Abstract
Understanding the structure–function relationship in proteins is a longstanding goal in molecular and computational biology. The development of structure-based parameters has helped to relate the structure with the function of a protein. Although several structural features have been reported in the literature, no single server can calculate a wide-ranging set of structure-based features from protein three-dimensional structures. In this work, we have developed a web-based tool, PDBparam, for computing more than 50 structure-based features for any given protein structure. These features are classified into four major categories: (i) interresidue interactions, which include short-, medium-, and long-range interactions, contact order, long-range order, total contact distance, contact number, and multiple contact index, (ii) secondary structure propensities such as α-helical propensity, β-sheet propensity, and propensity of amino acids to exist at various positions of α-helix and amino acid compositions in high B-value regions, (iii) physicochemical properties containing ionic interactions, hydrogen bond interactions, hydrophobic interactions, disulfide interactions, aromatic interactions, surrounding hydrophobicity, and buriedness, and (iv) identification of binding site residues in protein–protein, protein–nucleic acid, and protein–ligand complexes. The server can be freely accessed at http://www.iitm.ac.in/bioinfo/pdbparam/. We suggest the use of PDBparam as an effective tool for analyzing protein structures.
Keywords: protein three-dimensional structure, physicochemical properties, binding sites, secondary structure propensity
Introduction
It is widely accepted that the structure of a protein dictates its function.1 Most studies of protein structure and function rely on the analysis of the crystal structure of proteins. This is done by calculating various structure-based parameters, which have been developed to describe the folding, stability, and functions of proteins and their complexes, such as the nature of interactions among the amino acid residues and the surrounding solvent molecules, the preferred amino acid residues in the protein environment, the location of residues in the interior/surface of the protein, and the amino acid clusters.2
These parameters focus on specific aspects of the protein structure and are described in the literature. For instance, Lee and Richards3 developed the concept of solvent accessibility of amino acid residues. Chou and Fasman4 studied the secondary structures of proteins and deduced the propensity of amino acid residues present in α-helices, β-strands, and turns. Thornton’s group developed several algorithms for identifying ion pairs, hydrogen bonds, and catalytic sites in proteins.5–7 Manavalan and Ponnuswamy8 proposed the concept of surrounding hydrophobicity to characterize the hydrophobic behavior of amino acid residues in the protein environment. Plaxco et al.9 analyzed the contacts between amino acid residues and developed the concept of contact order (CO) to relate the folding rates of two-state proteins. Gromiha and Selvaraj10 considered contacts that are close in space but far away in the sequence and proposed long-range order (LRO) as a parameter for understanding protein-folding rates. This concept was refined by developing multiple contact index, ie, residues having multiple contacts in two- and three-state proteins.11
Methods are also available to identify binding site residues in protein complexes based on distances between atoms, energetic contributions, and changes in accessible surface area upon binding.12–14 Many standalone programs and online servers (such as DSSP,15 NACCESS,16 HYDROPRO,17 HYDRONMR,18 GETAREA,19 SCide,20 ContPro,21 CAPTURE,22 HBPLUS,23 CALCOM,24 PSAP,25 and SBPS26) are available to calculate various structural parameters. For instance, DSSP15 provides information on the secondary structure and accessible surface area of each amino acid residue in a protein. CALCOM is used to locate residues in the interior and surface based on the distance between the residues and the calculated center of mass of the given protein or peptide chain.24 Tina et al.27 developed a server, protein interactions calculator, to calculate the center of mass, hydrogen bond interactions, hydrophobic interactions, aromatic–aromatic interactions, aromatic–sulfur interactions, and cation–π interactions. Kozma et al.28 developed a server to obtain the contact map for any given protein. Magyar et al.29 utilized the concept of surrounding hydrophobicity, LRO, stabilization center, and conservation scores to identify the stabilizing residues in protein structures. ExPASy30 is a collection of tools on various bioinformatic aspects including proteomics, genomics, structural bioinformatics, and systems biology. PDBsum31 provides pictorial analyses of several structural features of proteins, DNA, and ligands, as well as the interactions between them.
Although a number of structural parameters have been described in the literature and can be calculated using various servers and standalone programs, no single server exists to calculate a diverse set of parameters and provide the output in a standard format. Hence, we have developed a web server, PDB-param (http://www.iitm.ac.in/bioinfo/pdbparam/), to calculate the following four distinct groups of properties: (i) physicochemical properties, (ii) secondary structure propensities, (iii) interresidue interactions, and (iv) identification of binding site residues in protein–DNA/RNA, protein–ligand, and protein–protein complexes. The server and the properties calculated are explained later.
