Abstract
The study details characterization of Newcastle disease virus (NDV) isolates recovered from commercial poultry flocks (chicken) and wild birds (crane) of India during the time period from 1989 to 2013. Phylogenetic analysis revealed that most of the NDV isolates belongs to class II, genotype XIIIa and a chicken isolate (108/BAREILLY/AD-IVRI/91) was of genotype VI, where it showed diversity of 3 % from the other viruses belonging to same genotype. Another chicken isolate (75/RAMPUR/AD-IVRI/89) grouped in genotype III and showed 4 % diversity with viruses of genotype III. The crane origin NDV identified as of genotype II corresponding to the vaccine virus. This appears to be the first report about existence of genotype XIIIa and its ancestral viruses are circulating in India for the last two decades in different species of birds. Furthermore, genetically distinct viruses belonging to genotypes II, III and VI are also circulating in India.
Keywords: Newcastle disease virus, Poultry, Phylogenetic analysis, India, Fusion protein, Genetic diversity
Newcastle disease (ND) is an economically important disease affecting poultry industry across the globe [2]. It is caused by Newcastle disease virus (NDV) belonging to the genus Avulavirus in the family Paramyxoviridae under the order Mononegavirales (ICTV, 2012). This enveloped virus has a negative-sense single-stranded genome of approximately 15 kb which codes for six proteins [13]. The NDV in general may broadly be categorized into three major pathotypes depending on the severity of the disease or virulence as velogenic, mesogenic and lentogenic. The most common methods of pathotyping of NDV involve determination of intracerebral pathogenicity index (ICPI) in day old chicks and determination of amino acid motif at fusion protein (F) gene cleavage site [12]. The serotypes APMV-1 is found to be associated with naturally occurring NDV infections of economic importance [11]. The serotype APMV-1 has two distinct clades: class I and II, further class II is subdivided into 18 recognized genotypes [5]. The outbreaks of NDV in different parts of the world are associated with genotype VII of class II NDV isolates [7–10]. The recent vNDV belongs to a new sub-genotype VIIi, VIIh, XIIIa and XIIIb [10]. In our previous study, NDV in peafowl documented the presence of VIIi, XIIIa related viruses which is causing fifth panzootic outbreak in countries like Pakistan, Israel and Indonesia [4]. In the present study intent was to explore the genetic diversity among NDVs circulating in poultry flocks (chicken) and wild birds (crane) of India, during 1989–2013.
Twelve archived NDV isolates of 11 chicken and one crane origin, maintained in Avian Diseases Section, Division of Pathology, Indian Veterinary Research Institute, Izatnagar, Uttar Pradesh, India were used for the study purpose (Table 1). The freeze dried NDV isolates were reconstituted in one ml of 1 % phosphate buffered saline (PBS, pH 7.4). For virus propagation, inoculums were prepared as per standard protocol [12]. The reconstituted 0.2 ml of inoculums was inoculated into 11 day-old specific pathogen free (SPF) embryonated chicken eggs (Venkateshwara Hatcheries Private Limited, Pune, India) through allantoic route and the eggs were incubated at 37 °C till death or maximum period of 120 h, whichever was earlier. The embryos were candled every day and the dead embryos were chilled at 4 °C for overnight, and the allantoic fluids were tested for hemagglutination (HA) activity (3 OIE, 2012). After 120 h, all the remaining live embryos were chilled at 4 °C for overnight and then allantoic fluids were harvested and tested for HA activity. The allantoic fluids that were found negative by HA were further passaged at least once in embryonated chicken eggs. Hemagglutination inhibition (HI) [12] test with LaSota specific serum (known HI titre of log2 HI > 3) was also performed to confirm the presence of NDV in the allantoic fluids. The allantoic fluids harvested were also subjected to reverse transcription-polymerase chain reaction (RT-PCR) testing for detection of RNA of NDV. Total RNA was extracted from the allantoic fluids using TRIzolR reagent (Invitrogen, USA) following manufacturer’s instructions. The extracted RNA was used to synthesize cDNA with the help of random hexamers (MBI Fermentas, USA), Reverse transcription (RT) for first strand synthesis was carried out by using Revert aid H minus (MMuLV-RT) (MBI Fermentas, USA) and RT-PCR was performed with the primers targeted to amplify F gene of NDV including the cleavage site. The primer sequences used were Forward-5′GCAGCTGCAGGGATTGTGGT3′, Reverse- 5′TCTTTGAGCAGGAGGATGTTG3′ and the expected product size was 356 bp [14]. The PCR products were analysed by electrophoresis in 1.5 % agarose gel stained with ethidium bromide (0.5 µg/ml) (Fig. 1). The RT-PCR amplified products were sequenced (Two replicates using forward and reverse primer) with help of commercial sequencing centre (BioServe, India). The sequence data of these twelve NDV isolates was subjected to blast analysis with the help of NCBI BLAST tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and compared with the NDV sequences available in GenBank. The phylogenetic analysis based on the 114 nucleotide sequences of F gene in the highly variable region including cleavage site was performed in MEGA6 software by using the maximum likelihood method based on the General Time Reversible model, and Gamma distributed with invariable sites (G+I). The codon positions 1st, 2nd, 3rd, and non-coding sequences were included and the positions containing gaps and missing data were eliminated [5, 10].
