Abstract
Enzymes are continually evolving in response to environmental pressures. In order to increase enzyme fitness, amino acid substitutions can occur leading to a changing function or an increased stability. These evolutionary drivers determine the activity of an enzyme and its success in future generations in response to changing conditions such as environmental stressors or to improve physiological function allowing continual persistence of the enzyme. With recent warning reports on antibiotic resistance and multidrug resistant bacterial infections, understanding the evolution of β-lactamase enzymes, which are a large contributor to antibiotic resistance, is increasingly important. Here, we investigated a variant of the SHV β-lactamase identified from a clinical isolate of Escherichia coli in 2011 (SHV-129, G238S-E240K-R275L-N276D) to identify the first instance of a global suppressor substitution in the SHV β-lactamase family. We have used this enzyme to show that several evolutionary principles are conserved in different class A β-lactamases, such as active site mutations reducing stability and requiring compensating suppressor substitutions in order to ensure evolutionary persistence of a given β-lactamase. However, the pathway taken by a given β-lactamase in order to reach its evolutionary peak under a given set of conditions is likely different. We also provide further evidence for a conserved stabilizing substitution among class A β-lactamases, the back to consensus M182T substitution. In addition to expanding the spectrum of β-lactamase activity to include the hydrolysis of cefepime, the amino acid substitutions found in SHV-129 provide the enzyme with an excess of stability, which expands the evolutionary landscape of this enzyme and may result in further evolution to potentially include resistance to carbapenems or β-lactamase inhibitors.
Keywords: global suppressors, antibiotic resistance, protein evolution, β-lactamase
Introduction
Generally, amino acid substitutions that occur in the active site of β-lactamases leading to a change in the hydrolysis profile in response to antibiotic challenge are thought to be the main evolutionary mechanism of these enzymes. These substitutions provide the ability to hydrolyze different β-lactams or to be resistant to inactivation by β-lactamase inhibitors. A second important mechanism of enzyme evolution was also identified: substitutions that do not directly affect interactions with substrates or inhibitors, but stabilize the protein (i.e., global suppressors) (Huang and Palzkill 1997; Orencia et al. 2001; Sideraki et al. 2001; Wang et al. 2002; Bershtein et al. 2006; Kather et al. 2008; Marciano et al. 2008; Brown et al. 2010; Dellus-Gur et al. 2013). Global suppressor substitutions can increase the ability of the enzymes to acquire additional substitutions (Orencia et al. 2001; Wang et al. 2002; Bershtein et al. 2006; Dellus-Gur et al. 2013). These changes can also allow the protein to remain functional in challenging conditions (e.g., in increasing antibiotic concentration, during the activation of the SOS system; in changing pH; and with increasing intracellular concentration of salt) (Bershtein et al. 2006).
A paradigm of protein evolution has developed where there is a “trade-off” between active site amino acid substitutions that change the function and activity of the enzyme and stabilizing substitutions that do not affect the protein function, but allow additional mutations to accumulate (Huang and Palzkill 1997; Orencia et al. 2001; Sideraki et al. 2001; Wang et al. 2002; Bershtein et al. 2006; Kather et al. 2008; Marciano et al. 2008; Brown et al. 2010; Thomas et al. 2010; Salverda et al. 2011; Goldsmith and Tawfik 2012; Dellus-Gur et al. 2013; Wellner et al. 2013; Tawfik 2014; Toth-Petroczy and Tawfik 2014). This theory proposes that every protein has a particular “threshold robustness,” which is a margin of excess stability that can be lost with the accumulation of amino acid substitutions that affect substrate specificity without losing fitness of the overall protein (Bershtein et al. 2006). The evolution of these substitutions is dependent on epistasis, or the interactions of one amino acid change with another and its effect on fitness (Bershtein et al. 2006; Salverda et al. 2011; Goldsmith and Tawfik 2012; Dellus-Gur et al. 2013; Wellner et al. 2013; Toth-Petroczy and Tawfik 2014). This epistatic relationship can affect the type and order of mutation accumulation and determine the direction that evolution proceeds (Salverda et al. 2011; Goldsmith and Tawfik 2012; Wellner et al. 2013; Toth-Petroczy and Tawfik 2014). Understanding the relationship of amino acid substitutions not only to overall protein function but also to each other is key to predicting the evolution of β-lactamases.
Several amino acid substitutions have been shown to confer a global suppressor phenotype in the Temoneira (TEM) β-lactamase family (Huang and Palzkill 1997; Sideraki et al. 2001; Wang et al. 2002; Kather et al. 2008; Marciano et al. 2008; Brown et al. 2010) and this type of substitution has been found in other enzymes including the Staphylococcal nuclease, phage P22, the p53 tumor suppressor, Saccharomyces cerevisiae cytochrome c, and bacterial lysozyme (Matthews et al. 1987; Doyle et al. 1996; Aramli and Teschke 1999; Odell et al. 2013). However, the presence of these substitutions in β-lactamases besides the TEM enzyme is unknown. One of the best characterized global suppressor substitutions in TEM, a back-to-consensus substitution, M182T, was identified after its appearance in multiple enzymes in combination with other amino acid substitutions, but never alone (Jacoby and Bush 2014). The Thr at position 182 represents a “back to consensus” substitution for TEM as most class A β-lactamases have 182T; consensus substitutions are also commonly stabilizing for enzymes (Risso et al. 2015).
Although TEM has frequently been used as a model enzyme for the study of evolution, it is unclear whether the principles found in TEM apply to other β-lactamases. SulfHydryl Variable (SHV) is a β-lactamase that was discovered in 1982 and is one of the most common extended-spectrum β-lactamases (ESBLs) observed clinically (Bush 2008). SHV and TEM are members of the same class of β-lactamase (class A), which describes both their structure and the wild-type function of predominant penicillinase activity (Jacoby and Bush 2014). Although the secondary structure and active sites of SHV and TEM are very similar and these enzymes share 68% sequence identity (fig. 1), there are several differences in the enzymes that may affect their evolution. Global suppressor or stabilizing substitutions commonly occur outside the active site of enzymes (Marciano et al. 2008; Brown et al. 2010). The various differences between SHV and TEM outside of the active site may contribute to differences in stabilization effects in these enzymes and to what amino acids are selected at a given position in each. Thus far, this question has not been studied to our knowledge.
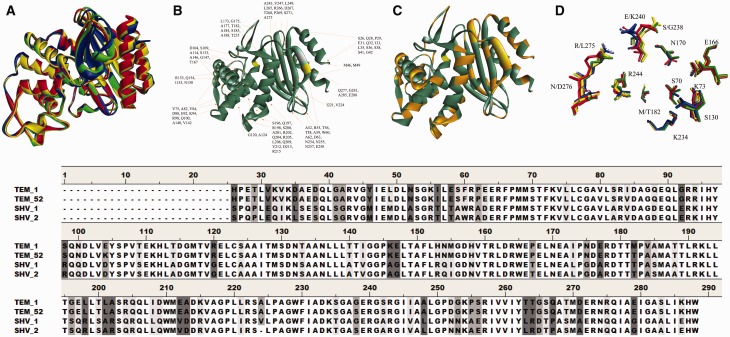
Fig. 1.
(A) Overlay of the ribbon structure of TEM-1 (protein database (PDB) ID: 1ZG4, yellow), TEM-52 (PDB ID: 1M40, red), SHV-1 (PDB ID: 1SHV, green), and SHV-2 (PDB ID: 1N9B, blue). (B) Ribbon diagram of SHV-1 (PDB ID: 1SHV) with amino acid differences with TEM-1 identified, S70 is in yellow. (C) Ribbon diagram of SHV-1 (PDB ID: 1SHV) with the location of amino acid differences with TEM-1 marked in orange. (D) Active site overlay of TEM-1 (PDB ID: 1ZG4, yellow), TEM-52 (PDB ID: 1M40, red), SHV-1 (PDB ID: 1SHV, green), and SHV-2 (PDB ID: 1N9B, blue). The amino acids studied in this article are also shown. Overall, the active site amino acids are very similar between these proteins. The amino acid alignment of these four β-lactamases is also shown below with shading at the site of amino acid differences illustrated in B and C.
