The biosynthesis of many different types of antibiotics and other secondary metabolites is regulated by phosphate. Production of these valuable compounds occurs only under phosphate-limiting nutritional conditions. In a few cases, there is evidence showing that the negative phosphate control is exerted at the transcriptional level. Recently, it was shown that phosphate control of antibiotic biosynthesis in Streptomyces lividans and Streptomyces coelicolor is mediated by the two-component PhoR-PhoP system that also controls the alkaline phosphatase gene (phoA). The PhoR protein is a standard membrane sensor kinase, whereas PhoP is a member of the DNA-binding response regulators. In Escherichia coli and Bacillus subtilis, the phosphorylated PhoP protein (PhoP∼P) activates, in response to phosphate starvation, expression of the pho regulon genes by binding to consensus phosphate boxes in the promoter regions (PHO boxes). Expression of phoA in S. lividans is induced by PhoP∼P, and mutants lacking phoP (or phoR and phoP) do not form PhoA. These mutants overproduce large amounts of actinorhodin and undecylprodigiosin. No consensus PHO boxes occur in the upstream region of phosphate-regulated secondary metabolism genes. However, pathway-specific activator proteins (ActII-open reading frame 4 [ORF4], RedD, CcaR, and DnrI) are known to bind to these regions. In S. coelicolor, actII-orf4 is positively regulated by the AfsS protein, which, in turn, is induced by the phosphorylated AfsR protein. It is likely that the PhoR-PhoP system exerts its action on actinorhodin and undecylprodigiosin by a cascade mechanism mediated by AfsR and AfsS. Directed phoR-phoP gene disruption will be very useful for the construction of tailored phosphate-deregulated strains overproducing valuable secondary metabolites.
PHOSPHATE CONTROL OF ANTIBIOTIC BIOSYNTHESIS: HISTORICAL PERSPECTIVE
The negative control exerted by inorganic phosphate on the biosynthesis of antibiotics and other secondary metabolites has been known for many years. Over the last four decades, an impressive number of antibiotics and secondary metabolites have been shown to be regulated negatively by phosphate. These include streptomycin, oxytetracycline, clavulanic acid, tylosin, echinomycin, cephalosporin, cephamycin C, and thienamycin, among many other secondary metabolites (23, 27, 32), but, surprisingly, the molecular mechanism of phosphate control has remained obscure (30) in spite of its basic and industrial relevance. It is interesting that inorganic phosphate in the culture medium controls the synthesis of a large number of secondary metabolites belonging to different biosynthetic groups such as, for example, macrolides, tetracyclines, anthracyclines, polyether compounds, aminoglycosides, and amino acid-derived metabolites such as clavulanic acid, among others (10, 11, 15, 16, 25). Why are all these compounds repressed by high concentrations of inorganic phosphate? From a biosynthetic point of view, these groups of metabolites have very little in common, except that they all are dispensable “secondary” metabolites. The negative effect exerted by inorganic phosphate on the biosynthesis of secondary metabolites is observed in a wide range of microorganisms, including proteobacteria, gram-positive bacteria (e.g., actinomycetes), and filamentous fungi, and probably has a wide ecological role. Martín and Demain proposed that phosphate control is used as a mechanism that triggers secondary metabolite biosynthesis when phosphate in the environment is depleted and, therefore, growth of the microorganisms cannot proceed at a normal rate (28). When the phosphate concentration in the culture medium decreases below a threshold level, bacteria increase their production of a variety of metabolites that might serve as direct antagonists to other microorganisms (48) or as biochemical cross talk signals (17, 38, 51) to enhance survival under harsh nutritional conditions (26).
A number of scientific studies published since the 1970s describe the negative effect of high concentrations of phosphate on the production of a variety of secondary metabolites (reviewed in reference 27). However, most of these reports are basic accounts of the overall phosphate effect on the control of antibiotic production and were made before the gene clusters encoding the biosynthesis of secondary metabolites were sequenced. These early studies were helpful in designing medium composition to avoid phosphate control, thereby favoring the production of secondary metabolites that are expressed at very low levels.
