Abstract
Campylobacter jejuni expresses two hemoglobins, each of which exhibits a heme pocket and structural signatures in common with vertebrate and plant globins. One of these, designated Cgb, is homologous to Vgb from Vitreoscilla stercoraria and does not possess the reductase domain seen in the flavohemoglobins. A Cgb-deficient mutant of C. jejuni was hypersensitive to nitrosating agents (S-nitrosoglutathione [GSNO] or sodium nitroprusside) and a nitric oxide-releasing compound (spermine NONOate). The sensitivity of the Cgb-deficient mutant to methyl viologen, hydrogen peroxide, and organic peroxides, however, was the same as for the wild type. Consistent with the protective role of Cgb against NO-related stress, cgb expression was minimal in standard laboratory media but strongly and specifically induced after exposure to nitrosative stress. In contrast, the expression of Cgb was independent of aeration and the presence of superoxide. In the absence of preinduction by exposure to nitrosative stress, no difference was seen in the degree of respiratory inhibition by NO or the half-life of the NO signal when cells of the wild type and the cgb mutant were compared. However, cells expressing GSNO-upregulated levels of Cgb exhibited robust NO consumption and respiration that was relatively NO insensitive compared to the respiration of the cgb mutant. Based on similar studies in Campylobacter coli, we also propose an identical role for Cgb in this closely related species. We conclude that, unlike the archetypal single-domain globin Vgb, Cgb forms a specific and inducible defense against NO and nitrosating agents.
Campylobacter jejuni and Campylobacter coli are recognized as the predominant agents of bacterial gastrointestinal disease worldwide (18). The main symptoms of infection are fever, severe abdominal pain, and diarrhea. Although this acute phase is normally self-limiting, serious sequelae may include Guillain-Barré syndrome (32).
NO and its reaction products, peroxynitrite in particular, have strong bactericidal activities and during infection campylobacters are likely to encounter these agents from a variety of sources. Invasion of the epithelial mucosa is considered to play an important role during Campylobacter infection (14, 33), and NO and/or its redox products form a key component of the inducible defense of intestinal cells against microbial infection (52). As a result of this antimicrobial mechanism, NO synthesis is markedly increased in patients with infective gastroenteritis (15) and, accordingly, during infection campylobacters are likely to be exposed to significant amounts of NO in the gut. Campylobacters may be exposed to NO in the stomach since the chemical generation of NO in this organ can occur as a consequence of microbial nitrite production in the mouth (10, 29). It has even been suggested that generation of NO in the stomach represents a separate and yet powerful defense against gut pathogens (10, 11). Consequently, although nothing is known of the interaction of campylobacters with NO, resistance to this agent and it redox products are likely to be critical during colonization and infection.
The most fully understood mechanisms for detoxification of NO involve the inducible bacterial flavohemoglobin (Hmp) of Escherichia coli (44) and flavorubredoxin (19). In the presence of oxygen, Hmp detoxifies NO by acting as an NO dioxygenase (20, 23, 54, 55) and affords protection of respiration (47). Under anaerobic conditions, in the absence of Hmp activity, the flavorubredoxin serves as an oxygen-independent NO reductase (19). Hmp may also function to repair NO-damaged lipid membranes, since the purified protein has recently been shown to possess both alkyl hydroperoxide reductase activity and lipid-binding properties (3, 4). However, whether these activities are physiologically relevant has not yet been confirmed by in vivo studies.
The N-terminal domain of Hmp is homologous to the globin family (reviewed in references 43, 54, and 55), having a single protoheme and highly conserved heme pocket, whereas the C-terminal domain has binding sites for FAD and NAD(P)H. Many other microorganisms possess proteins having clear homology to Hmp. These bacterial hemoglobins may be classified into three broad groups (54, 55): the small single-domain globins such as Vgb (49), the flavohemoglobins exemplified by Hmp, and the truncated globins that are 20 to 40 amino acids shorter than vertebrate hemoglobins (40, 51). The functions of the single-domain globins are not clear. Vgb, the archetypal single-domain globin from Vitreoscilla stercoraria has been implicated in oxygen storage, delivery, or reduction based upon its induced expression at low-oxygen tensions (9), the absence of an induced response to nitrosative stress (17), the ability to enhance microaerobic growth when expressed in E. coli (2), and its binding to cytochrome bo′ (36). However, Vgb may also have an alternative functional role since a recombinant chimeric version of this globin, which carries a flavoreductase domain, can alleviate nitrosative stress in E. coli (27). We provide here the first conclusive evidence for a single-domain globin that performs an NO scavenging and detoxification function in the organism from which it originated, namely, Campylobacter. We also demonstrate that the globin is uniquely and specifically induced by nitrosative stress and not by oxygen limitation, or superoxide stress as is the case for other globins.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains and plasmids used in the present study are listed in Table 1. C. jejuni NCTC 11168 and C. jejuni NCTC 11828 were obtained from the National Collection of Type Cultures (NCTC; PHLS, London, United Kingdom). The naturally transformable strain C. coli UA585 was a gift from D. E. Taylor (University of Alberta) and has been described previously (50). E. coli DH5α was obtained from Life Technologies. Campylobacters were grown at 37 or 42°C when indicated in Mueller-Hinton (MH) broth (Oxoid)or on MH agar containing kanamycin (50 μg ml−1), tetracycline (10 μg ml−1), or chloramphenicol (10 μg ml−1) when necessary. Microaerophilic conditions were generated by using the CampyGen (Oxoid) gas generating kit or a MACS-VA500 microaerophilic work station (10% O2, 10% CO2, and 80% N2) from Don Whitley Scientific, Ltd. For cloning experiments, E. coli DH5α was grown at 37°C in Luria-Bertani (LB) broth or agar containing ampicillin (100 μg ml−1), kanamycin (50 μg ml−1), or tetracycline (10 μg ml−1) as required.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| C. jejuni | ||
| NCTC 11168 | Parental strain | NCTC |
| NCTC 11828 | Source of astA reporter gene | NCTC |
| CJCGB01 | 11168 cgb::Kanr | This study |
| CJA01 | 11168 ahpC::Cmr | This study |
| CJAC01 | 11168 cgb::Kanr, ahpC::Cmr | This study |
| C. coli strains | ||
| UA585 | Parental strain | 50 |
| CCCGB01 | UA585 cgb::Kanr | This study |
| CCCF1 | UA585::pKE56 (cgb-astA fusion strain) | This study |
| CCSF1 | UA585::pKE58 (sodB-astA fusion strain) | This study |
| E. coli DH5α | F− φ80dlacZ ΔM15 | Life Technologies |
| Plasmids | ||
| pCR2.1 | Cloning vector; Apr Kanr | Invitrogen |
| pBAD-TOPO | Cloning vector; Apr | Invitrogen |
| pET16b | N-terminal His tag fusion expression vector; Apr | Novagen |
| pSP105 | C. coli integrative vector; Tetr | 8 |
| pJMK30 | C. coli Kanr cassette in pUC19 | 1 |
| pAV103 | Cmr cassette cloned into unique EcoRV site of ahpC | 1 |
| pKE1 | 1,187-bp PCR product containing cgb in pCR2.1 | This study |
| pKE2 | 1,187-bp PCR product containing cgb in pBAD-TOPO | This study |
| pKE18 | Kanr cassette cloned into a unique PCR generated BglII site in cgb in pKE2 | This study |
| pKE54 | pSP105 with cgb promoter fragment | This study |
| pKE55 | pSP105 with sodB promoter fragment | This study |
| pKE56 | pSP105 with a cgb-astA transcriptional fusion | This study |
| pKE58 | pSP105 with a sodB-astA transcriptional fusion | This study |
Cmr, chloramphericol resistance; Apr, ampicillin resistance; Tetr, tetracycline resistance.
Generation of C. jejuni and C. coli mutants.
Genomic DNA was isolated from all campylobacters by using guanidinium thiocyanate (42). The putative bacterial hemoglobin gene (Cj1586) from C. jejuni NCTC 11168 (accession no. AL111168 [37]) was amplified from genomic DNA by using PCR and the oligonucleotide primers HMP1 (5′-CACTACAAGTACTCCATCACAAGA-3′) and HMP6 (5′-GGATCCGTCAAAGATAATGAACTGACCTTAACCA-3′). The PCR conditions have been described previously (12). The 1,187-bp DNA fragment generated was cloned into the TA cloning vectors pBAD-TOPO (Invitrogen) and pCR2.1 (Invitrogen) to generate pKE2 and pKE1, respectively.
