Abstract
Viral encephalopathy and retinopathy otherwise known as viral nervous necrosis (VNN) is a neuropathological condition affecting more than 50 fish species worldwide, mostly marine. Different PCR protocols with specific primers were reported from many countries for confirmation of VNN in fishes. In the present study, two pairs of primers were designed and evaluated for the diagnosis of clinical and subclinical cases of infections from field. These primers designated as BARL-F1/BARL-R1 amplified a 902 bp product in the variable region (T4) of the coat protein gene by first step PCR. Nested PCR primers BARL-F2/BARL-R2 amplified a fragment of 313 bp. The results were comparable with other commonly used primer sets such as F2/R3 and RG668f/RG919r primers. These new primers could detect betanodavirus in standard reference samples containing low, moderate and high viral load. Known positive and negative control samples of fish also revealed a predictive value of 100 % by RT-PCR diagnosis.
Electronic supplementary material
The online version of this article (doi:10.1007/s13337-016-0313-0) contains supplementary material, which is available to authorized users.
Keywords: Viral nervous necrosis, Viral encephalopathy and retinopathy, Betanodavirus, Mortality, Diagnosis, Nested RT-PCR
Introduction
Viral encephalopathy and retinopathy (VER) otherwise known as viral nervous necrosis (VNN) is a neuropathological condition affecting more than 50 fish species, mostly marine across the world [19, 27]. The disease is caused by Betanodavirus of the family Nodaviridae which are icosahedral, non enveloped viruses with a diameter of about 25 nm and a bipartite positive-sense RNA genome. Betanodaviruses have been classified into five major genotypes: red spotted grouper nervous necrosis virus (RGNNV), striped jack nervous necrosis virus (SJNNV), tiger puffer nervous necrosis virus (TPNNV), barfin flounder nervous necrosis virus (BFNNV) and turbot nodavirus (TNV) based on the variable region (T4) of the viral capsid protein (CP) gene [23].
In India, the betanodavirus infection has been reported in wild and hatchery/farm grown marine fishes and considered as an emerging fish disease affecting finfish species of aquaculture potential [1, 3, 5, 15]. Generic classifications based on partial or full coat protein gene sequences of all these investigations revealed that the nucleotide sequences showed up to 98 % similarity to the RGNNV genotype. Specific clinical signs include neurological abnormalities like erratic or aberrant swimming behavior and lethargy. Distinct vacuolization of the nervous tissue (brain, retina) on histological examination was observed. Mortality may be rapid, generally during early larvae and juvenile fish and may reach up to 100 % of the fish stock within 2–3 days. Its bipartite genome consists of two positive RNA strands; RNA1 (3.1 kb) codes for viral RdRP (protein A) while the RNA2 (1.4 kb) codes for coat protein [18]. A sub genomic RNA3 synthesized from RNA1 during early viral replication codes for protein B2 [31] which acts against host RNA interference [9, 13]. The virus causes acute as well as latent infections [19]. Biological and environmental stress factors may induce latent form of the infection to acute infection with disease signs [20] and/or facilitate vertical/horizontal transmission of the virus [6, 16]. The first described PCR protocol using F2/R3 primers for betanodavirus [24] is now accepted as the gold standard for its confirmatory diagnosis [26]. Several workers opined that F2/R3 primers are not perfectly adapted to efficient amplification of every betanodavirus isolates [4, 8], more so for RGNNV due to wide range of host species and diversity of isolates. Diagnostics with specific objectives such as health management of broodstock and confirmation of acute VNN were developed in many countries [10, 11, 32] but information on diagnosis of latent infection is scanty. In this paper, an attempt was made for a robust universal test for amplifying and genotyping betanodaviruses, by designing two pairs of primer sets in variable region of the viral capsid protein gene and evaluation of RT-PCR protocol for early diagnosis of betanodavirus in fish. The efficiency of these primers was compared with that of F2/R3 [24] and RG668f/RG919r [22] primers using a range of samples originated from diverse fish species besides a reference strain in cell culture.
