Abstract
Dysregulation of the EGFR signaling axis enhances bone metastases in many solid cancers. However, the relevant downstream effector signals in this axis are unclear. miR-1 was recently shown to function as a tumor suppressor in prostate cancer cells, where its expression correlated with reduced metastatic potential. In this study, we demonstrated a role for EGFR translocation in regulating transcription of miR-1-1, which directly targets expression of TWIST1. Consistent with these findings, we observed decreased miR-1 levels that correlated with enhanced expression of activated EGFR and TWIST1 in a cohort of human prostate cancer specimens and additional datasets. Our findings support a model in which nuclear EGFR acts as a transcriptional repressor to constrain the tumor-suppressive role of miR-1 and sustain oncogenic activation of TWIST1, thereby leading to accelerated bone metastasis.
Introduction
Evidence suggests that acquisition of androgen independence may be due to upregulation of growth factor receptor signaling pathways, principally the EGFR, making it an attractive target for therapeutic interventions (1). Enhanced EGFR expression is correlated with disease relapse and progression to androgen independence in many cases of prostate cancer (1). EGFR modulates pathways that are implicated in androgen-independent proliferation and survival as well as invasion and metastasis to the bone (2). Although targeting the membrane-bound EGFR showed some benefit, there is an emerging need to further explore targetable signaling components involved in the nuclear EGFR signaling network (3). Our study showed that the main function of nuclear EGFR appears to be as an upstream factor for various oncogenic genes including TWIST1. Given the central role of EGFR in prostate bone metastasis, further characterization of targets of EGFR-regulated genes is of interest (4).
TWIST1 is a highly conserved basic helix-loop-helix transcription factor and putative oncogene that is upregulated in many types of aggressive solid tumors, including a subset of advanced prostate cancers (5). In human prostate cancer, upregulation of TWIST1 was positively correlated with Gleason grades (6). In contrast, downregulation of TWIST1 in androgen-independent prostate cancer cells increased their sensitivity to anticancer drugs and reduced their migratory and invasive abilities (6). Interestingly, nuclear EGFR cooperates with STAT3 and induces a TWIST1-mediated malignant transformation of breast cancer cells (7). Therefore, establishing a functional link between EGFR and TWIST1 activities is potentially clinically significant for selecting therapies and monitoring responses to them (8). However, it has not been established whether an EGFR-dependent transcriptional mechanism influences TWIST1 activity in prostate cancer. We hypothesized that the EGFR may act on TWIST1 by affecting the expression of the miRNA (miR), miR-1, which acts as a putative tumor suppressor in prostate cancer cells.
miRs play many roles in biologic processes of cancer cells with respect to the post-transcriptional regulation of target genes (9). Altered expressions of miRs also cause loss of tumor-suppressive or gain of oncogenic activities that affect tumor progression (10). miR-1 is the most downregulated miR in prostate tumors compared with noncancerous prostate tissues (11). Moreover, miR-1 was implicated as a candidate tumor suppressor and prognostic marker for prostate cancer (12, 13). We previously showed that miR-1 is an essential factor regulating aspects of tumorigenesis and progression using the aggressive Pten/Tp53 null prostate cancer mouse model system (14). Importantly, our previous report showed that the growth and invasive capability of highly metastatic PC3 cells were impaired by ectopic miR-1 expression (14).
In the current study, our in vitro and in vivo data showed that activated EGFR enhanced prostate cancer progression and bone metastasis via downregulating miR-1 and activating TWIST1. To address the mechanism underlying the effects on tumor progression, we demonstrated that miR-1 is directly and transcriptionally regulated by nuclear EGFR. We identified miR-1 targets in the 3′-UTR of TWIST1 that can lead to TWIST1 downregulation at both mRNA and protein levels. We confirmed our findings in patient tissue samples from prostate cancers with low miR-1 expression by showing a positive correlation with enhanced TWIST1. Our studies support a model that EGFR facilitates tumor malignancy through EGFR-dependent reduction of miR-1 to disrupt the inhibitory effects of miR-1–dependent post-transcriptional regulation of TWIST1 and enhance TWIST1 activities. This study provides an example of EGFR signaling being linked to downstream activation of TWIST1 through a molecular mechanism by miRs.
Materials and Methods
Cell culture
DU145, PC3, LNCap, and 22Rv1 human prostate cancer cell lines were obtained from ATCC. The cell lines were authenticated within 6 months before use according to the provider's recommendations. All the cells were tested and negative for mycoplasma contamination. The metastatic RasB1 cell line was provided by Dr. Kathleen Kelly (NCI/NIH, Bethesda, MD). This cell line was characterized and used to study molecular mechanisms of prostate cancer metastasis previously in multiple peer-reviewed articles (15–20). Cells expressing miR-1 or the control miR were generated as described previously (14, 20). EGFR was subcloned into the pFUGW lentiviral vector and an IRES-mCherry reporter with a puromycin-selectable marker. Stable EGFR-expressing cell lines were established by FACS sorting of mCherry-positive cells. LNCap, 22Rv1, PC3, DU145, and RasB1 cell lines were cultured in RPMI-1640 medium supplemented with 10% FCS. Transient transfections were carried out using Lipofectamine RNAiMAX (Invitrogen). The dose of the EGF was 100 ng/mL in a serum-free condition. The dose of the EGFR inhibitor was 10 nmol/L for CI1033.
Migration and invasion assay
Invasion assays were conducted using 106 cells that had invaded Matrigel-coated Transwells in response to EGF (100 ng/mL). After 6 hours, Transwells were fixed and stained with a 0.5% crystal violet fixative solution for 15 minutes. Invaded cells on the underside of the membrane were counted and quantified with five medium-power fields for each replicate. The migration assay used Transwells without Matrigel, and cells were fixed and stained as described in the invasion assay.
Promoter analysis and FACS analysis
A promoter functional analysis using FACS and the relative median fluorescent intensity (MFI) value were measured as previously described (14). Cells were treated with or without the EGF (100 ng/mL) and CI1033 (10 nmol/L) for 24 hours. The MFI value for RFP was measured by FACS using FACSDiva software and normalized to the value of the vehicle as previously described (21). Predictions of transcription factor-binding sites within the promoter regions were adopted from the AliBaba 2.1 program.
miRNA luciferase assay
Cells were transfected with 1 µg of human TWIST1 3′UTR reporter and 1 µg of precursor miRs encoding a control or the miR-1 precursor. Cell extracts were prepared 24 hours after EGF (100 ng/mL) or CI1033 (10 nmol/L) treatment, and luciferase (FL) and Renilla (RL) activities were measured using Dual Luciferase Reporter Assay System (Promega). RL activities were calculated as mean ± SEM after normalization to FL activities. Three independent experiments were done in triplicate. The miR-binding sites on human TWIST1 3′UTR were determined using the Computational Biology Center, Memorial Sloan-Kettering Cancer Center (MSKCC) website (microRNA.org) and the Bioinformatics and Research Computing, Whitehead Institute for Biomedical Research (TargetScan.org).
Tissue samples
The clinical samples used 32 independent primary prostate tumors were collected from Taipei Medical University Joint human biological database, Taiwan. Tissue samples were obtained and used according to protocols approved by Taipei Medical University-Joint Institutional Review Board (approval no.: 201311034). The study was conducted according to the Declaration of Helsinki principles.