A novel SHV variant, SHV-129, was identified in 2012 (Lascols et al. 2012) and found to contain two well-characterized ESBL substitutions (G238S and E240K) in addition to two substitutions that were not previously observed in clinical isolates containing SHV enzymes; these were formerly characterized as global suppressor substitutions in TEM (R275L and N276D) (Kather et al. 2008; Brown et al. 2010). Interestingly, this is the first complex SHV variant to emerge clinically (Jacoby and Bush 2014). The N276D substitution was also characterized as leading to clavulanic acid resistance in SHV in laboratory studies, but its stabilization effects were not appreciated (Drawz et al. 2009). The appearance of the R275L and N276D substitutions in a bacterial isolate containing SHV-129 led us to hypothesize that 1) these substitutions would result in a different hydrolysis profile of bacteria containing this enzyme, and 2) these amino acid substitutions were serving a role as global suppressors to offset destabilization due to the G238S and E240K substitutions. We used this isolate as an entry into the study of stabilizing substitutions in the SHV β-lactamase and to begin to understand the universal principles of global suppressors and stabilizers in class A β-lactamases. We also wanted to explore the effects of making the T182M substitution in SHV and whether a Met at position 182 would destabilize this β-lactamase. Our investigation revealed that 1) the substitution N276D had a major impact on SHV-mediated β-lactam resistance, 2) there are differences in the role of R275L in TEM and SHV, and 3) that a Thr at position 182 is universally important in the stabilization of class A β-lactamases providing further evidence about the stabilizing effects of consensus amino acids.
Results and Discussion
Mutagenesis
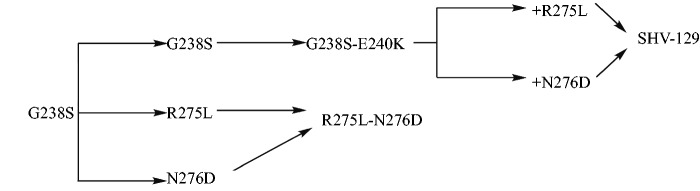
We developed a mutagenesis scheme in order to mimic the hypothesized evolutionary pathway of SHV-129 (fig. 2). SHV-2 (G238S) and SHV-5 (G238S-E240K) were engineered first as they have been identified repeatedly both alone and in combination in clinical isolates containing blaSHV variant enzymes since the 1980s (Jacoby and Bush 2014). We also constructed all variants as single substitutions (E240K, R275L, and N276D) in addition to the single substitution T182M to create a variant of SHV that resembles wild-type TEM at this position. We then made triple mutants on the pathway to SHV-129 by designing G238S-E240K-R275L and G238S-E240K-N276D. We also engineered the double mutant R275L-N276D to determine if stabilization effects by either single substitution were additive.
Fig. 2.
Proposed evolutionary mutagenesis scheme for SHV-129 including a separate path testing only our two hypothesized suppressor substitutions (R275L and N276D). G238S is SHV-2, G238S-E240K is SHV-5, and G238S-E240K-R275L-N276D is SHV-129.
Whole Cell Assays and Resistance Phenotype
Whole cell assays were performed to determine the resistance profile of all variants (table 1). Generally, in enzymes expressed in Escherichia coli DH10B, we found that resistance to ceftazidime predominated when compared with resistance to β-lactamase inhibitors as ampicillin–clavulanic acid resistance was lost when N276D was combined with any other substitution (alone the minimum inhibitory concentration [MIC] is 16 mg/l but with other substitutions it is 0.5–4 mg/l). Unlike in TEM where it leads to inhibitor resistant (IR) properties (Kather et al. 2008; Brown et al. 2010), the R275L substitution had no effect on the resistance of SHV expressed in an isogenic E. coli background.
Table 1.
MICs in mg/l of Variants Leading to SHV-129 with a Variety of β-Lactams and Inhibitors.
| AMP | THIN | CAZ | TAX | FEP | AZT | AMP-CLAVa | AMP-SULa | AMP-TAZOa | |
|---|---|---|---|---|---|---|---|---|---|
| Escherichia coli DH10B empty | 0.5 | 4–8 | <0.06 | <0.06 | <0.06 | <0.06 | 0.12 | 0.12 | 0.12 |
| SHV-1 | >16,384 | 256 | 4 | 0.12 | 1 | 0.12 | 8 | 512 | 64 |
| SHV-2 (G238S) | 4,096 | >512 | 16 | 16–32 | 4 | 1 | 0.5 | 16 | 8 |
| E240K | >16,384 | 128 | 16 | 0.12 | 0.5 | 0.12–0.25 | 4 | 256 | 64 |
| SHV-5 (G238S-E240K) | 1,024 | >512 | >32 | 32 | 4 | 32 | 0.5 | 16 | 8 |
| R275L | >16,384 | 256 | 4 | 0.12 | 0.12 | 0.12 | 2 | 128 | 16 |
| N276D | 16,384 | 32 | 4 | 0.12 | 1 | <0.06 | 16 | 256 | 64 |
| R275L-N276D | 8,192 | 32 | 4 | <0.06 | 1 | <0.06 | 4 | 128 | 32 |
| G238S-E240K-R275L | 2,048–4,096 | >512 | >32 | 16–32 | 8 | >32 | 0.25 | 8 | 8 |
| G238S-E240K-N276D | 2,048–4,096 | >512 | >32 | 16 | 8 | >32 | 0.5 | 8 | 8 |
| SHV-129 (G238S-E240K-R275L-N276D) | 2,048–4,096 | 512 | >32 | 4 | 16 | 32 | 0.5 | 8 | 8 |
| T182M | 8,192–16,384 | 32 | 0.12 | <0.06 | 0.12 | 0.12 | 0.5 | ND | ND |
Note.—AMP = ampicillin; THIN = cephalothin; CAZ = ceftazidime; FEP = cefepime; AZT = aztreonam; CLAV = clavulanic acid; SUL = sulbactam; TAZO = tazobactam; ND = not determined.
aInhibitor held constant at 4 mg/l and ampicillin concentration increased in doubling dilutions.
SHV-129 expressed in E. coli DH10B showed elevated resistance to expanded-spectrum cephalosporins (ceftazidime, cefepime, cefotaxime) compared with SHV-1, but lower MICs against inhibitor combinations, suggesting that it displays a true ESBL phenotype with inhibitor “hypersusceptibility” (Kalp et al. 2009).
In comparison with the two proposed ancestor enzymes, SHV-2 and SHV-5, SHV-129 expressed in E. coli DH10B displayed a lower cefotaxime MIC (4 vs. 16–32 mg/l) but a higher cefepime MIC (16 vs. 4 mg/l). The MIC for E. coli DH10B expressing SHV-129 against ceftazidime is the same as E. coli expressing SHV-5 at >32 mg/l. The two triple mutants (G238S-E240K-R275L and G238S-E240K-N276D) showed the same level of resistance when expressed in E. coli DH10B. These variants displayed the same ceftazidime MIC as E. coli DH10B expressing SHV-5 and SHV-129; cefepime resistance was intermediate at 8 mg/l. The elevation in cefepime MIC for the mutants may be due to a change in the β-lactamase structure and/or hydrogen bonding that only occurs when 3–4 of these substitutions are present, or it may be due to stabilization of the enzyme with a R275L and/or N276D substitution in SHV leading to increased protein production. Neither of the R275L or N276D single substitutions in an isogenic background increases cefepime resistance; in fact, the MIC for R275L alone in E. coli is reduced for cefepime (0.12 vs. 1 mg/l for SHV-1), which indicates that stabilization and increased active protein production may be leading to the cefepime resistance phenotype. Additionally, R275L-N276D in combination also does not increase cefepime resistance when expressed in E. coli (MIC of 1 mg/l).
The singly substituted E240K variant expressed in E. coli DH10B showed an unexpected phenotype as it has an elevated ceftazidime MIC (16 mg/l) without affecting resistance to any other β-lactam or inhibitor combinations. Interestingly, this single substitution is not prevalent in clinical isolates (Bush 2008; Doi et al. 2013). One explanation for this may be that the E240K substitution is highly destabilizing and therefore not favored compared with the SHV-2 or G238S-E240K combination through natural selection (as discussed in Introduction, for an enzyme to be selected it must be not only more fit than the wild type, but also more fit than other variants) (Majiduddin and Palzkill 2003). This hypothesis is further probed by assessing steady-state protein expression levels and protein stability.
