Abstract
Evolutionary changes in cis-regulatory elements are thought to play a key role in morphological and physiological diversity across animals. Many conserved noncoding elements (CNEs) function as cis-regulatory elements, controlling gene expression levels in different biological contexts. However, determining specific associations between CNEs and related phenotypes is a challenging task. Here, we present a computational “reverse genomics” approach that predicts the phenotypic functions of human CNEs. We identify thousands of human CNEs that were lost in at least two independent mammalian lineages (IL-CNEs), and match their evolutionary profiles against a diverse set of phenotypes recently annotated across multiple mammalian species. We identify 2,759 compelling associations between human CNEs and a diverse set of mammalian phenotypes. We discuss multiple CNEs, including a predicted ear element near BMP7, a pelvic CNE in FBN1, a brain morphology element in UBE4B, and an aquatic adaptation forelimb CNE near EGR2, and provide a full list of our predictions. As more genomes are sequenced and more traits are annotated across species, we expect our method to facilitate the interpretation of noncoding mutations in human disease and expedite the discovery of individual CNEs that play key roles in human evolution and development.
Keywords: reverse genomics, conserved noncoding elements, genotype–phenotype matching, mammals
Introduction
Advancements in comparative genomics, along with the increased availability of whole genome sequences, have led to the identification of many noncoding genomic regions evolving under strong purifying selection (Lindblad-Toh et al. 2011). These conserved noncoding elements (CNEs) often act as cis-regulatory elements of nearby genes in specific tissues and time points, and some have already been shown to be associated with vertebrate development and transcriptional regulation (Bejerano et al. 2004; Woolfe et al. 2005; Lowe et al. 2011). In humans, a large fraction of common trait and disease variation is noncoding (Hindorff et al. 2009), and a growing number of cis-regulatory mutations have been implicated in human disease (Ragvin et al. 2010; Wasserman et al. 2010). In fact, over 80% of single nucleotide polymorphisms found to be associated with diseases through genome-wide association studies are noncoding (Hindorff et al. 2009). Although CNE loss events during mammalian evolution are relatively rare (McLean and Bejerano 2008), a recent report highlighted hundreds of CNEs that have been lost in at least two independent mammalian lineages (Hiller, Schaar, Bejerano, et al. 2012). Moreover, evolutionary changes in CNEs have been demonstrated to play key roles in the remarkable morphological, physiological, and behavioral diversity observed across species (Wray 2007; Carroll 2008). Evidence supporting phenotypic modifications via gains and losses in cis-regulatory regions has been collected over the past decades in many different species, including yeast (Tuch et al. 2008), flies (Bradley et al. 2010), fish (Chan et al. 2010), mammals (Schmidt et al. 2010), and humans (McLean et al. 2011).
Despite vast increases over the last few years in the amount of available genomic data, determining the function of cis-regulatory elements has remained a challenging task, as cross-species sequence differences are confounded by millions of nonsignificant mutations and genomic changes. Another major challenge in phenotype–genotype mapping is pleiotropy, where a genomic element is involved in multiple unrelated functions and can be active in an even greater number of contexts without contributing (or disturbing) function. Recent efforts, such as the ENCODE project (Ecker et al. 2012), have focused on identifying active elements in the human and mouse genomes by measuring patterns of transcription factor (TF) binding and histone modification marks across different cell lines and developmental time points. Although these maps have become a valuable resource for identifying active cis-regulatory elements and generating new hypotheses about their roles, they are limited to a subset of cell types and conditions, and it is unclear which of these biochemically active events actually contribute to gene regulation or are important for evolutionary fitness (Pennacchio et al. 2013). As a result, directly linking biochemically active cis-regulatory elements with specific phenotypic adaptation or disease is yet difficult.