Materials and methods
Primer design and evaluation
Two sets of primer pair (BARL-F1/BARL-R1 and BARL-F2/BARL-R2) were designed (Table 1) by alignment of betanodavirus RNA2 gene sequences (n = 28) from all five distinct genotypes (NC003449, NC013459, AY324870, NC013461 and AJ608266) using the program Clustal W (MegAlign program version 5.03 of the DNASTAR package) and two consensus sequences targeting the coat protein gene (RNA2) were chosen for nested PCR. The performance of these new sets of primers were evaluated using fish that displayed clinical signs of VNN infection, reference strain of the betanodavirus (CSIRO, Australia) and fish samples without apparent clinical signs (persistent or negative for VNN infection) and compared with a set of published primers [22, 24].
Table 1.
Primers used in the PCR reaction to detect betanodavirus
| Primer code | Sequence (5′… 3′) | Product (bp) | Application/reference (s) |
|---|---|---|---|
| BARL-F1 | ACGCAAAGGTGAGAAGAAA | 902 | I step PCR (this study) |
| BARL-R1 | GTCCCAGATGCCCCA | ||
| BARL-F2 | AACTGACAACGACCACACCTT | 313 | Nested PCR (this study) |
| BARL-R2 | TGTGGAAAGGGAATCGTTG | ||
| F2 | CGT GTC AGT CAT GTG TCG CT | 430 | I step PCR [24]a |
| R3 | CGA GTC AAC ACG GGT GAA GA | ||
| RG668f | ACC TGA GGA GAC TAC CGC TC | 280 | Nested PCR [22] |
| RG919r | CAG CGA AAC CAG CCT GCA GG |
aOIE primer
Fish samples
Subclinical samples
Fish samples (Lates calcarifer, Mugil cephalus, Chanos chanos, Liza parsia, Liza tade and Eleutheronema tetradactylus) were collected from finfish hatchery/farms and wild habitats located in Chennai (Tamil Nadu) and Kakdwip (West Bengal) along the east-coast of India (Table 2). The target organs (brain and eyeball) were excised using a sterile scalpel and were collected in RNAlater Tissue Protect Tubes (QIAGEN) or TRIzol® Reagent (Invitrogen) for molecular diagnosis.
Table 2.
Results of RT-PCR for betanodavirus for fish samples collected from wild and culture facilities during April 2013–December 2014
| Species | Disease status | Origin | No. examined | No. positive by OIE primers | No. positive by newly designed primers |
|---|---|---|---|---|---|
| Lates calcarifer | Clinical | Farm/hatchery | 4 | 4 | 4 |
| Sub clinical | Wild | 3 | 1 | 1 | |
| Mugil cephalus | Sub clinical | Farm | 11 | 1 | 1 |
| Sub clinical | Wild | 1 | – | – | |
| Chanos chanos | Sub clinical | Farm | 4 | 2 | 2 |
| Sub clinical | Wild | 9 | – | – | |
| Liza parsia | Sub clinical | Farm | 1 | – | – |
| Sub clinical | Wild | 6 | 1 | 1 | |
| Others Liza tade, Eleutheronema tetradactylus |
Sub clinical | Farm | 0 | – | – |
| Sub clinical | Wild | 1 | – | – | |
| Total | – | – | 40 | 9 | 9 |
Clinical samples
A pool of Asian seabass (L. calcarifer) larval samples with known history of VNN outbreak in a hatchery, preserved at −80 °C was used in this study as clinical samples (BARL-LC01). A portion of the target organs from clinical and sub clinical samples were collected aseptically for virus isolation studies by inoculation in susceptible cell line (seabass spleen cell line-SISS) for confirmation, and for routine histopathological studies in 10 % neutral buffered formalin.
Virus-free fish samples (negative control)
A healthy stock of Asian seabass larvae (28 dph) collected earlier from a hatchery and stored at −80 °C was used as negative control (n = 4).