The MICs for the T182M variant expressed in E. coli DH10B were severely impaired for nonpenicillin β-lactams and β-lactamase inhibitor combinations. This could either be due to catalytic impairment of this enzyme or due to the predicted instability. We further probe this phenotype through steady-state expression and stability measurements as well.
Kinetics
We next tested the kinetic parameters of SHV-129 and SHV-1 for β-lactams and β-lactamase inhibitors to understand the resistance that has evolved in this enzyme compared with its evolutionary precursors (table 2). We compared these values with SHV-2 and SHV-5 (Kalp et al. 2009). Generally, SHV-129 shows similar kinetic properties as SHV-2 and SHV-5 with observed hydrolysis of cefotaxime and ceftazidime and the values for catalytic efficiency mirror the observed whole cell assays in table 1. Additionally, SHV-129 is better able to hydrolyze cefepime (higher kcat/Km than the three precursor enzymes SHV-1, SHV-2, and SHV-5). SHV-129 is also hypersusceptible to β-lactamase inhibitors with a low Ki app for tazobactam, sulbactam, and clavulanic acid (0.44, 1.6, and 1.3 µM, respectively).
Table 2.
Enzyme Kinetics for β-Lactams and β-Lactamase Inhibitors by Several SHV Variant β-Lactamases.
| β-Lactam or Inhibitor | β-Lactamase |
|||||
|---|---|---|---|---|---|---|
| SHV-1 | SHV-2b | SHV-5b | SHV-129 | |||
| NCF | Km | 25 ± 5 | 5 | 5 | 4.6 ± 1.1 | |
| kcat | 348 ± 21 | 101 | 52 | 93.1 ± 9 | ||
| kcat/Km | 14 ± 0.2 | 20.2 | 10.4 | 20.4 ± 2 | ||
| AMP | Km | 152 ± 33 | 12 | 10 | 46.8 ± 24 | |
| kcat | 2,802 ± 214 | 206 | 122 | 22.8 ± 11 | ||
| kcat/Km | 18 ± 0.2 | 17 | 12 | 0.5 ± 0.7 | ||
| PIP | Km | 75 ± 12 | ND | ND | 25 ± 9 | |
| kcat | 1,179 ± 65 | ND | ND | 168 ± 4 | ||
| kcat/Km | 16 ± 0.6 | ND | ND | 7 ± 0.4 | ||
| THIN | Km | 49 ± 11 | ND | ND | 12.1 ± 3.7 | |
| kcat | 43 ± 3 | ND | ND | 26 ± 1 | ||
| kcat/Km | 0.9 ± 0.3 | ND | ND | 2.2 ± 0.3 | ||
| TAX | Km | NA | 18 | 4 | 26.7 ± 5.5 | |
| kcat | NA | 11 | 6 | 4.8 ± 3.4 | ||
| kcat/Km | ≤0.001c | 0.6 | 1.5 | 0.2 ± 0.5 | ||
| CAZ | Km | NA | ND | 17 | 24 ± 3 | |
| kcat | NA | ND | 2.6 | 3.1 ± 1.5 | ||
| kcat/Km | ≤2 × 10−5,c | ND | 0.15 | 0.13 ± 0.5 | ||
| FEP | Km | NA | NA | NA | 52 ± 35 | |
| kcat | NA | NA | NA | 4.5 ± 0.5 | ||
| kcat/Km | ≤0.004c | ≤0.008c | ≤0.03c | 0.09 ± 0.01 | ||
| TAZO | Ki | 0.44 | ND | ND | 0.04 | |
| SUL | Ki | 1.6 | ND | ND | 0.4 | |
| CLAV | Ki | 1.3 | ND | ND | 0.4 | |
Note.—NCF = nitrocefin; AMP ampicillin; PIP = piperacillin; THIN = cephalothin; TAX = cefotaxime; CAZ = ceftazidime; FEP = cefepime; TAZO = tazobactam; SUL = sulbactam; CLAV = clavulanic acid; ND = not determined in the article; NA = not able to determine the rate of hydrolysis and affinity. Km (µM), kcat (/s), kcat/Km (/µM/s).
aSHV-2 is G238S, SHV-5 is G238S-E240K, and SHV-129 is G238S-E240K-R275L-N276D.
bValues are adapted with permission from Kalp et. al. (2009).
cEstimate of upper limit of kcat/Km calculated as described in Methods.
SHV-129 has a lower kcat for ampicillin compared with SHV-1, SHV-2, and SHV-5 leading to a reduced catalytic efficiency. For expanded-spectrum cephalosporins, SHV-129 has a similar catalytic efficiency as SHV-2 for cefotaxime and as SHV-5 for ceftazidime. The catalytic efficiency of SHV-129 for cefepime is 10-fold lower than it is for ceftazidime, but still significantly higher than for the SHV-1, SHV-2, or SHV-5 variants. Overall, the lower catalytic efficiencies of SHV-129 compared with SHV-1, -2, and -5 for various substrates are due to both changes in kcat or Km. For substrates with a smaller R2 group but a larger R1 group (ampicillin, piperacillin, and cephalothin) the kcat is lower. However, for substrates with larger R1 and R2 groups (ceftazidime and cefotaxime), the Km is higher. The N276D substitution in SHV was previously proposed to restrict the location of R244, which impairs carboxylate recognition (Giakkoupi et al. 1999; Drawz et al. 2009). Perhaps in SHV-129 the restriction of R244 leads to this higher Km by impairing recognition of this same group. However, this is not in accordance with our inhibitor susceptibility. Overall, it seems that the active site of SHV-129 is optimized for the hydrolysis of third and fourth generation cephalosporins, which makes it less like the penicillinase enzymes of the typical TEM and SHV class A β-lactamases.
Steady-State Protein Expression
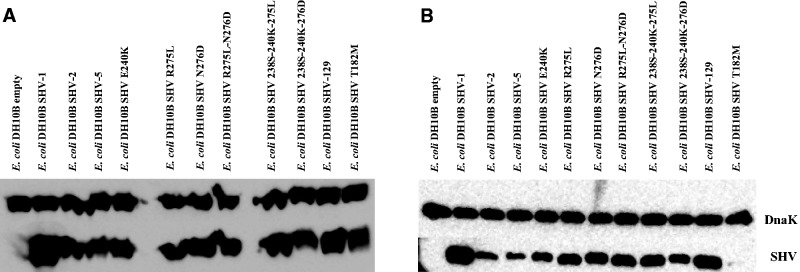
Immunoblots were examined for the steady-state expression level of the variants in whole cell and crude periplasmic preparations collected at OD600 = 0.8 (optical density) (fig. 3). We performed these experiments to determine if there was reduced stability due to bacterial proteolysis of different enzyme variants or if there was evidence of improper expression due to the formation of inclusion bodies. The whole cell immunoblot (fig. 3A) showed the highest expression of SHV-1 compared with all the variants. SHV-5 is expressed at a lower level in comparison with the other proteins. However, all the β-lactamase enzymes seem to be expressed well in the whole cell immunoblot.
Fig. 3.
(A) Immunoblot of variants leading to SHV-129 showing the various expression levels of these proteins in whole cell bacterial preparations. (B) Immunoblot of variants leading to SHV-129 showing the various expression levels of these proteins in soluble fraction preparations. SHV-2 is G238S, SHV-5 is G238S-E240K, and SHV-129 is G238S-E240K-R275L-N276D.