Animal studies
To analyze tumorigenesis, 5-week-old male nude mice (NCI/NIH) were injected intracardially with 105 tumor cells, and metastases were monitored by bioluminescent imaging (BLI) as previously described (14). Bone metastases were evaluated on magnified (×3) radiographs taken with a Faxitron MX-20 (Faxitron Bioptics). Each bone metastasis was scored based on the following criteria: 0, no metastasis; 1, bone lesion covering <1/4 of the bone width; 2, bone lesion involving 1/4~1/2 of the bone width; 3, bone lesion across 1/2~3/4 of the bone width; and 4, bone lesion of >3/4 of the bone width. The bone metastasis score for each mouse represented the sum of scores of all bone lesions from four limbs. For survival studies, mice were euthanized when one of the following situations applied: 10% loss of body weight, paralysis, or head tilting.
Statistical analysis
In vivo animal results and clinical outcome analyses are expressed as plots showing the median and box boundaries extending between the 25th and 75th percentiles, with whiskers down to the minimum and up to the maximum value. All in vitro data are presented as mean ·SEM. Statistical calculations were performed with GraphPad Prism (GraphPad Software, Inc.) analytical tools. Differences between individual groups were analyzed by a one- or two-way ANOVA test. Bonferroni post test was used for comparisons among three or more groups. The log-rank test was used for the survival curve analysis. P values of <0.05 were considered statistically significant.
Results
EGFR signaling regulates miR-1 expression levels
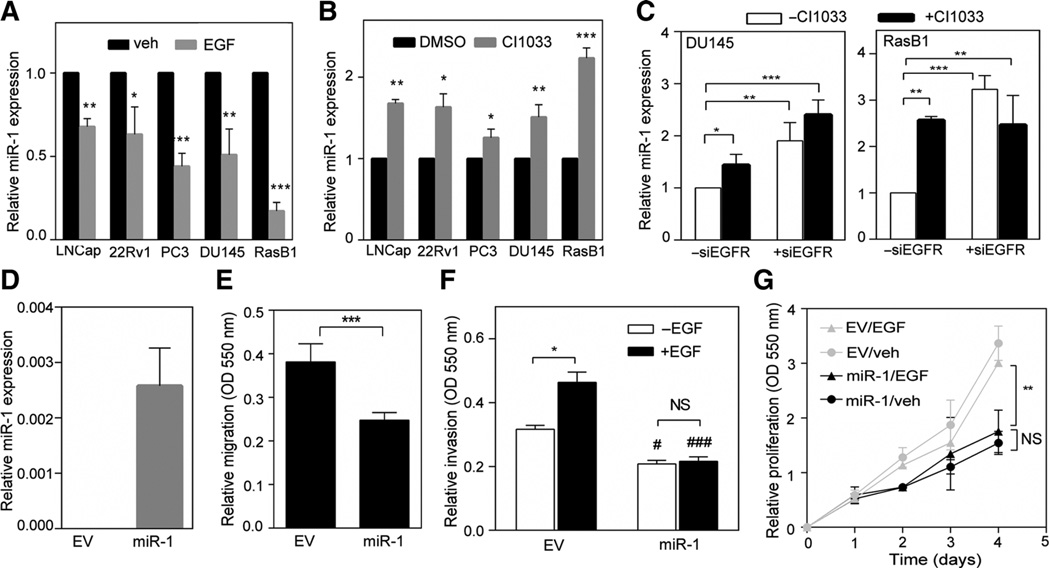
To study the molecular mechanisms involved in the relationship between EGFR pathway dysregulation and miR-1 in advanced prostate cancer, we first investigated miR-1 levels in two clinical datasets from the MSKCC (22) and The Cancer Genome Atlas (TCGA) that, respectively, contained 111 and 372 samples. We found enrichment of gene sets that exhibited downregulated EGFR signaling in cancers expressing higher levels of miR-1 by a Gene Set Enrichment Analysis (GSEA; Supplementary Fig. S1A and S1B). To further confirm whether miR-1 has an inverse correlation with EGFR signaling in prostate cancer progression, we divided specimens into two groups of up- and downregulated EGFR signaling-responsive signature expressions based on a measure of relative mRNA expression using z-scores. An analysis of mean expression confirmed that higher miR-1 was significantly expressed in tissues with downregulated EGFR signaling signatures (Supplementary Fig. S1C and S1D). Because EGFR signaling is crucial to the malignant progression of prostate cancer (1), it is possible that EGFR signaling reduces miR-1 and promotes aggressive prostate phenotypes. Indeed, EGF treatment markedly decreased miR-1 expression in prostate cancer cells (Fig. 1A), whereas inhibition of EGFR signaling with EGFR inhibitor treatment increased miR-1 levels (Fig. 1B). Consistent with the effect of EGFR inhibitor treatment, EGFR siRNA also resulted in an increase in miR-1 in prostate cancer cells (Fig. 1C). We next examined endogenous EGFR and miR-1 levels in different prostate cancer cell lines. As shown in Supplementary Fig. S2A and S2B, there was an inverse relationship between endogenous EGFR and miR-1 in the metastatic cells, PC3 and RasB1 (15). To further clarify the role of miR-1 in the metastatic prostate cancer cell line, miR-1 was overexpressed in RasB1 cells (Fig. 1D). Consistent with our previous results (14), cells with ectopically expressed miR-1 exhibited decreased migration (Fig. 1E). Moreover, miR-1– overexpressing cells did not exhibit enhanced invasion (Fig. 1F) or proliferation (Fig. 1G) in response to the EGF, supporting the tumor-suppressive role of miR-1 in prostate cancer cells. In addition, we did not observe enhanced migration or invasiveness abilities in EGF-treated AR-positive cells (Supplementary Fig. S2C). We concluded that the reduced miR-1 in response to the EGF in AR-positive cells was insufficient to drive the malignant phenotype. This is discussed below in the "Discussion" section.
Figure 1.
EGFR signaling activation is correlated with reduced miR-1 expression. A and B, expressions of miR-1 levels after EGF (A) and CI1033 (B) treatment in various prostate cancer cell lines. C, expressions of miR-1 levels after EGFR siRNA and CI1033 treatment in DU145 and RasB1 cells. Data are presented as mean ± SEM of separate treatments or transfections; n = 3. D, miR-1 levels of RasB1 cells expressing empty vector (EV) or miR-1 precursor construct. E and F, cellular migration (E) and invasion (F) of RasB1 cells expressing EV or miR-1 precursor following EGF treatment. Data are presented as mean ± SEM; n = 5. #, EV vs. miR-1. #, P < 0.01; ###, P < 0.001. G, growth curves of RasB1 cells expressing EV or miR-1 precursor following EGF treatment; n = 6. *, P < 0.05; **, P < 0.01; ***, P < 0.001. NS, nonsignificant.