Table 1.
The NDV isolates used in this study, their GenBank accession number, F gene cleavage site amino acid sequence, host, place and year of collection
| S. no. | Accession no. | Isolate name | F gene cleavage site amino acid (112–117) | Host | Place | Year |
|---|---|---|---|---|---|---|
| 1 | KJ627784 | IVRI/91 | RRQKRF | Chicken | Bareilly | 1991 |
| 2 | KJ627783 | IVRI/92 | RRQKRF | Chicken | Bareilly | 1992 |
| 3 | KJ627782 | 123/KATHUA/AD-IVRI/92 | RRQKRF | Chicken | Kathua | 1992 |
| 4 | KJ627781 | 1/chicken/IVRI/13 | RRQRRF | Chicken | Bareilly | 2013 |
| 5 | KJ627780 | 2/chicken/IVRI/13 | GRQGRL | Chicken | Bareilly | 2013 |
| 6 | KJ627779 | 3/chicken/IVRI/13 | GRQGRL | Chicken | Bareilly | 2013 |
| 7 | KJ627778 | 4/chicken/IVRI/13 | GRQGRL | Chicken | Bareilly | 2013 |
| 8 | KJ627777 | 75/RAMPUR/AD-IVRI/89 | RRQRRF | Chicken | Rampur | 1989 |
| 9 | KJ627776 | 108/BAREILLY/AD-IVRI/91 | RRQKRF | Chicken | Bareilly | 1991 |
| 10 | KJ627775 | 127/FAIZABAD/AD-IVRI/92 | RRQKRF | Chicken | Faizabad | 1992 |
| 11 | KJ627774 | 129/FAIZABAD/AD-IVRI/92 | RRQKRF | Chicken | Faizabad | 1992 |
| 12 | KJ627773 | Crane/ADS/IVRI/13 | GKQGRL | Crane | Bareilly | 2013 |
| 13 | KJ398400 | NDV/Peafowl/Haryana/IVRI-037/12a | RRQKRF | Peafowl | Haryana | 2012 |
| 14 | KJ398401 | NDV/Peafowl/UP/IVRI-024/12a | RRQRRF | Peafowl | Uttar Pradesh | 2012 |
| 15 | KJ398399 | NDV/Peafowl/Delhi/IVRI-0022/12a | RRQKRF | Peafowl | Delhi | 2012 |
aIsolates pertaining to previous study [4]
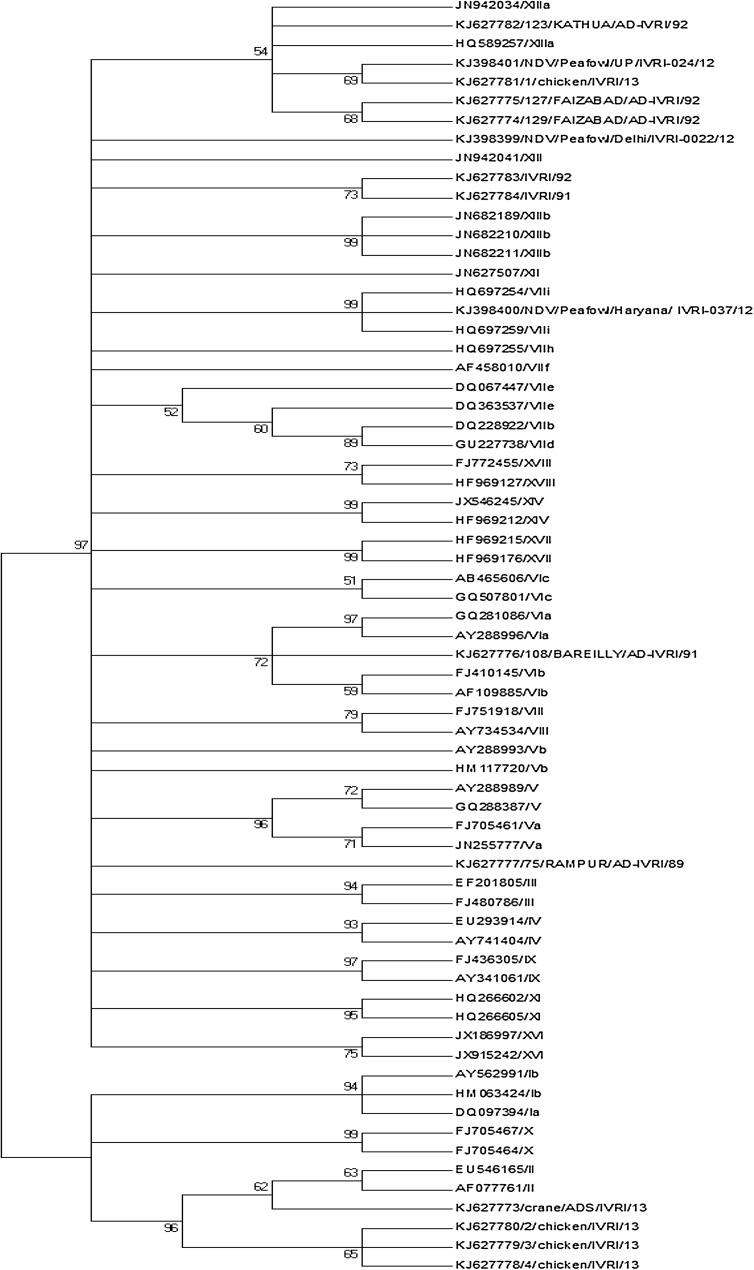
Fig. 1.
The phylogenetic analysis of NDV isolates based on the 114 nucleotide sequence F gene in the highly variable region including cleavage site—performed in MEGA6 by using the maximum likelihood method based on the General Time Reversible model, and Gamma distributed with invariable sites (G+I). The codon positions 1st, 2nd, 3rd, and non-coding were included. All positions containing gaps and missing data were eliminated
