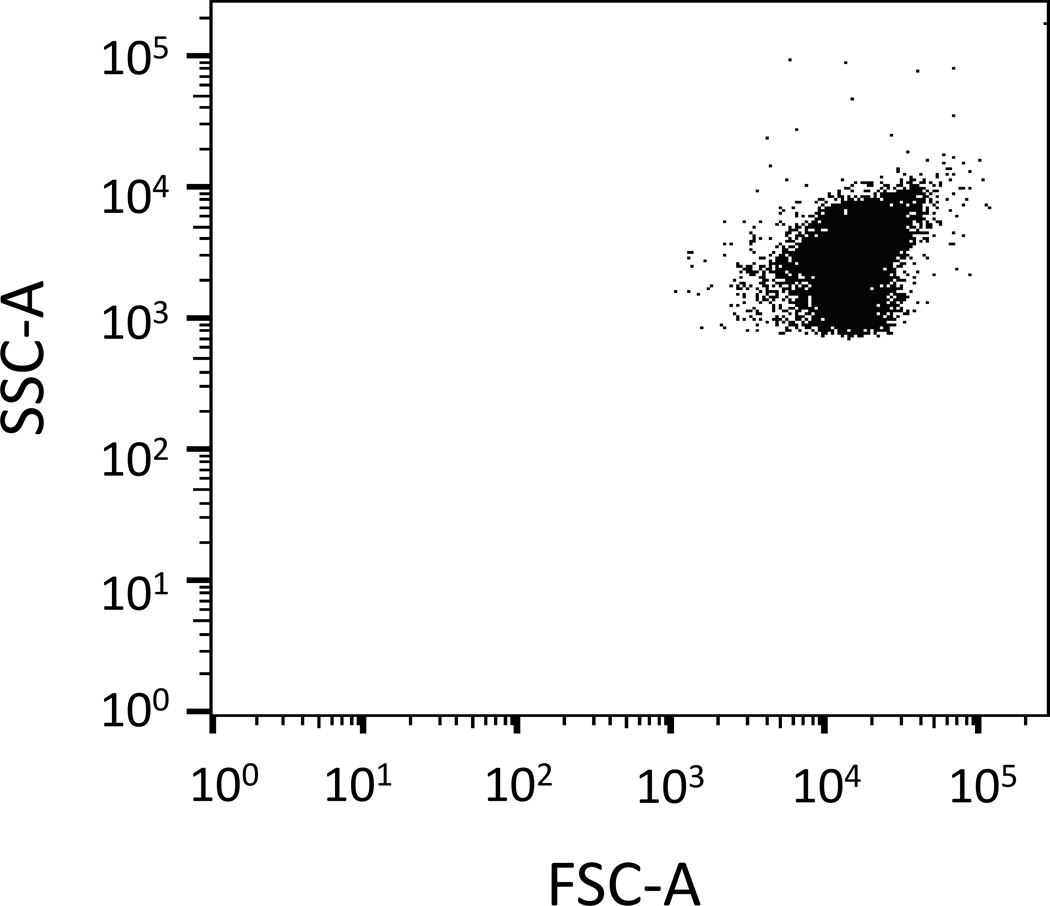
Place a tube of live, non-fluorescent, exponential phase E. coli cells (e.g., unstained, non-FP expressing) on the system and adjust FSC and SSC PMT voltage settings so that cells are on scale and electronic noise remains low (Notes 12 and 13) (Figure 3). Make sure sample concentration is optimized for your FACS system setup. We have found that cell concentrations ~ 107/mL or less work well (Note 14).
Rinse the sample uptake line with clean dI water and re-sample the PBS. Confirm that there is little (normally less than 100 events/sec) signal contribution from electronic noise.
Remove PBS tube and run non-fluorescent E. coli cells (negative control: unstained, non-FP expressing) to evaluate cellular autofluorescence. Adjust fluorescence parameter PMT voltage setting(s) to place the signal from this negative control sample toward the lower end of the scale for each fluorescent signal to be detected.
Run positive controls (see Section 3.3) to check if sorter can detect the fluorescence signals. Adjust the fluorescence parameter PMT voltage settings if positive signal is off scale (too bright). Rerun negative control sample at new PMT voltage setting.