EGFR expression reconstitutes metastasis in miR-1–expressing cells
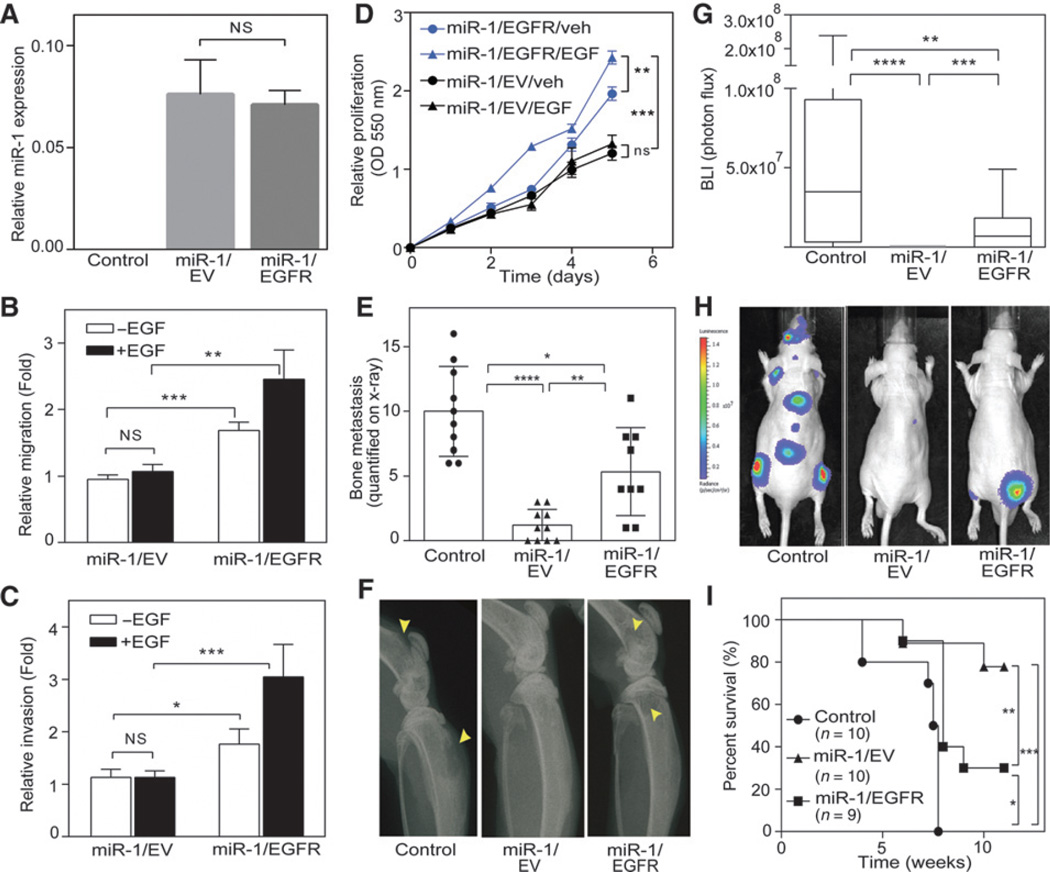
To study the role of the EGFR, we then asked whether reconstitution of EGFR levels in miR-1–expressing cells induces the malignant phenotype. miR-1 expression levels remained elevated after stable EGFR overexpression (Fig. 2A). Reconstitution of EGFR in the miR-1–expressing cells showed significantly enhanced migration (Fig. 2B), invasiveness (Fig. 2C), and proliferation (Fig. 2D) in both the presence and absence of EGF stimulation. These data confirm the dominant role of the EGFR in prostate cancer cells. Next, we evaluated the effects of EGFR overexpression in cells harboring miR-1 in vivo. The parental RasB1 control group mostly showed higher bone metastasis by x-ray and BLI (Fig. 2E–H) and lower survival rates (Fig. 2I) compared with the group expressing miR-1 alone or coexpressing miR-1 and EGFR. Moreover, reconstituted EGFR in the presence of miR-1 resulted in bone metastases characterized by mixed osteolytic and osteoblastic elements in mice (Supplementary Fig. S2D). Taken together, these data suggest that EGFR is a physiologically important factor that functions in the development of prostatic bone metastasis. Robust EGFR expression alone was sufficient to reverse the inhibitory effects of high levels of miR-1, suggesting that EGFR is a dominant factor in determining the bone metastatic phenotype. These data are consistent with a mechanism by which activated EGFR regulates target gene expressions as an upstream inducer.
Figure 2.
EGFR expression reconstitutes malignancy and bone metastasis in miR-1–expressing prostate cancer cells. A, miR-1 levels in parental RasB1 (control), RasB1/miR-1 (miR-1/EV), and EGFR-rescued RasB1/miR-1 (miR-1/EGFR) cells. B and C, cellular migration (B) and invasion (C) of RasB1/miR-1 cells expressing empty vector (EV) or EGFR cDNA vector following EGF treatment. D, in vitro growth rate of RasB1/miR-1 cells expressing EGFR or EV following EGF treatment; n = 6 per group. E, bone metastasis scores for each mouse in tumor-bearing mice inoculated with control (n = 10), miR-1/EV (n = 10), or miR-1/EGFR (n = 9) cells. F, representative radiographic images of bone metastases in mice from E. Bone metastases are indicated by arrows. Yellow arrows, osteolysis. G, BLI signal of bone metastasis per mouse for mice bearing tumor cells described in E at week 5. H, representative BLIs in mice from G. I, survival rate of tumor-bearing mice from E. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
EGFR translocation to nuclei
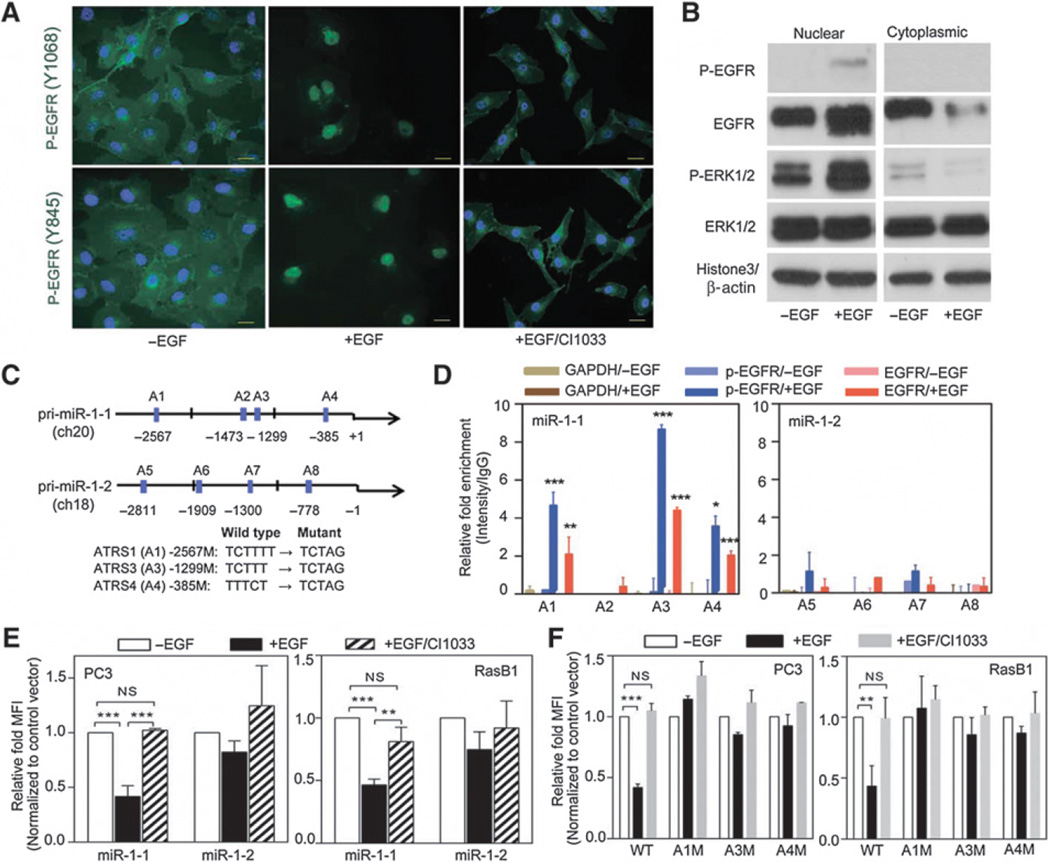
To address the molecular mechanism by which activated EGFR regulates the metastatic capability of prostate cancer cells, we focused on searching for target substrates that might be involved in promoting malignancy through a transcriptional regulator. In our prostate cancer model, miR-1 decreased as evidenced by monitored levels following EGFR activation (Fig. 1A), indicating a functional role of EGFR in regulating miR-1 expression. On the basis of the effects of EGFR translocation observed in various solid cancers (23–26), we reasoned that the miR-1 level is determined by the nuclear-translocated EGFR in prostate cancer. To test this possibility, we examined the presence of nuclear EGFR in prostate cancer cells by IF labeling. RasB1 and PC3 metastatic prostate cancer cells exhibited increased expression of nuclear EGFR with EGF stimulation; however, higher membrane and cytoplasmic EGFR levels were found in cells treated with an EGFR inhibitor (CI1033; Supplementary Fig. S3A). Increased nuclear EGFR translocation was shown in serum-starved cells with EGF stimulation using two phosphorylated (p)-EGFR antibodies, but CI1033 inhibited EGFR translocation (Fig. 3A). Moreover, FFPE cell blocks from RasB1 and PC3 cells confirmed that the EGFR inhibitor indeed prevented translocation of nuclear p-EGFR (Supplementary Fig. S3B). Comparable with the effects with EGF, we also observed enhanced EGFR signal transduction as measured by increased p-EGFR and p-ERK1/2 in nuclear extracts of cells (Fig. 3B). Although cytoplasmic expression of p-EGFR was not detected (Fig. 3B), the ratio of total EGFR to internal controls from the nuclear and cytoplasmic fractions in response to EGF confirmed EGFR translocation downstream of EGF (Supplementary Fig. S3C). The translocated EGFR also altered the levels of p-ERK1/2 in the cytoplasmic fraction (Fig. 3B), and the ratio of p-ERK1/2 to ERK1/2 from the nuclear and cytoplasmic fractions was also changed in response to EGF (Supplementary Fig. S3D). Our results show that as in many other cancers, activated EGFR is translocated to nuclei in prostate cancer cells.