PHOSPHATE CONTROL OF ANTIBIOTIC BIOSYNTHESIS AT THE TRANSCRIPTIONAL LEVEL
The phosphate control signals of antibiotic biosynthesis, i.e., the biochemical intermediates involved in the signal transduction cascade, have long remained largely unknown. These signals appear to be integrated with the inputs produced by other sensors of environmental or nutritional stress, e.g., carbon or nitrogen limitation (7). Phosphate control of the biosynthesis of secondary metabolites is exerted at the transcriptional and posttranscriptional (antibiotic synthases activity) levels (reviewed in references 23 and 30). In at least two cases, it is clearly established that phosphate control of the biosynthesis of the secondary metabolites candicidin and oxytetracycline is exerted at the transcriptional level (3, 34). Expression of the pabS gene of S. griseus, which encodes the candicidin precursor-forming enzyme p-aminobenzoic acid (PABA) synthase (3), and expression of the tetracycline biosynthesis genes otcC, otcX, and otcY (34) are observed only under phosphate-limiting conditions. otcC encodes anhydrotetracycline oxygenase, and otcY codes for the tetracycline polyketide synthase, whereas otcX is a gene divergent to otcC encoding a protein of unknown function (19).
The more advanced studies on phosphate control of secondary metabolites have been done with the model actinomycetes S. coelicolor and S. lividans (11, 16). The synthesis of the S. coelicolor pigmented secondary metabolites actinorhodin and undecylprodigiosin is also negatively controlled by high phosphate concentrations (11, 16). Actinorhodin is synthesized by a type II polyketide synthetase, whereas undecylprodigiosin is a pyrrolic compound derived from proline. Surprisingly, no transcriptional studies were available until recently (see below) on the molecular mechanism of phosphate control of the expression of the actinorhodin and undecylprodigiosin genes.
Genes encoding secondary metabolites are frequently linked in clusters (5, 29). Phosphate control appears to repress the expression of entire clusters of antibiotic biosynthesis genes, e.g., genes encoding the activating enzyme PABA-coenzyme A ligase and a thioesterase, in addition to PABA synthase in the candicidin gene cluster (6, 9) or the complete pimaricin gene cluster (2). A question that remains unanswered is whether each promoter corresponding to phosphate-sensitive genes contains a phosphate box or whether there is a master gene encoding a regulatory protein involved in the activation of antibiotic biosynthetic genes that in turn is controlled by the inorganic phosphate level.
ARE THERE PHO BOXES IN PHOSPHATE-REGULATED GENES IN STREPTOMYCES SPECIES?
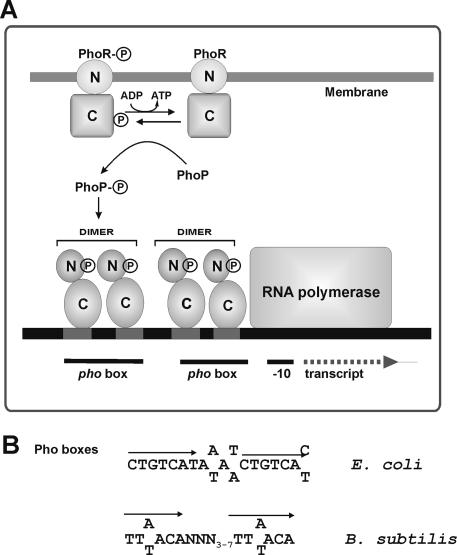
In E. coli and B. subtilis, about 30 genes belonging to the pho regulon are controlled by the two-component PhoR-PhoB (named PhoR-PhoP in B. subtilis) system (18, 46). More genes of the pho regulon have been identified by proteomics and transcriptional studies of B. subtilis (1) and by microarray analysis of Corynebacterium glutamicum (21).