Virus strain and viral propagation (positive control)
The strain used in this study designated as BARL-LC02 belongs to RGNNV genotype was originally isolated from seabass during an acute disease outbreak in a rearing facility. Larval tissue homogenate was treated with antibiotics (1000 IU ml−1 penicillin, 100 µg ml−1 streptomycin, 500 µg ml−1 gentamycin and 10 µg ml−1 amphotericin B) filtered using 0.22 micron syringe filter [2]. SISS cell line from Asian seabass spleen (provided by Dr. Sahul Hameed, C. Abdul Hakeem College, Melvisharam, India) was used for viral propagation and were grown in L-15 medium containing 10 % foetal bovine serum [28], 100 µl virus suspension inoculated in overnight cell monolayer and after an hour of viral adsorption, the inoculum was removed and fresh maintenance L-15 medium containing 5 % foetal bovine serum was added and incubated at 28 °C up to a maximum of two weeks. When cytopathic effect (CPE) becomes extensive, fluids were harvested after freeze thawing for three times and clarified by centrifugation at 3000×g for 15 min at 4 °C and stored at −80 °C until use. Virus titration was conducted on monolayers of cells in 96 well plates using 10 fold serial dilutions in triplicate. The 50 % of the tissue culture infective dose (TCID50/ml) was calculated as described by Reed and Muench [30]. The titre of the culture supernatant was 103 TCID50/ml at 4 days post infection (dpi) and 108 TCID50/ml at 7 dpi. The culture supernatants were subjected to RNA extraction and used as positive control for further analyses by reverse transcription nested polymerase chain reaction (RT-nPCR).
Reference standard (betanodavirus positive)
A non-infectious sample of ethanol precipitated (80 % v/v) tissue culture supernatants from active virus cultures with low, moderate and high dose of betanodavirus provided by CSIRO, Australia was used for standardizing the sensitivity of the new primer sets.
RT-nPCR and sequence analysis
Total RNA was extracted from target tissues (clinical and sub clinical), cell culture supernatants (field isolate of BARL-LC02) and the betanodavirus reference strain from CSIRO using TRIzol™ Reagent (Invitrogen, USA) following the manufacturer’s protocol. The purity and quantity of the extracted RNA was carried out using a NanoSpectrophotometer (Implen, Germany) at 260 nm. Reverse transcription was carried out using iScript cDNA synthesis kit (BioRad) in 10 μl reaction as per the manufacturer’s instructions. The synthesized cDNA was subjected to PCR amplification using the newly designed first and nested PCR primer sets. Following optimized thermal cycling conditions for first step PCR was used: 95 °C initial denaturation for 5 min followed by 40 cycles of 1 min denaturation at 95 °C, 1 min annealing at 60 °C and 1 min extension at 72 °C and final extension at 72 °C for 5 min. The optimized thermal cycling profile for nested PCR was 95 °C initial denaturation for 5 min followed by 40 cycles of 30 s denaturation at 95 °C, 30 s annealing at 60 °C and 30 s extension at 72 °C and final extension at 72 °C for 5 min. PCR was carried out in a final volume of 25 μl reaction mixture containing Tris- HCl (pH 8.5), 1.5 mM MgCl2, 0.2 % Tween 20, 0.4 mM dNTPs, 100 pM of forward and reverse primer, 1U Taq DNA Polymerase, 50–100 ng of template DNA and inert dye and stabilizer (Ampliqon, Denmark). The amplification was performed in gradient thermal cycler (Eppendorf, Germany). The amplified PCR products were resolved in 1.5 % TBE agarose gel stained with 0.5 µg/ml ethidium bromide. The identities of the first step PCR products (903 bp) were confirmed by nucleotide sequencing (First Base Laboratories, Malaysia) and NCBI BLAST analysis.