The cgb gene was mutated by using the inverse PCR protocol described by Wren et al. (53), which deleted 2 bp of the open reading frame and introduced a unique BglII restriction site. Oligonucleotide primers HMP7 (5′-gcaacaAGATCTAAGCTTTTGGTTGTTCTCCTGAAATT-3′) and HMP8 (5′-gcaacaAGATCTGCAATGGCGATTTTAATGGCGGCT-3′) containing unique restriction sites for BglII (underlined) and clamp sequences and not complementary to C. jejuni DNA (lowercase) were used. The PCR fragment, generated by using a two-stage cycling program (12), was digested with BglII and self-ligated. The resulting plasmid was digested with BglII, and a kanamycin resistance (Kanr) cassette with BamHI ends from pJMK30 (J. Ketley, University of Leicester) was inserted. This step generated the suicide plasmid pKE18, which was introduced into C. jejuni NCTC 11168 by electroporation (12) and into C. coli UA585 by natural transformation (50).
The gene encoding AhpC (Cj0334) was disrupted by insertional mutagenesis with a chloramphenicol gene by using plasmid pAV103 as described previously (1). The insertion was confirmed by PCR, and the single ahpC and double cgb ahpC mutants were designated CJA01 and CJAC01, respectively.
Sequencing of the C. coli UA585 cgb gene.
The cgb gene from C. coli UA585 was amplified by PCR with primers HMP1 and HMP3 (5′-GGATCCTGTTGATGATACGCTTATAGATGA-3′) and cloned into the pCR2.1-TOPO TA cloning vector (Invitrogen). Nucleotide sequence analysis was carried out by the dideoxy chain termination method by using a CEQ dye terminator cycle sequencing quick start kit on a CEQ 2000 sequencer (Beckman Coulter).
Reverse transcriptase PCR.
RNA was extracted from NCTC 11168 and CJCGB01 by using Qiagen RNeasy and RNAprotect as described by the manufacturer. Briefly, for each extraction, 2 ml of culture was transferred into 4 ml of RNA protect reagent, followed by incubation at room temperature for 5 min. The samples were next centrifuged at 6,000 × g for 10 min, the supernatant was discarded, and the cells were lysed after the addition of 100 μl of Tris-EDTA buffer containing 1 mg of lysozyme (Sigma) ml−1. The RNA was purified by using the RNeasy kit (Qiagen). To remove contaminating DNA, 1 μg of total RNA was treated with DNA-free kit (Ambion) according to the manufacturer's instructions.
First-strand cDNA synthesis was performed with 100 ng of total RNA with random hexamers (250 ng; Invitrogen) and Superscript III RT (Invitrogen). PCR amplification consisted of 35 cycles of 45 s of denaturation at 94°C, 30 s of annealing at 55°C, and 1 min of extension at 72°C. The oligonucleotides used to amplify Cj1585c were the forward primer RED6 (5′-TACCGTTGAACCTGCCTTTC-3′) and the reverse primer RED5 (5′-CGGAATGTGCTGTGGAACTA-3′), and for Cj1587c we used the forward primer ABC1 (5′-ATCTGCAACACTTGCCCATT-3′) and the reverse primer ABC2 (5′-AATGCAAAGGCGCGTAGTA-3′). The PCR products were analyzed by gel electrophoresis on 0.8% agarose.
Oxidative and nitrosative stress tolerance assays.
S-Nitrosoglutathione (GSNO) was obtained from Sigma or synthesized according to the method of Hart (22) and kindly donated by M. N. Hughes. Concentrations of GSNO in stock solutions were determined from absorbance at 334 nm by using a molar absorption coefficient of 900 M−1 cm−1 (35). Cumene hydroperoxide, methyl viologen, and sodium nitroprusside (SNP) were also obtained from Sigma. The NO-releasing agent spermine NONOate (Calbiochem) was also used.
The sensitivity of strains to a range of oxidative and nitrosative stress-inducing factors was initially assayed in a plate diffusion assay using the method described by Baillon et al. (1). For viability assays, C. jejuni strains were grown microaerobically overnight at 37°C to form a lawn on MH agar. The lawns were harvested in MH broth and adjusted to an optical density at 600 nm (OD600) of 0.35. A total of 100 μl of the cell suspension was inoculated into 50 ml of MH broth, and 180 μl of this diluted cell suspension was dispensed into 96-well microtiter plates. The plates were incubated microaerobically and shaken horizontally (175 rpm) at 37°C for 1 h. After 1 h, 20 μl of GSNO or SNP solutions were added to the wells in duplicate to give final concentrations of 0.2, 0.4, 0.8, 1.6, and 2 mM. Spermine NONOate was also used but at lower concentrations. In addition, 20-μl aliquots of MH broth were added to other wells as controls. Viable counts were carried out after a further 1 h of incubation under the conditions described above. All assays were repeated three times.
Growth of C. jejuni in the presence of GSNO.
Wild-type NCTC 11186 and the cgb mutant CJCGB01 were grown in MH liquid medium with vancomycin (10 μg ml−1) in the microaerophilic work station at 40°C overnight before they were inoculated at 1% to 10 ml of MH broth (without vancomycin) in 50-ml flasks containing different concentrations of GSNO. The cultures were shaken (100 rpm), and growth was monitored by measuring the ODs of samples at various time points.
Measuring gene expression by using cgb-astA and sodB-astA reporter gene fusions.
The promoter regions of cgb and sodB (45) were amplified from C. coli UA585 by using PCR and oligonucleotide primer pairs CGB3 (5′-GGTACCCACTACAAGTACTCCATCACAAGA-3′)-CGB4 (5′-CCCGGGCTGCCATTAAAATTGCCATAGCTAAGG-3′) and SOD3 (5′-GGTACCGCTACAATATTTTGTCAAACTACT-3′)-SOD4 (5′-CCCGGGATCTTCAGTGATAGGTGTAGCTGC-3′). The restriction sites for KpnI and SmaI, incorporated into the primers to facilitate subcloning, are indicated by underlining. PCR, under conditions described previously (12), was used to generate 627- and 521-bp fragments derived from cgb and sodB, respectively, that were cloned into pCR2.1. Inserts were isolated as SmaI and KpnI fragments and cloned into the integrational Campylobacter vector pSP105 (8) that had been digested with the same restriction enzymes. This generated the vectors pKE54 and pKE55 containing the promoter regions of the cgb and sodB genes, respectively. A promoterless arylsulfatase gene (56) including the Shine-Dalgarno sequence was amplified from genomic DNA derived from C. jejuni NCTC 11828 by using PCR and the oligonucleotide primers AST1 (5′-CCCGGGTTAAAGGATTGATCATGAGACTTAG-3′) and AST2 (5′-CTGCAGAGAAAATGCTTCTATTACACTATT-3′) with restriction sites for SmaI and PstI (underlined). The PCR cycling conditions used are described previously (12). The 2,094-bp DNA fragment generated in this manner was cloned into pCR2.1 and then excised by using SmaI and PstI. This was then ligated into the site created by digestion of the vectors pKE54 and 55 with SmaI and then PstI to generate the integrational vectors pKE56 (cgb-astA transcriptional fusion) and pKE58 (sodB-astA). These were transformed into the naturally competent strain C. coli UA585 as described previously (50). Transformants were recovered after incubation under microaerophilic conditions at 37°C on MH agar plates containing tetracycline (10 μg ml−1).
Overnight cultures of the C. coli strains containing astA transcriptional fusions were grown microaerobically at 37°C in 10 ml of MH broth or 10 ml of MH broth containing tetracycline (10 μg ml−1), respectively. These cultures were adjusted to OD600 of 0.3, and each strain was inoculated (100 μl) into flasks containing 100 ml of MH broth and incubated microaerobically at 37°C for 16 h with shaking (125 rpm). Samples were taken to record viable counts, the OD, and arylsulfatase activity (see below), after which GSNO was added to flasks to give final concentrations of 0.05, 0.1, and 0.25 mM. Arylsulfatase activity was also measured after addition of 0.005, 0.01, and 0.05 mM SNP and 1, 5, and 10 μM methyl viologen. Samples were taken at regular intervals.
Arylsulfatase enzyme assay.