Run additional controls to verify that RSG reports on metabolic activity (see Section 3.3.1) (Figure 2A), or FP is not degraded during the time frame of the experiment (see Section 3.3.2) (representative images of this control can be found in Figure S2 of reference (10)).
Run test samples to be sorted (Note 15). Assign sort gates via acquisition software, insert clean collection tubes, and begin sorting (Note 16).
To evaluate the purity of post-sort fractions, rinse the sample lines well with clean dI water or PBS after sorting and then run a small amount of each quantile. To ensure proper analysis, be sure to rinse well between each sample (Note 17).
Figure 3. SSC-A vs. FSC-A dot-plot.
FSC and SSC PMT voltage settings are adjusted so that exponential phase E. coli cells (unstained, non-FP expressing) are on SSC-A and FSC-A scales. Each dot represents a cell.
3.5 Culturability and persistence assays on sorted samples
Following sorting, bacterial cells from their respective quantiles are suspended in sheath fluid, which is usually PBS, and these cell suspensions will be used to measure both culturability and the abundance of persisters within the quantiles (Note 18).
3.5.1. Persistence and culturability assays on sorted fractions
-
After the total number of cells (T) have been isolated from each quantile, mix the collected sample with an equal volume of 2x-concentrated fresh LB medium in a test tube (Notes 19 and 20). If the total volume (V) will be >2mL, we recommend using a 50mL Falcon tube for treatment to ensure proper aeration of the sample.
For controls:- Dilute approximately T number of cells from the culture (without sorting) into V volume of 2x-concentrated LB media+PBS mixture (mixed at equal volumes) to analyze the effects of flow through the sorter on persister levels.
- Dilute approximately T number of cells from the culture (without sorting) into V volume of fresh 1× LB media to examine the effects of media+PBS mixture when compared to media only on persister levels.
- Collect T number of cells using FACS (from the entire population without segregating) and mix the collected sample at equal volume with 2x-concentrated LB medium to analyze the effects of segregation on persister levels (Note 21).
Remove 10µL from the samples and dilute into 90µL PBS. Plate serial 10-fold dilutions of these untreated samples onto LB agar plates. CFUs from this sample will enumerate the number of cells in the cultures before treatment (t=0h).
Add the appropriate volume of freshly prepared 100x-concentrated antibiotic to the samples (Note 3).
Incubate the samples at 37°C with shaking at 250rpm.
Remove the samples from the shaker at desired time points.
If the volume of sample is less than 2mL, transfer sample to a microcentrifuge tube and go to step 8.
If the volume of the sample is greater than 2mL, transfer sample to a 15mL Falcon tube. Spin at 4,000rpm for 15 mins. Remove all but 1mL supernatant. Resuspend the pellet in the 1mL and transfer to a microcentrifuge tube.
Spin at 15,000rpm for 3 mins. Remove all but 100µL supernatant. Resuspend the pellet in the 100µL.
Add 900µL PBS.
Repeat steps 8 and 9 until the antibiotic concentration is below the MIC. Resuspend the pellet in the remaining 100µL PBS, resulting in a concentrated sample.
Plate serial dilutions of the concentrated samples on LB agar plates.
In order to increase the limit of detection, plate the remaining 80µL of the concentrated samples onto another agar plate.
Incubate plates at 37°C for 16h and count the CFUs by taking into account the concentration factor.
Repeat Steps 1–13 for unstained cells (control of RSG staining) and un-induced cells (control of FP expression) (Note 22).
Acknowledgments
This work was supported by the Department of the Army under award number W81XWH-12-2-0138 and the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number R21AI105342. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
Footnotes
The number of test tubes depends on the duration of treatment. For each time point, one test tube is used. We have found that 5 hours of treatment results in biphasic killing of E. coli growing in LB (4). We re-iterate that one must ensure that the duration of antibiotic treatment is long enough to measure CFUs within the second regimen of a biphasic kill curve.
Agar plates should be dried 1–2 days at room temperature to ensure that 10µL spots do not run together when plated.