Figure 3.
Nuclear EGFR directly binds to pri-miR-1-1 promoter and regulates miR-1 transcription. A, immunofluorescent staining of RasB1 cells with antibodies for p-EGFR (Y1068 or Y845) following EGF treatment. Nuclei were visualized with DAPI staining (blue). Scale bars, 50 µm. B, immunoblotting of nuclear (left) and cytoplasmic (right) cell extracts from RasB1 cells following EGF treatment. C, schematic of the predicted ATRSs in the pri-miR-1-1 and pri-miR-1-2 stem-loop promoters. D, ChIP analyses of predicted ATRSs in the pri-miR-1-1 and pri-miR-1-2 promoter regions of RasB1 cells following EGF treatment. Enrichment of each protein at each site is given as a percentage of the total input, which was then normalized to each IgG. E, promoter analyses of PC3 and RasB1 cells transiently transfected with the pri-miR-1-1 or pri-miR-1-2-RFP reporter following EGF and CI1033 treatment. Relative fold of MFI is given as normalization to a control vector. F, promoter analyses of PC3 and RasB1 cells transiently transfected with the wild-type or mutated pri-miR-1-1-RFP reporter following EGF and CI1033 treatment. Data are presented as mean ± SEM; n = 3. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
The primary miR-1-1 stem loop promoter is the target of the nuclear EGFR
A report showed that nuclear EGFR binds specific genomic sequences, including AT-rich minimal consensus sequences (ATRS) that promote transcriptional regulation (23). To investigate how EGFR signaling transcriptionally regulates miR-1 expression, we examined the putative promoter region upstream of the human primary miR-1-1 (pri-miR-1-1) and miR-1-2 (pri-miR-1-2) transcripts encoding miR-1, respectively, located on chromosomes 20 and 18. Following a sequence analysis, we identified four ATRSs within both promoter regions (Fig. 3C). To test whether EGF directly mediates the binding of nuclear EGFR to either pri-miR-1-1 or pri-miR-1-2, we performed a chromatin immunoprecipitation (ChIP) assay on RasB1 cells following quantitative PCR analyses. Significant increases in nuclear EGFR-binding signals were found at ARTS1, ARTS3, and ARTS4 after EGF treatment using a p-EGFR or EGFR antibody (Fig. 3D). Moreover, we performed the ChIP assay in the AR-positive prostate cancer cell line, 22RV1, and a similar p-EGFR- or EGFR-binding signal was found of pri-miR-1-1 after EGF treatment (Supplementary Fig. S3E). In addition, we performed a promoter assay to examine whether the ATRSs in the promoter region are functional. The reporter assays showed significantly decreased primiR-1-1 promoter activity in cells treated with EGF; however, treatment of cells with an EGFR inhibitor disrupted EGF-repressed promoter activity in wild-type pri-miR-1-1 (Fig. 3E). Furthermore, we characterized the specificity of nuclear EGFR binding in the promoter region of pri-miR-1-1 by introducing mutations (Fig. 3C). Mutations at ARTS1, ARTS3, and ARTS4 abolished EGF-reduced reporter activity in a promoter assay (Fig. 3F), indicating the specificity of nuclear EGFR downregulation on pri-miR-1-1. These data are consistent with a mechanism whereby nuclear EGFR regulates pri-miR-1-1 transcription through a physical interaction between nuclear EGFR complex and promoter region of pri-miR-1-1. Our findings validate this cellular system by recapitulating the effects of EGFR translocation through which nuclear EGFR can inhibit miR-1 functions.
Increased nuclear EGFR and decreased miR-1 are correlated with clinical outcomes
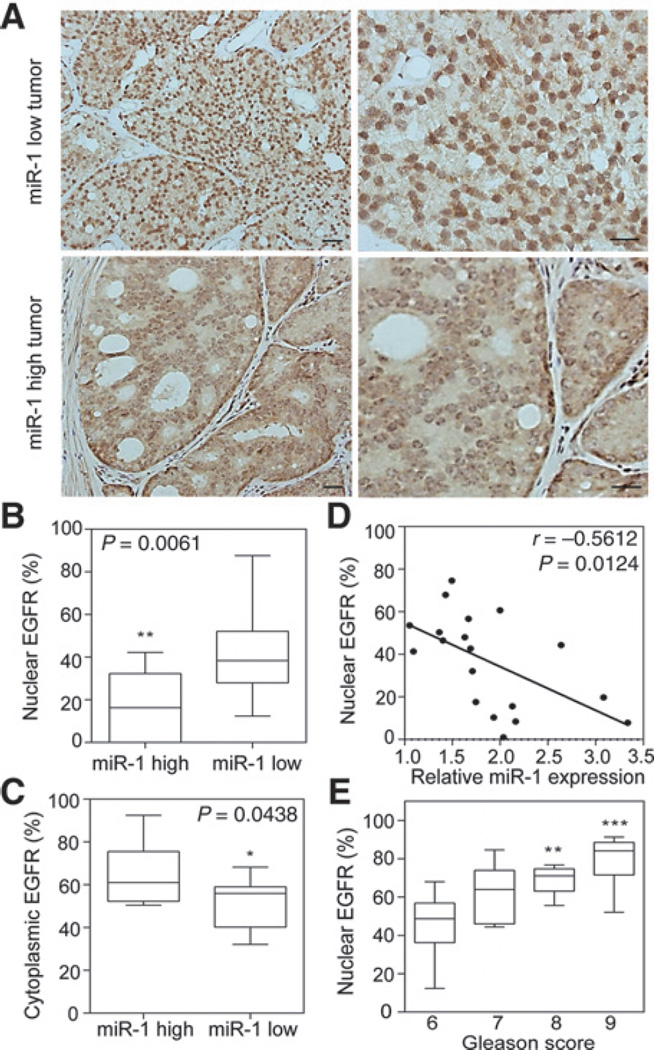
Next, we validated our in vitro results by examining nuclear EGFR expression in metastatic prostate tumors by an IHC analysis. p-EGFR nuclear expression was enhanced in prostate cancer samples with lower miR-1 expression levels (Fig. 4A, top, 4B), which is consistent with our finding that miR-1 levels are reduced following nuclear EGFR activation. Tumor tissues with higher miR-1 levels had strong cytoplasmic EGFR labeling compared with tissue samples with lower miR-1 expression (Fig. 4A, bottom, 4C). Importantly, the correlation was consistent between nuclear EGFR and mean miR-1 expressions according to a Pearson coefficient analysis, which showed that samples with high miR-1 levels had lower nuclear EGFR expression (Fig. 4D). Moreover, we also noted that a number of cases of tumors with high Gleason grades also expressed nuclear EGFR (Fig. 4E). These results are consistent with our molecular mechanism whereby hyperactive EGFR signaling reduces miR-1 expression, resulting in activation of nuclear EGFR function and enhancement of the clinical malignant potential.