The virulent and avirulent NDV have the sequence 112R/K-R-Q/K/R-K/R-R-F117 and 112G/E-K/R-Q-G/E-R-L117, respectively in fusion protein (F) gene cleavage site [12]. The F gene cleavage site amino acid motif revealed that eight chicken isolates were virulent NDV and four isolates including crane isolates belonged to lentogenic NDV (Table 1). The phylogenetic relationship of these viruses is presented in Fig. 1. The newly defined sub-genotype XIIIa was reported from Iran, South Africa and India [10]. This study reports that the 123/KATHUA/AD-IVRI/92, 1/chicken/IVRI/13, 127/FAIZABAD/AD-IVRI/92 and 129/FAIZABAD/AD-IVRI/92 isolates belonged to NDV class II, genotype XIIIa. The 123/KATHUA/AD-IVRI/92 isolate showed 100 % identity with UP1-93/Gallus gallus/India (AJ249529), ZA C1868/95/UK, ZW 3422/95/UK, ZA 360/95/UK, VRDC/NDV/F/N1/poultry/Maharashtra/India/2004 (KJ621041), Bareilly/chicken/2006 (KF727980) and NDV/Chicken/Bareilly/01/10 (KJ577585) isolates. The 123/KATHUA/AD-IVRI/92 virus or viruses like this were detected in different years in India in 1993, 2004, 2006 and 2010 in chicken, suggesting that this type of NDV circulates in Indian chicken for the last two decades without genetic change. The 1/chicken/IVRI/13 and NDV/Peafowl/UP/IVRI-024/12 isolates revealed 100 % identity with each other and had same identity with PiAD18/Guinea fowl/India (AY581303) and WB1-94/Gallus gallus/India (AJ249530) isolates. This finding suggests that 1/chicken/IVRI/13 virus is well adopted in all these three species (chickens, Guinea fowl and peafowl) and also this virus circulates in India for the last two decades.