Results
Fish samples and clinical history and gross pathology
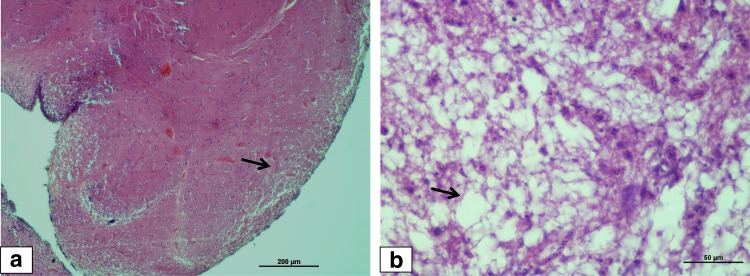
In the present study, a group of clinical, subclinical, healthy fish samples of different fish species were used for evaluation two pairs of newly designed primers for the diagnosis of betanodavirus (Table 2). The disease status and the presence of fish nodavirus in these groups were ascertained by the known history of the disease, histopathological studies and confirmation by a RT-nPCR targeting T4 region of coat protein gene (RNA2) as per the earlier published reports (22,24). The primers designed by Nishizawa et al. [24] are the most widely used OIE primers across the world, with capability to detect all genotypes of betanodavirus. Of the nine samples tested positive by published primers above, four samples were L. calcarifer collected from clinical disease outbreaks at different time intervals in a hatchery, and another five from different fish species without any apparent clinical signs of VNN infection. Histologically, severe vacuolation in the brain and retinal tissues of the eye was characteristically seen as the most important and reliable diagnostic feature of VNN in fish. Fish samples from clinical disease outbreak revealed mild necrosis and vacuolization in the brain (Fig. 1).
Fig. 1.
Histological section of brain of VNN affected Asian seabass larvae (code 01), H&E stain. a Peripheral vacuolation in the brain (×10); b severe vacuolations with coalescence of the vacuoles seen scattered in the brain tissue (arrow), H&E ×40
A strain of betanodavirus designated as BARL-LC02 (positive control) cultured in SISS cell line was used as positive control used in this study. BARL-LC02 grew well in this cell line at 28 °C with progression of cytopathic effect by vacuolation of cells, cellular aggregation and rounding up of cells and clumps. The titre of the culture supernatant was 103 TCID50/ml at 4 days post infection (dpi) and 108 TCID50/ml at 7 dpi. Healthy seabass larvae (28 dph) tested earlier for VNN and stored at −80 °C was used as negative control.
Reverse transcriptase nested PCR and sequence analysis
All the forty fish samples belonging to six fish species from wild and culture facility were tested by RT-nPCR for betanodavirus infection using published primer sets as well as the newly designed primers (Table 2). Nine samples showed positive for betanodavirus by RT-nPCR using both sets of primers. The RT-PCR amplification using the new primer pair BARL-F1/BARL-R1 for first-step gave an amplified product of 902 bp, while BARL-F2/BARL-R2 primer pair yielded a 313 bp nested PCR product. No PCR artifacts or host fish genomes were amplified under the specified thermal cycling conditions.
The new primers were specific and amplified targeted product of betanodavirus coat protein gene in all four samples of L. calcarifer from clinical disease outbreaks and five samples of different fish species with no history of disease signs (Figs. 2, 3) at par with the F2/R3 primers (data not shown). The reference strain of betanodavirus (CSIRO, Australia) containing low (20 copies), moderate (100 copies) and heavy infection (200 copies) of virus also tested positive without any non-specific bands (Fig. 4). However, F2/R3 primers (first step PCR) could not amplify target product in cell culture supernatants (positive control) containing low (103 TCID50/ml) viral titres in this study (Fig. 5a), while they were amplified with the new primer sets used in this work for both first and second step PCR (Fig. 5b). No non-specific amplification of host fish genome was observed by using virus-free seabass larvae (negative control).
Fig. 2.
Evaluation of the primers using VNN affected seabass larvae from clinical outbreak of disease (lanes 1–6 first step PCR using primer set BARL-F1/BARL-R1 and lanes 7–12 nested PCR using primer set BARL-F2/BARL-R2)—these samples were tested positive by OIE primer (F2/R3). M DNA ladder, 1 Asian seabass larvae—code 01, 2 Asian seabass larvae—code 02, 3 Asian seabass larvae—code 03, 4 reference strain (CSIRO), 5 negative control, 6 positive control, 7 Asian seabass larvae—code 01, 8 Asian seabass larvae—code 02, 9 Asian seabass larvae—code 03, 10 reference strain (CSIRO), 11 negative control, 12 positive control
Fig. 3.