Treat samples with a concentration of antibiotic that is many-fold higher than the minimum inhibitory concentration (MIC) of the strain. The MIC of the strain may be determined by a broth dilution method (30) or an agar method (31). We have determined the MIC of our strains to be ~0.075–0.15µg/mL OFL and 1.5–3µg/mL AMP. Our antibiotic treatment concentrations for persister assays are 5µg/mL OFL or 200µg/mL AMP.
It is optimal to have a volume of exactly 100µL, not simply an approximation. Therefore, we recommend measuring the amount of liquid with the pipet tip, and adjusting the volume.
We generally count spots containing 10–100 CFUs (32).
Note that since we use sterile-filtered PBS as a sheath fluid in FACS, sorted samples are suspended in PBS, and antibiotic treatments are done in a mixture of PBS and 2xLB media. We have found that treatment in the PBS+2xLB mixture results in persister fractions comparable to those of samples treated in 1xLB for exponential phase cultures (OD600~0.1), and therefore, our preliminary experiments were done using 1xLB. However, one may wish to perform the preliminary experiments in Section 3.2.1 by treating in a PBS+2xLB mixture, so that the conditions more accurately represent those that are used with FACS.
A sample of sheath fluid can be taken from the sheath fluid stream directly above the waste catcher on the front of the instrument and placed into culture medium for incubation to confirm the cleanliness of the system.
We perform sorting with a FACSVantage SE w/DiVa (BD Biosciences, San Jose, CA) with a 70µm nozzle at 16 psi and the following settings: frequency 32.3, amplitude 27.5, phase 10, drop delay 14.75. Purity precision sort mode is used. All system settings are unique to each FACS instrument and sort setup should be optimized and tested for your specific system prior to conducting all sort experiments.
FSC and SSC are measurements taken from the amount of laser light scattered from the interrogating laser beam as each particle (cell, debris, or aggregates of cells) passes through. FSC is affected more by the cross sectional area and refractive index of the cell, whereas SSC is related to the granularity or internal complexity of a cell (24). Using log scale for FSC and SSC is helpful when looking at small particles such as bacteria.
One may also wish to create one or more doublet discrimination plots. Doublets (two cells stuck together) and/or aggregates of cells can be a confounding factor in FACS. Pulse processing analysis allows one to reduce the likelihood of doublets/clumps in subsequent analysis plots by gating on discrimination plots (FSC-W vs. FSC-A and/or SSC-W vs. SSC-A). Each particle passing through the laser beam creates a peak pulse in all activated detection parameters. The width (W) signal displays the duration of the peak pulse; the height (H) signal, the maximum light; and the area (A) signal, the total light detected. Since the E. coli samples we used had insignificant cell aggregation, we did not cover this section in detail. See reference (24) for specifics.
To minimize the electronic noise signal detected, set the SSC threshold value slightly above the minimum allowable value for your system and lower the FSC & SSC PMT voltage values until event rate falls below 100 events per second. In systems that allow dual threshold parameters one may also wish to activate the FSC threshold as well to better address the elimination of system noise from the analysis.
One may use 1µm sized beads, which are similar in size to E. coli, rather than cells to adjust the PMT voltages and make sure that the sorter detects the signal from these events. In order to create the bead sample, dilute 5µl Fluoresbrite Plain YG 1 micron beads or equivalent in 2mL PBS. Filter bead suspension through a 35µm mesh cell strainer to remove any aggregates.
Observe the events/second of the cells or beads alone. As one adjusts the FSC and SSC voltages, the event rate will spike (often to tens of thousands events/sec) when noise is detected. Lower the voltage(s) until noise signal disappears and event rate decreases.
Samples that are too concentrated can cause difficulty in system setup and performance. If fluorescence signal drifts while running (moves from high to low to high again), remove sample and dilute.
Data files of all test and control samples should be recorded to generate flow diagrams. Once PMT voltage values are optimized using control samples, all test samples should be recorded using the same voltage values.
Sort into appropriate culture medium or simply collect droplets containing sorted cells into empty tubes. We sort at room temperature, but some sorters are equipped with temperature control options for both the sample and collection tubes if needed.