Arylsulfatase activity in whole cells was measured by a modification of the methods described by Henderson and Milazzo (24) and by Delisle and Milazzo (7). Routinely, 10 ml of culture was centrifuged at 3,500 rpm for 15 min, and cells resuspended in 1 ml of 0.1 M Tris-HCl buffer (pH 7.0) and kept on ice for 10 min. One milliliter of 20 mM p-nitrophenyl sulfate (Sigma) in 0.1 M Tris-HCl (pH 7.0) was added to the cells, followed by incubation at 37°C for 1 h. The reaction was terminated by centrifugation at 13,000 rpm for 5 min. The amount of liberated free p-nitrophenol was determined spectrophotometrically at 410 nm from 1 ml of the supernatant by using a Helios Alpha spectrophotometer (Unicam) with a 1-cm path length. The results are expressed as a ratio of micrograms of p-nitrophenol (determined by applying the Beer Lambert Law) to OD600/hour.
Purification of Cgb, preparation of antibodies, and immunoblotting.
The purification of the Cgb protein will be described in detail elsewhere. In brief, Cgb was overexpressed in the pET16b vector (from Novagen) and purified with DEAE-Sepharose, butyl-Toyopearl 650s hydrophobic, and Superdex 200 gel filtration columns. Antibody to Cgb was raised essentially as described before for the E. coli hemoglobin (47), except that test bleeds were performed after the third and fourth injections. The animals were bled out after a fifth injection. In preparation for immunoblotting, C. jejuni cultures were grown in MH medium at 42°C with shaking at 110 rpm in the microaerophilic work station. After 10 h, 6 ml of culture was supplemented with 4 ml of fresh MH medium in 20 ml of Sterilin tubes and grown for a further 1 h. GSNO was added to final concentrations of 0.05, 0.1, or 0.25 mM; cells were harvested at different time points after the addition of chloramphenicol (100 μg ml−1), washed with Tris-buffered saline buffer without phenol red (46), and stored at −70°C overnight. Cells were then resuspended in Tris-buffered saline (75 μl), and an equal volume of acid-washed glass beads (25 μl, 106 μm and finer, G-4649; Sigma) was added. The samples were vigorously vortexed for four periods of 1 min, with chilling on ice during the intervals. The disrupted cell suspension was centrifuged at 13,800 × g and assayed for protein with the Bio-Rad protein assay kit and bovine serum albumin as the standard. Then, 10 μg of each sample was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The anti-Cgb antibody was diluted 2,000-fold for use with a 2,000-fold dilution of peroxidase-conjugated monoclonal anti-rabbit immunoglobulin G (γ-chain specific, clone RG-96, A-1949; Sigma) as the secondary antibody. Western blots and detection by using enhanced chemiluminescence (Amersham Biosciences) were done as described previously (47). Blot intensities were quantified by scanning the film with a Syngene Gene Genius gel documentation and analysis system.
To study the regulation of Cgb by aeration, cells were grown overnight in MH broth with vancomycin (10 μg ml−1) before being subcultured (3%) into 5, 10, 25, 35, or 45 ml of MH broth in 50-ml flasks and grown in the microaerophilic work station at 40°C with shaking at 110 rpm. After 5 h of growth, chloramphenicol (100 μg ml−1) was added to stop further protein synthesis. Cells were harvested after a further 10 min of incubation. Western blotting was performed as described above, except that more protein (45 μg) was loaded to each lane.
Inhibition of respiration by nitric oxide and consumption of NO by C. jejuni.
Next, 4 ml of an overnight culture of Campylobacter in MH broth with vancomycin (10 μg ml−1) was inoculated into 150 ml of MH broth in 250-ml baffled flasks, and the cells were incubated for 5 h in the microaerophilic work station at 40°C with shaking at 110 rpm. GSNO was then added to a final concentration of 0.05 mM. At this point the OD600 was between 0.16 and 0.20. At this concentration of GSNO, the growth of neither the mutant nor the wild type was inhibited, but the expression of Cgb was induced. The cells were further incubated for 1 h before harvesting and washed in 50 mM morpholinepropanesulfonic acid (MOPS) plus 50 mM NaCl (pH 7.0) (MOPS buffer). Cell respiration was measured with an oxygen electrode as described by Stevanin et al. (48). Sodium formate (2 mM) used as respiratory substrate was incubated with MOPS buffer before the cells were added. NO solution was used to inhibit the respiration. Alternatively, NO solution was added before the addition of cells in order to measure NO consumption by cells.
Adherence and invasion of Caco-2 cells and nitrite assay.
Caco-2 cells (ECACC) were grown as monolayers in minimal essential Eagle medium (Sigma). After infection of these with campylobacters, adhesion, invasion, and intracellular survival was assessed as described previously (12). As a stable nonvolatile breakdown product of NO, nitrite can be used to measure NO production in human colon epithelial cells (52). Nitrite levels in cell culture bathing medium were measured by the Griess reagent system (Promega). Caco-2 cells were seeded into six-well tissue culture trays (12) and infected with C. jejuni NCTC 11168 and CJCGB01. Wells containing Caco-2 cells without Campylobacter and others containing minimum essential Eagle medium with C. jejuni NCTC 11168 and CJCGB01 alone were used as controls. All were incubated for 3 h at 37°C in 5% CO2, after which 1 ml of cell culture bathing medium was transferred to 1.5-ml microfuge tubes. Tubes were centrifuged at 13,000 rpm for 3 min to precipitate the cells, and nitrite levels were measured according to the manufacturer's instructions by using 50 μl of supernatant. Absorbance was assayed at 540 nm and was compared to a standard curve by using sodium nitrite.
RESULTS
Identification of two globins in C. jejuni.
Analysis of the genome sequence of C. jejuni NCTC 11168 (37) reveals coding sequences for two hemoglobin-like proteins. Cj0465c is of the truncated type (data not shown) as described by Pesce et al. (40). The second, Cj1586, which we designate Cgb (for Campylobacter globin), was shown to have 42% amino acid identity to Vgb from V. stercoraria (Fig. 1a). Cgb also showed 39, 34 and 33% identity to the heme domain in flavohemoglobins from B. subtilis, Salmonella enterica serovar Typhimurium, and E. coli, respectively, but does not possess the binding sites for FAD and NAD(P)H seen in the two-domain globins.
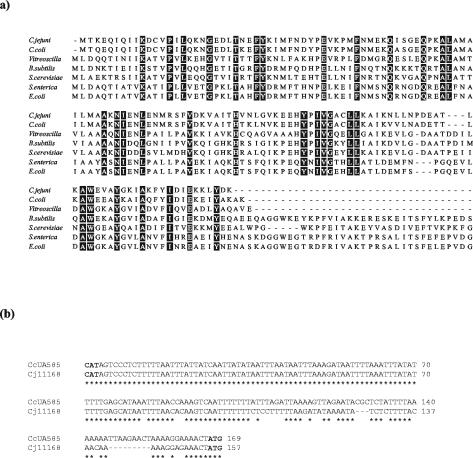
FIG. 1.
(a) Comparison of Cgb from C. jejuni NCTC 11168 and C. coli UA585 with hemoglobins from other bacterial and yeast species. Hemoglobin sequences from C. jejuni NCTC 11168 (SwissProt Q9PM89), C. coli UA585, V. stercoraria (sp P04252), B. subtilis (sp P49852), S. cerevisiae (SwissProt [sp] P39676), S. enterica serovar Typhimurium (sp P26353), and E. coli (sp P24232) from BLASTP results were aligned by using the CLUSTAL W algorithm (www.ebi.ac.uk). Alignments for the last four species are truncated. The amino acid residues that are identical in all sequences are shaded in black. (b) Alignment of intergenic nucleotide sequences from C. jejuni NCTC 11168 and C. coli UA585. CAT and ATG code for methionine for the start of the Cj1585c putative oxidoreductase and of the Cj1586 putative bacterial hemoglobin Cgb, respectively. Identical nucleotides in C. coli UA585 and C. jejuni NCTC 11168 are shown by an asterisk.
Despite the similarity between the sequences of Cgb and the heme domains of flavohemoglobins, there are some striking differences. In flavohemoglobins, the interaction between the heme- and flavin-binding domains occurs mainly through helices H and CE. A salt bridge is formed between the domains through Lys-84 that is conserved in Hmp and other globins (13) but not in Cgb. This suggests that Cgb may not interact with the same kind of reductase. The dimerization domain of Vgb is also not conserved in Cgb, which is consistent with the fact that Cgb elutes as a monomer after gel filtration (G. Wu and R. K. Poole, unpublished data).