Evaluation of the primers using sub-clinically affected fishes (lanes 1–7 first step PCR using primer set BARL-F1/BARL-R1 and lanes 8–14 nested PCR using primer set BARL-F2/BARL-R2)—these samples were tested positive by OIE primer (F2/R3). M DNA ladder, 1 M. cephalus (code-15), 2 L. calcarifer (code-27), 3 L. calcarifer (code-28), 4 C. chanos (code-07), 5 negative control, 6 positive control, 7 M. cephalus (code-15), 8 L. calcarifer (code-27), 9 L. calcarifer (code-28), 10 C. chanos (code-07), 11 negative control, 12 positive control
Fig. 4.
Detection of betanodavirus positive reference strain (CSIRO, Australia) containing low, moderate and heavy infection using the newly designed primers (lane M molecular marker (100 bp), lanes 1–3 Positive control (reference sample) containing low, moderate and high level of betanodavirus by first step PCR using primer set BARL-F1/BARL-R1, lanes 4–6 same samples by nested PCR using primer set BARL-F2/BARL-R2
Fig. 5.

a Betanodavirus detection using published primers F2/R3 for first step (top) and RG668f/RG919r for nested PCR (bottom); b newly designed primers BARL-F1/BARL-R1 for first step (top) and BARL-F2/BARL-R2 for nested PCR (bottom) showing specific products; lane M DNA ladder (100 bp), lane N negative control, lanes 1–3 cell culture samples C1–C3 (positive control), lanes 4–7 healthy Asian seabass larval samples S1–S4 (known VNN-negative control)
The sequences of the PCR product obtained in this study were analysed using NCBI BLAST and found to have >90 % sequence homology to the nucleotide sequences of fish nodavirus isolated from other Asian countries. Further, the first step PCR product sequences confirmed the specificity of the amplified products as that of coat protein gene of betanodavirus with sequence homology to RGNNV genotype.
Discussion
Molecular methods have played a pivotal role in detecting and characterizing betanodavirus, and RT-PCR has emerged as a powerful, rapid and sensitive method for detection of VNN. Recently, two conventional PCR based methods have been used to amplify and genotype betanodaviruses by sequencing. First one targets a region of the polymerase gene carried by RNA1 as a source of genetic information that is useful for identification of an isolate. The second method, more versatile based on the amplification of the T4 region of RNA2 [24] has been widely used on a large number of isolates in numerous studies. High genetic variation within the T4 region occasionally leads to mismatches between primers and their targets and consequently the sensitivity [8, 25]. However, these primers were designed before the large genetic diversity of betanodavirus was appreciated and is still considered as gold standard for diagnostic purpose. Subsequently, several diagnostic protocols have been applied to detect various virus strains, mostly on confirmation of the virus once an outbreak occurs [7, 8, 10–13, 21, 29, 32, 33]. Among molecular methods, quantitative PCR seems to be efficient for this purpose, considering its sensitivity, speed and the possible control of contamination. Unfortunately, the significant genetic diversity of betanodaviruses is a serious difficulty in developing universal primers and hence not yet advantageous for accurate genotyping [4]. Epidemiological studies till date revealed only a single genotype (RGNNV) in India (3,5,15) with varying degree of sensitivity of F2/R3 primers, especially in asymptomatic wild fish samples. A similar observation has been made by Bigarre et al. [4] indicating false positive PCR products in gilthead, Sparus aurata. The present study demonstrated the suitability of newly designed RT-nPCR primers for routine disease diagnosis using different types of biological samples from infected fish samples. The results were comparable to commonly used primers used for VNN diagnostics in India and may be used as a management tool for screening hatchery samples, grow-out monitoring, quarantine and biosecurity programme in finfish aquaculture sector.
Availability of precise and sensitive diagnostics for betanodavirus is important, both as diagnostic tools and for morphological studies of the virus. It is inevitable in case of latent (persistent) betanodavirus infection where the disease condition is asymptomatic and an acute disease predisposes with right combination of biological and environmental conditions. Hence, confirmatory diagnosis of viral diseases has been always a problem in field leading to poor documentation of fish mortalities and scarce epidemiological data.