In contrast, the soluble fraction immunoblot (fig. 3B) demonstrated differences in expression levels indicating that the enzymes are either unstable and being degraded during the purification process or that the enzymes are not being properly folded, and inclusion bodies are formed that are removed during the preparation. Figure 3B shows that SHV-2 (G238S), SHV-5 (G238S-E240K), and SHV E240K seemed to be present at the lowest amount in the soluble fractions in comparison with SHV-1 and the other SHV variants. In contrast, SHV-129 (G238S-E240K-R275L-N276D) has a high level of protein expression on the immunoblot. Therefore, R275L and N276D either separately or together restored expression and/or proper folding of SHV enzymes containing the G238S and/or E240K substitutions indicating that one or both of the R275L and/or N276D substitutions may serve as a global suppressor. Strikingly, the T182M protein is barely detected in the soluble fraction immunoblot. This indicates that there is a severe destabilizing effect when a methionine is present at this position, which is similar to results observed with TEM (Huang and Palzkill 1997; Orencia et al. 2001; Sideraki et al. 2001; Kather et al. 2008; Brown et al. 2010). Therefore, the consensus sequence of class A β-lactamases with a Thr at position 182 is likely very important in the stability of these enzymes and the Met at position 182 of TEM-1 does seem to be an “evolutionary aberration” with an unclear purpose as the evolution of TEM also seems to favor a Thr at position 182 for its stabilizing effects (Jacoby and Bush 2014). Further exploration of the stability and folding of the SHV variants was then performed through circular dichroism (CD) and thermal denaturation.
Protein Structure and Stability
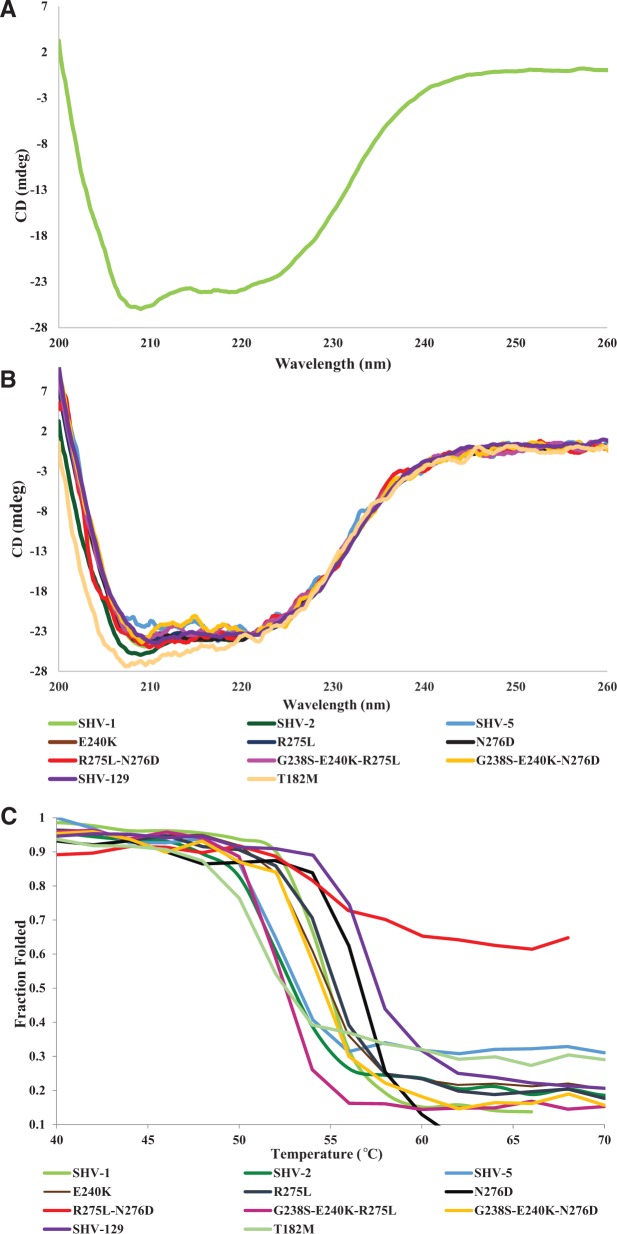
In SHV-1, the percentage of various secondary structure elements can be visualized through CD (fig. 4A) (Greenfield and Fasman 1969). The α-helical moieties of SHV-1 predominate with negative peaks at 221 and 208 nm (Greenfield and Fasman 1969). The β-sheet domains of SHV-1 appear as a negative peak at 214 nm (Greenfield and Fasman 1969). In figure 4B, we observe the secondary structures of the SHV variant enzymes. Overall, the secondary structures are very similar between the variant enzymes. This is to be expected as single amino acid substitutions are unlikely to lead to an unfolding or change in secondary structure unless a proline residue, which would destabilize a helix, was added. However, one difference is in the T182M structure: The 214-nm α-helical peak is larger for this enzyme than for the other variants. One hypothesis for the stabilization of the M182T substitution in TEM is that it allows an additional hydrogen bond between the domains of the β-lactamase (Huang and Palzkill 1997; Orencia et al. 2001). The loss of an interdomain hydrogen bond with the methionine substitution may lead to our observed secondary structure changes with the T182M substitution in addition to destabilization of the enzyme. This change in structure may also affect the folding of the T182M SHV variant, which results in reduced stability and lower expression observed in the periplasmic immunoblot in figure 3B.
Fig. 4.
(A) Overall CD structure of SHV-1 showing the secondary structure elements of this wild-type β-lactamase. (B) Overall CD structure of the various SHV enzymes showing similarities in α-helical and β-sheet structure with the exception of the T182M variant. (C) Melting curves of the various SHV enzymes showing stabilization of the SHV-129 enzyme and destabilization of T182M, SHV-2 (G238S), and SHV-5 (G238S-E240K) β-lactamases. The R275L-N276D protein does not completely unfold.
The thermal denaturation curves (fig. 4C) allowed us to calculate a melting temperature for each variant (table 3). From these results, several of our hypotheses of the role of these substitutions are supported (as discussed in the Introduction). If we compare the stability of SHV-1 (as it is the wild-type enzyme) with the other purified β-lactamases, we can determine changes in the stability of these variants. We found that the T182M substitution is significantly destabilizing (a Tm of 50.4 °C for the variant vs. 54.5 °C for SHV-1). This corresponds to the major changes in the MICs for the T182M variant of SHV, particularly for cephalosporins and inhibitor combinations. We also see destabilization of SHV-2 and SHV-5 (melting temperatures of 52 °C and 51.6 °C, respectively). Unexpectedly, the E240K substitution seems to have only a minor effect on stability with the melting temperature of SHV-5 slightly lower than SHV-2 and the E240K melting temperature (53.9 °C) slightly lower than the SHV-1 melting temperature (54.5 °C). We expected the E240K substitution to be more destabilizing as it is generally not observed in ESBL SHV variant enzymes in the absence of a second substitution (Jacoby and Bush 2014).
Table 3.
Melting Temperatures of β-Lactamase Variants Leading to SHV-129.
| Tm (°C) | ΔTm (°C)b | |
|---|---|---|
| SHV-1 | 54.5 ± 0.2 | — |
| SHV-2 (G238S) | 52.0 ± 0.4 | −2.5 |
| E240K | 53.9 ± 0.8 | −0.6 |
| SHV-5 (G238S-E240K) | 51.6 ± 0.2 | −2.9 |
| R275L | 54.6 ± 1.0 | +0.1 |
| N276D | 56.3 ± 0.7 | +1.8 |
| R275L-N276D | NDa | |
| G238S-E240K-R275L | 52.2 ± 0.3 | +0.6 |
| G238S-E240K-N276D | 54.2 ± 0.3 | +2.6 |
| SHV-129 (G238S-E240K-R275L-N276D) | 57.3 ± 1.2 | +2.8 (compared with SHV-1) |
| +5.1 (compared with G238S-E240K-R275L) | ||
| +3.1 (compared with G238S-E240K-N276D) | ||
| T182M | 50.4 ± 0.5 | −4.1 |
aValue unable to be determined due to incomplete denaturation of the protein.
bΔTm reported relative to SHV-1 for single variants and relative to SHV-5 for triple variants.