PCR was used to clone cgb from C. coli and C. jejuni. Sequence analysis of the C. coli cgb gene (GenBank no. AY321511) showed that the corresponding C. coli protein was closely related to Cgb (87% amino acid identity) (Fig. 1a).
Generation of Cgb-deficient mutants of C. jejuni and C. coli.
The gene encoding cgb was disrupted in both C. jejuni and C. coli by insertion of a Kanr cassette, at a position 165 bp downstream from the ATG start codon, by using the suicide plasmid pKE18 (see Materials and Methods). Confirmation that the genomic copy of cgb was disrupted was obtained by Southern blot analysis (data not shown) and the corresponding mutants designated CJCGB01 (C. jejuni) and CCCGB01 (C. coli). In C. jejuni, mutants that had the Kanr cassette in opposite orientations behaved identically in all tests (data not shown), suggesting that the phenotype caused by the cgb mutation was not due to polar effects generated by the antibiotic resistance marker. Furthermore, the insertion of the Kanr cassette did not effect expression the genes immediately proximal to cgb (Cj1585c and Cj1587c) as determined by reverse transcriptase PCR (data not shown).
Cgb confers resistance to nitrosative stress but does not provide protection against superoxide or peroxides.
The role of the C. jejuni Cgb was investigated by comparison of nitrosative stress resistance in the wild-type and cgb mutant strains. Diffusion assays on solid medium revealed that the cgb mutant of C. jejuni was hypersensitive to GSNO (a nitrosating agent) with a significantly greater zone of killing compared to the wild type (P = 0.002; two-tailed t test assuming equal variance). However, sensitivities to methyl viologen, hydrogen peroxide, and cumene hydroperoxide were equivalent to the wild-type (nonsignificant t test; Table 2).
TABLE 2.
Resistance and sensitivity of C. jejuni strains to agents of oxidative and nitrosative stress
| Stress inducer | Initial concn on disk | Mean diam of disk inhibition zone (mm)a ± SEM (P) for C. jejuni:
|
|||
|---|---|---|---|---|---|
| NCTC 11168 | CJCGB01 (cgb mutant) | CJA01 (ahpC mutant) | CJAC01 (ahpC cgb mutant) | ||
| Methyl viologen | 3% (wt/vol) | 37.2 ± 0.8 | 36.4 ± 0.6 (0.40) | ND | ND |
| Cumene hydroperoxide | 3% (wt/vol) | 43.9 ± 2.1 | 42.6 ± 0.7 (0.56) | 59.0 ± 0.6 (0.001*) | 62.3 ± 1.5 (0.002*) |
| Hydrogen peroxide | 3% (wt/vol) | 38.7 ± 2.5 | 37.2 ± 0.5 (0.58) | 45.0 ± 1.0 (0.16) | 40.0 ± 0.6 (0.16) |
| S-Nitrosoglutathione | 100 mM | 9.2 ± 0.9 | 18.4 ± 2.4 (0.002*) | 10.0 ± 0.6 (0.29) | ND |
Values are expressed as mean diameter of the zone of killing. Data shown are from nine replicates. Significant P values, where P < 0.05; are indicated by an asterisk. P values are a result of t test, assuming an equal variance (two tailed) comparing the wild type with each mutant. ND, not determined.
Hmp has been reported to function as an alkyl hydroperoxide reductase (4). To eliminate the possibility that the major iron-regulated alkyl hydroperoxide reductase (AhpC) present in C. jejuni might mask any contribution of Cgb to peroxide resistance, single ahpC and double ahpC cgb mutants were constructed. The AhpC-deficient mutant (CJA01) showed increased sensitivity to cumene hydroperoxides (Table 2) as expected (1), but no additional sensitivity to GSNO. Furthermore, neither the single cgb mutant nor the ahpC cgb double mutant (CJAC01) exhibited additional sensitivity to cumene hydroperoxide.
Contribution of Cgb to nitrosative stress resistance in C. jejuni and C. coli.
The effect of the cgb mutation on nitrosative stress resistance was probed further by exposing cells to a variety of nitrosating agents (GSNO or SNP) and a nitric oxide releaser (spermine NONOate) and assessing viability. The cgb mutant was markedly less resistant to all of these agents compared to the wild type (Fig. 2). The difference in viability varied markedly, however, depending on the nature of the compound and its concentration. For example, when GSNO was used, both the mutant and parental strains showed very little reduction in viability at up to 0.4 mM GSNO. In contrast, exposure to 0.8 mM GSNO reduced the viability of the mutant ∼102-fold, while counts of the parental strain were not affected at these concentrations. In the presence of spermine NONOate, the wild type showed no decrease in viability at concentrations of up to 0.2 mM, whereas the cgb mutant began to lose viability at NONOate concentrations lower than 0.05 mM. At 0.2 mM, the difference in viability between the two strains was >104-fold. The C. coli Cgb-deficient mutant also displayed a similar sensitivity to GSNO and SNP (data not shown), suggesting that Cgb is a crucial defense against nitrosative stress in at least two Campylobacter species. To confirm that the sensitivity of the cgb mutant to GSNO and spermine NONOate reflected a difference in nitrosative stress resistance and not a changed tolerance to spermine or glutathione, the sensitivity of the wild-type and mutant to these agents was assessed by using the disk diffusion assay. When 10 μl of 100 mM glutathione or spermine was used in this assay, no zones of inhibition were produced for either the wild type or the mutant (data not shown).
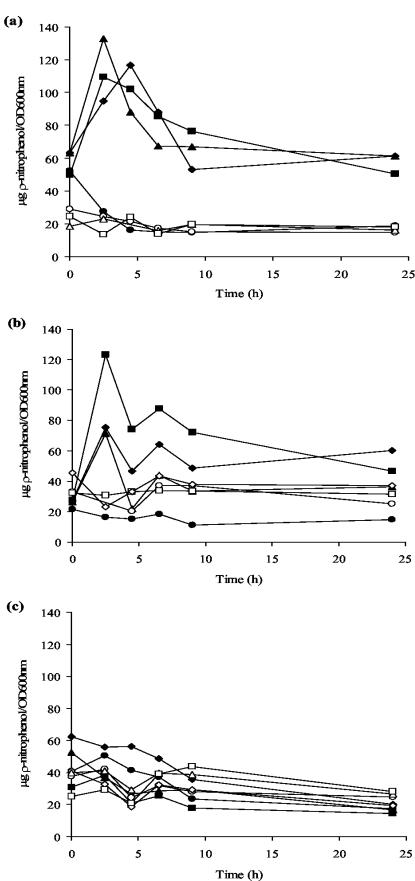
FIG. 2.
Effects of nitrosative stress on the viability of C. jejuni. Growing cultures of C. jejuni NCTC 11168 (▴) and CJCGB01 (○) were challenged with the indicated concentrations of GSNO (a), SNP (b), and spermine NONOate (c) for 1 h and then plated on MH agar. The results are a mean of two independent experiments. Bars indicate one standard error.
The effect of a deficiency in Cgb on growth in liquid medium in the presence of GSNO was also assessed. In the absence of GSNO and in the presence of 0.05 mM GSNO, the cgb mutant and the wild type grew similarly, whereas growth of the mutant, but not the wild type, was almost completely inhibited in the presence of a 0.2 mM concentration of this agent (data not shown).
Construction of cgb-astA and sodB-astA reporter gene fusions in C. coli.
A novel Campylobacter reporter gene system, based on arylsulfatase, has recently been described (25). This has the advantage that expression of astA can be detected when the reporter gene is present in single copy and that expression can be readily monitored on agar plates by using chromogenic substrates (56), allowing regulatory mutants to be detected. In strain NCTC 11168, astA is present only as a pseudogene (Cj0866) that is not expressed (37) and, consequently, this strain provides a suitable background for this system. Nevertheless, when a copy of the astA was integrated into cgb, no expression of the reporter gene could be detected (data not shown). Consequently, cgb-astA gene fusions were next introduced into C. coli UA585 to determine whether astA expression could be assessed in this background and to provide information on this closely related species. Integrable vectors capable of generating transcriptional fusions via single crossover recombination events in C. coli have been described previously (8). In the present study, these vectors were modified to include an arylsulfatase reporter gene (see Materials and Methods). The integration of the reporter gene fusions into the chromosome was confirmed by Southern hybridization analysis with three different labeled probes (data not shown). Strains containing sodB (control) and cgb fusions to astA were designated CCSF1 and CCCF1, respectively.