Materials and Methods
A brief description of the properties under the four categories (physicochemical properties, secondary structure propensities, interresidue interactions, and binding site residues in protein complexes) is provided in this section.
Interresidue interactions
For the past three decades, studies on the mechanism of protein folding and stability have focused on interresidue interactions.32 Interactions between amino acid residues of the protein and with the surrounding solvent molecules play an important role in the formation of stable secondary structures and a unique tertiary structure for the protein. These interactions are usually noncovalent and include hydrogen bonds, ion pairs, van der Waals interactions, and hydrophobic interactions. In fact, parameters such as CO and LRO show a very strong correlation with the folding rate of small proteins.9,10
Short-, medium-, and long-range interactions
For a given residue, the surrounding residues within a sphere of 8 Å radius are analyzed in terms of their sequence position. Residues within a distance of two residues from the central residue are considered to contribute to short-range interactions, those within a window between three and four residues to medium-range interactions and those more than four residues apart to long-range interactions.
Number of contacts (8/14 Å, Cα/Cβ atoms)
The contacts between amino acid residues in the crystal structure are computed with cutoffs of 8 and 14 Å using Cα or Cβ atoms, as reported widely in literature.32
Contact order
This parameter reflects the relative importance of local and nonlocal contacts to the native structure of a protein.9 It is defined as
where N is the total number of contacts, ΔSij is the sequence separation between two contacting residues i and j, and L is the total number of residues in the protein.
Long-range order
LRO is derived from long-range contacts (contacts between two residues that are close in space and far in the sequence) in the protein structure.10 It is defined as
where i and j are the two contacting residues within a distance of 8 Å, and N represents the total number of residues in the protein.
Total contact distance
A new parameter total contact distance was developed by taking the product of CO and LRO. This parameter shows good correlation with the folding rates of proteins.33
Multiple contact index
It considers the distance between amino acid residues in protein structure, residue separation at the sequence level, and the number of residues that have multiple contacts.11 Multiple contact index has been derived separately for two- and three-state proteins.
Two-state proteins:
Three-state proteins:
where nci is the number of contacts for each residue, and rij is the distance between the residues i and j.
Propensities
Propensities indicate the preference of amino acid residues for different secondary structures. The propensities listed in PDBparam are given below.
α-Helical, β-strand, and coil tendencies
The α-helical propensities can be computed by taking into account the frequency of amino acids in these regions.
i varies from 1 to 20, number of amino acid residues. Similar equations have been used to compute strand and coil propensities.
Frequency of occurrence in β-bends
Certain segments in the polypeptide chain help in bringing the distant residues into close proximity during the folding process. For example, β-bends34 allow hydrogen bonds to form between the C = O group of residue i and NH group of residue (i + 3).
Criteria to occur in β-bends:
Distance between Cα(i) to Cα(i + 3) carbon atoms should be less than 7 Å.
The (i + 1)th or (i + 2)th residue is not in an α-helix.
Amino acid compositions in turns
An open turn exists in a protein if the distance between to carbon atoms is <5.7 Å.35 Turns are usually present where a strand of β-sheet reverses itself to form the next antiparallel strand or keep the helices, β-sheets, and random coils in a compact globular form and are thus used to predict protein structure.
Normalized frequency of helix
Helical regions are divided into three zones35: the first three residues represent the N- helix, the last three represent the C-helix, and the residues in the middle represent the M-helix. The amino acid frequency in each helical zone divided by the total frequency (in the entire protein) constitutes normalized frequency.
Propensity to form multiple contact index
The frequency of occurrence of amino acid residues that form multiple contacts (fmc) and in the protein as a whole (ft) is computed.11 The propensity, Pmc can be calculated as follows:
where i represents each of the 20 amino acid residues.
Amino acid composition in high B-value regions
Temperature factors (ie, B-values) provide a measure of the degree of uncertainty in the position of an atom due to thermal motion and/or positional disorder. Analyzing B-values provides insights into protein flexibility and protein dynamics. The B-values at Cα atoms are normalized and residues with B-values greater than Bmean + 0.5 × Bσ are labeled as high B-value residues.36
Physicochemical properties of proteins
Center of mass
The center of mass can be used to define constraints in predicting protein tertiary structures to assess the global shape of the protein partners in protein–protein complexes and to measure their distance.24 It is given by
where xi is the X coordinate of the atom i and mi is the atomic mass. The Y and Z coordinates of the center of mass can be calculated using a corresponding formula.