Transfer a small amount of each sorted fraction to a clean tube for reanalysis to avoid any risk of mixing sorted samples. Also be advised that any electronic noise contribution from the system will lower the sort fraction purity values displayed. Compare sort check data to PBS only sample to draw conclusions as to the level of success obtained for each sorted fraction.
The FACS procedure might affect the culturability of the cells. Therefore, once sorting parameters (such as pressure and flow rate) have been optimized, the culturability of cells being sorted should be checked by plating a number of cells immediately after sorting. We identified that more than 80% of the cells from exponentially growing cultures in LB sorted with FACS with the indicated parameters in Note 8 can form CFUs.
Under our conditions, adding 2x-concentrated LB to the cell samples sorted into PBS does not change persister fractions from samples treated in 1x-LB.
If the cells are sorted into a large volume of PBS, one may wish to remove the excessive PBS by centrifugation.
We have used a two-tailed t-test to perform a pairwise comparison of the persister fractions that result from the sort quantiles (10). We have already confirmed that the CFU measurements performed for persister assays were normally distributed by using a larger sample data set and the Anderson-Darling and Shapiro-Wilk tests (16).
References
- 1.Amato SM, Fazen CH, Henry TC, et al. The role of metabolism in bacterial persistence. Front Microbiol. 2014;5:70. doi: 10.3389/fmicb.2014.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kint CI, Verstraeten N, Fauvart M, et al. New-found fundamentals of bacterial persistence. Trends Microbiol. 2012;20(12):577–585. doi: 10.1016/j.tim.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 3.Balaban NQ, Merrin J, Chait R, et al. Bacterial persistence as a phenotypic switch. Science. 2004;305(5690):1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 4.Lewis K. Persister Cells. In: Gottesman S, Harwood CS, editors. Annual Review of Microbiology. Vol. 64. Palo Alto: Annual Reviews; 2010. pp. 357–372. [DOI] [PubMed] [Google Scholar]
- 5.Lewis K. Persister cells, dormancy and infectious disease. Nat Rev Microbiol. 2007;5(1):48–56. doi: 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- 6.Fauvart M, De Groote VN, Michiels J. Role of persister cells in chronic infections: clinical relevance and perspectives on anti-persister therapies. J Med Microbiol. 2011;60(Pt 6):699–709. doi: 10.1099/jmm.0.030932-0. [DOI] [PubMed] [Google Scholar]
- 7.Joers A, Kaldalu N, Tenson T. The Frequency of Persisters in Escherichia coli Reflects the Kinetics of Awakening from Dormancy. J Bacteriol. 2010;192(13):3379–3384. doi: 10.1128/JB.00056-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roostalu J, Jõers A, Luidalepp H, et al. Cell division in Escherichia coli cultures monitored at single cell resolution. BMC Microbiol. 2008;8:68. doi: 10.1186/1471-2180-8-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orman MA, Brynildsen MP. Establishment of a method to rapidly assay bacterial persister metabolism. Antimicrob Agents Chemother. 2013;57(9):4398–4409. doi: 10.1128/AAC.00372-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Orman MA, Brynildsen MP. Dormancy is not necessary or sufficient for bacterial persistence. Antimicrob Agents Chemother. 2013;57(7):3230–3239. doi: 10.1128/AAC.00243-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wakamoto Y, Dhar N, Chait R, et al. Dynamic persistence of antibiotic-stressed mycobacteria. Science. 2013;339(6115):91–95. doi: 10.1126/science.1229858. [DOI] [PubMed] [Google Scholar]
- 12.Oliver JD. The viable but nonculturable state in bacteria. J Microbiol. 2005;43(Spec No):93–100. [PubMed] [Google Scholar]