Induction of cgb expression by nitrosative stress.
Strains containing the reporter gene fusion were grown for 16 h, after which GSNO, SNP, or methyl viologen was added. The parental strain of C. coli does not possess a copy of astA and, when it was assessed for expression, no AstA activity was detected (data not shown). In contrast, a basal level of astA expression was detected when CCCF1 and CCSF1 were grown in MH broth with no additions. This level remained constant throughout the growth experiment at ca. 20 to 30 U (Fig. 3a).
FIG. 3.
Cgb expression, as monitored from transcriptional astA fusions, is induced by nitrosative stress in C. coli. Cells of CCSF1 sodB-astA (open symbols) and CCCF1 cgb-astA (solid symbols) were grown with no additions (○, •) or in the presence of GSNO at 0.05 mM (♦), 0.1 mM (▪, □), or 0.25 mM (▴, ▵) (a); SNP at 0.005 mM (♦, ⋄), 0.01 mM (▪, □), or 0.05 mM (▴) (b); and methyl viologen at 1 μM (♦, ⋄), 5 μM (▴, ▵), or 10 μM (▪, □) (c). The agents were added at time zero, and arylsulfatase activity was assessed and is expressed as micrograms of p-nitrophenol released/hour/OD600. The data shown are representative of the results obtained from three independent experiments.
After the addition of 0.1 and 0.25 mM GSNO, expression from the cgb-astA fusion showed a dramatic increase, reaching a peak of expression after 2.5 h. Similarly, the addition of 0.05 mM GSNO induced expression, but in this case the peak was after 4.5 h. At the peak, induced levels of expression from the cgb-astA fusions were 4-, 5-, and 7-fold higher than the expression levels seen in MH broth alone (Fig. 3a). In contrast, no induction of arylsulfatase activity was detected when the same experiment was repeated with the sodB-astA fusion strain CCSF1. Increased cgb-astA activity was also seen when SNP was added to the media. In this case, the peak in expression from the cgb-astA fusion occurred at 2.5 h when 0.05, 0.01, or 0.005 mM SNP was used, with levels of induction at this point being 4-, 8-, and 5-fold higher, respectively, than that measured from the cgb-astA fusion in the absence of SNP (Fig. 3b). No induction was seen when either CCCF1 or CCSF1 were exposed to methyl viologen (Fig. 3c), suggesting that Cgb expression responds specifically to nitrosative stress.
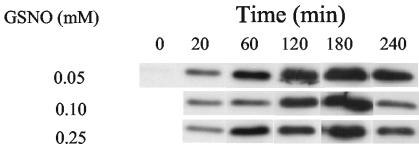
Purification of Cgb allowed preparation of a polyclonal antibody for immunoblotting of C. jejuni extracts. In cultures not challenged with GSNO, Cgb was undetectable (Fig. 4). However, even after the addition of only 0.05 mM GSNO, a clear band, corresponding to a protein of molecular mass 16 kDa, was detectable, which increased in intensity over 3 h. A similar pattern of induction was seen after the addition of 0.1 or 0.25 mM GSNO (Fig. 4). No Cgb was detected when the culture was incubated for the same time in the absence of GSNO. The possibility that the expression of Cgb was regulated by oxygen was checked by growing C. jejuni in a microaerobic atmosphere, but at different volumes (5, 10, 25, 35, and 45 ml) of medium in 50-ml conical flasks. Such variation of culture volume has been shown previously to be reflected in the oxygen transfer rate (KLa) from the gas to liquid phase (21). Since Cgb was present at a very low level under noninduced conditions, higher amounts of soluble protein (45 μg) were used for Western blotting. No difference was seen under different aeration conditions (results not shown).
FIG. 4.
GSNO induces the expression of Cgb in C. jejuni. Different concentrations of GSNO were added to growing cultures of C. jejuni. The expression of Cgb was detected by using the anti-Cgb antibody. Each lane of the 15% SDS gels was loaded with 10 μg of protein extract.
Cgb protects the respiration of C. jejuni from inhibition by nitric oxide.
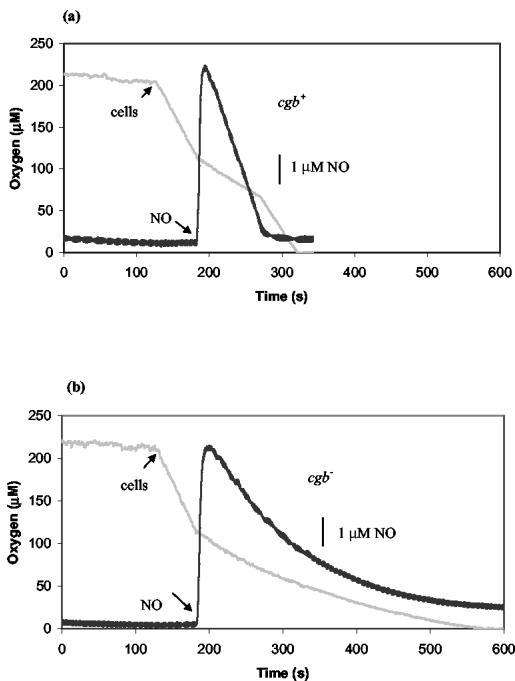
Respiratory chains are well-known targets of nitric oxide. Cells were incubated in a dual-electrode apparatus (see Materials and Methods), and the effects of adding NO on respiration were assessed. Using cells grown under noninduced conditions, no difference was seen between the wild-type and the cgb mutant in terms of the degree and the period of respiratory inhibition at various NO concentrations. The half-lives of NO signals were identical in both the mutant and the wild-type (data not shown). To induce Cgb expression, 0.05 mM GSNO, which had no effect on the growth of either the cgb mutant or the wild type and yet significantly induced Cgb expression, was used. Both the mutant and the wild type were grown to mid-exponential phase (5 h) before 0.05 mM GSNO was added to the cultures, followed by incubation for a further hour. The effect of NO on cell respiration was then assessed. Figure 5 shows that the addition of 5 μM NO to a respiring suspension of wild-type cells (oxygen tension, ∼110 μM; respiration rate, 1.6 μM s−1) immediately inhibited respiration by ca. 60 to 70%. However, inhibition was transient and respiration resumed at the initial rate after about 90 s. The period of respiratory inhibition coincided precisely with the time during which NO was detectable in the vessel (Fig. 5a). In contrast, addition of NO to Cgb-deficient cells caused a continuous decline in the rate of respiration until all O2 was exhausted; NO was detectable throughout the experiment (Fig. 5b). The half-lives of NO were typically 58 and 131 s for the wild type and the cgb mutant, respectively. In control experiments without NO addition, the respiration of both wild-type and cgb cells was linear down to very low O2 tensions. When lower NO concentrations were used, cell respiration of the mutant resumed at low O2 tension (not shown). These patterns of O2 and NO consumption were reproducible in three to six experiments done on cells from each of two independent growths.
FIG. 5.
Inhibition of respiration by NO. Sodium formate (2 mM) was incubated with MOPS buffer at 37°C until the traces were stable. Cells (160 μg of cell protein) were added to initiate respiration of C. jejuni NCTC 11168 (a) and CJCGB01 (b). NO (5 μΜ), indicated by the black line, was added when the oxygen tension (gray line) was ∼110 μM. The experiments were repeated twice, and the assay was repeated three to six times, yielding essentially the same results.
An alternative method was also used to assay the consumption of NO by campylobacters. Sodium formate (2 mM) was first incubated with MOPS buffer before NO (5 μM) was added. When NO signals reached the maximum, cells were added to consume NO. The results were essentially the same as seen in Fig. 6 except that, at the higher oxygen concentration, NO signals disappeared faster in both strains. The half-life of NO was 50 s in the wild type and 67 s in the mutant. In a control without cells, the half-life was 117 s.
FIG. 6.