Radius of gyration
The radius of gyration describes the compactness of the protein. It is calculated as follows:
where mi denotes the mass of each atom, COM denotes the center of mass of protein, and xi represents the atomic coordinate.
Surrounding hydrophobicity
The sum of hydrophobic indices assigned to the residues that appear within a distance of 8 Å from the central residue8 can be used to characterize the hydrophobic behavior of each amino acid residue in the protein environment. It is defined as
where nij is the total number of surrounding residues of type j around the ith residue of the protein, and hj is the hydrophobicity index (kcal/mol) obtained from thermodynamic transfer experiments.37,38
Gain in surrounding hydrophobicity of a residue
For a given amino acid, the increase in surrounding hydrophobicity as the protein transitions from its unfolded state to its native (ie, folded) state represents the enrichment in the hydrophobic property of that residue. To compute the gain in surrounding hydrophobicity39 for each residue in the protein molecule, it is assumed that the fully extended chain conformation is the unfolded reference state.
Surrounding hydrophobicity in the unfolded
The average gain ratio in surrounding hydrophobicity is given by
where H f and H u denote the hydrophobic index of the jth residue in the folded state and unfolded state of the protein, respectively.
Surface hydrophobicity
This is computed from the protein crystal structure by considering the hydrophobic contribution of exposed amino acid residues. Surface hydrophobicity38 is given by
where si is the solvent accessible surface area occupied by the ith residue, ψi is the hydrophobicity value assigned to the residue, and sp is the solvent accessible surface area of protein.
Hydrophobic accessible area
It is calculated as the solvent accessible surface area of the hydrophobic residues on the protein surface.40 We considered Ala, Val, Leu, Ile, Met, Phe, and Pro as the hydrophobic residues to calculate the hydrophobic accessible area.
Accessible surface area for the native protein
The accessible surface area (ASA) for the native protein is calculated as the sum of the accessible surface area of each residue present in the protein, which is obtained from DSSP.15
Buriedness
The buriedness2 of each residue is calculated as the ratio of number of residues in the interior of the protein and the total number of residues in the protein.
Mean area buried on transfer
The mean area buried on transfer41 is given by difference in the accessible area in the unfolded and folded states of the protein.
where A0 and <A> represent the accessible areas in unfolded and folded states of protein, respectively.
Mean fractional area loss
During the process of folding, the nonpolar residues avoid contact with solvent molecules and are buried inside the protein. The area lost when a residue is buried is proportional to its hydrophobic contribution. This is termed as solvent accessible reduction ratio41 or mean fractional area loss, denoted as <RA>:
where A 0 and <A> represent the accessible areas in unfolded and folded states of protein, respectively.
Normalized flexibility parameters (B-values)
This parameter can be computed from the temperature factors extracted from the PDB for the N, Cα, C, and O atoms. Based on the deviation of B-value from the mean, each residue was classified as flexible or rigid. The normalized B-values42 were determined for each residue type, ie, when surrounded by none, one, or two rigid neighbors.
Noncovalent interactions
Several interactions (hydrophobic, hydrogen bond, ionic, aromatic, cation–π, and disulfide bonds) have been described in terms of the amino acid residues involved and the distance between two specific amino acid residues. The details of the amino acid residues in each interaction along with the distances27 are given in Table 1.
Table 1.
Distance criteria for noncovalent interactions and disulfide bonds.
| NAME OF THE INTERACTIONS | INTERACTING RESIDUES | DISTANCE CRITERIA |
|---|---|---|
| Disulfide | Pair of cysteines | 2.2Å |
| Ionic Interactions | (R,K) with (D,E,H) | 6.0 Å |
| Hydrophobic interactions | A,V,L,I,M,F,W,P,Y | 5.0 Å |
| Hydrogen bond interactions | Donor-acceptor distance cut-off (O and N) Donor-acceptor distance cut-off (sulfur) |
3.5Å 4.0Å |
| Aromatic-Aromatic interactions | Pairs of phenyl ring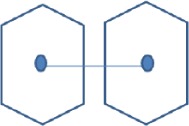
|
4.5 to 7.0 Å |
| Aromatic-sulfur interactions | Sulfur atoms of C, M and thearomatic rings of F,Y,W | 5.3 Å |
| Cation-π interactions | Cationic side chain (Lys or Arg) is near an aromatic side chain (Phe, Tyr, or Trp) | 6.0 Å |
Hydrophobic-free energy
The hydrophobic-free energy43 is expressed as
where Ai(folded) and Ai(unfolded) represent the accessible surface areas of each atom in the folded and unfolded (extended) states of the protein, respectively.