Cockatoo/India/7847/1982 was considered as an ancestral strain for sub-genotype XIIIa [10]. The NDV/Peafowl/Delhi/IVRI-0022/12, IVRI/92 and IVRI/91 isolates were closely related to NDV class II, genotype XIIIa ancestral virus (Cockatoo/India/7847/1982). The IVRI/92 and IVRI/91 isolates showed 100 % identity with each other and had the same identity with NP1-93 (AJ249526) isolate from India, and 99 % identity with Cockatoo/India/7847/1982 isolate. This finding suggests that NDV genotype XIIIa ancestral Cockatoo/India/7847/1982 virus related viruses are circulating in Indian peafowl and chickens.
Genotypes VI emerged after 1960 and considered as “late” genotype [3]. During the 1980s, viruses of genotype VI were responsible for the third panzootic [6]. The 108/BAREILLY/AD-IVRI/91 isolate belonged to NDV class II, genotype VI. This isolate showed 97 % identity with AV 1573/92 120/90/United Arab Emirates (AY135748), Pi07/AD/01/INDIA (AJ781075) and Pi04/AD/94/INDIA (AJ781072) isolates, and 95 % identity with Pigeon/New York/44407/1984 (JN872173), Pigeon/Maryland/11936/1985 (JN967788), PPMV-1/Maryland/1984 (FJ410147) and PPMV-1/New York/1984 (FJ410145) isolates. Even though this virus belonged to genotype VI, it had minimum 3 % diversity with VI sub-genotypes in NCBI blast analysis.
Viruses belongs to the genotypes II, III and IV were responsible for the first panzootic between 1920s and 1960s [1]. The 75/RAMPUR/AD-IVRI/89 isolate belonged to NDV class II, genotype III. This isolate showed 98 % identity with UP3/2006/chicken/India (FJ665433) isolate, 96 % identity with IT-7/60/Italy (EU604256) and POK/70/Russia (AJ243388), isolates and 95 % identity with Mukteswar (JF950509) and H/Ph (FJ687488) isolates. Even though this virus belonged to genotype III, it showed minimum 4 % diversity with genotypes III viruses in NCBI blast analysis.
The 2/chicken/IVRI/13, 3/chicken/IVRI/13, 4/chicken/IVRI/13, Crane/ADS/IVRI/13 isolate belonged to NDV class II, genotype II. Chicken isolates showed 100 % identity with NDV vaccine strain F. The flock may be vaccinated with live F strain of vaccine hence it might be detected in RT-PCR and sequencing. Another finding is that crane origin virus showed 97 % identity with LaSota vaccine strain. Hence, this virus needs to be validated further as a vaccine candidate for wild birds to control NDV in wild population. In conclusion, most of the NDV isolates of the present study belong to NDV class II, genotype XIIIa and its ancestral virus. Genetically distinct virus belongings to genotype VI and III are circulating in poultry and wild birds of India. The crane origin NDV showed identity to vaccine virus hence may have potential value to be used as a vaccine candidate for wild birds which needs validation. Further intensive monitoring and surveillance of NDV in commercial and wild birds is warranted to establish genetic diversity among NDVs existing/circulating in Indian poultry population, for adopting proper and timely controlling measures in growing commercial poultry sector and wild birds also.
Acknowledgments
The authors are highly thankful to the institute’s authority for providing necessary facilities to carry out the research work.