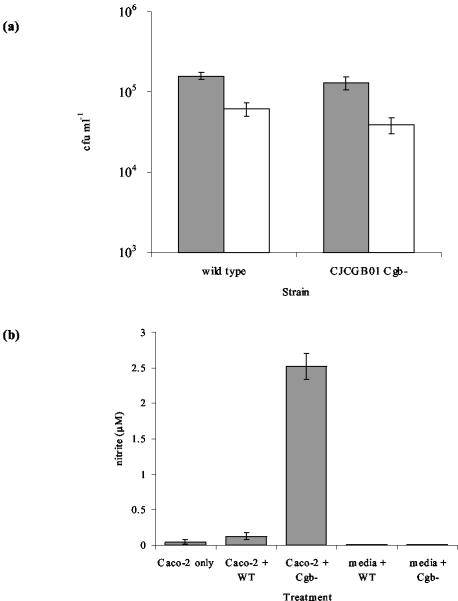
(a) Adherence to and invasion of Caco-2 cells by C. jejuni NCTC 11168 and CJCGB01. The numbers of adhered and invaded cells (gray shaded bars) and invasive cells only (open bar) were determined by viable counts after 3 h. The results are a mean ± the standard error of three replicates. (b) Nitrite production by Caco-2 cells in response to C. jejuni infection. Nitrite production was measured after 3 h of infection with C. jejuni NCTC 11168 and CJCGB01. Values are means ± the standard error of 16 replicates for Caco-2 cells and 4 replicates for media.
Adherence and invasion of Caco-2 cells and nitrite assays.
Enteroinvasive bacteria induce NO production in human colon epithelial cells, and consequently NO and/or its redox products form part of the defense of intestinal cells against microbial infection (52). We therefore assessed the ability of the Cgb-deficient mutant to invade Caco-2 cells and to induce NO production, as measured by monitoring its stable end product nitrite (52). During the 3-h adherence and invasion assay, the wild type and the cgb mutant showed similar abilities to adhere to (P = 0.36) and invade (P = 0.21) epithelial cells (Fig. 6). NO production was not detected in the cells of C. jejuni exposed to tissue culture medium alone. When Caco-2 cells were exposed to wild-type C. jejuni, a modest increase in NO production was detected. However, when the eukaryotic cells were exposed to the cgb mutant, the level of NO production was 50-fold higher than with uninfected Caco-2 cells and 20-fold greater than with Caco-2 cells infected with wild-type bacteria. This suggests that C. jejuni cells that are deficient in Cgb are unable to reduce the levels of NO produced upon infection of Caco-2 cells, whereas the wild-type bacteria can do so.
Intracellular survival was also assessed 24 h after the infection of Caco-2 cells. Although the number of intracellular bacteria had decreased compared to survival at 3 h, numbers of the wild type (1,100 ± 800) and cgb mutant (2,200 ± 1,300) were not significantly different at this time point.
DISCUSSION
The two-domain flavohemoglobins exhibit NO-consuming activity (20, 23), and mutants deficient in these globins exhibit increased sensitivity to NO and nitrosative stress (6, 20, 31, 47). The functions of the single-domain globins and truncated globins are, however, less clear. The archetypal Vitreoscilla single-domain globin Vgb has been implicated in oxygen storage, delivery, or reduction based upon (i) its induced expression at low-oxygen tensions (9), (ii) the ability to enhance microaerobic growth when heterologously expressed in E. coli (2), and (iii) its binding to cytochrome bo′ (36). More recently, however, a recombinant form of Vgb carrying a heterologous reductase domain has been shown to metabolize NO and confer nitrosative stress protection to E. coli (27). In fact, several single-domain globins from a number of species, including C. jejuni, have recently been reported to alleviate nitrosative stress when heterologously expressed in E. coli (16). In addition, although the HbO truncated globin from Mycobacterium tuberculosis may be important in oxygen sequestration (39), the truncated HbN globins from M. tuberculosis and Mycobacterium bovis have also been shown to scavenge and detoxify NO (34, 38).
In order to investigate the role of the single-domain globin Cgb in the physiology of campylobacters, cgb-deficient mutants of C. jejuni and C. coli were generated. Cgb-deficient cells had levels of resistance comparable to the levels of resistance to methyl viologen, hydrogen peroxide, and organic peroxides, as did wild-type cells. Thus, it seems unlikely that Cgb plays a major role in the resistance of campylobacters to oxidative stresses. In addition, Cgb does not appear to have a physiological role as an alkyl hydroperoxide reductase in C. jejuni at least, as has been suggested for Hmp (4), since the cgb mutation did not confer sensitivity to organic peroxides even in a strain in which the iron-regulated alkyl hydroperoxide reductase, AhpC, was not present.
An important finding of the present study is that cgb mutants showed increased sensitivity to the NO-releasing compound spermine NONOate, as well as to the nitrosating agents GSNO and SNP. All of these compounds are widely used to generate nitrosative stress. However, with SNP, NO release occurs only after nitrosation, i.e., the transfer of the NO+ group to a nucleophilic receptor (43) and, although GSNO may undergo homolysis to generate the oxidized thiol RS. and NO, it can also act as a transnitrosating agent. In contrast, spermine NONOate directly liberates NO with a specified half-life (230 min; Calbiochem data) and can be used to study the effect of NO per se. Since the cgb mutant was hypersensitive to this agent, it might be concluded that Cgb protects campylobacters directly from NO but, in the presence of O2, the NO liberated might act as a nitrosating agent. Thus, care needs to be exercised in interpreting the results with different nitrosative agents. Since Salmonella strains defective in AhpC are hypersusceptible to reactive nitrogen intermediates (5), C. jejuni CJA01 (AhpC−) was also assessed for sensitivity to GSNO. However, in a disk diffusion assay, the ahpC mutant showed no increased sensitivity to this compound.
It is not possible to complement mutations in these strains of Campylobacter by plasmid-borne copies of cgb, since plasmids containing DNA homologous to the genome are unstable in these host backgrounds. Nevertheless, the nitrosative stress-sensitive phenotype is due solely to the inability of cells to express cgb alone for several reasons. First, the Kanr cassette used contains its own promoter but lacks a transcriptional terminator, and when the resistance cassette is inserted with the same transcriptional polarity as the mutated gene, mutations have been shown to be nonpolar on downstream genes (26). In addition, cgb mutants had the same phenotype irrespective of the orientation of the Kanr cassette. Second, the two neighboring open reading frames, Cj1585c and Cj1587c, which encode a putative oxidoreductase and putative ABC transporter, respectively, are expressed in the opposite direction to cgb and on the complementary strand. Furthermore, the insertion of the Kanr cassette did not affect expression of the genes immediately proximal to cgb, as determined by reverse transcriptase PCR, and polar effects can, therefore, be ruled out.
In other bacteria, the expression of two-domain globins is modulated by a number of environmental conditions. In E. coli, Hmp expression is induced by both NO (44) and oxidative stress (30), whereas in B. subtilis Hmp expression is induced by anaerobiosis and nitrite (28). Less information is available concerning the regulation of the single-domain globins, such as Cgb or the truncated globins, although Vgb is expressed in response to oxygen limitation (9). Recent work did not demonstrate the induction of Vgb by nitrosative stress (17). The present study is the first, to our knowledge, to demonstrate upregulation of a single-domain hemoglobin (i.e., non-flavo-hemoglobin) in response to nitrosative stress. For example, the presence of GSNO and SNP stimulated expression of the cgb-astA reporter gene fusion by 4- to 8-fold, whereas expression from a control sodB-astA fusion was not affected by the presence of either of these compounds. The astA gene fusion vectors could be used only in C. coli. Nevertheless, the expression of Cgb in C. jejuni was clearly induced under similar conditions when Western blotting was used to assess its expression. Given the fact that the intergenic region between cgb and Cj1585c is highly conserved, particularly in the region proximal to Cj1585c (Fig. 1b), this finding is not unexpected. Since Cgb expression is not regulated by aeration, we suggest that Cgb is specifically responsible for dealing with nitrosative stress.
NO reacts with many cellular targets, such as FeS-containing proteins, and inhibits respiratory chains. Using uninduced cells, no difference was seen between the wild type and the cgb mutant in terms of the severity and the degree of respiratory inhibition at various NO concentrations. The half-lives of NO signals were identical in both the mutant and the wild type. These results are not surprising given that Cgb was not expressed significantly without induction. However, when the expression of Cgb was induced, it significantly contributed to the protection of respiration and the consumption of nitric oxide. The fact that Cgb-deficient cells consumed NO at a faster rate than measured for its decay in the buffer control suggests that there may be additional mechanisms of NO detoxification in campylobacters. However, this activity is not due to a flavorubredoxin (19) since genes encoding this function are not present in the genome sequence (37).