The solvent accessible surface areas of all the atoms in the folded state were computed using the program NAC-CESS.16 The extended state ASA of the atom was obtained from literature. They are in the form of a Gly–X–Gly (where X is the amino acid) sequence in a typical extended conformation. σi (atomic solvation parameters) for the five classes of atoms (namely, carbon, neutral nitrogen and oxygen, charged nitrogen, charged oxygen, and sulfur) are determined by a least-squares fit of above equation. The σi values are C: 12.02, N/O: −5.86, N+: −19.46, O−: −34.98, and S: 35.51 (in units of cal/mol Å2).43
Free energy due to disulfide interactions
The free energy due to disulfide interactions is calculated using the formula:
where Nss is the number of disulfide bonds in the protein.
Hydrogen bond interactions
It is classified into the following three main categories: main chain–main chain, main chain–side chain, and side chain–side chain interactions. These interactions are calculated using HBPLUS,23 a hydrogen bond calculation program.
Identification of binding sites in protein–DNA/RNA and protein–protein complexes
Protein–DNA interactions play a key role in many vital processes, including regulation of gene expression, DNA replication and repair, and packaging. The binding sites for a protein–DNA/RNA complex can be identified using the following distance criteria12: an amino acid residue within a protein is designated as a binding site residue if its side chain or backbone atoms are within a cutoff distance (eg, 3.5 Å) from any atom in DNA/RNA.44–46 The binding sites for protein–protein complexes were also computed using the distance criteria between different chains present in the protein.
Server Description and Implementation
The PDBparam server can calculate more than 50 parameters from the three-dimensional structure of a protein. Each parameter has been treated as a separate module, and the script has been written using perl. The perl-CGI scripts are used to render the HTML web pages. The PDBparam server works with the PDB file as input and provides the computed results in a single output page. The output can be downloaded as a PDF file. The results for all the parameters were cross-checked manually with several structures of proteins and their complexes. Furthermore, the documentation has been provided for all the parameters listed in PDBparam on the website. It is linked with other online tools available in the literature. The utility of the server is described with a few examples.
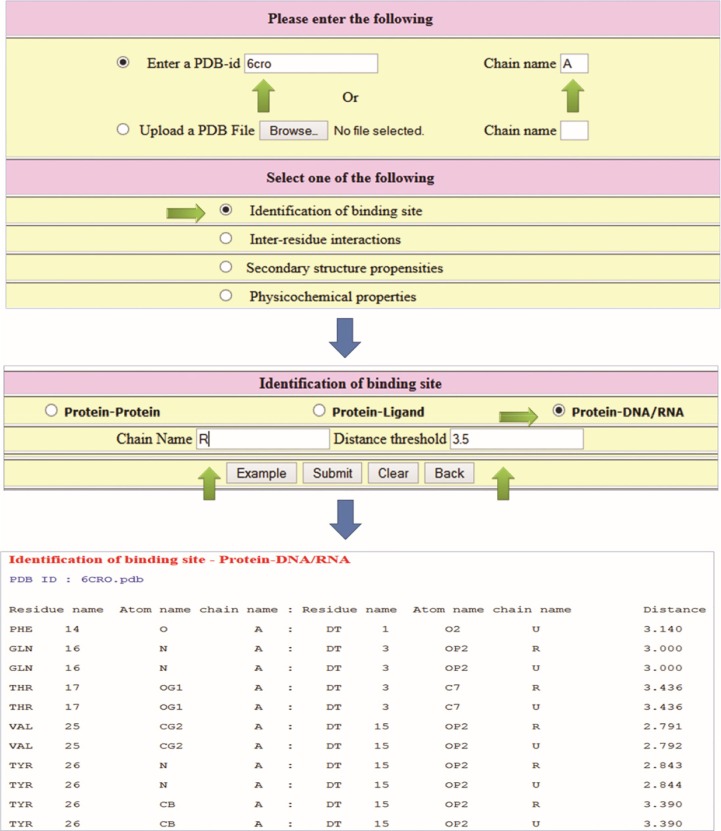
Example 1: Identify the binding site residues in a protein–DNA complex (PDB code: 6CRO) using the distance cutoff of 3.5 Å.
Steps:
Enter the PDB code and chain (optional; case sensitive); eg, PDB code: 6CRO.
Check “identification of binding site” and submit.
In the new page, check protein–DNA/RNA.
Give the distance (default cutoff is 3.5 Å).