In E. coli, the sensor protein kinase PhoR self-phosphorylates under conditions of phosphate starvation (forming PhoR∼P) that transfers its phosphate group to the dephosphorylated PhoB. The phosphorylated PhoB activates expression of about 30 phosphate-regulated genes by binding to the PHO boxes located in the 5′ region of these genes (46). Expression of phoA and other members of the phosphate regulon takes place under phosphate-limiting conditions when the PhoB transcriptional activator is available in its phosphorylated form (PhoB∼P). The mechanism is essentially identical in B. subtilis (18, 40). PHO boxes in E. coli consist of 18 nucleotides (C/T)TGTCATA(A/T)A(A/T)CTGTCA(T/C) formed by two direct repeats of 7 nucleotides (C/T)TGTCAT separated by four adenines or thymines. This sequence has been found, with minor changes, in the promoters of a variety of phosphate-controlled genes in different proteobacteria (30, 46) (Fig. 1).
FIG. 1.
Phosphate boxes and signal transduction pathway from PhoR to PhoP. (A) PhoR and PhoP consist of the following two domains: C, the carboxyl-terminal domain, and N, the amino-terminal domain. The circled P corresponds to a phosphate group. Each pho box is formed by two elements arranged in a direct repeat separated by three to seven nucleotides. Some promoters contain one pho box, whereas others contain more than one. Two pho boxes are shown in this figure. This model is based on the information available for B. subtilis (24), S. coelicolor, and S. lividans (43). (B) Nucleotide sequences of the pho boxes of E. coli and B. subtilis.
Similar PHO boxes, although with a different consensus sequence, occur in B. subtilis. Each box consists of two hexanucleotides [TT(A/T)ACA] repeated in tandem and separated by three to seven nucleotides (24), in contrast to E. coli, for which the separation of the two repeats in each PHO box is always of four nucleotides (Fig. 1).
The full genome sequence of S. coelicolor (4) and S. avermitilis (20, 39) is now available, and some others will soon be accessible. Several research groups have provided evidence showing that the production of methylenomycin, actinorhodin, and undecylprodigiosin in cultures of S. coelicolor is reduced by high phosphate concentrations (11, 16). Until a few years ago, it was unclear whether standard PHO boxes might occur in the upstream regions of antibiotic biosynthesis genes (23). However, repeated searches to find standard PHO boxes in the upstream region of act (for actinorhodin) or red (for undecylprodigiosin) genes have been unsuccessful. Similarly, no consensus PHO boxes have been found in the upstream region of the phosphate-regulated ppk gene of S. lividans (8).
Expression of the act and red genes is regulated by the specific transcriptional activators ActII-ORF4 (12, 14) and RedD (37, 44). Similarly, expression of the daunorubicin gene cluster in Streptomyces peucetius is controlled by the transcriptional activator DnrI (45). The activator proteins ActII-ORF4, RedD, DnrI, and other pathway-specific regulators, such as CcaR in Streptomyces clavuligerus (41), belong to the group of SARP (Streptomyces antibiotic regulatory protein) regulators (50) that form part of the OmpR superfamily (31, 35, 41). It is possible that the expression of the transcriptional activators ActII-ORF4 and RedD is controlled by phosphate, but again, the upstream regions of these genes lack classical PHO boxes. Initial evidence suggests, however, that the phosphate effect is mediated by a different sequence in the GC-rich promoters of Streptomyces species than in E. coli.
THE STREPTOMYCES PhoR-PhoP SYSTEM IS INVOLVED IN PHOSPHATE CONTROL OF ACTINORHODIN AND UNDECYLPRODIGIOSIN
After the cloning and characterization of the extracellular alkaline phosphatase (phoA) gene of S. griseus (36), we found three putative phosphatase genes in the S. coelicolor genome. One of them, phoA (the putative extracellular alkaline phosphatase gene) showed 71% identity to S. griseus phoA, identified unequivocally by sequencing of the amino-terminal end of the purified extracellular alkaline phosphatase (36). The biochemical characteristics of PhoB are unknown and PhoC corresponds to a phosphodiesterase.