Linking human CNEs with specific phenotypic traits by analyzing their orthologous regions in other species may provide an alternative approach for annotating adaptive cis-regulatory elements. A conserved genomic region encoding a particular trait may show clear signs of erosion in species where the trait has been altered or lost (e.g., due to inactivating mutations) if sufficient evolutionary time has elapsed to allow neutral selection to erode the DNA sequences. Trait loss in independent lineages (i.e., in two or more unrelated taxa) should result in neutral drift in all trait-related regions (in the trait-loss lineages), regardless of which element in the independent lineages was initially affected by a trait-inactivating mutation (fig. 1A). Thus, a matching evolutionary pattern between an independently lost trait and an independently eroding genomic region violates the expected evolutionary conservation more than once, and suggests a functional relationship between the phenotype and the genotype. Based on this principle, Hiller et al. devised a “forward genomics” approach—named for its similarity to forward genetics—to identify protein-coding genes most likely to be responsible for a given independently lost trait (Hiller, Schaar, Indjeian, et al. 2012). Given phenotypic evolutionary conservation patterns, they were able to link loss of vitamin C synthesis with the inactivation of the gene Gulo, and low biliary phospholipid levels in guinea pig and horse with the inactivation of the gene ABCB4 (Hiller, Schaar, Indjeian, et al. 2012). Recently, the forward genomics approach has been used in comparative evolutionary analysis of a previously undescribed lncRNA (long non-coding RNA) in a transcriptional study of neuronal progenitors in the human cortex (Johnson et al. 2015).
Fig. 1.
(A) An ancestral mammalian phenotype is encoded by a set of genomic regions (such as CNEs) that are required for that trait. Species or clade-specific trait inactivation events may occur via inactivating mutations in any of the genomic regions related to the trait (a schematic “phenotree” is shown, where a phenotype across species is projected on the phylogenetic tree). As a consequence of trait inactivation, all related genomic regions switch from purifying to neutral selection, resulting in decay of the genomic region over time, and matching evolutionary profiles of the eroding elements and lost trait. Independent loss patterns are in general far less common than single-clade loss patterns, and are expected to match independent CNE inactivation events in the mammalian phylogeny with higher specificity. (B) A proposed “reverse genomics” approach to link human IL-CNEs to a large data set of scored phenotypes by matching the evolutionary patterns for many IL-CNEs (projected onto one-dimensional vectors) against the evolutionary patterns of many mammalian traits.
CNEs are thought to be less pleiotropic than the genes they regulate (Carroll 2008). Their inactivation is thus less deleterious, and it is anticipated that CNE loss events are more common than gene loss events (Carroll 2005). In this article, we hypothesized that some evolutionary phenotypic modifications could map to CNE losses, and we examined our hypothesis using a large phenotypic data set from a recent study that scores thousands of diverse anatomical and physiological traits across 86 extant and extinct mammalian species (O’Leary et al. 2013). Specifically, we present an inverted phenotype–genotype mapping approach called “reverse genomics”—akin to “reverse genetics”—that examines thousands of human CNEs, one at a time, and tries to match them against hundreds of possible phenotypic roles each CNE could play. We first identify thousands of human CNEs that have been independently lost (IL-CNEs) twice or more during placental mammal evolution. Next, we trace the evolutionary histories of human CNEs and mammalian traits in order to predict phenotype–genotype pairs that are likely to be functionally related on the basis of shared independent evolutionary patterns that persist in both (fig. 1B). In this way, we identify a total of 2,759 candidate trait-CNE associations in a diverse set of traits. By assigning IL-CNEs to nearby genes, we demonstrate that this set is enriched for agreement between the CNE matched traits and the functions of the neighboring genes. We discuss multiple examples of such associations, and provide a comprehensive list of predictions.
Results
Identifying Independent CNE Loss Patterns in Placental Mammal Phylogenies
We analyzed a set of placental mammalian CNEs anchored in the human genome (assembly GRCh37/hg19) to identify losses of CNEs in two or more independent lineages. We started with a set of human elements at least 50 bp in length that are highly conserved across mammals (Siepel et al. 2005), excluding all protein-coding or other known or predicted transcribed regions (see Materials and Methods). To ensure the robustness of our phenotype–genotype matches later in our process, we removed regions not conserved in at least 7 of the 19 sequenced and phenotypically characterized placental mammals (fig. 2B and supplementary table S1, Supplementary Material online). Our conservative CNE set contained 266,116 elements with a mean length of 174 bp and a maximum length of 2,191 bp, covering 1.5% of the human genome (supplementary table S4, Supplementary Material online).