Click on submit.
Figure 1 shows the relevant items to be checked, the required information, and the output. The output contains information on the residue name, residue number, atom name, and chain name of both protein and DNA and the distance between the atoms. These residues are identified as binding sites. We have also provided options to display the structure of the complex, highlighting the binding site residues.
Figure 1.
Steps to identify the binding sites in a protein–DNA complex.
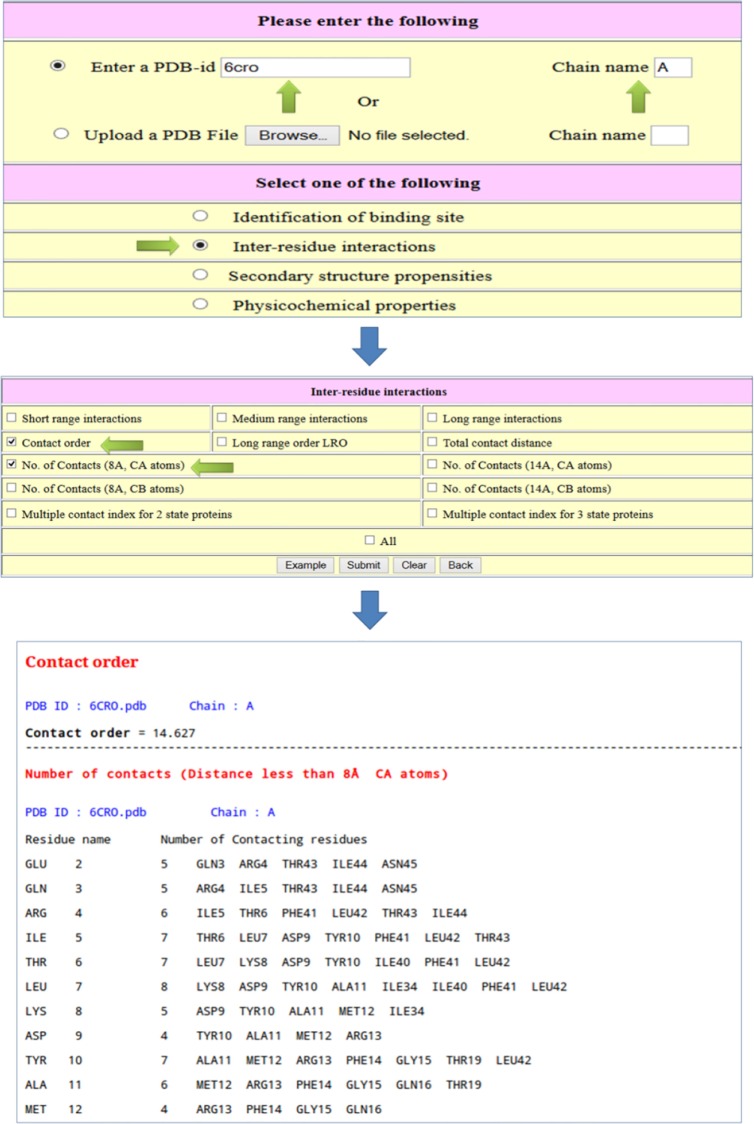
Example 2: Calculate the CO of the protein, 6CRO (A chain), and the number of contacts for all the residues using Cα atoms within the limit of 8 Å.
Steps:
Enter the PDB code and chain (optional; case sensitive).
Check “interresidue interactions” and submit.
In the new page, check “contact order and number of contacts (8 Å, CA atoms)”.
Click on submit.
Figure 2 shows the relevant items for computing the CO and number of contacts and the output. The output displays the CO for the protein and the number of contacts for all the residues with residue name and number. The contacting residues are also shown in the output.
Figure 2.
Example to compute the contact order of a protein and the number of contacts for all the amino acid residues in a protein.
Availability of PDBparam
PDBparam is freely available at http://www.iitm.ac.in/bioinfo/pdbparam.