- 13.Keren I, Shah D, Spoering A, et al. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J Bacteriol. 2004;186(24):8172–8180. doi: 10.1128/JB.186.24.8172-8180.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shah D, Zhang Z, Khodursky A, et al. Persisters: a distinct physiological state of E. coli. BMC Microbiol. 2006;6:53. doi: 10.1186/1471-2180-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Canas-Duarte SJ, Restrepo S, Pedraza JM. Novel protocol for persister cells isolation. PLoS One. 2014;9(2):e88660. doi: 10.1371/journal.pone.0088660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amato SM, Orman MA, Brynildsen MP. Metabolic Control of Persister Formation in Escherichia coli. Mol Cell. 2013;50(4):475–487. doi: 10.1016/j.molcel.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 17.Vega NM, Allison KR, Khalil AS, et al. Signaling-mediated bacterial persister formation. Nat Chem Biol. 2012;8(5):431–433. doi: 10.1038/nchembio.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allison KR, Brynildsen MP, Collins JJ. Heterogeneous bacterial persisters and engineering approaches to eliminate them. Curr Opin Microbiol. 2011;14(5):593–598. doi: 10.1016/j.mib.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keren I, Kaldalu N, Spoering A, et al. Persister cells and tolerance to antimicrobials. FEMS Microbiol Let. 2004;230(1):13–18. doi: 10.1016/S0378-1097(03)00856-5. [DOI] [PubMed] [Google Scholar]
- 20.Kalyuzhnaya MG, Lidstrom ME, Chistoserdova L. Real-time detection of actively metabolizing microbes by redox sensing as applied to methylotroph populations in Lake Washington. ISME J. 2008;2(7):696–706. doi: 10.1038/ismej.2008.32. [DOI] [PubMed] [Google Scholar]
- 21.Ullrich S, Karrasch B, Hoppe HG, et al. Toxic effects on bacterial metabolism of the redox dye 5-cyano-2,3-ditolyl tetrazolium chloride. Appl Environ Microbiol. 1996;62(12):4587–4593. doi: 10.1128/aem.62.12.4587-4593.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gray DR, Yue S, Chueng CY, et al. Bacterial vitality detected by a novel fluorogenic redox dye using flow cytometry. Abstracts of the General Meeting of the American Society for Microbiology. 2005;105:331. [Google Scholar]
- 23.Arnold LW, Lannigan J. Practical issues in high-speed cell sorting. Curr Protoc Cytom. 2010;Chapter 1(Unit 1.24):1–30. doi: 10.1002/0471142956.cy0124s51. [DOI] [PubMed] [Google Scholar]
- 24.Givan A. Flow Cytometry: First Principles. Second. New York: Wiley-Liss; 2001. [Google Scholar]
- 25.Shapiro H. Practical Flow Cytometry. Fourth. New York: Wiley-Liss; 2003. [Google Scholar]
- 26.Ormerod M. Flow Cytometry: A Practical Approach. New York: Oxford University Press; 2000. [Google Scholar]
- 27.Holmes KL, Fontes B, Hogarth P, et al. International Society for the Advancement of Cytometry cell sorter biosafety standards. Cytom A. 2014;85(5):434–453. doi: 10.1002/cyto.a.22454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmid I, Nicholson JK, Giorgi JV, et al. Biosafety guidelines for sorting of unfixed cells. Cytometry. 1997;28(2):99–117. doi: 10.1002/(sici)1097-0320(19970601)28:2<99::aid-cyto2>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 29.Schmid I, Lambert C, Ambrozak D, et al. International Society for Analytical Cytology biosafety standard for sorting of unfixed cells. Cytom A. 2007;71(6):414–437. doi: 10.1002/cyto.a.20390. [DOI] [PubMed] [Google Scholar]
- 30.Ericsson HM, Sherris JC. Antibiotic sensitivity testing. Report of an international collaborative study. Acta Pathol Microbiol Scand B Microbiol Immunol. 1971;217(Suppl 217) 1+ [PubMed] [Google Scholar]
- 31.Andrews JM. Determination of minimum inhibitory concentrations. J Antimicrob Chemother. 2001;48(Suppl 1):5–16. doi: 10.1093/jac/48.suppl_1.5. [DOI] [PubMed] [Google Scholar]
- 32.Kohanski MA, Dwyer DJ, Hayete B, et al. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130(5):797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 33.Baba T, Ara T, Hasegawa M, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100050. 2006 0008. [DOI] [PMC free article] [PubMed] [Google Scholar]