Figure 4.
Induction of nuclear EGFR is correlated with lower miR-1 and associated with increased Gleason scores in prostate cancer. A, IHC staining with an antibody specific for p-EGFR (Y1068) in prostate cancer tissue sections with different miR-1 levels. Scale bars, 100 µm. B and C, percentage of nuclear EGFR (B) and cytoplasmic EGFR (C) in two groups of tissue samples based on miR-1 levels (n = 10 per group). D, inverse correlation of relative miR-1 expression to the percentage of nuclear EGFR in prostate samples (n = 20). Significance was determined by the Gaussian population (Pearson) and a two-tailed test. E, correlation of nuclear EGFR expression percentage with Gleason scores according to prostate cancer clinical annotation (n = 20). * vs. GGS6. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
EGFR signaling regulates TWIST1 expression in advanced prostate cancer
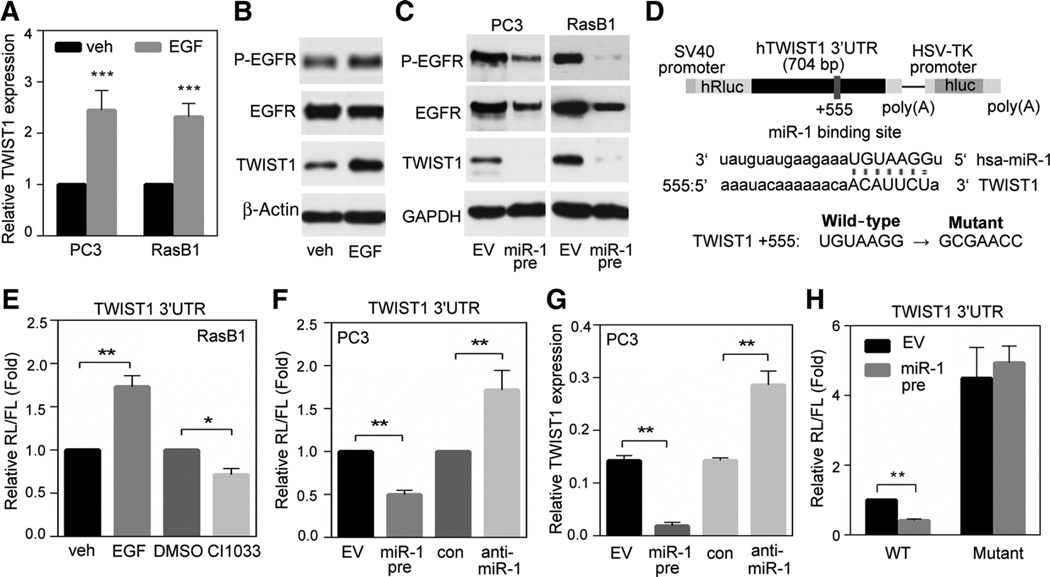
TWIST1 is expressed at high levels in prostate cancers (6) identified as having EGFR activation (7). Although the molecular mechanisms involved in this effect are still under investigation, the specific relationship of TWIST1 and EGFR pathway in advanced prostate cancer remains unclear. Compared with normal tissues, analyses of the actual intensity of mean expressions in a clinical prostate database (22) showed elevated TWIST1 expression in primary tumors and a further increase in metastatic samples (Supplementary Fig. S4A). To further confirm the association between TWIST1 and EGFR signaling, we examined gene expression profiles using two prostate cancer datasets, MSKCC and TCGA. Indeed, after performing the GSEA, we observed that tissues expressing lower TWIST1 were enriched in an EGFR signaling-downregulated gene set (Supplementary Fig. S4B and S4C). Our results support the idea that increased TWIST1 expression is a downstream event of EGFR signaling pathway in prostate cancer. We tested the relationship between EGFR signaling and TWIST1 levels in clinical samples via a z-score analysis and observed a significant increase in upregulated EGFR-signaling gene signatures in samples with high TWIST1 expression (Supplementary Fig. S4D). Interestingly, an analysis of summed z-scores also confirmed that miR-1 was significantly expressed at low levels in prostate tissues with upregulated EGFR signaling (Supplementary Fig. S4E). We hypothesized that miR-1 may be a negative regulator of TWIST1, following EGFR signaling that reduces miR-1 to ensure persistent TWIST1 function and promote malignant phenotypes. Although there was no significant change in vimentin levels or morphology in response to EGF (Supplementary Fig. S4F and S4G), we found that levels of TWIST1 and p-EGFR increased following EGF stimulation (Fig. 5A and B) and were reduced by the overexpression of miR-1 in PC3 and RasB1 cells (Fig. 5C). These findings are consistent with our dataset studies showing that the activated EGFR upregulates TWIST1 expression in advanced prostate cancer.
Figure 5.
Activation of EGFR signaling results in upregulation of TWIST1 through reduced miR-1–mediated specific targeting to the 3′UTR of TWIST1. A, TWIST1 levels in PC3 and RasB1 cells following EGF treatment. B, immunoblotting of extracts from RasB1 cells following EGF treatment. C, immunoblotting of extracts from PC3 and RasB1 cells with stable expression of miR-1 precursor or empty vector (EV). D, schematic of the predicted miR-1–binding site (+555) in the full-length 3′UTR reporter constructs of wild-type and mutated TWIST1. SV40 and HSV-TK, promoters. E, normalized reporter activity in RasB1 cells following EGF or CI1033 treatment. F, normalized reporter activity in PC3 cells treated with agents that regulate miR-1 levels. G, endogenous levels of TWIST1 in cells treated with agents that regulate miR-1 levels. H, normalized reporter activity of the TWIST1 3′UTR containing wild-type or mutated miR-1–binding sites in RasB1 cells with transient expression of miR-1 precursor or EV. Data are presented as mean ± SEM; n = 5. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
miR-1 directly targets the 3′UTR of TWIST1 and regulates TWIST1 mRNA stability
To address the molecular mechanism whereby miR-1 affects TWIST1 expression in tumor cells, we found that TWIST1 is a potential target with a predicted miR-1–binding site in the 3′UTR region of the mRNA transcript (Fig. 5D). Using a bicistronic luciferase assay system with firefly luciferase (FL) as the internal control, we combined Renilla luciferase (RL) and the 3′UTR of TWIST1 in a single transcriptional unit and monitored luciferase activities. After normalization (RL/FL), we showed that the reporter activity increased following EGF treatment and was reduced after treatment with CI1033 (Fig. 5E). In addition, an exogenously expressing miR-1 precursor reduced the reporter activity, whereas miR-1 depletion increased the signal (Fig. 5F). Moreover, we observed that miR-1 downregulated TWIST1 mRNA levels similar to that found in the reporter assay (Fig. 5G vs. F), which is consistent with the finding that miR-1 targets the 3′UTR of TWIST1, leading to its downregulation. Next, we asked whether the predicted miR-1–binding site in the 3′UTR of TWIST1 provides specificity by monitoring luciferase activities of the reporter construct containing individual mutations at the miR-1 target site (Fig. 5D). As shown in Fig. 5H, we demonstrated that a mutation at the +555-binding site conferred resistance to the inhibitory effects of miR-1, supporting a physical interaction between miR-1 and the TWIST1 3′UTR. Therefore, miR-1 may act as a negative regulator of TWIST1.