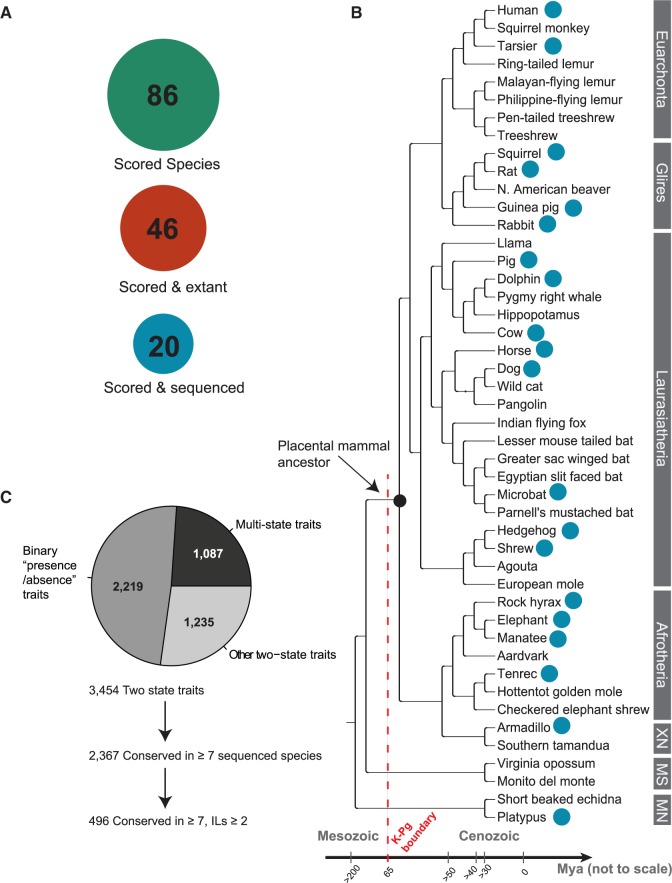
Fig. 2.
Phenotypic database from Morphobank scoring morphological and physiological mammalian traits across species. (A) A phenomic matrix curated in a recent study (O’Leary et al. 2013) that scores over 3,400 “two state” and presence/absence traits across 86 species, of which 46 are extant mammals. For 20 mammals (19 placental mammals and platypus), whole genome sequences are available. (B) Sequenced placental mammal phylogenetic tree highlighting the 19 + 1 phenotyped mammals used in the reverse genomics screen. Each major placental clade is sampled, and the screened species span over 60 My of evolution. XN = Xenarthra; MS = Marsupialia; MN = Monotremata. (C) A subset of 496 independently modified traits (ILs) is extracted based on the annotation level of the trait across the extant sequenced species, and its conservation pattern across the placental mammal phylogeny.
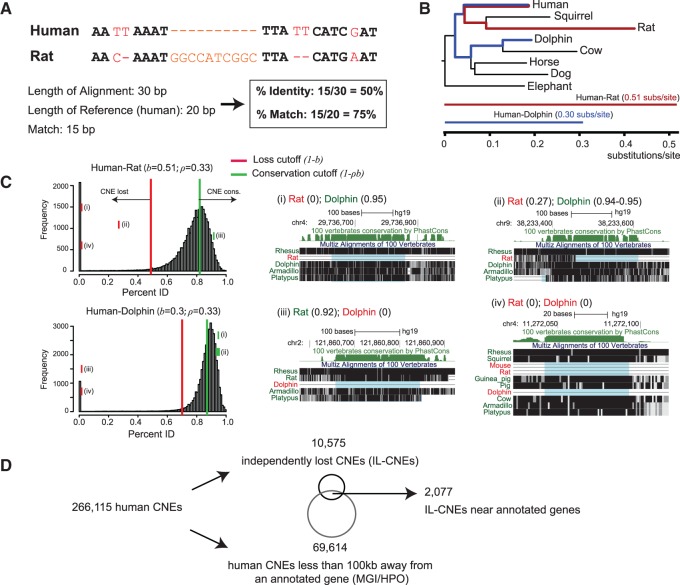
Using a pairwise alignment between human CNEs and their orthologous locations in other genomes (Kent et al. 2003), we determined if a CNE is conserved or lost in a given mammalian species. Specifically, we compute two quantities for the orthologous genomic region: 1) percent identity, defined as the number of matches divided by the total alignment length and 2) percent match, defined as the number of matches divided by the total number of bases in the reference genome (fig. 3A). Percent identity penalizes insertions (by length) in the orthologous region, while percent match ignores insertions. As percent identity is always less than or equal to percent match, these two quantities can be thought of as lower and upper bounds, respectively, on the CNE similarity between human and a query species. We derived two species-specific thresholds based on the evolutionary distance from human (fig. 3B; see Materials and Methods)—the conservation threshold and the loss threshold—that define whether a given CNE is considered conserved or lost. We call a human CNE “lost” in a species if its percent match (i.e, upper bound for similarity) is less than the loss threshold, and call it “conserved” if its percent identity (i.e., lower bound for similarity) is greater than the conserved threshold; otherwise, its state is labeled as “unknown” (fig. 3C). Our approach does not require full CNE deletion, which allows us to identify partial deletion and excessive substitution events as losses. Moreover, because the functional effects of large insertions remain unclear (e.g., an insertion could eliminate regulatory activity by disrupting a TF binding site or by increasing the spacing between synergistic binding sites [Guturu et al. 2013], or have negligible impact on enhancer activity [Smith et al. 2013]), we only call an element conserved if its percent identity—which considers insertions significantly deleterious—is high, and only called an element lost if its percent match—which ignores insertions—is low.