In other bacterial pathogens, the ability of the cell to detoxify NO and its congeners, through the action of globins, has been correlated with virulence. For example, Salmonella hmp mutants survive phagocytosis by peripheral monocyte-derived macrophages relatively poorly compared to wild-type cells (48). Consequently, Cgb is likely to play some role during colonization and infection. Here, we have shown that, although cells deficient in Cgb are able to invade and survive intracellularly in Caco-2 cells with the same efficiency as the wild-type cells, the levels of NO produced by epithelial cells are much higher in cells infected with the cgb mutant. Since NO production was not detected in cells of C. jejuni incubated in tissue culture medium alone, this NO must be derived from the eukaryotic cells, suggesting that Cgb is required for the efficient removal of NO. Although this difference in NO detoxification was not manifested in differences in invasion potential, this may be due to the fact that other resistance mechanisms, such as the detoxification of superoxide by superoxide dismutase (41), are also involved in intracellular survival.
In conclusion, the data presented here demonstrate for the first time that (i) the single-domain globin Cgb, unlike Vgb, plays a key role in detoxification of NO and related compounds and that (ii) consumption of and protection from NO is inducible and that this is a consequence of the specific induction of Cgb expression by nitrosative stress. Future studies concerning Cgb and its regulators are likely to confirm the importance of this globin in Campylobacter physiology.
Acknowledgments
This study was supported by BBSRC grants BFP11346 and D18368 to R.K.P., S.F.P., and G.W. and by a Wellcome Trust Vacation Scholarship to N.J.G.
We thank Simon Smith (University of Sheffield Antibody Resource Centre) for preparation of antibody, Martin Hughes (King's College London) for helpful advice on NO chemistry and provision of GSNO, and H. Corker for critical reading of the manuscript.
REFERENCES
- 1.Baillon, M. L., A. H. van Vliet, J. M. Ketley, C. Constantinidou, and C. W. Penn. 1999. An iron-regulated alkyl hydroperoxide reductase (AhpC) confers aerotolerance and oxidative stress resistance to the microaerophilic pathogen Campylobacter jejuni. J. Bacteriol. 181:4798-4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bollinger C. J., J. E. Bailey, and P. T. Kallio. 2001. Novel hemoglobins to enhance microaerobic growth and substrate utilization in Escherichia coli. Biotechnol. Prog. 17:798-808. [DOI] [PubMed] [Google Scholar]
- 3.Bonamore, A., A. Farina, M. Gattoni, E. Schininà, A. Bellelli, and A. Boffi. 2003. Interaction with membrane lipids and heme ligand binding properties of Escherichia coli flavohemoglobin. Biochemistry 20:5792-5801. [DOI] [PubMed] [Google Scholar]
- 4.Bonamore, A., P. Gentili, A. Ilari, M. E. Schinina, and A. Boffi. 2003. Escherichia coli flavohemoglobin is an efficient alkylhydroperoxide reductase. J. Biol. Chem. 278:22272-22277. [DOI] [PubMed] [Google Scholar]
- 5.Chen, L., Q. W. Xie, and C. Nathan. 1998. Alkyl hydroperoxide reductase subunit C (AhpC) protects bacterial and human cells against reactive nitrogen intermediates. Mol. Cell 1:795-805. [DOI] [PubMed] [Google Scholar]
- 6.Crawford, M. J., and D. E. Goldberg, D. E. 1998. Role for the Salmonella flavohemoglobin in protection from nitric oxide. J. Biol. Chem. 273:12543-12547. [DOI] [PubMed] [Google Scholar]
- 7.Delisle, G. J., and F. H. Milazzo. 1972. Characterization of arylsulfatase isoenzymes from Pseudomonas aeruginosa. Can. J. Microbiol. 18:561-568. [DOI] [PubMed] [Google Scholar]
- 8.Dickinson, J. H., K. A. Grant, and S. F. Park. 1995. Targeted and random mutagenesis of the Campylobacter coli chromosome with integrational plasmid vectors. Curr. Microbiol. 31:92-96. [DOI] [PubMed] [Google Scholar]
- 9.Dikshit, K. L., D. Spaulding, A. Braun, and D. A. Webster. 1989. Oxygen inhibition of globin gene transcription and bacterial haemoglobin synthesis in Vitreoscilla. J. Gen. Microbiol. 135:2601-2609. [DOI] [PubMed] [Google Scholar]
- 10.Duncan, C., H. Dougall, P. Johnston, S. Green, R. Brogan, C. Leifert, L. Smith, M. Golden, and N. Benjamin. 1995. Chemical generation of nitric oxide in the mouth from the enterosalivary circulation of dietary nitrate. Nat. Med. 1:546-551. [DOI] [PubMed] [Google Scholar]
- 11.Dykhuizen, R. S., R. Frazer, C. Duncan, C. C. Smith, M. Golden, N. Benjamin, and C. Leifert. 1996. Antimicrobial effect of acidified nitrite on gut pathogens: importance of dietary nitrate in host defense. Antimicrob. Agents Chemother. 40:1422-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elvers, K. T., and S. F. Park. 2002. Quorum sensing in Campylobacter jejuni: detection of a luxS encoded signaling molecule. Microbiology 148:1475-1481. [DOI] [PubMed] [Google Scholar]
- 13.Ermler, U., R. A. Siddiqui, R. Cramm, and B. Friedrich. 1995. Crystal structure of the flavohemoglobin from Alcaligenes eutrophus at 1.75 Å resolution. EMBO J. 14:6067-6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Everest, P. H., H. Goossens, J. P. Butzler, D. Lloyd, S. Knutton, J. M. Ketley, and P. H. Williams. 1992. Differentiated Caco-2 cells as a model for enteric invasion by Campylobacter jejuni and C. coli. J. Med. Microbiol. 37:319-325. [DOI] [PubMed] [Google Scholar]
- 15.Forte, P., R. S. Dykhuizen, E. Milne, A. McKenzie, C. C. Smith, and N. Benjamin. 1999. Nitric oxide synthesis in patients with infective gastroenteritis. Gut 45:355-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frey, A. D., J. Farres, C. J. Bollinger, and P. T. Kallio. 2002. Bacterial hemoglobins and flavohaemoglobins for alleviation of nitrosative stress in Escherichia coli. Appl. Environ. Microbiol. 68:4835-4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frey, A. D., T. Koskenkorya, and P. T. Kallio. 2003. Vitreoscilla hemoglobin promoter is not responsive to nitrosative and oxidative stress in Escherichia coli. FEMS Microbiol. Lett. 224:127-132. [DOI] [PubMed] [Google Scholar]
- 18.Friedman, C. R., J. Neimann, H. C. Wegener, and R. V. Tauxe. 2000. Epidemiology of Campylobacter jejuni infections in the United States and other industrialized nations, p. 121-138. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. ASM Press, Washington, D.C.