In recent years, VNN as a disease entity has been recognized as an emerging disease in finfish culture with increasing number of host species across ecosystem barriers, marine to freshwater [5, 14, 17]. With the increasing diversification of finfish aquaculture and consequent transfer of live organisms for culture, new strains and genotypes of betanodaviruses are likely to emerge in future. Considering the highly destructive nature of betanodavirus, it is increasingly crucial to prevent the introduction of virus in farms/hatcheries by screening for healthy stock and elucidating promptly the source of virus in case of outbreaks. Cell culture and immunological tests are efficient for diagnostics, with inherent limitation of speed and strain identification. Among molecular methods, quantitative PCR seems efficient for this purpose, considering its sensitivity and speed, but need sophisticated equipments. In this context, the newly designed RT-nPCR primers could serve as additional tool for routine disease diagnosis, epidemiology, quarantine and biosecurity programme in Indian finfish aquaculture.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The authors are grateful to the Director, Central Institute of Brackishwater Aquaculture (Chennai) for providing necessary facilities. The authors also thank Australian National Quality Assurance Program (Australia) and Commonwealth Scientific and Industrial Research Organisation (Australia) for providing a positive reference sample of betanodavirus (non-infectious) under a joint Regional Proficiency Testing Programme for Aquatic Animal Disease Laboratories in Asia, coordinated by Network of Aquaculture Centres in Asia Pacific (Bangkok) during 2013–2014.
References
- 1.Azad IS, Shekhar MS, Thirunavukkarasu AR, Poornima M, Kailasam M, Rajan JJS, Ali SA, Abraham M, Ravichandran P. Nodavirus infection causes mortalities in hatchery produced larvae of Lates calcarifer: first report from India. Dis Aquat Org. 2005;63:113–118. doi: 10.3354/dao063113. [DOI] [PubMed] [Google Scholar]
- 2.Bandin I, Olveira JG, Borrego JJ, Dopazo CP, Barja JL. Susceptibility of the fish cell line SAF-1 to betanodavirus. J Fish Dis. 2006;29:633–636. doi: 10.1111/j.1365-2761.2006.00757.x. [DOI] [PubMed] [Google Scholar]
- 3.Banerjee D, Hamod MA, Suresh T, Karunasagar I. Isolation and characterization of a nodavirus associated with mass mortality in Asian seabass (Lates calcarifer) from the west coast of India. Virus Dis. 2014 doi: 10.1007/s13337-014-0226-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bigarre L, Baud M, Cabon J, Crenn K, Castric J. New PCR probes for detection and genotyping of piscine betanodavirus. J Fish Dis. 2010;33:907–912. doi: 10.1111/j.1365-2761.2010.01188.x. [DOI] [PubMed] [Google Scholar]
- 5.Binesh CP, Jithendran KP. Genetic characterization of betanodavirus isolates from Asian seabass Lates calcarifer (Bloch) in India. Arch Virol. 2013;158:1543–1547. doi: 10.1007/s00705-012-1554-x. [DOI] [PubMed] [Google Scholar]
- 6.Breuil G, Pepin JFP, Boscher S, Thiery R. Experimental vertical transmission of nodavirus from broodfish to eggs and larvae of the sea bass, Dicentrarchus labrax (L.) J Fish Dis. 2002;25:697–702. doi: 10.1046/j.1365-2761.2002.00406.x. [DOI] [Google Scholar]
- 7.Comps M, Raymond JC. Virus-like particles in the retina of the sea-bream, Sparus aurata. Bull Eur Assoc Fish Pathol. 1996;16:161–163. [Google Scholar]