We observed that the N276D substitution was stabilizing (Tm of 56.3 °C compared with 54.5 °C for SHV-1). This substitution was able to act as a suppressor substitution in combination with G238S-E240K (melting temperature of 54.2 °C). Unexpectedly, the R275L substitution did not seem to be stabilizing on its own (54.6 °C) or in combination with G238S-E240K (52.2 °C). Therefore, the R275L substitution is unlikely to be a global suppressor substitution. However, when the R275L and N276D substitutions are combined in SHV-129 further stabilization is observed (melting temperature of 57.3 °C). Therefore, R275L seems to be a stabilizing substitution, but it is not a global suppressor, because it requires a certain protein sequence background (perhaps N276D) for stabilizing effects to be observed in SHV. We conclude that this is an example of sign epistasis, where the positive effects of the R275L substitution are dependent on a particular genetic background. The R275L-N276D double substitution melting curve lends some support to this as the β-lactamase does not completely unfold. Therefore, a formal melting temperature cannot be assigned for this variant, but a lack of unfolding indicates that this variant is likely very stabilized relative to the other β-lactamase enzymes that were measured. From these results, we posit that the R275L and N276D substitutions evolved in the SHV β-lactamase mainly as stabilizing substitutions with a contribution of cefepime resistance.
In order to further analyze the stability and movement of the variant proteins, we performed a B factor analysis of several crystallized enzyme variants, SHV-1 (PDB ID: 1SHV), SHV-2 (PDB ID: 1N9B), TEM-1 (PDB ID: 1ZG4), and a “SHV-2-like” TEM, TEM-52 (PDB ID: 1M40). B factors provide an estimate of the amount that an atom deviates from its average position in a crystal, which can be used to make comparisons of the molecular flexibility of a given amino acid or region of the protein (Dellus-Gur et al. 2015). Unfortunately, SHV-129 and SHV-5 have not been successfully captured by X-ray crystallography, so we cannot make further conclusions about the effects of E240K, R275L, and N276D on amino acid movement in SHV.
Our B factor analysis (supplementary fig. S1, Supplementary Material online) is similar to previously published studies on the TEM β-lactamase (Dellus-Gur et al. 2015). We found that the Ω-loop (R164-D179) is highly flexible (highly positive B factor). The G238 loop is also highly flexible (G238-A243). The major notable difference we observe when comparing SHV-1 with SHV-2 (G238S) is that the G238 loop is more mobile in SHV-2. This is likely part of the explanation behind the increased hydrolysis of cefotaxime by this enzyme in addition to its wider more shallow active site (Nukaga et al. 2003). Additionally, the increased mobility of this loop in SHV-2 may attribute to its lower thermal stability.
We also looked at the flexibility of individual amino acids important in the hydrolytic pathway of class A β-lactamases (S70, K73, S130, E166, N170, K234) (Drawz and Bonomo 2010). Generally, S70, S130, and K234 were not very mobile in any of the structures and no differences were observed in our comparisons among the enzymes. These results are key in the enzymatic function of the β-lactamase and a relatively fixed position may be favorable. In contrast, E166 and N170 were more flexible in SHV compared with TEM and in SHV-2 N170 was more mobile than in SHV-1. E166 and N170 are known to anchor a key water molecule in the deacylation pathway of β-lactamases and the mobility of these residues may also have implications for the enzyme function (Kuzin et al. 1999; Nukaga et al. 2003). We also found differences in the mobility of K73; in SHV-1 and TEM-1, this residue was not mobile. However, in TEM-52 and SHV-2, the mobility of K73 was increased, a higher movement of this residue may also have important implications for the ESBL activity of these variant enzymes.
The increased mobility of several areas of the enzyme in SHV-2 compared with SHV-1 likely contribute to the expanded hydrolytic profile and reduced stability of SHV-2. An X-ray crystal structure of SHV-5 and of enzymes with R275L and N276D will be required in order to provide information of the mobility effects of E240K, R275L, and N276D. Generally, the mobility of SHV and TEM is similar throughout the enzyme with the only major differences observed outside of the active site area (besides the differences in E166 and N170 mentioned above). The mobility similarities between these enzymes are important in allowing the hydrolysis of a similar spectrum of β-lactams. However, the differences in mobility that we observe outside of the active site may provide a partial explanation for the differences in stabilizing substitutions in TEM and SHV. The largest difference is in the area of amino acids 116–125, which is much less mobile in SHV than in TEM. This rigid portion of the scaffold in SHV likely contributes to the overall stability of the enzyme and affects the result of amino acid substitutions differently than the more mobile area of TEM.
Competition Experiments
The blaSHV-5 variant was compared with the blaSHV-129 variant in competition experiments using E. coli Ara+ and Ara− strains because SHV-5 is the most likely ESBL ancestor of the SHV-129 enzyme. Competition experiments did not clearly demonstrate the favorability of one bla variant over another.
Implications for Evolutionary Potential of β-Lactamases
Production of β-lactamase enzymes is one of the major mechanisms leading to antibiotic resistance in bacteria. Therefore, understanding how these enzymes evolve in response to the introduction and use of antibiotics is essential in the continual fight against antibiotic-resistant infections. The TEM β-lactamase has been used in the past as a model in order to understand the evolution of β-lactamases (Huang and Palzkill 1997; Sideraki et al. 2001; Majiduddin and Palzkill 2003; Bershtein et al. 2006; Marciano et al. 2008; Brown et al. 2010; Salverda et al. 2011; Goldsmith and Tawfik 2012; Dellus-Gur et al. 2013; Wellner et al. 2013; Tawfik 2014; Toth-Petroczy and Tawfik 2014). One computer simulation predicted that the principles discovered in the probing of the evolution of TEM could be applied to the evolution of SHV, the β-lactamase with 68% protein identity to TEM (fig. 1) (Bloom 2014). However, experimental evidence thus far does not exist for this hypothesis.
Here, we use a naturally occurring SHV variant enzyme, SHV-129 (G238S-E240K-R275L-N276D), to begin to understand the universal principles and differences in evolution of β-lactamase enzymes. From our comprehensive studies, we found that the results observed in TEM cannot be universally applied to SHV. In particular, the R275L substitution, which is a global suppressor in TEM does not show this property in the SHV background. However, the R275L substitution shows positive sign epistasis with N276D in SHV resulting in greater stability of the enzyme when these substitutions occur together. Additionally, the R275L substitution does not result in β-lactamase inhibitor resistance in SHV in contrast to TEM. However, N276D has a similar role in both SHV and TEM. This amino acid substitution functions both as a global suppressor and as an inhibitor resistant substitution in these two divergent enzymes. Additionally, the G238S substitution has a similar role in both enzymes leading to an ESBL phenotype and destabilization. In fact, even the result of the G238S substitution is similar in both enzymes leading to a more open conformation of the enzyme with easier access of large substrates (such as cefotaxime) to the active site as well as an increased flexibility of the loop containing G238 in both enzymes (Nukaga et al. 2003; Dellus-Gur et al. 2015). In contrast, the E240K substitution has different stability effects in each enzyme. In TEM, E240K has been postulated as stabilizing (Salverda et al. 2010). However, here we find that E240K is neutral in terms of thermal denaturation of SHV. The results of R275L are also different in both enzymes. In TEM, R275L is a global suppressor believed to improve the packing effects of the protein by removing the partially charged arginine residue (Brown et al. 2010). The effects of substituting position 275 in SHV had not been previously studied, but here we find that the global suppressor phenotype does not occur with the R275L substitution in SHV. However, in combination with N276D, the R275L substitution does result in favorable thermodynamics of the enzyme with an increased melting temperature of SHV when these two amino acid substitutions occur together. This may explain why the only substitution at position 275 in a clinical isolate of SHV (SHV-129) occurs in the background of N276D, while both isolates of TEM with substitutions at position 275 (TEM-38 and TEM-45) do not have the N276D background (Jacoby and Bush 2014). The differences in the effects of these substitutions illustrate that the trajectory each β-lactamase takes in its evolution may differ depending on the underlying amino acid sequence of the protein.
Despite the differences in the roles of particular amino acids in these two enzymes, the underlying principles of their evolution seem to be universal. Active site amino acid substitutions that change the activity of the enzyme (such as G238S and E240K) occur at a cost in terms of enzyme stability. Therefore, in order for the β-lactamase to continue to adapt to novel antibiotic challenges, stabilizing substitutions must be acquired. These stabilizing substitutions can occur in the form of global suppressors (N276D) or in the form of substitutions that exhibit epistasis (R275L). However, without the acquisition of a stabilizing substitution, the enzyme cannot further evolve without the risk of unfolding under physiologic conditions. Therefore, in the description and characterization of β-lactamases, it is important to identify not only how the enzymes change phenotypically in their resistance pattern, but also in their stability as this provides essential information about how future generations of the enzyme may look. This universal theme of destabilization and compensation in the evolutionary trajectory of β-lactamase enzymes in response to antibiotic challenge is likely also important in other evolutionary processes occurring in a longer timescale.