- 19.Gardner, A. M., R. A. Helmick, and P. R. Gardner. 2002. Flavorubredoxin, an inducible catalyst for nitric oxide reduction and detoxification in Escherichia coli. J. Biol. Chem. 277:8172-8177. [DOI] [PubMed] [Google Scholar]
- 20.Gardner, P. R., A. M. Gardner, L. A. Martin, and A. L. Salzman. 1998. Nitric oxide dioxygenase: an enzymic function for flavohemoglobin. Proc. Natl. Acad. Sci. USA 95:10378-10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gil, A., R. G. Kroll, and R. K. Poole. 1992. The cytochrome composition of the meat spoilage bacterium Brochothrix thermosphacta: identification of cytochrome a3- and d-type terminal oxidases under various conditions. Arch. Microbiol. 158:226-233. [DOI] [PubMed] [Google Scholar]
- 22.Hart, T. W. 1985. Some observations concerning the S-nitroso and S-phenylsulphonyl derivatives of l-cysteine and glutathione. Tetra. Lett. 26:2013-2016. [Google Scholar]
- 23.Hausladen, A., A. J. Gow, and J. S. Stamler. 1998. Nitrosative stress: metabolic pathway involving the flavohemoglobin. Proc. Natl. Acad. Sci. USA 95:14100-14105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henderson, M. J., and F. H. Milazzo. 1979. Arylsulfatase in Salmonella typhimurium: detection and influence of carbon source and tyramine on its synthesis. J. Bacteriol. 139:80-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hendrixson, D. R., and V. J. DiRita. 2003. Transcription of σ54-dependent but not σ28-dependent flagellar genes in Campylobacter jejuni is associated with formation of the flagellar secretory apparatus. Mol. Microbiol. 50:687-702. [DOI] [PubMed] [Google Scholar]
- 26.Hickey, T. E., A. L. McVeigh, D. A. Scott, R. E. Michielutti, A. Bixby, S. A. Carroll, A. L. Bourgeois, and P. Guerry. 2000. Campylobacter jejuni cytolethal distending toxin mediates release of interleukin-8 from intestinal epithelial cells. Infect. Immun. 68:6535-6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaur, R., R. Pathania, V. Sharma, S. C. Mande, and K. L. Dikshit. 2002. Chimeric Vitreoscilla hemoglobin (VHb) carrying a flavoreductase domain relieves nitrosative stress in Escherichia coli: new insight into the functional role of VHb. Appl. Environ. Microbiol. 68:152-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.LaCelle, M., M. Kumano, K. Kurita, K. Yamane, P. Zuber, and M. M. Nakano. 1996. Oxygen-controlled regulation of the flavohemoglobin gene in Bacillus subtilis. J. Bacteriol. 178:3803-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, H., C. Duncan, J. Townend, K. Killham, L. M. Smith, P. Johnston, R. Dykhuizen, D. Kelly, M. Golden, N. Benjamin, and C. Leifert. 1997. Nitrate-reducing bacteria on rat tongues. Appl. Environ. Microbiol. 63:924-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Membrillo-Hernández, J., S. O. Kim, G. M. Cook, and R. K. Poole. 1997. Paraquat regulation of hmp (flavohemoglobin) gene expression in Escherichia coli K-12 is SoxRS independent but modulated by sigma S. J. Bacteriol. 179:3164-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Membrillo-Hernández, J., M. D. Coopamah, M. F. Anjum, T. M. Stevanin, A. Kelly, M. N. Hughes, and R. K. Poole. 1999. The flavohemoglobin of Escherichia coli confers resistance to a nitrosating agent, a “nitric oxide releaser,” and paraquat and is essential for transcriptional responses to oxidative stress. J. Biol. Chem. 274:748-754. [DOI] [PubMed] [Google Scholar]
- 32.Nachamkin, I., B. M. Allos, and T. Ho. 1998. Campylobacter species and Guillain-Barré syndrome. Clin. Microbiol. Rev. 11:555-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newell, D. G., and A. Pearson. 1984. The invasion of epithelial cell lines and the intestinal epithelium of infant mice by Campylobacter jejuni/coli. J. Diarrhoeal Dis. 2:19-26. [PubMed] [Google Scholar]
- 34.Ouellet, H., Y. Ouellet, C. Richard, M. Labarre, B. Wittenberg, J. Wittenberg, and M. Guertin. 2002. Truncated hemoglobin HbN protects Mycobacterium bovis from nitric oxide. Proc. Natl. Acad. Sci. USA 99:5902-5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park, J. K., and P. Kostka. 1997. Fluorometric detection of biological S-nitrosothiols. Anal. Biochem. 249:61-66. [DOI] [PubMed] [Google Scholar]
- 36.Park, K. W., K. J. Kim, A. J. Howard, B. C. Stark, and D. A. Webster. 2002. Vitreoscilla hemoglobin binds to subunit I of cytochrome bo ubiquinol oxidases. J. Biol. Chem. 277:33334-33337. [DOI] [PubMed] [Google Scholar]
- 37.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M.-A. Rajandream, K. M. Rutherford, A. H. M. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 38.Pathania, R., N. K. Navani, A. M. Gardner, P. R. Gardner, and K. L. Dikshit. 2002. Nitric oxide scavenging and detoxification by the Mycobacterium tuberculosis haemoglobin, HbN, in Escherichia coli. Mol. Microbiol. 45:1303-1314. [DOI] [PubMed] [Google Scholar]
- 39.Pathania, R., N. K. Navani, G. Rajamohan, and K. L. Dikshit. 2002. Mycobacterium tuberculosis hemoglobin HbO associates with membranes and stimulates cellular respiration of recombinant Escherichia coli. J. Biol. Chem. 277:15293-15302. [DOI] [PubMed] [Google Scholar]
- 40.Pesce, A., M. Couture, S. Dewilde, M. Guertin, K. Yamauchi, P. Ascenzi, L. Moens, and M. Bolognesi. 2000. A novel two-over-two alpha-helical sandwich fold is characteristic of the truncated hemoglobin family. EMBO J. 19:2424-2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pesci, E. C., D. L. Cottle, and C. L. Pickett. 1994. Genetic, enzymatic, and pathogenic studies of the iron superoxide dismutase of Campylobacter jejuni. Infect. Immun. 62:2687-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pitcher, D. G., N. A. Saunders, and R. J. Owen. 1989. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 8:51-156. [Google Scholar]
- 43.Poole, R. K., and M. N. Hughes. 2000. New functions for the ancient globin family: bacterial responses to nitric oxide and nitrosative stress. Mol. Microbiol. 36:775-783. [DOI] [PubMed] [Google Scholar]
- 44.Poole, R. K., M. F. Anjum, J. Membrillo-Hernandez, S. O. Kim, M. N. Hughes, and V. Stewart. 1996. Nitric oxide, nitrite, and Fnr regulation of hmp (flavohemoglobin) gene expression in Escherichia coli K-12. J. Bacteriol. 178:5487-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Purdy, D., and S. F. Park. 1994. Cloning, nucleotide sequence, and characterization of a gene encoding superoxide dismutase from Campylobacter jejuni and Campylobacter coli. Microbiology 140:1203-1208. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook, J., E. F. Fritsch, and T. Maniatis (ed.). 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, New York, N.Y.
- 47.Stevanin, T. M., N. Ioannidis., C. E. Mills, S. O. Kim, M. N. Hughes, and R. K. Poole. 2000. Flavohemoglobin Hmp affords inducible protection for Escherichia coli respiration, catalyzed by cytochromes bo′ or bd, from nitric oxide. J. Biol. Chem. 275:35868-35875. [DOI] [PubMed] [Google Scholar]
- 48.Stevanin, T. M., R. K. Poole, E. A. Demoncheaux, and R. C. Read. 2002. Flavohemoglobin Hmp protects Salmonella enterica serovar Typhimurium from nitric oxide-related killing by human macrophages. Infect. Immun. 70:4399-4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wakabayashi, S., H. Matsubara, and D. A. Webster. 1986. Primary sequence of a dimeric bacterial haemoglobin from Vitreoscilla. Nature 322:481-483. [DOI] [PubMed] [Google Scholar]
- 50.Wang, Y., and D. E. Taylor. 1990. Natural transformation in Campylobacter species. J. Bacteriol. 172:949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wittenberg, J. B., M. Bolognesi, B. A. Wittenberg, and M. Guertin. 2002. Truncated hemoglobins: a new family of hemoglobins widely distributed in bacteria, unicellular eukaryotes, and plants. J. Biol. Chem. 277:871-874. [DOI] [PubMed] [Google Scholar]
- 52.Witthoft, T., L. Eckmann, J. M. Kim, and M. F. Kagnoff. 1998. Enteroinvasive bacteria directly activate expression of iNOS and NO production in human colon epithelial cells. Am. J. Physiol. 275:G564-G571. [DOI] [PubMed] [Google Scholar]
- 53.Wren, B. W., J. Henderson, and J. M. Ketley. 1993. A PCR-based strategy for the rapid construction of defined bacterial deletion mutants. BioTechniques 16:994-996. [PubMed] [Google Scholar]
- 54.Wu, G., L. M. Wainwright, and R. K. Poole. 2003. Microbial globins. Adv. Microbial Physiol. 47:257-300. [DOI] [PubMed] [Google Scholar]
- 55.Wu, G., L. M. Wainwright, J. Membrillo-Hernández, and R. K. Poole. Bacterial haemoglobins: old proteins with “new” functions? Roles in respiratory and nitric oxide metabolism. In D. Zannoni (ed.), Respiration in Archaea and Bacteria, in press. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 56.Yao, R., and P. Guerry, P. 1996. Molecular cloning and site-specific mutagenesis of a gene involved in arylsulfatase production in Campylobacter jejuni. J. Bacteriol. 178:3335-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]