Recently, the phoR-phoP systems of S. coelicolor and S. lividans were cloned, and their role in actinorhodin and undecylprodigiosin biosynthesis in S. lividans was studied by gene disruption and gene replacement (43). The phoR-phoP cluster has also been identified and disrupted in the S. avermitilis genome (H. Ikeda, J. F. Martín, and S. Omura, unpublished data). In Streptomyces species, PhoR shows all of the characteristics of a transmembrane sensor protein, whereas PhoP is a member of the DNA-binding OmpR family (35). PhoR (426 amino acids; 45.4 kDa in S. coelicolor) has two hydrophobic, membrane-spanning regions in the N-terminal domain and a large extramembrane domain that may serve as a sensor of environmental signals (35, 43). In B. subtilis the cytoplasmic kinase domain of PhoR is sufficient for the low phosphate expression of the pho regulon genes (42). The S. coelicolor and S. lividans 24.7-kDa PhoP protein belonging to the OmpR family contains a DNA-binding domain in the carboxyl-terminal region (residues 190 to 201) (43).
The S. lividans phoR-phoP deletion mutants (named ΔphoP and ΔphoRP) lacking either PhoP or both PhoR and PhoP proteins are unable to synthesize extracellular alkaline phosphatase, as shown by Western blot analysis. The formation of alkaline phosphatase was restored by complementation of the deletion mutants with phoR-phoP, confirming the involvement (positive effect) of the two-component system in the phosphate control of phoA in this actinomycete (43). A very interesting finding is the observation that mutants with phoR-phoP deletions overproduce large amounts of actinorhodin and undecylprodigiosin in a manner that is partially insensitive to phosphate (1 to 10 mM) control.
Phosphate control of the biosynthesis of actinorhodin and undecylprodigiosin was restored by complementation of the Δpho mutants with the phoR-phoP cluster (43). These results clearly indicate that expression of either primary metabolism genes such as phoA or genes involved in the biosynthesis of secondary metabolites such as actinorhodin and undecylprodigiosin are under the phosphate control mechanism mediated by PhoR-PhoP.
THE PhoR-PhoP ACTION ON ACTINORHODIN AND UNDECYLPRODIGIOSIN MAY BE MEDIATED BY THE PATHWAY-SPECIFIC REGULATORS
It is possible that there is a phosphate control mechanism of antibiotic gene expression different from that recognizing the classical PHO box sequences. It is important to note that, whereas PhoP exerts a positive regulation on expression of the phoA gene, as occurs in other bacteria, expression of S. lividans or S. coelicolor pigments and antibiotics is regulated negatively, i.e., inactivation of the response regulator PhoP or deletion of the entire PhoR-PhoP system results in overexpression of actinorhodin and undecylprodigiosin. This suggests that the modes of regulation of the alkaline phosphatase and secondary metabolite genes are different and that the PhoR-PhoP action on antibiotic gene expression may be mediated through other DNA-binding proteins, e.g., pathway-specific regulators.
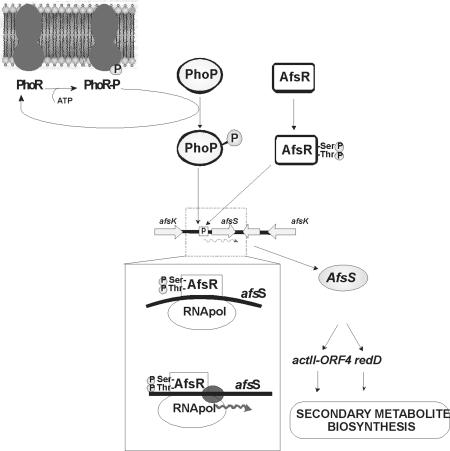
In S. lividans and S. coelicolor, there is a small (63-amino-acid) protein, AfsS, encoded by a short ORF located in the 3′ region immediately downstream of the regulatory gene AfsR. AfsS is a rare protein (thus far, it is found only in a few Streptomyces species) that contains three repeats of the 11-amino-acid sequence TXXDHMPXXPA (where X represents any amino acid) (33). Overexpression of AfsS in S. lividans (or S. coelicolor) leads to an overproduction of actinorhodin and undecylprodigiosin, even in mutants disrupted in AfsR. Recently, Lee et al. (22) have shown that the afsS gene is the target for the AfsR regulator, a widespread transcription factor that has ATPase activity (47). S. lividans has an afsS analogue named afsR2 (49). Floriano and Bibb (13) reported that AfsS protein stimulates the production of actinorhodin by enhancing the transcription of the pathway-specific regulator ActII-ORF4 and that the same occurs in S. lividans when afsR2 was overexpressed.