Conclusions
Stabilizing substitutions that evolve to increase the threshold robustness of the SHV β-lactamase in order to enhance its ability to acquire additional destabilizing active site substitutions are present in the novel SHV-129 β-lactamase. Particularly, in enzymes highly prone to evolution in response to drug challenge, such as β-lactamases, these stabilizing substitutions are centrally important to allowing continual mutation accumulation. N276D is the first global suppressor substitution identified in the SHV β-lactamase. Although this substitution was previously characterized as an IR amino acid substitution (Drawz et al. 2009), it had not been observed in a clinical isolate and its stability was not explored (Jacoby and Bush 2014). In SHV-129, this substitution does not seem to serve an IR role as the final enzyme is susceptible to clavulanic acid inhibition with a low Ki.
The R275L substitution may be a stabilizing suppressor that is not “global” and only able to stabilize the protein in combination with N276D. The limitations of other stabilizing substitutions have been reported (Marciano et al. 2008). The stabilizing properties of L201P and M182T were not universal for all destabilizing substitutions. The combination of R275L and N276D in SHV-129 also expands the hydrolysis spectrum of the enzyme to include cefepime. Structural characterization through molecular modeling or X-ray crystallography is required to determine if active site changes are responsible for this phenotype. However, we propose that the increased stability of the SHV-129 β-lactamase permits the hydrolysis of cefepime. Perhaps an alternative conformation of the enzyme is stabilized that allows cefepime hydrolysis in SHV-129 compared with SHV-5. Future structural comparisons of these enzymes in the presence and absence of β-lactam substrates will be important in elucidating the enzyme conformation and explaining its stability.
In addition to studying the effects of amino acid substitutions in terms of substrate activity, stabilizing properties must also be considered. Here, we have shown that the SHV-129 β-lactamase has likely evolved from SHV-5 probably due to the increasing clinical use of cefepime. Since its introduction in 1994, we have observed a similar increase in cefepime resistance in Pseudomonas aeruginosa (Endimiani et al. 2008; Akhabue et al. 2011). However, as a consequence of this evolution, SHV-129 now has an excess of stability. We are concerned that this clinically acquired β-lactamase, SHV-129, represents an intermediate to a more highly evolved SHV variant. SHV-129 now has an excess of stability; it is possible that an IR substitution which confers resistance to clavulanic acid, such as S130G or K234R, both of which are destabilizing (Sun et al. 2004; Winkler et al. 2013) could be acquired in SHV-129 and lead to an enzyme variant with both ESBL and IR properties. This “gain of function” would be a significant threat to our antibiotic armamentarium as SHV is one of the most common plasmid-encoded β-lactamases (Bush 2008). Most concerning would be substitutions that increase turnover of carbapenems or confer resistance to new β-lactamase inhibitors in SHV combined with global suppressors. These changes can forebode a truly ominous β-lactamase that can become readily widespread in Gram-negative bacteria.
Materials and Methods
bla Gene Mutagenesis
Site-directed mutagenesis was performed on the blaSHV-1 gene in the pBC SK (−) phagemid as previously described (Drawz et al. 2009; Winkler et al. 2013). The SHV-2 (G238S), SHV-5 (G238S-E240K), and N276D substitutions were already present in the blaSHV gene in the pBC SK (−) phagemid and previously published (Nukaga et al. 2003; Drawz et al. 2009; Kalp et al. 2009). We also engineered the T182M substitution as it converts the wild-type sequence of SHV-1 (Thr) to what is observed in TEM (Met) and the M182T substitution is a common global suppressor in TEM (Huang and Palzkill 1997; Sideraki et al. 2001; Kather et al. 2008; Brown et al. 2010). Gene changes corresponding to the desired nucleotide substitutions were confirmed by gene sequencing (McLab, South San Francisco, CA) and the plasmids were transformed into E. coli DH10B for further experimentation.
Whole Cell Assays
The susceptibility of E. coli containing each bla variant was determined using whole cell assays or MIC measurements. These were performed by the agar dilution method previously described and according to Clinical and Laboratory Standards Institute (CLSI) guidelines (CLSI 2014; Winkler et al. 2015).
Protein Expression
An immunoblot was used to visualize the levels of protein expression among the different variants in a standard E. coli pBC SK (−) background on whole cell preparations as previously described (Hujer et al. 2002; Winkler et al. 2013). This technique was also used to view the levels of protein expression among the different variants in a standard E. coli pBC SK (−) background soluble fraction preparations. The soluble fractions were isolated as previously described (Winkler et al. 2015). The α-SHV-1 antibody was previously characterized (Hujer et al. 2002). After transfer to a nitrocellulose membrane, blocking, and incubations with primary and secondary antibodies, the immunoblot was developed with the Prime ECL kit from Amersham. All immunoblots were performed in triplicate with one representative experiment shown.
We included the loading control E. coli DnaK in both immunoblots. It is easily identified on the immunoblot of the whole cell preparation as well as the soluble fractions as the higher molecular mass protein.
Protein Purification
SHV-1, SHV-2, R275L, E240K, T182M, and SHV-129 were purified to homogeneity from E. coli DH10B containing the respective blaSHV genes as previously reported (Winkler et al. 2013). SHV-5, G238S-E240K-R275L, and G238S-E240K-N276D were cloned into the pGEX-6P-2 vector, transformed into E. coli Origami 2 DE3 cells, and the proteins were expressed with isopropyl β-d-1-thiogalactopyranoside (IPTG) induction. The N276D variant was purified from E. coli BL21DE3 cells containing blaSHV N276D as previously described (Drawz et al. 2009). For plasmids in E. coli DH10B, cells were grown overnight at 37 °C with shaking in super optimal broth with 50 mg/l of chloramphenicol for plasmid maintenance and then cells were pelleted and the supernatant was discarded. For the other cells and plasmids, bacteria were grown to an OD600 = 0.6 at 37 °C with shaking. Then, 1 mM of IPTG was added and cells were grown for an additional 3 h with shaking at 37 °C. After 3 h of growth, cells were pelleted and the supernatant was discarded. All cell pellets were then frozen overnight at −20 °C.
For bla genes expressed in E. coli DH10B, frozen pellets were thawed and E. coli DH10B was resuspended in Tris–HCl pH 8.0 with the addition of benzonuclease, ethylenediaminetetraacetic acid (EDTA), MgSO4, and lysozyme as previously described (Winkler et al. 2013). The pellet of E. coli cells expressing N276D protein was resuspended in Tris–HCl pH 8.0 with the addition of lysozyme, benzonuclease, and EDTA as previously described (Drawz et al. 2009). After incubations with the various reagents, the bacterial debris was pelleted and separated. The supernatants were collected and run on a pIEF gel apparatus and then gel filtration chromatography as previously described (Drawz et al. 2009; Winkler et al. 2013). Protein was identified as >95% pure through sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) chromatography.
The frozen pellets of E. coli cells with pGEX-6P-2 plasmids containing bla genes were resuspended in phosphate-buffered saline (PBS) with the addition of benzonuclease, EDTA, and lysozyme as previously described (Winkler et al. 2013). After pelleting to remove the bacterial debris, the supernatant was run over a 5 ml GSTrapFF GST column (GE Healthcare), then the gel filtration column to remove residual TEM β-lactamase from the pGEX-6P-2 plasmid. The GST tag was cleaved from the protein using an overnight incubation with PreScission Protease at 4 °C. Then, the cleaved protein was isolated using the GSTrapFF GST column and verified as >95% pure with SDS-PAGE electrophoresis.
β-Lactam and β-Lactamase Inhibitor Kinetics
Kinetics with β-lactams and β-lactamase inhibitors were conducted as previously described (Drawz et al. 2009; Levitt et al. 2012; Winkler et al. 2013). A full panel of kinetic measurements was performed with the SHV-129 β-lactamase. Extinction coefficients were Δε235 = −900 M/cm for ampicillin, Δε262 = −7,660 M/cm for cephalothin, Δε482 = −17,400 M/cm for nitrocefin, Δε262 = −7,250 M/cm for cefotaxime, Δε256 = −7,600 M/cm for ceftazidime, Δε260 = 750 M/cm for cefepime, and Δε318 = −640 M/cm for aztreonam (Levitt et al. 2012).