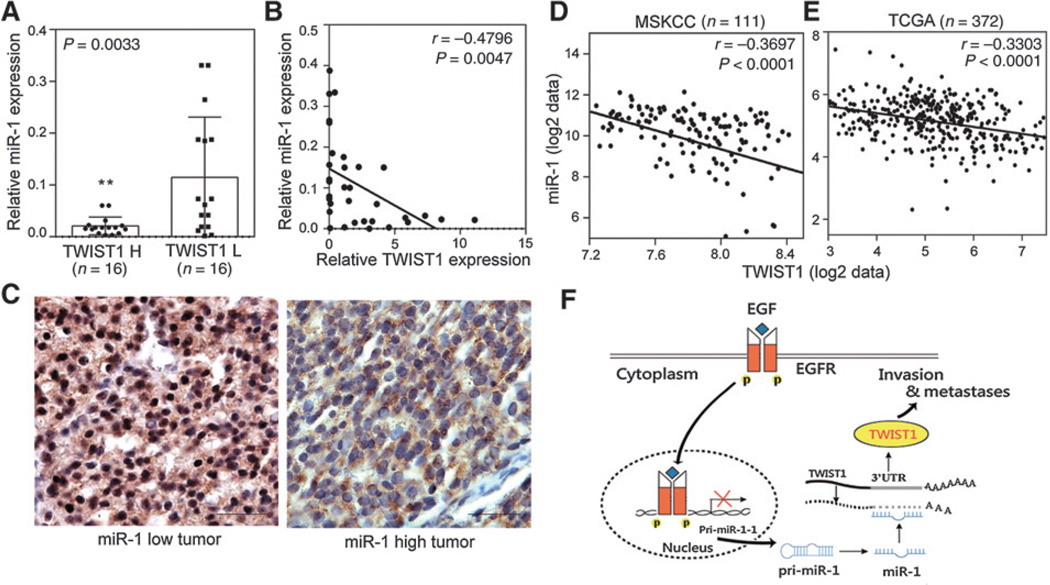
miR-1 and TWIST1 levels are inversely correlated in clinical prostate cancer specimens
To further investigate the inverse relation between miR-1 and TWIST1 in human prostate cancer tissues, we analyzed 32 independent prostate tumors collected from Taipei Medical University Joint human biological database. Samples were divided into two groups of "low" and "high" TWIST1 expression based on qRT-PCR analyses. An analysis of the variance confirmed that miR-1 was differentially expressed between the low- and high-expression groups, where tissues with higher levels of TWIST1 expression had lower miR-1 expression levels (Fig. 6A and B). We performed an IHC analysis in distant metastatic tumors to examine TWIST1 protein levels. We found that TWIST1 staining was enriched in nuclei of prostate cancer samples with low miR-1 expression levels (Fig. 6C, left) compared with samples with high miR-1 levels (Fig. 6C, right), consistent with our finding that a reduction in TWIST1 is miR-1 dependent (Fig. 5). Importantly, it is consistent with the inverse correlation between miR-1 and TWIST1 expressions in two clinical datasets (Fig. 6D and E). We tested the relationship of TWIST1 and miR-1 expressions with prostate cancer clinical outcomes in the dataset, and the observation that patient samples with induced TWIST1 and reduced miR-1 had higher pathology grades (Supplementary Fig. S5A and S5B), and patients with higher TWIST1 and lower miR-1 expressions also had lower survival rates (Supplementary Fig. S5C and S5D). Moreover, TWIST1 expression can be detected by IHC in metastatic lesions harvested from parental RasB1 and RasB1-miR-1/EGFR cell bearing mice (Supplementary Fig. S5E). Ectopic miR-1 alone in RasB1 cells did not form tumors and bone metastasis (Supplementary Fig. S5E). These results suggested that EGFR activation in vivo decreases miR-1 in turn increases TWIST1 expression. Taken together, our results support the idea that reduced miR-1 levels promote oncogenic properties of TWIST1 and highlight a regulatory network where the induction of TWIST1 is linked to inactivation of miR-1 in advanced prostate cancer.
Figure 6.
Establishment of inverse correlation between miR-1 and TWIST1 levels in metastatic prostate cancer patients. A, levels of miR-1 in two groups of tissue samples based on TWIST1 levels (n = 16 per group). B, inverse correlation of relative miR-1 expression to relative TWIST1 mRNA expression in prostate cancer samples (n = 32). Significance was determined by Gaussian population (Pearson) and two-tailed test. C, IHC staining with an antibody specific for TWIST1 in prostate cancer tissue sections with different miR-1 levels. Scale bars, 100 µm. D and E, inverse correlation of mean miR-1 expression to mean TWIST1 mRNA expression in two prostate cancer datasets, MSKCC (D) and TCGA (E). Significance was determined by Gaussian population (Pearson) and two-tailed test. F, model for EGFR signaling regulation of miR-1 and TWIST1 functions leading to bone metastases. EGF stimulates EGFR functions through a nuclear EGFR translocation pathway to reduce pri-miR-1-1 transcription. miR-1 can disrupt the oncogenic effects of TWIST1 through targeting the 3′UTR of TWIST1 mRNA. Inducible phosphorylation of EGFR expression disrupts the inhibitory effects of miR-1 and promotes TWIST1 activity and bone metastases. * vs. TWIST L. **, P < 0.01.
Our results show that nuclear EGFR serves as a suppressor of miR-1 expression. As shown in Fig. 6F, our model suggests that activated EGFR represses miR-1 transcription in prostate cancer, which in turn, reduces the negative regulatory ability of one of miR-1 targets, TWIST1. Activated TWIST1 then enhances the invasive and metastatic capabilities of prostate cancer cells.
Discussion
In the current study, we provide evidence for a molecular mechanism linking miR-1 and EGFR regulation to malignant phenotypes observed in advanced prostate cancers. We showed that EGFR expression reverses ectopic miR-1–mediated inhibition of invasiveness and bone metastasis (Fig. 2), implying that EGFR levels are dominant in determining the metastatic potential of prostate cancer cells. Our results support a model whereby nuclear EGFR might function as a transcriptional repressor to directly modulate miR-1 expression (Fig. 3). Although other groups observed binding of EGFR to chromatin (23), our study is the first to demonstrate that EGFR can bind to specific sequences of primary miR stem-loop promoter regions in order to suppress transcription. Overexpression of EGFR was shown in a majority of cases of prostate cancer (27), supporting our idea that EGFR signaling plays an important role in repressing miR-1 in prostate cancer. Moreover, our results showed that nuclear EGFR-dependent signatures and metastatic disease are correlated with decreased miR-1 levels as observed in clinical samples (Fig. 4). Our findings raise the interesting idea that miRs might be general targets of many transmembrane receptors, as evidenced by our results showing the regulation of miR-1 by EGFR in prostate cancer and the involvement of TWIST1 in facilitating bone metastasis by EGFR signaling networks. These data are consistent with previous studies, which implied that miR-1 acts as a potential tumor suppressor (11–13), and TWIST1 acts as an oncogene (5, 6) of prostate cancer. We demonstrated that TWIST1 and miR-1 levels are modulated by EGFR activation in a metastatic prostate cancer model and that miR-1 directly and negatively contributes to the post-transcriptional regulation of TWIST1 in experimental models (Fig. 5). It was shown that nuclear EGFR cooperates with STAT3 and induces a TWIST1-mediated malignant transformation of breast cancer cells (7). Nuclear EGFR might not only target miR-1, but it may also mediate other pathways to induce TWIST1 and promote malignant phenotypes of prostate cancer cells. Importantly, we showed that TWIST1 expression was inversely correlated with miR-1 levels in clinical samples (Fig. 6). Our current study suggests two molecular mechanisms whereby high EGFR output contributes to reduced miR-1 and increased TWIST1 levels.