A possible cascade mechanism involves a negative effect of phosphorylated PhoP on the expression of afsS, resulting in downregulation of actII-ORF4 and redD expression (Fig. 2). Due to their low concentration, ActII-ORF4 and RedD would be unable to induce the actinorhodin and undecylprodigiosin biosynthetic genes. Null phoR-phoP mutants lack the PhoP response regulator and therefore would be unable to repress the afs gene, leading to constitutive formation of AfsS, resulting in induction of actII-ORF4 and redD and finally in actinorhodin and undecylprodigiosin overproduction.
FIG. 2.
Proposed cascade mechanism involved in phosphate control of actinorhodin and undecylprodigiosin biosynthesis. The involvement of the two-component system PhoR-PhoP has been confirmed (see the text). The cascade action of AfsR mediated by AfsS on actII-ORF4 and redD pathway-specific regulators is based on the results of Umeyama and coworkers (47). The inset shows the binding of phosphorylated AfsR to the afsS promoter.
FUTURE PERSPECTIVES
The recent identification of a large number of structural and regulatory genes involved in antibiotic biosynthesis permits analysis of the mechanism of PhoR-PhoP-mediated global control in other Streptomyces species, some of which produce antibiotics, antitumor agents, immunosuppressants, and many other biologically active compounds of high commercial interest. In all of these cases, phosphate represses the biosynthesis of these compounds, and a proper understanding of the control mechanism in these actinomycetes will allow targeted derepression of antibiotic production.
Acknowledgments
This work was supported by a grant from the Ministry of Education and Science of Spain CICYT Bio2003 01489).
I thank P. Liras, A. Sola, and A. Rodriguez for valuable discussions.
REFERENCES
- 1.Antelmann, H., C. Scharf, and M. Hecker. 2000. Phosphate starvation-inducible proteins of Bacillus subtilis: proteomics and transcriptional analysis. J. Bacteriol. 182:4478-4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aparicio, J. F., R. Fouces, M. V. Mendes, N. Olivera, and J. F. Martín. 2000. A complex multienzyme system encoded by five polyketide synthase genes is involved in the biosynthesis of the 26-membered polyene macrolide pimaricin in Streptomyces natalensis. Chem. Biol. 7:895-905. [DOI] [PubMed] [Google Scholar]
- 3.Asturias, J. A., P. Liras, and J. F. Martín. 1990. Phosphate control of pabS gene transcription during candicidin biosynthesis. Gene 93:79-84. [DOI] [PubMed] [Google Scholar]
- 4.Bentley, S. D., K. F. Chater, A. M. Cerdeño-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 5.Bibb, M. J. 1996. The regulation of antibiotic production in Streptomyces coelicolor A3(2). Microbiology 142:1335-1344. [DOI] [PubMed] [Google Scholar]
- 6.Campelo, A. B., and J. A. Gil. 2002. The candicidin gene cluster from Streptomyces griseus IMRU 3570. Microbiology 48:51-59. [DOI] [PubMed] [Google Scholar]
- 7.Chater, K. F., and M. J. Bibb. 1996. Regulation of bacterial antibiotic production, p. 59-105. In H. Kleinkauf and H. von Doren (ed.), Products of secondary metabolism. WCH, Weinheim, Germany.