For β-lactam substrates, steady-state kinetics were measured on an Agilent 8453 photospectrometer at room temperature in 10 mM PBS using variable amounts of enzyme depending on the substrate. Values for Km and kcat were calculated according to the Henri–Michaelis–Menten equation (eq. 1) using EnzFitter and equation 2 as previously described (Levitt et al. 2012; Winkler et al. 2013).
| (1) |
| (2) |
For several substrates that could not be fully characterized due to the low hydrolytic activity of the β-lactamases, we estimated an upper limit of kcat/Km according to equation 3 using the initial velocity of hydrolysis measured at the same wavelengths as described above for cefotaxime, ceftazidime, and cefepime (Papp-Wallace et al. 2010).
| (3) |
In equation 3, [E] represents the concentration of enzyme used and [S] represents the concentration of the respective substrate.
Steady-state kinetics for β-lactamase inhibitors were determined as competition assays varying inhibitor concentration against a constant concentration of nitrocefin (100 µM) as previously described (Winkler et al. 2013). The reciprocal velocities (1/v) at various inhibitor concentrations [I] were plotted versus the inhibitor concentration to a simple linear equation using Excel. The slope was then corrected for the affinity of nitrocefin to obtain the Ki app representing the dissociation constants of the Michaelis–Menten complex.
| (4) |
Circular Dichroism
CD was performed on a Jasco J-815 spectrometer using quartz cells with a 0.1 cm path length on all purified proteins to determine any structural differences using a previously published method (Winkler et al. 2013). Thermal denaturation curves were also measured using a Peltier effect temperature controller between 22 °C and 70 °C with a heating rate of 2 °C/min for all the purified enzymes as previously described (Winkler et al. 2013). Raw equilibrium data were normalized to the fraction of unfolded protein (fu). A van’t Hoff plot (ln Keq vs. 1/T) was constructed at λ = 214 nm to calculate the melting temperature (Tm), van’t Hoff enthalpy (ΔHVH), and the entropy of unfolding (ΔSu). The van’t Hoff enthalpy was defined as the slope of the plot times−R (R = ideal gas constant) and the y-intercept of the plot was defined as ΔSu/R. Equations 5, 6, and 7 were used to derive these values and calculate Tm:
| (5) |
| (6) |
| (7) |
The temperatures are reported with an error as determined by the standard deviation of three independent experiments. As has been observed with TEM, the melting curves were irreversible due to the precipitation of the β-lactamase because of the high quantity of enzyme required for the Tm measurement (Brown et al. 2010). Therefore, the reported values represent apparent Tm as the standard for β-lactamase melting point determination.
B Factor Determination
The log2 normalized B factors for each amino acid in SHV-1 (PDB ID: 1SHV), SHV-2 (PDB ID: 1N9B), TEM-1 (PDB ID: 1ZG4), and TEM-52 (PDB ID: 1M40) were calculated from their respective X-ray crystallography structures according to previously published methods (Dellus-Gur et al. 2015).
Competition Assays
Competition experiments were performed using E. coli B Ara+ and Ara− strains that were a kind gift from Dr. Timothy Palzkill (Marciano et al. 2007). These strains were electroporated with blaSHV-5 or blaSHV-129 on the pBC SK (−) phagemid. E. coli B Ara+ containing blaSHV-5 and E. coli B Ara− containing blaSHV-129 were grown separately overnight in Lysogeny Broth (LB) supplemented with chloramphenicol at 37 °C with shaking. The following morning, the OD600 of each culture was determined, then the cultures were each diluted to OD600 = 0.025 and mixed at a 1:1 ratio in 5 ml of LB supplemented with chloramphenicol for a final OD600 = 0.05. Cells were diluted and plated on tetrazolium and arabinose agar plates at the initial time point and then at OD600 = 0.1, 0.2, 0.4, 0.6, 0.8, 1.0, 1.5, and 24 h. Red and white bacterial cells were then counted for each OD to determine the number of Ara−and Ara+ cells, respectively. The reverse experiment with E. coli B Ara+ containing blaSHV-129 and E. coli B Ara− containing blaSHV-5 was also performed. Each version of the experiment was performed in triplicate.
Supplementary Material
Supplementary figure S1 is available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
This work was supported by the National Institutes of Health and the National Institute of Allergy and Infectious Diseases of the National Institutes of Health, and the Cleveland Department of Veterans Affairs. Individual funding was provided by the National Institutes of Health (grant T32-GM007250 to Case Western Reserve University, M.L.W.), the Veterans Affairs Merit Review Program (R.A.B.), the National Institutes of Health (grants AI072219-05 and AI063517-07 to R.A.B.), and the Geriatric Research Education and Clinical Center VISN 10 (R.A.B.). We would like to thank Christine Lascols for identifying the Escherichia coli isolate containing the SHV-129 enzyme and Steven M. Marshall and Andrea M. Hujer for initial genetic characterization of the isolate. We would like to thank Dr Krisztina M. Papp-Wallace for guidance in kinetics experiments and Magdalena A. Taracila for assistance in CD and B factor measurements.
References
- Akhabue E, Synnestvedt M, Weiner MG, Bilker WB, Lautenbach E. 2011. Cefepime-resistant Pseudomonas aeruginosa. Emerg Infect Dis. 17:1037–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aramli LA, Teschke CM. 1999. Single amino acid substitutions globally suppress the folding defects of temperature-sensitive folding mutants of phage P22 coat protein. J Biol Chem. 274:22217–22224. [DOI] [PubMed] [Google Scholar]
- Bershtein S, Segal M, Bekerman R, Tokuriki N, Tawfik DS. 2006. Robustness-epistasis link shapes the fitness landscape of a randomly drifting protein. Nature 444:929–932. [DOI] [PubMed] [Google Scholar]
- Bloom JD. 2014. An experimentally informed evolutionary model improves phylogenetic fit to divergent lactamase homologs. Mol Biol Evol. 31:2753–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NG, Pennington JM, Huang W, Ayvaz T, Palzkill T. 2010. Multiple global suppressors of protein stability defects facilitate the evolution of extended-spectrum TEM β-lactamases. J Mol Biol. 404:832–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush K. 2008. Extended-spectrum β-lactamases in North America, 1987–2006. Clin Microbiol Infect. 14(Suppl. 1):134–143. [DOI] [PubMed] [Google Scholar]
- CLSI. 2014. Performance standards for antimicrobial susceptibility testing; twenty-fourth informational supplement. Wayne (PA): Clinical and Laboratory Standards Institute; CLSI document M100-S24. [Google Scholar]
- Dellus-Gur E, Elias M, Caselli E, Prati F, Salverda ML, de Visser JA, Fraser JS, Tawfik DS. 2015. Negative epistasis and evolvability in TEM-1 β-lactamase—the thin line between an enzyme’s conformational freedom and disorder. J Mol Biol. 427:2396–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellus-Gur E, Toth-Petroczy A, Elias E, Tawfik DS. 2013. What makes a protein fold amenable to functional innovation? Fold polarity and stability trade-offs. J Mol Biol. 425:2609–2621. [DOI] [PubMed] [Google Scholar]
- Doi Y, Park YS, Rivera JI, Adams-Haduch JM, Hingwe A, Sordillo EM, Lewis JS, 2nd, Howard WJ, Johnson LE, Polsky B, et al. 2013. Community-associated extended-spectrum β-lactamase-producing Escherichia coli infection in the United States. Clin Infect Dis. 56:641–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle DF, Waldner JC, Parikh S, Alcazar-Roman L, Pielak GJ. 1996. Changing the transition state for protein (Un) folding. Biochemistry 35:7403–7411. [DOI] [PubMed] [Google Scholar]