- 8.Dalla Valle L, Zanella L, Patarnello P, Paolucci L, Belvedere P, Colombo L. Development of a sensitive diagnostic assay for fish nervous necrosis virus based on RT-PCR plus nested PCR. J Fish Dis. 2001;23:312–327. [Google Scholar]
- 9.Fenner BJ, Thiagarajan R, Chua HK, Kwang J. Betanodavirus B2 is an RNA interference antagonist that facilitates intracellular viral RNA accumulation. J Virol. 2006;80:85–94. doi: 10.1128/JVI.80.1.85-94.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gagne N, Johnson SC, Cook-Versloot M, MacKinnon AM, Olivier G. Molecular detection and characterization of nodavirus in several marine fish species from the northeastern Atlantic. Dis Aquat Org. 2004;62:181–189. doi: 10.3354/dao062181. [DOI] [PubMed] [Google Scholar]
- 11.Gomez DK, Sato J, Mushiake K, Isshiki T, Okinaka Y, Nakai T. PCR-based detection of betanodaviruses from cultured and wild marine fish with no clinical signs. J Fish Dis. 2004;27:603–608. doi: 10.1111/j.1365-2761.2004.00577.x. [DOI] [PubMed] [Google Scholar]
- 12.Huang B, Tan C, Chang SF, Munday B, Mathew JA, Ngoh GH, Kwang J. Detection of nodavirus in barramundi, Lates calcarifer (Bloch) using recombinant coat protein-based ELISA and RT-PCR. J Fish Dis. 2001;24:135–141. doi: 10.1046/j.1365-2761.2001.00270.x. [DOI] [Google Scholar]
- 13.Iwamoto T, Mise K, Takeda A, Okinaka Y, Mori KI, Arimoto M, Okuno T, Nakai T. Characterization of Striped jack nervous necrosis virus subgenomic RNA3 and biological activities of its encoded protein B2. J Gen Virol. 2005;86:2807–2816. doi: 10.1099/vir.0.80902-0. [DOI] [PubMed] [Google Scholar]
- 14.Jithendran KP, Shekhar MS, Kannappan S, Azad IS. Nodavirus infection in freshwater ornamental fishes in India—diagnostic histopathology and nested RT-PCR. Asian Fish Sci. 2011;24:12–19. [Google Scholar]
- 15.John KR, George MR, Jeyatha B, Saravanakumar R, Sundar P, Jithendran KP, Koppang EO. Isolation and characterization of Indian betanodavirus strain from infected farm-reared Asian seabass Lates calcarifer (Bloch, 1790) juveniles. Aquac Res. 2014;45:1481–1488. doi: 10.1111/are.12095. [DOI] [Google Scholar]
- 16.Kai YH, Su HM, Tai KT, Chi SC. Vaccination of grouper broodfish (Epinephelus tukula) reduces the risk of vertical transmission by nervous necrosis virus. Vaccine. 2010;28:996–1001. doi: 10.1016/j.vaccine.2009.10.132. [DOI] [PubMed] [Google Scholar]
- 17.Keawcharoen J, Techangamsuwan S, Ponpornpisit A, Lombardini ED, Patchimasiri T, Pirarat N. Genetic characterization of a betanodavirus isolated from a clinical disease outbreak in farm-raised tilapia Oreochromis niloticus (L.) in Thailand. J Fish Dis. 2015;38:49–54. doi: 10.1111/jfd.12200. [DOI] [PubMed] [Google Scholar]
- 18.Mori K, Nakai T, Muroga K, Arimoto M, Mushiake K, Furusawa I. Properties of a new virus belonging to nodaviridae found in larval striped jack (Pseudocaranx dentex) with nervous necrosis. Virology. 1992;187:368–371. doi: 10.1016/0042-6822(92)90329-N. [DOI] [PubMed] [Google Scholar]
- 19.Munday BL, Kwang J, Moody N. Betanodavirus infections in teleost fish: a review. J Fish Dis. 2002;25:127–142. doi: 10.1046/j.1365-2761.2002.00350.x. [DOI] [Google Scholar]
- 20.Mushiake K, Nishizawa T, Nakai T, Furusawa I, Muroga K. Control of VNN in striped jack: selection of spawners based on the detection of SJNNV gene by polymerase chain reaction (PCR) Fish Pathol. 1994;29:177–182. doi: 10.3147/jsfp.29.177. [DOI] [Google Scholar]