Applications
PDBparam computes various structure-based parameters on interresidue interactions, amino acid propensities, physicochemical properties, and binding sites. This information can be used to understand the structure and functions of proteins and their complexes. The contacts between amino acid residues in protein structures provide data on the location of amino acid residues and preferred contacts in the protein environment, which can be used to comprehend protein folding and predict protein structures.32 The topological parameters, such as CO, LRO, total contact distance, and multiple contact distance, are helpful in understanding protein-folding rates and folding kinetics.9–11 Specific physicochemical interactions between amino acid residues in protein structures, such as cation–π, aromatic clusters, and hydrogen bonds, reveal the importance of these interactions inproteinstability.27 The combination of secondary structure and solvent accessibility is useful in identifying functionally important residues in proteins.15,16 Furthermore, the identification of binding sites in protein–protein, protein–nucleic acid, and protein–ligand complexes can be effectively used to compute the binding propensity and affinity and understand the recognition mechanism of protein complexes.46–51
PDBparam can be used to compute important parameters for any specific protein, providing deep insights into its structure–function relationship. It can also be used for large-scale analysis of different types of proteins to explore potential interactions and contacts, which will provide insights on the similarities and differences crucial to understanding the function.
Conclusion
The PDBparam server can calculate more than 50 parameters from the three-dimensional structure of a protein, classified into the following four categories: physicochemical properties, interresidue interactions, secondary structure propensities, and identification of binding sites in protein–DNA/RNA and protein–protein complexes. All the parameters have been coded using perl. Furthermore, perl-CGI scripts are used to render the HTML web pages. Detailed documentation for the protein properties and links of other available web servers related to such properties are provided, in order to enhance the user’s ease of access.
Acknowledgments
We thank the Bioinformatics Facility, Department of Biotechnology, and IIT Madras for computational facilities.
Footnotes
ACADEMIC EDITOR: J. T. Efird, Associate Editor
PEER REVIEW: Five peer reviewers contributed to the peer review report. Reviewers’ reports totaled 824 words, excluding any confidential comments to the academic editor.
FUNDING: This study was funded by the Department of Science and Technology, Government of India to MMG (SR/SO/BB-0036/2011). The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE). Provenance: the authors were invited to submit this paper.
Author Contributions
Conceived and designed the study: MMG, DV. Web server development: AA, AMT, RN. Discussions: AA, AMT, RN, SJ, DV, MMG. Wrote the first draft of the article: AA, MMG. Contributed to the writing of the article: AMT, RN, SJ, DV. All the authors reviewed and approved the final article.
REFERENCES
- 1.Branden C, Tooze J. Introduction to Protein Structure. New York: Garland Science; 1999. pp. 13–34. [Google Scholar]
- 2.Gromiha MM. Protein Bioinformatics: From Sequence to Function. Cambridge, MA: Academic Press; 2010. [Google Scholar]
- 3.Lee B, Richards FM. The interpretation of protein structures: estimation of static accessibility. J Mol Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 4.Chou PY, Fasman GD. Prediction of protein conformation. Biochemistry. 1974;13:222–245. doi: 10.1021/bi00699a002. [DOI] [PubMed] [Google Scholar]
- 5.Barlow DJ, Thornton JM. Ion-pairs in proteins. J Mol Biol. 1983;168:867–85. doi: 10.1016/s0022-2836(83)80079-5. [DOI] [PubMed] [Google Scholar]
- 6.McDonald IK, Thornton JM. Satisfying hydrogen bonding potential in proteins. J Mol Biol. 1984;238:777–93. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- 7.Furnham N, Holliday GL, de Beer TA, Jacobsen JO, Pearson WR, Thornton JM. The Catalytic Site Atlas 2.0: cataloging catalytic sites and residues identified in enzymes. Nucleic Acids Res. 2014;42:D485–9. doi: 10.1093/nar/gkt1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manavalan P, Ponnuswamy PK. Hydrophobic character of amino acid residues in globular proteins. Nature. 1978;275:673–4. doi: 10.1038/275673a0. [DOI] [PubMed] [Google Scholar]