- Drawz SM, Bethel CR, Hujer KM, Hurless KN, Distler AM, Caselli E, Prati F, Bonomo RA. 2009. The role of a second-shell residue in modifying substrate and inhibitor interactions in the SHV β-lactamase: a study of Ambler position Asn276. Biochemistry 48:4557–4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drawz SM, Bonomo RA. 2010. Three decades of β-lactamase inhibitors. Clin Microbiol Rev. 23:160–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endimiani A, Perez F, Bonomo RA. 2008. Cefepime: a reappraisal in an era of increasing antimicrobial resistance. Expert Rev Anti Infect Ther. 6:805–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giakkoupi P, Tzelepi E, Legakis NJ, Tzouvelekis LS. 1999. Aspartic acid for asparagine substitution at position 276 reduces susceptibility to mechanism-based inhibitors in SHV-1 and SHV-5 β-lactamases. J Antimicrob Chemother. 43:23–29. [DOI] [PubMed] [Google Scholar]
- Goldsmith M, Tawfik DS. 2012. Directed enzyme evolution: beyond the low-hanging fruit. Curr Opin Struct Biol. 22:406–412. [DOI] [PubMed] [Google Scholar]
- Greenfield N, Fasman GD. 1969. Computed circular dichroism spectra for the evaluation of protein conformation. Biochemistry 8:4108–4116. [DOI] [PubMed] [Google Scholar]
- Huang W, Palzkill T. 1997. A natural polymorphism in β-lactamase is a global suppressor. Proc Natl Acad Sci U S A. 94:8801–8806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hujer AM, Page MG, Helfand MS, Yeiser B, Bonomo RA. 2002. Development of a sensitive and specific enzyme-linked immunosorbent assay for detecting and quantifying CMY-2 and SHV β-lactamases. J Clin Microbiol. 40:1947–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby G, Bush K. 2014. ß-Lactamase classification and amino acid sequences for TEM, SHV and OXA extended-spectrum and inhibitor resistant enzymes. Available from: http://www.lahey.org/Studies/. [Google Scholar]
- Kalp M, Bethel CR, Bonomo RA, Carey PR. 2009. Why the extended-spectrum β-lactamases SHV-2 and SHV-5 are “hypersusceptible” to mechanism-based inhibitors. Biochemistry 48:9912–9920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kather I, Jakob RP, Dobbek H, Schmid FX. 2008. Increased folding stability of TEM-1 β-lactamase by in vitro selection. J Mol Biol. 383:238–251. [DOI] [PubMed] [Google Scholar]
- Kuzin AP, Nukaga M, Nukaga Y, Hujer AM, Bonomo RA, Knox JR. 1999. Structure of the SHV-1 β-lactamase. Biochemistry 38:5720–5727. [DOI] [PubMed] [Google Scholar]
- Lascols C, Hackel M, Hujer AM, Marshall SH, Bouchillon SK, Hoban DJ, Hawser SP, Badal RE, Bonomo RA. 2012. Using nucleic acid microarrays to perform molecular epidemiology and detect novel β-lactamases: a snapshot of extended-spectrum β-lactamases throughout the world. J Clin Microbiol. 50:1632–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt PS, Papp-Wallace KM, Taracila MA, Hujer AM, Winkler ML, Smith KM, Xu Y, Harris ME, Bonomo RA. 2012. Exploring the role of a conserved class A residue in the Omega-Loop of KPC-2 β-lactamase: a mechanism for ceftazidime hydrolysis. J Biol Chem. 287:31783–31793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majiduddin FK, Palzkill T. 2003. An analysis of why highly similar enzymes evolve differently. Genetics 163:457–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciano DC, Karkouti OY, Palzkill T. 2007. A fitness cost associated with the antibiotic resistance enzyme SME-1 β-lactamase. Genetics 176:2381–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciano DC, Pennington JM, Wang X, Wang J, Chen Y, Thomas VL, Shoichet BK, Palzkill T. 2008. Genetic and structural characterization of an L201P global suppressor substitution in TEM-1 β-lactamase. J Mol Biol. 384:151–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews BW, Nicholson H, Becktel WJ. 1987. Enhanced protein thermostability from site-directed mutations that decrease the entropy of unfolding. Proc Natl Acad Sci U S A. 84:6663–6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nukaga M, Mayama K, Hujer AM, Bonomo RA, Knox JR. 2003. Ultrahigh resolution structure of a class A β-lactamase: on the mechanism and specificity of the extended-spectrum SHV-2 enzyme. J Mol Biol. 328:289–301. [DOI] [PubMed] [Google Scholar]
- Odell AF, Odell LR, Askham JM, Alogheli H, Ponnambalam S, Hollstein M. 2013. A novel p53 mutant found in iatrogenic urothelial cancers is dysfunctional and can be rescued by a second-site global suppressor mutation. J Biol Chem. 288:16704–16714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orencia MC, Yoon JS, Ness JE, Stemmer WP, Stevens RC. 2001. Predicting the emergence of antibiotic resistance by directed evolution and structural analysis. Nat Struct Biol. 8:238–242. [DOI] [PubMed] [Google Scholar]
- Papp-Wallace KM, Taracila M, Wallace CJ, Hujer KM, Bethel CR, Hornick JM, Bonomo RA. 2010. Elucidating the role of Trp105 in the KPC-2 β-lactamase. Protein Sci. 19:1714–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risso VA, Manssour-Triedo F, Delgado-Delgado A, Arco R, Barroso-delJesus A, Ingles-Prieto A, Godoy-Ruiz R, Gavira JA, Gaucher EA, Ibarra-Molero B, et al. 2015. Mutational studies on resurrected ancestral proteins reveal conservation of site-specific amino acid preferences throughout evolutionary history. Mol Biol Evol. 32:440–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salverda ML, De Visser JA, Barlow M. 2010. Natural evolution of TEM-1 β-lactamase: experimental reconstruction and clinical relevance. FEMS Microbiol Rev. 34:1015–1036. [DOI] [PubMed] [Google Scholar]
- Salverda ML, Dellus E, Gorter FA, Debets AJ, van der Oost J, Hoekstra RF, Tawfik DS, de Visser JA. 2011. Initial mutations direct alternative pathways of protein evolution. PLoS Genet. 7:e1001321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sideraki V, Huang W, Palzkill T, Gilbert HF. 2001. A secondary drug resistance mutation of TEM-1 β-lactamase that suppresses misfolding and aggregation. Proc Natl Acad Sci U S A. 98:283–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Bethel CR, Bonomo RA, Knox JR. 2004. Inhibitor-resistant class A β-lactamases: consequences of the Ser130-to-Gly mutation seen in Apo and tazobactam structures of the SHV-1 variant. Biochemistry 43:14111–14117. [DOI] [PubMed] [Google Scholar]
- Tawfik DS. 2014. Accuracy-rate tradeoffs: how do enzymes meet demands of selectivity and catalytic efficiency? Curr Opin Chem Biol. 21:73–80. [DOI] [PubMed] [Google Scholar]
- Thomas VL, McReynolds AC, Shoichet BK. 2010. Structural bases for stability-function tradeoffs in antibiotic resistance. J Mol Biol. 396:47–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth-Petroczy A, Tawfik DS. 2014. The robustness and innovability of protein folds. Curr Opin Struct Biol. 26:131–138. [DOI] [PubMed] [Google Scholar]
- Wang X, Minasov G, Shoichet BK. 2002. Evolution of an antibiotic resistance enzyme constrained by stability and activity trade-offs. J Mol Biol. 320:85–95. [DOI] [PubMed] [Google Scholar]
- Wellner A, Raitses Gurevich M, Tawfik DS. 2013. Mechanisms of protein sequence divergence and incompatibility. PLoS Genet. 9:e1003665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler ML, Papp-Wallace KM, Bonomo RA. 2015. Activity of ceftazidime/avibactam against isogenic strains of Escherichia coli containing KPC and SHV β-lactamases with single amino acid substitutions in the Omega-loop. J Antimicrob Chemother. 70:2279–2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler ML, Rodkey EA, Taracila MA, Drawz SM, Bethel CR, Papp-Wallace KM, Smith KM, Xu Y, Dwulit-Smith JR, Romagnoli C, et al. 2013. Design and exploration of novel boronic acid inhibitors reveals important interactions with a clavulanic acid-resistant sulfhydryl-variable (SHV) β-lactamase. J Med Chem. 56:1084–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.