- 8.Chouayekh, H., and M. J. Virolle. 2002. The polyphosphate kinase plays a negative role in the control of antibiotic production in Streptomyces lividans. Mol. Microbiol. 43:919-930. [DOI] [PubMed] [Google Scholar]
- 9.Criado, L. M., J. F. Martín, and J. A. Gil. 1993. The pabS gene of Streptomyces griseus, encoding p-aminobenzoic acid synthase, is located between genes possibly involved in candicidin biosynthesis. Gene 126:135-139. [DOI] [PubMed] [Google Scholar]
- 10.Dekeva, M. L., J. A. Titus, and W. R. Strohl. 1985. Nutrient effects on anthracycline production by Streptomyces peucetius in a defined medium. Can. J. Microbiol. 31:287-294. [DOI] [PubMed] [Google Scholar]
- 11.Doull, J. L., and L. C. Vining. 1990. Nutritional control of actinorhodin production by Streptomyces coelicolor A3(2): suppressive effects of nitrogen and phosphate. Appl. Microbiol. Biotechnol. 32:449-454. [DOI] [PubMed] [Google Scholar]
- 12.Fernández-Moreno, M. A., J. L. Caballero, D. A. Hopwood, and F. Malpartida. 1991. The act cluster contains regulatory and antibiotic export genes, direct targets for translational control by the bldA transfer RNA gene of Streptomyces. Cell 66:769-780. [DOI] [PubMed] [Google Scholar]
- 13.Floriano, B., and M. Bibb. 1996. afsR is a pleiotropic but conditionally required regulatory gene for antibiotic production in Streptomyces coelicolor A3(2). Mol. Microbiol. 21:385-396. [DOI] [PubMed] [Google Scholar]
- 14.Gramajo, H. C., E. Takano, and M. J. Bibb. 1993. Stationary-phase production of the antibiotic actinorhodin in Streptomyces coelicolor A3(2) is transcriptionally regulated. Mol. Microbiol. 7:837-845. [DOI] [PubMed] [Google Scholar]
- 15.Hobbs, G., C. M. Frazer, D. C. J. Gardner, F. Flett, and S. G. Oliver. 1990. Pigmented antibiotic production by Streptomyces coelicolor A3(2): kinetics and the influence of nutrients. J. Gen. Microbiol. 136:2291-2296. [Google Scholar]
- 16.Hobbs, G., A. I. C. Obanye, J. Petty, J. C. Mason, E. Barrat, D. C. J. Gardner, F. Flett, C. P. Smith, P. Broda, and S. G. Oliver. 1992. An integrated approach to studying regulation of production of the antibiotic methylenomycin by Streptomyces coelicolor A3(2). J. Bacteriol. 174:1487-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horinouchi, S., and T. Beppu. 1992. Autoregulatory factors and communication in actinomycetes. Annu. Rev. Microbiol. 46:377-398. [DOI] [PubMed] [Google Scholar]
- 18.Hulett, F. M. 1996. The signal transduction network for Pho regulation. Mol. Microbiol. 19:933-939. [DOI] [PubMed] [Google Scholar]
- 19.Hunter, I. S., and R. A. Hill. 1997. Tetracyclines: chemistry and molecular genetics of their formation, p. 659-682. In W. Strohl (ed.), Biotechnology of industrial antibiotics. Marcel Decker, Inc., New York, N.Y.