References
- 1.Alexander DJ. Newcastle disease. Br Poult Sci. 2001;42:5–22. doi: 10.1080/713655022. [DOI] [PubMed] [Google Scholar]
- 2.Cattoli G, Susta L, Terregino C, Brown C. Newcastle disease: a review of field recognition and current methods of laboratory detection. J Vet Diagn Invest. 2011;23(4):637–656. doi: 10.1177/1040638711407887. [DOI] [PubMed] [Google Scholar]
- 3.Czegledi A, Ujvari D, Somogyi E, Wehmann E, Werner O, Lomniczi B. Third genome size category of avian paramyxovirus serotype 1 (Newcastle disease virus) and evolutionary implications. Virus Res. 2006;120:36–48. doi: 10.1016/j.virusres.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 4.Desingu PA. Studies on characterization and pathogenicity of Indian Peafowl-origin Newcastle disease virus isolates. Submitted in partial fulfillment of the requirement for the Degree of Master of Veterinary Science in Veterinary Pathology to Deemed University Indian Veterinary Research Institute, Izatnagar (U.P.), India. 2013 (Thesis).
- 5.Diel DG, da Silva LHA, Liu H, Wang Z, Miller PJ, Afonso CL. Genetic diversity of avian paramyxovirus type 1: proposal for a unified nomenclature and classification system of Newcastle disease virus genotypes. Infect Genet Evol. 2012;12:1770–1779. doi: 10.1016/j.meegid.2012.07.012. [DOI] [PubMed] [Google Scholar]
- 6.Kaleta EF, Alexander DJ, Russell PH. The first isolation of the avian pmv-1 virus responsible for the current panzootic in pigeons? Avian Pathol. 1985;14:553–557. doi: 10.1080/03079458508436258. [DOI] [PubMed] [Google Scholar]
- 7.Liu H, Wang Z, Wu Y, Zheng D, Sun C, Bi D, Zuo Y, Xu T. Molecular epidemiological analysis of Newcastle disease virus isolated in China in 2005. J Virol Methods. 2007;140:206–211. doi: 10.1016/j.jviromet.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 8.Miller PJ, Kim LM, Ip HS, Afonso CL. Evolutionary dynamics of Newcastle disease virus. Virology. 2009;391:64–72. doi: 10.1016/j.virol.2009.05.033. [DOI] [PubMed] [Google Scholar]
- 9.Miller PJ, Decanini EL, Afonso CL. Newcastle disease: evolution of genotypes and the related diagnostic challenges. Infect Genet Evol. 2010;10:26–35. doi: 10.1016/j.meegid.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 10.Miller PJ, Haddas R, Simanov L, Lublin A, Rehmani SF, Wajid A, Bibi T, Khan TA, Yaqub T, Setiyaningsih S, Afonso CL. Identification of new sub-genotypes of virulent Newcastle disease virus with potential panzootic features. Infect Genet Evol. 2015;29:216–229. doi: 10.1016/j.meegid.2014.10.032. [DOI] [PubMed] [Google Scholar]
- 11.Nayak B, Dias FM, Kumar S, Paldurai A, Collins PL, Samal SK. Avianparamyxovirus serotypes 2-9 (APMV-2-9) vary in the ability to induce protective immunity in chickens against challenge with virulent Newcastle disease virus (APMV-1) Vaccine. 2012;30:2220–2227. doi: 10.1016/j.vaccine.2011.12.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.OIE. OIE Terrestrial Manual 2012. Newcastle Disease, Chapter 2.3.14, 2012.
- 13.Tirumurugaan KG, Kapgate S, Vinupriya MK, Vijayarani K, Kumanan K, Elankumaran S. Genotypic and pathotypic characterization of Newcastle disease viruses from India. PLoS One. 2011;6:28414. doi: 10.1371/journal.pone.0028414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toyoda T, Sakaguchi T, Hirota H, Gotoh B, Kuma K, Miyata T, Nagai Y. Newcastle disease virus evolution II. Lack of gene recombination in generating virulent and avirulent strains. Virology. 1989;169:273–282. doi: 10.1016/0042-6822(89)90152-9. [DOI] [PubMed] [Google Scholar]