- 21.Nguyen HD, Mushiake K, Nakai T, Muroga K. Tissue distribution of striped jack nervous necrosis virus (SJNNV) in adult striped jack. Dis Aquat Org. 1997;28:87–91. doi: 10.3354/dao028087. [DOI] [Google Scholar]
- 22.Nishioka T, Mori K, Sugaya T, Tezuka N, Takebe T, Imaizumi H, Kumon K, Masuma S, Nakai T. Involvement of viral nervous necrosis in larval mortality of hatchery-reared Pacific bluefin tuna Thunnus olientalis. Fish Pathol. 2010;45:69–72. doi: 10.3147/jsfp.45.69. [DOI] [Google Scholar]
- 23.Nishizawa T, Furuhashi M, Nagai T, Nakai T, Muroga K. Genomic classification of fish nodaviruses by molecular phylogenetic analysis of the coat protein gene. Appl Environ Microbiol. 1997;63:1633–1636. doi: 10.1128/aem.63.4.1633-1636.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishizawa T, Mori K, Nakai T, Furusawa I, Muroga K. Polymerase chain reaction (PCR) amplification of RNA of striped jack nervous necrosis virus. Dis Aquat Org. 1994;18:103–107. doi: 10.3354/dao018103. [DOI] [Google Scholar]
- 25.Nishizawa T, Muroga K, Arimoto M. Failure of the polymerase chain reaction (PCR) method to detect striped jack nervous necrosis virus (SJNNV) in Striped Jack Pseudocaranx dentex selected as spawners. J Aquat Anim Health. 1996;8:332–334. doi: 10.1577/1548-8667(1996)008<0332:FOTPCR>2.3.CO;2. [DOI] [Google Scholar]
- 26.OIE-World Organisation for Animal Health . Viral encephalopathy and retinopathy. In: OIE, editor. Manual of diagnostic tests for aquatic animals, chapter 2.3.11. Paris: Office International des Epizooties; 2013. [Google Scholar]
- 27.Panzarin V, Fusaro A, Monne I, Cappellozza E, Patarnello P, Bovo G, Capua I, Holmes EC, Cattoli G. Molecular epidemiology and evolutionary dynamics of betanodavirus in southern Europe. Infect Genet Evol. 2012;12:63–70. doi: 10.1016/j.meegid.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 28.Parameswaran V, Rajesh Kumar S, Ishaq Ahmed VP, Sahul Hameed AS. A fish nodavirus associated with mass mortality in hatchery-reared Asian seabass, Lates calcarifer. Aquaculture. 2008;275:366–369. doi: 10.1016/j.aquaculture.2008.01.023. [DOI] [Google Scholar]
- 29.Peducasse S, Castic J, Thiery R, Jeffroy J, Le Ven A, Baudin Laurencin F. Comparative study of viral encephalopathy and retinopathy in juvenile seabass Dicentrarchus labrax infected in different ways. Dis Aquat Org. 1999;36:11–20. doi: 10.3354/dao036011. [DOI] [PubMed] [Google Scholar]
- 30.Reed LJ, Muench M. A simple method of estimating fifty per cent end points. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 31.Sommerset I, Nerland AH. Complete sequence of RNA1 and subgenomic RNA3 of Atlantic halibut nodavirus (AHNV) Dis Aquat Org. 2004;58:117–125. doi: 10.3354/dao058117. [DOI] [PubMed] [Google Scholar]
- 32.Thiery R, Raymond JC, Castric J. Natural outbreak of viral encephalopathy and retinopathy in juvenile seabass, Dicentrarchus labrax: study by nested reverse transcriptase-polymerase chain reaction. Virus Res. 1999;63:11–17. doi: 10.1016/S0168-1702(99)00053-2. [DOI] [PubMed] [Google Scholar]
- 33.Thiery R, Cozien J, de Boisseson C, Kerbart-Boscher S, Nevarez I. Genomic classification of new betanodavirus isolates by phylogenetic analysis of the coat protein gene suggests a low host-fish species specificity. J Gen Virol. 2004;85:3079–3087. doi: 10.1099/vir.0.80264-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.