- 20.Ikeda, H., J. Ishikawa, A. Hanamoto, M. Shinose, H. Kikuchi, T. Shiba, Y. Sakaki, M. Hattori, and S. Omura. 2003. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 21:526-531. [DOI] [PubMed] [Google Scholar]
- 21.Ishige, T., M. Krause, M. Bott, V. F. Wendisch, and H. Sahm. 2003. The phosphate starvation stimulon of Corynebacterium glutamicum determined by DNA microarray analyses. J. Bacteriol. 185:4519-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee, P.-C., T. Umeyama, and S. Horinouchi. 2002. afsS is a target of AfsR, a transcriptional factor with ATPase activity that globally controls secondary metabolism in Streptomyces coelicolor A3(2). Mol. Microbiol. 43:1413-1430. [DOI] [PubMed] [Google Scholar]
- 23.Liras, P., J. A. Asturias, and J. F. Martín. 1990. Phosphate control sequences involved in transcriptional regulation of antibiotic biosynthesis. Trends Biotechnol. 8:184-189. [DOI] [PubMed] [Google Scholar]
- 24.Liu, W., and F. M. Hulett. 1998. Comparison of PhoP binding to the tuaA promoter with PhoP binding to other Pho-regulon promoters establishes a Bacillus subtilis Pho core binding site. Microbiology 144:1443-1450. [DOI] [PubMed] [Google Scholar]
- 25.Lounes, A., A. Lebrihi, C. Benslimane, G. Lefebvre, and P. Germain. 1996. Regulation of spiramycin synthesis in Streptomyces ambofaciens: effects of glucose and inorganic phosphate. Appl. Microbiol. Biotechnol. 45:204-211. [DOI] [PubMed] [Google Scholar]
- 26.Mapplestone, R. A., M. J. Stone, and D. H. Williams. 1992. The evolutionary role of secondary metabolites: a review. Gene 115:151-157. [DOI] [PubMed] [Google Scholar]
- 27.Martín, J. F. 1989. Molecular mechanism for the control by phosphate of the biosynthesis of antibiotic and secondary metabolites, p. 213-237. In S. Shapiro (ed.), Regulation of secondary metabolism in actinomycetes. CRC Press, Inc., Boca Raton, Fla.
- 28.Martín, J. F., and A. Demain. 1980. Control of antibiotic biosynthesis. Microbiol. Rev. 44:230-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martín, J. F., and P. Liras. 1989. Organization and expression of genes involved in the biosynthesis of antibiotics and other secondary metabolites. Annu. Rev. Microbiol. 43:173-206. [DOI] [PubMed] [Google Scholar]
- 30.Martín, J. F., A. T. Marcos, A. Martín, J. A. Asturias, and P. Liras. 1994. Phosphate control of antibiotic biosynthesis at the transcriptional level, p. 140-147. In A. Torriani-Gorini, E. Yagil, and S. Silver (ed.), Phosphate in microoganisms. ASM Press, Washington, D.C.
- 31.Martínez-Hackert, E., and A. M. Stock. 1997. Structural relationships in the OmpR family of winged-helix transcription factors. J. Mol. Biol. 269:301-312. [DOI] [PubMed] [Google Scholar]
- 32.Masuma, R., Y. Tanaka, H. Tanaka, and S. Omura. 1986. Production of nanomycin and other antibiotics by phosphate-depressed fermentation using phosphate-trapping agents. J. Antibiot. 39:1557-1564. [DOI] [PubMed] [Google Scholar]
- 33.Matsumoto, A., H. Ishizuka, T. Beppu, and S. Horinouchi. 1995. Involvement of a small ORF downstream of the afsR gene in the regulation of secondary metabolism in Streptomyces coelicolor A3(2). Actinomycetologica 9:37-43. [Google Scholar]
- 34.McDowall, K., A. Thamchaipenet, and I. S. Hunter. 1999. Phosphate control of oxytetracycline production by Streptomyces rimosus is at the level of transcription from promoters overlapped by tandem repeats similar to those of the DNA-binding sites of the OmpR family. J. Bacteriol. 181:3025-3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mizuno, T., and I. Tanaka. 1997. Structure of the DNA-binding domain of the OmpR family of response regulators. Mol. Microbiol. 24:665-667. [DOI] [PubMed] [Google Scholar]