Trafficking of EGFR from cytoplasmic membranes to nuclei is well documented. EGF stimulation of EGFR causes endocytosis of EGFR and an interaction with importin β via its tripartite nuclear localization sequence (28). Consistent with our study, high levels of EGFR indeed existed in nuclei upon EGF treatment (Figs. 3A and B and Supplementary Fig. S3A–S3C). Interestingly, a number of studies suggested that the nuclear counterpart of EGFR is likely the phosphorylated form of full-length receptor (23, 29–31). This idea was also supported by our results that showed that p-EGFR (Y1068 and Y845) was the predominant form observed in nuclei (Figs. 3A and Supplementary Fig. S3B). Our results indicated that EGFR can function as a transcription factor, which directly represses the tumor-suppressive role of miR-1. Interestingly, we did not observe enhanced migration or invasiveness abilities in AR-positive cell lines (Supplementary Fig. S2C). It is supported by the previous study that loss of androgen regulation of EGFR increased proliferation and survival, as well as invasion and metastasis to the bone in prostate cancer (2). Moreover, we showed that AR-positive cells express relatively higher miR-1 compared with AR-negative cells (Supplementary Fig. S2A). We concluded that the reduced miR-1 in response to EGF in AR-positive cells was insufficient to produce a malignant phenotype. Of interest, independent prostate cancer studies suggested a rationale for the correlation between TWIST1 activity and the bone metastatic phenotype (32, 33). Our results showed that cells with high EGFR signaling exhibited increased TWIST1 (Fig. 5A and B) and decreased miR-1 levels (Fig. 1A). Importantly, the activity of a TWIST1 3′-UTR reporter increased following activation of EGFR (Fig. 5E), demonstrating that post-transcriptional regulation of TWIST1 was correlated with EGFR-dependent miR-1 levels.
It was shown that TWIST but not Snail is required for Drosophila miR-1 expression in the mesoderm during muscle development (34). The tissue-specific miRNA and transcription factor regulatory network were identified by several reports (35–37), indicating that tissue-specific patterns across different tissues and species play different roles in regulating gene expressions. Our study highlights a mechanism by which prostate tumors can modulate miR-1 and TWIST1 activities leading to malignant phenotypes observed when patients relapse and progress after failing standard therapies. Data accumulated through the combined use of in vitro and in vivo systems support our idea that nuclear EGFR-miR-1 andmiR-1-TWIST1 regulatory networks are both relevant to clinical outcome analyses. High levels of nuclear EGFR are observed in many cancers, and it is well documented that nuclear EGFR signaling networks also play a vital role in many solid tumors, including prostate cancer (3). In addition, miR-1 was shown to be among the most downregulated miRs in prostate tumors compared with normal prostate tissues (13). Finally, upregulation of TWIST1 in prostate cancer was linked to the acquisition of malignant traits and may be an important therapeutic target (6). Herein, we provide a molecular explanation linking high EGFR levels in prostate cancer cells with decreased miR-1 and elevated TWIST1 signaling. These conclusions were further supported by our analyses of clinical prostate cancer specimens, which showed that miR-1 expression was mutually correlated with increased nuclear EGFR (Fig. 4) and TWIST1 (Fig. 6). miR-1 represents a novel regulatory target of EGFR that can be exploited in order to promote EGFR-mediated tumorigenic activities. Therefore, the ability to modulate miR-1 activity and/or activities of miR-1 targets, such as TWIST1, might open up new therapeutic avenues to overcome EGFR-dependent prostate bone metastasis.
Supplementary Material
Acknowledgments
Grant Support
This work was jointly supported by grants from the Ministry of Science and Technology (MOST103-2314-B-038-051) and the National Health Research Institutes (NHRI-EX104-10308BC) of Taiwan.
Footnotes
Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Authors' Contributions
Conception and design: Y.-S. Chang, Y.-N. Liu
Development of methodology: Y.-S. Chang, J.J. Yin, J. Huang, Y.-N. Liu
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): Y.-S. Chang, W.-Y. Chen, J.J. Yin, H. Tillman, J. Huang, Y.-N. Liu
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): Y.-S. Chang, W.-Y. Chen, J.J. Yin, H. Tillman, J. Huang, Y.-N. Liu
Writing, review, and/or revision of the manuscript: Y.-S. Chang, J.J. Yin, H. Tillman, Y.-N. Liu
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): Y.-S. Chang, W.-Y. Chen, Y.-N. Liu
Study supervision: Y.-N. Liu
The authors thank Dr. Ji-Hshiung Chen (Tzu Chi University, Taiwan) for reading the article and for his comments and helpful suggestions.
References
- 1.Di Lorenzo G, Tortora G, D'Armiento FP, De Rosa G, Staibano S, Autorino R, et al. Expression of epidermal growth factor receptor correlates with disease relapse and progression to androgen-independence in human prostate cancer. Clin Cancer Res. 2002;8:3438–3444. [PubMed] [Google Scholar]
- 2.Traish AM, Morgentaler A. Epidermal growth factor receptor expression escapes androgen regulation in prostate cancer: a potential molecular switch for tumour growth. Br J Cancer. 2009;101:1949–1956. doi: 10.1038/sj.bjc.6605376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brand TM, Iida M, Li C, Wheeler DL. The nuclear epidermal growth factor receptor signaling network and its role in cancer. Discov Med. 2011;12:419–432. [PMC free article] [PubMed] [Google Scholar]
- 4.Maugeri-Sacca M, Coppola V, De Maria R, Bonci D. Functional role of microRNAs in prostate cancer and therapeutic opportunities. Crit Rev Oncog. 2013;18:303–315. doi: 10.1615/critrevoncog.2013007206. [DOI] [PubMed] [Google Scholar]
- 5.Qin Q, Xu Y, He T, Qin C, Xu J. Normal and disease-related biological functions of Twist1 and underlying molecular mechanisms. Cell Res. 2012;22:90–106. doi: 10.1038/cr.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwok WK, Ling MT, Lee TW, Lau TC, Zhou C, Zhang X, et al. Up-regulation of TWIST in prostate cancer and its implication as a therapeutic target. Cancer Res. 2005;65:5153–5162. doi: 10.1158/0008-5472.CAN-04-3785. [DOI] [PubMed] [Google Scholar]
- 7.Lo HW, Hsu SC, Xia W, Cao X, Shih JY, Wei Y, et al. Epidermal growth factor receptor cooperates with signal transducer and activator of transcription 3 to induce epithelial-mesenchymal transition in cancer cells via up-regulation of TWIST gene expression. Cancer Res. 2007;67:9066–9076. doi: 10.1158/0008-5472.CAN-07-0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pala A, Karpel-Massler G, Kast RE, Wirtz CR, Halatsch ME. Epidermal to mesenchymal transition and failure of EGFR-targeted therapy in glioblastoma. Cancers (Basel) 2012;4:523–530. doi: 10.3390/cancers4020523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicoloso MS, Spizzo R, Shimizu M, Rossi S, Calin GA. MicroRNAs–the micro steering wheel of tumour metastases. Nat Rev Cancer. 2009;9:293–302. doi: 10.1038/nrc2619. [DOI] [PubMed] [Google Scholar]