- 9.Plaxco KW, Simons KT, Baker D. Contact order, transition state placement and the refolding rates of single domain proteins. J Mol Biol. 1998;277:985–94. doi: 10.1006/jmbi.1998.1645. [DOI] [PubMed] [Google Scholar]
- 10.Gromiha MM, Selvaraj S. Comparison between long-range interactions and contact order in determining the folding rate of two-state proteins: application of long-range order to folding rate prediction. J Mol Biol. 2001;310:27–32. doi: 10.1006/jmbi.2001.4775. [DOI] [PubMed] [Google Scholar]
- 11.Gromiha MM. Multiple contact network is a key determinant to protein folding rates. J Chem Inf Model. 2009;49:1130–5. doi: 10.1021/ci800440x. [DOI] [PubMed] [Google Scholar]
- 12.Ahmad S, Gromiha MM, Sarai A. Analysis and prediction of DNA-binding proteins and their binding residues based on composition, sequence and structural information. Bioinformatics. 2004;20:477–86. doi: 10.1093/bioinformatics/btg432. [DOI] [PubMed] [Google Scholar]
- 13.Tjong H, Zhou HX. DISPLAR: an accurate method for predicting DNA-binding sites on protein surfaces. Nucleic Acids Res. 2007;35:1465–77. doi: 10.1093/nar/gkm008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gromiha MM, Fukui K. Scoring function based approach for locating binding sites and understanding the recognition mechanism of protein-DNA complexes. J Chem Inf Model. 2011;51:721–9. doi: 10.1021/ci1003703. [DOI] [PubMed] [Google Scholar]
- 15.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 16.Hubbard SJ, Thornton JM. Biochemistry and Molecular Biology. University College London; 1993. Naccess [Computer Program] Available at: http://www.bioinf.manchester.ac.uk/naccess/ [Google Scholar]
- 17.Ortega A, Amorós D, García de la Torre J. Prediction of hydrodynamic and other solution properties of rigid proteins from atomic- and residue-level models. Biophys J. 2011;101:892–8. doi: 10.1016/j.bpj.2011.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.García de la Torre J, Huertas ML, Carrasco B. HYDRONMR: prediction of NMR relaxation of globular proteins from atomic-level structures and hydrodynamic calculations. J Magn Reson. 2000;147:138–46. doi: 10.1006/jmre.2000.2170. [DOI] [PubMed] [Google Scholar]
- 19.Fraczkiewicz R, Braun W. Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. J Comput Chem. 1998;19:319–33. [Google Scholar]
- 20.Dosztanyi Z, Magyar C, Tusnády G, Simon I. SCide: identification of stabilization centers in proteins. Bioinformatics. 2003;19:899–900. doi: 10.1093/bioinformatics/btg110. [DOI] [PubMed] [Google Scholar]
- 21.Firoz A, Malik A, Afzal O, Jha V. ContPro: a web tool for calculating amino acid contact distances in protein from 3D-structures at different distance threshold. Bioinformation. 2010;5:55–7. doi: 10.6026/97320630005055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gallivan JP, Dougherty DA. Cation-π interactions in structural biology. Proc Natl Acad Sci. 1999;96:9459–64. doi: 10.1073/pnas.96.17.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDonald IK, Naylor D, Jones D, Thornton JM. HBPLUS [Computer Program] Department of Biochemistry and Molecular Biology, University College London. 1993. Available at: http://www.ebi.ac.uk/thornton-srv/software/HBPLUS/
- 24.Costantini S, Paladino A, Facchiano AM. CALCOM: software for calculating the center of mass of proteins. Bioinformation. 2008;2:271–2. doi: 10.6026/97320630002271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balamurugan B, Md Roshan MNA, Hameed BS, et al. PSAP: protein structure analysis package. J Appl Crystallogr. 2007;40:773–7. [Google Scholar]
- 26.Gurusaran M, Shankar M, Nagarajan R, Helliwell JR, Sekar K. Do we see what we should see? Describing non-covalent interactions in protein structures including precision. IUCrJ. 2015;1:74–81. doi: 10.1107/S2052252513031485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tina KG, Bhadra R, Srinivasan N. PIC: protein interactions calculator. Nucleic Acids Res. 2007;35:473–6. doi: 10.1093/nar/gkm423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozma D, Simon I, Tusnády GE. CMWeb: an interactive on-line tool for analysing residue-residue contacts and contact prediction methods. Nucleic Acids Res. 2012;40:W329–33. doi: 10.1093/nar/gks488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Magyar C, Gromiha MM, Pujadas G, Tusnády GE, Simon I. SRide: a server for identifying stabilizing residues in proteins. Nucleic Acids Res. 2005;33:W303–5. doi: 10.1093/nar/gki409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Artimo P, Jonnalagedda M, Arnold K, et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012;40:W597–603. doi: 10.1093/nar/gks400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Beer TA, Berka K, Thornton JM, Laskowski RA. PDBsum additions. Nucleic Acids Res. 2014;42:D292–6. doi: 10.1093/nar/gkt940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gromiha MM, Selvaraj S. Inter-residue interactions in protein folding and stability. Prog Biophys Mol Biol. 2004;86:235–77. doi: 10.1016/j.pbiomolbio.