- 36.Moura, R. S., J. F. Martín, A. Martín, and P. Liras. 2001. Substrate analysis and molecular cloning of the extracellular alkaline phosphatase of Streptomyces griseus. Microbiology 147:1525-1533. [DOI] [PubMed] [Google Scholar]
- 37.Narva, K. E., and J. S. Feitelson. 1990. Nucleotide sequence and transcriptional analysis of the redD locus of Streptomyces coelicolor A3(2). J. Bacteriol. 172:326-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nodwell, J. R., and R. Losick. 1998. Purification of an extracellular signaling molecule involved in production of the aerial mycelium by Streptomyces coelicolor. J. Bacteriol. 180:1334-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Omura, S., H. Ikeda, J. Ishikawa, A. Hanamoto, C. Takahashi, M. Shinose, Y. Takahashi, H. Horikawa, H. Nakazawa, T. Osonoe, H. Kikuchi, T. Shiba, Y. Sakaki, and M. Hattori. 2001. Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc. Natl. Acad. Sci. USA 98:12215-12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pragai, Z., and C. R. Harwood. 2002. Regulatory interactions between the Pho and σB-dependent general stress regulons of Bacillus subtilis. Microbiology 148:1593-1602. [DOI] [PubMed] [Google Scholar]
- 41.Santamarta, I., A. Rodríguez-García, R. Pérez-Redondo, J. F. Martín, and P. Liras. 2002. CcaR is an autoregulatory protein that binds to the ccaR and cefD-cmcI promoters of the cephamycin C-clavulanic acid cluster in Streptomyces clavuligerus. J. Bacteriol. 184:3106-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi, L., and F. M. Hulett. 1999. The cytoplasmic kinase domain of PhoR is sufficient for the low phosphate-inducible expression of pho regulon genes in Bacillus subtilis. Mol. Microbiol. 31:211-222. [DOI] [PubMed] [Google Scholar]
- 43.Sola-Landa, A., R. S. Moura, and J. F. Martín. 2003. The two-component PhoR-PhoP system controls both primary metabolism and secondary metabolite biosynthesis in Streptomyces lividans. Proc. Natl. Acad. Sci. USA 100:6133-6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takano, E., H. C. Gramajo, E. Strauch, N. Andres, J. White, and M. J. Bibb. 1992. Transcriptional regulation of the redD transcriptional activator gene accounts for growth-phase-dependent production of the antibiotic undecylprodigiosin in Streptomyces coelicolor A3(2). Mol. Microbiol. 6:2797-2804. [DOI] [PubMed] [Google Scholar]
- 45.Tang, L., A. Grimm, Y. X. Zhang, and C. R. Hutchinson. 1996. Purification and characterization of the DNA-binding protein DnrI, a transcriptional factor of daunorubicin biosynthesis in Streptomyces peucetius. Mol. Microbiol. 22:801-813. [DOI] [PubMed] [Google Scholar]
- 46.Torriani-Gorini, A. 1994. The Pho regulon of Escherichia coli, p. 1-4. In A. Torriani-Gorini, E. Yagil, and S. Silver (ed.), Phosphate in microorganisms. ASM Press, Washington, D.C.
- 47.Umeyama, T., P.-C. Lee, and S. Horinouchi. 2002. Protein serine/threonine kinases in signal transduction for secondary metabolism and morphogenesis in Streptomyces. Appl. Microbiol. Biotechnol. 59:419-425. [DOI] [PubMed] [Google Scholar]
- 48.Vining, L. C. 1992. Secondary metabolism, inventive evolution and biochemical diversity: a review. Gene 115:135-140. [DOI] [PubMed] [Google Scholar]
- 49.Vögtli, M., P. C. Chang, and S. N. Cohen. 1994. afsR2: a previously undetected gene encoding a 63-amino-acid protein that stimulates antibiotic production in Streptomyces lividans. Mol. Microbiol. 14:643-653. [DOI] [PubMed] [Google Scholar]
- 50.Wietzorrek, A., and M. J. Bibb. 1997. A novel family of proteins that regulates antibiotic production in Streptomyces appears to contain an OmpR-like DNA-binding fold. Mol. Microbiol. 25:1181-1184. [DOI] [PubMed] [Google Scholar]
- 51.Yamada, Y., and T. Nihira. 1999. Microbial hormones and microbial chemical ecology, p. 377-413. In K. Mori (ed.), Comprehensive natural products chemistry, vol. 8. Elsevier Scientific Publishers, Dordrecht, The Netherlands.