- 10.Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347–355. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ambs S, Prueitt RL, Yi M, Hudson RS, Howe TM, Petrocca F, et al. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res. 2008;68:6162–6170. doi: 10.1158/0008-5472.CAN-08-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hudson RS, Yi M, Esposito D, Watkins SK, Hurwitz AA, Yfantis HG, et al. MicroRNA-1 is a candidate tumor suppressor and prognostic marker in human prostate cancer. Nucleic Acids Res. 2012;40:3689–3703. doi: 10.1093/nar/gkr1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martens-Uzunova ES, Jalava SE, Dits NF, van Leenders GJ, Moller S, Trapman J, et al. Diagnostic and prognostic signatures from the small non-coding RNA transcriptome in prostate cancer. Oncogene. 2012;31:978–991. doi: 10.1038/onc.2011.304. [DOI] [PubMed] [Google Scholar]
- 14.Liu YN, Yin JJ, Abou-Kheir W, Hynes PG, Casey OM, Fang L, et al. MiR-1 and miR-200 inhibit EMT via Slug-dependent and tumorigenesis via Slug-independent mechanisms. Oncogene. 2013;32:296–306. doi: 10.1038/onc.2012.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yin JJ, Zhang L, Munasinghe J, Linnoila RI, Kelly K. Cediranib/AZD2171 inhibits bone and brain metastasis in a preclinical model of advanced prostate cancer. Cancer Res. 2010;70:8662–8673. doi: 10.1158/0008-5472.CAN-10-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yin J, Pollock C, Tracy K, Chock M, Martin P, Oberst M, et al. Activation of the RalGEF/Ral pathway promotes prostate cancer metastasis to bone. Mol Cell Biol. 2007;27:7538–7550. doi: 10.1128/MCB.00955-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siu MK, Abou-Kheir W, Yin JJ, Chang YS, Barrett B, Suau F, et al. Loss of EGFR signaling regulated miR-203 promotes prostate cancer bone metastasis and tyrosine kinase inhibitors resistance. Oncotarget. 2014;5:3770–3784. doi: 10.18632/oncotarget.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siu MK, Tsai YC, Chang YS, Yin JJ, Suau F, Chen WY, et al. Transforming growth factor-beta promotes prostate bone metastasis through induction of microRNA-96 and activation of the mTOR pathway. Oncogene. 2014 Dec 22; doi: 10.1038/onc.2014.414. [DOI] [PubMed] [Google Scholar]
- 19.Yin J, Liu YN, Tillman H, Barrett B, Hewitt S, Ylaya K, et al. AR-regulated TWEAK-FN14 pathway promotes prostate cancer bone metastasis. Cancer Res. 2014;74:4306–4317. doi: 10.1158/0008-5472.CAN-13-3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu YN, Yin J, Barrett B, Sheppard-Tillman H, Li D, Casey OM, et al. Loss of androgen-regulated MicroRNA 1 activates SRC and promotes prostate cancer bone metastasis. Mol Cell Biol. 2015;35:1940–1951. doi: 10.1128/MCB.00008-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Day CP, Carter J, Bonomi C, Esposito D, Crise B, Ortiz-Conde B, et al. Lentivirus-mediated bifunctional cell labeling for in vivo melanoma study. Pigment Cell Melanoma Res. 2009;22:283–295. doi: 10.1111/j.1755-148X.2009.00545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin SY, Makino K, Xia W, Matin A, Wen Y, Kwong KY, et al. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat Cell Biol. 2001;3:802–808. doi: 10.1038/ncb0901-802. [DOI] [PubMed] [Google Scholar]
- 24.Hanada N, Lo HW, Day CP, Pan Y, Nakajima Y, Hung MC. Co-regulation of B-Myb expression by E2F1 and EGF receptor. Mol Carcinog. 2006;45:10–17. doi: 10.1002/mc.20147. [DOI] [PubMed] [Google Scholar]
- 25.Lo HW, Cao X, Zhu H, Ali-Osman F. Cyclooxygenase-2 is a novel transcriptional target of the nuclear EGFR-STAT3 and EGFRvIII-STAT3 signaling axes. Mol Cancer Res. 2010;8:232–245. doi: 10.1158/1541-7786.MCR-09-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaganathan S, Yue P, Paladino DC, Bogdanovic J, Huo Q, Turkson J. A functional nuclear epidermal growth factor receptor, SRC and Stat3 heteromeric complex in pancreatic cancer cells. PLoS ONE. 2011;6:e19605. doi: 10.1371/journal.pone.0019605. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Schlomm T, Kirstein P, Iwers L, Daniel B, Steuber T, Walz J, et al. Clinical significance of epidermal growth factor receptor protein overexpression and gene copy number gains in prostate cancer. Clin Cancer Res. 2007;13:6579–6584. doi: 10.1158/1078-0432.CCR-07-1257. [DOI] [PubMed] [Google Scholar]
- 28.Lo HW, Ali-Seyed M, Wu Y, Bartholomeusz G, Hsu SC, Hung MC. Nuclear-cytoplasmic transport of EGFR involves receptor endocytosis, importin beta1 and CRM1. J Cell Biochem. 2006;98:1570–1583. doi: 10.1002/jcb.20876. [DOI] [PubMed] [Google Scholar]
- 29.Cao H, Lei ZM, Bian L, Rao CV. Functional nuclear epidermal growth factor receptors in human choriocarcinoma JEG-3 cells and normal human placenta. Endocrinology. 1995;136:3163–3172. doi: 10.1210/endo.136.7.7540549. [DOI] [PubMed] [Google Scholar]
- 30.Lo HW, Hsu SC, Ali-Seyed M, Gunduz M, Xia W, Wei Y, et al. Nuclear interaction of EGFR and STAT3 in the activation of the iNOS/NO pathway. Cancer Cell. 2005;7:575–589. doi: 10.1016/j.ccr.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 31.Lo HW, Xia W, Wei Y, Ali-Seyed M, Huang SF, Hung MC. Novel prognostic value of nuclear epidermal growth factor receptor in breast cancer. Cancer Res. 2005;65:338–348. [PubMed] [Google Scholar]
- 32.Yuen HF, Chua CW, Chan YP, Wong YC, Wang X, Chan KW. Significance of TWIST and E-cadherin expression in the metastatic progression of prostatic cancer. Histopathology. 2007;50:648–658. doi: 10.1111/j.1365-2559.2007.02665.x. [DOI] [PubMed] [Google Scholar]
- 33.Yuen HF, Kwok WK, Chan KK, Chua CW, Chan YP, Chu YY, et al. TWIST modulates prostate cancer cell-mediated bone cell activity and is upregulated by osteogenic induction. Carcinogenesis. 2008;29:1509–1518. doi: 10.1093/carcin/bgn105. [DOI] [PubMed] [Google Scholar]
- 34.Sokol NS, Ambros V. Mesodermally expressed Drosophila microRNA-1 is regulated by Twist and is required in muscles during larval growth. Genes Dev. 2005;19:2343–2354. doi: 10.1101/gad.1356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hobert O. Gene regulation by transcription factors and microRNAs. Science. 2008;319:1785–1786. doi: 10.1126/science.1151651. [DOI] [PubMed] [Google Scholar]
- 36.Re A, Cora D, Taverna D, Caselle M. Genome-wide survey of microRNA-transcription factor feed-forward regulatory circuits in human. Mol Biosyst. 2009;5:854–867. doi: 10.1039/b900177h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.