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 33.Zhou H, Zhou Y. Folding rate prediction using total contact distance. Biophys J. 2002;82:458–63. doi: 10.1016/S0006-3495(02)75410-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewis PN, Momany FA, Scheraga HA. Folding of polypeptide chains in proteins: a proposed mechanism for folding. Proc Natl Acad Sci. 1971;68:2293–7. doi: 10.1073/pnas.68.9.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crawford JL, Lipscomb WN, Schellman CG. The reverse turn as a polypeptide conformation in globular proteins. Proc Natl Acad Sci. 1973;70:538–42. doi: 10.1073/pnas.70.2.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parthasarathy S, Murthy MRN. Protein thermal stability: insights from atomic displacement parameters (B values) Protein Eng. 2000;13:9–13. doi: 10.1093/protein/13.1.9. [DOI] [PubMed] [Google Scholar]
- 37.Nozaki Y, Tanford C. The solubility of amino acids and two glycine peptides in aqueous ethanol and dioxane solutions establishment of a hydrophobicity scale. J Biol Chem. 1971;246:2211–7. [PubMed] [Google Scholar]
- 38.Tanford C. Contribution of hydrophobic interactions to the stability of the globular conformation of proteins. J Am Chem Soc. 1962;84:4240–7. [Google Scholar]
- 39.Ponnuswamy PK, Prabhakaran M, Manavalan P. Hydrophobic packing and spatial arrangement of amino acid residues in globular proteins. Biochim Biophys Acta. 1980;623:301–16. doi: 10.1016/0005-2795(80)90258-5. [DOI] [PubMed] [Google Scholar]
- 40.Mahn A, Lienqueo ME, Asenjo JA. Effect of surface hydrophobicity distribution on retention of ribonucleases in hydrophobic interaction chromatography. J Chromatogr A. 2004;1043:47–55. doi: 10.1016/j.chroma.2004.03.021. [DOI] [PubMed] [Google Scholar]
- 41.Rose GD, Geselowitz AR, Lesser GJ, Lee RH, Zehfus MH. Hydrophobicity of amino acid residues in globular proteins. Science. 1985;229:834–8. doi: 10.1126/science.4023714. [DOI] [PubMed] [Google Scholar]
- 42.Vihinen M, Torkkila E, Riikonen P. Accuracy of protein flexibility predictions. Proteins. 2004;19:141–9. doi: 10.1002/prot.340190207. [DOI] [PubMed] [Google Scholar]
- 43.Ponnuswamy PK, Gromiha MM. On the conformational stability of folded proteins. J Theor Biol. 1994;166:63–74. doi: 10.1006/jtbi.1994.1163. [DOI] [PubMed] [Google Scholar]
- 44.Nagarajan R, Ahmad S, Gromiha MM. Novel approach for selecting the best predictor for identifying the binding sites in DNA binding proteins. Nucleic Acids Res. 2013;41:7606–14. doi: 10.1093/nar/gkt544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagarajan R, Gromiha MM. Prediction of RNA binding residues: an extensive analysis based on structure and function to select the best predictor. PLoS One. 2014;9:e91140. doi: 10.1371/journal.pone.0091140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gromiha MM, Nagarajan R. Computational approaches for predicting the binding sites and understanding the recognition mechanism of protein-DNA complexes. Adv Protein Chem Struct Biol. 2013;91:65–99. doi: 10.1016/B978-0-12-411637-5.00003-2. [DOI] [PubMed] [Google Scholar]
- 47.Gromiha MM, Siebers JG, Selvaraj S, Kono H, Sarai A. Role of inter and intra-molecular interactions in protein-DNA recognition. Gene. 2005;364:108–13. doi: 10.1016/j.gene.2005.07.022. [DOI] [PubMed] [Google Scholar]
- 48.Nagarajan R, Chothani SP, Ramakrishnan C, Sekijima M, Gromiha MM. Structure based approach for understanding organism specific recognition of protein-RNA complexes. Biol Direct. 2015;10:8. doi: 10.1186/s13062-015-0039-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yugandhar K, Gromiha MM. Feature selection and classification of protein-protein complexes based on their binding affinities using machine learning approaches. Proteins. 2014;82:2088–96. doi: 10.1002/prot.24564. [DOI] [PubMed] [Google Scholar]
- 50.Yugandhar K, Gromiha MM. Protein-protein binding affinity prediction from amino acid sequence. Bioinformatics. 2014;30:3583–9. doi: 10.1093/bioinformatics/btu580. [DOI] [PubMed] [Google Scholar]
- 51.Yugandhar K, Gromiha MM. Computational approaches for predicting binding partners, interface residues and binding affinity of protein-protein complexes. Methods Mol Biol. 2016 doi: 10.1007/978-1-4939-6406-2_16. in press. [DOI] [PubMed] [Google Scholar]