Fig. 3.
A comparative genomic screen for identifying independently lost human CNEs. (A) Two quantities, percent identity and percent match, are computed over a pairwise alignment of a region between a reference (human, GRCh37/hg19) and a query species. (B) A mammalian phylogeny scaled by branch length between human and other mammals from which we derive (C) loss and conservation thresholds per species. A percent identity-percent match interval must fully reside above the conservation cutoff (%) or below loss cutoff (%) to be considered as “conserved” or “lost,” respectively. Histograms of human CNE percent identity are shown for rat and dolphin, with examples of lost CNEs (or partially lost) in one of the two species (i–iii), as well as a CNE that has been independently lost in both (iv). (D) From an initial set of 266,115 CNEs, we identify 10,575 IL-CNEs and 69,614 CNEs near (<100 kb) annotated genes. The intersection of the two derived sets (2,077 IL-CNEs) is the input for the reverse genomics screen.
Next, we identified human CNEs that were independently lost during placental mammalian evolution. We used a parsimony-based algorithm that, for a given rooted phylogenetic tree (in our case, a tree with 19 placental mammals; supplementary table S1, Supplementary Material online) with leaves annotated with either “1” or “0” (representing CNE “presence” or “absence,” respectively), infers the state of each ancestral node and computes the minimum number of loss events that are needed to explain the particular evolutionary pattern of ones and zeros. We restricted 0 to 1 transitions in accordance with Dollo’s irreversible evolution hypothesis (Gould 1970), which asserts that lost ancestral characters—in this case, ancestral DNA elements—cannot be restored. We extracted CNEs for which we identified at least two independent loss events in the eutherian tree (see Materials and Methods). From the initial set of 266,115 human CNEs, we extracted 10,575 independently lost CNEs (IL-CNEs), the majority of which contains exactly 2 independent losses (fig. 3D). We also applied the same method to trait phylogenetic trees to identify independently lost traits, as well as the matched trait-CNE phenotrees; both are discussed in more details below.
Hundreds of Anatomical and Physiological Traits Have Been Independently Modified during Placental Mammal Evolution
To discover interordinal relationships of living and fossil placental mammals and to date the origin of placental mammals relative to the Cretaceous-Paleogene boundary (a mass extinction event about 65 Ma), O’Leary et al. (2013) curated a phenomic matrix scoring 4,541 anatomical and physiological characters across 86 fossil and living mammals (fig. 2A). From this matrix, we extracted mammalian traits with patterns of independent evolutionary loss in extant species for which whole genome sequences are available. Specifically, of the extant phenotyped species in the matrix, 20 mammals including humans (19 placental mammals and platypus—for the complete list of assemblies and list of remaining species, see supplementary tables S1 and S2, Supplementary Material online) have whole genome sequences, and represent a wide phylogeny that spans over 60 My of placental mammal evolution (fig. 2B). There are 3,454 traits that are characterized by two possible states across species (e.g., “Brain–Cerebral cortex folding”: “smooth” or “multiple folds”), the majority of which are traits with binary presence/absence characterization (e.g., “Presence/absence of Omasum in the Digestive tract”) (fig. 2C).
Phylogenetically independent trait modification and losses (i.e., traits lost in at least two independent lineages) represent a subset of relatively rare evolutionary events which we hypothesized would yield fewer false positive genotype–phenotype associations. To infer the evolutionary patterns of independent gains and losses, we employed the same parsimony-based algorithm described above for CNEs (see Materials and Methods). To deal with the fact that the phenomic matrix contains missing annotations, we restricted our analysis to traits for which at least one of the two possible trait states is conserved across at least 7 of the 19 phenotyped placental mammals (2,367 of the 3,454 traits), and further identified a subset of 496 traits with at least 2 independent losses flanked by species conserving the original trait state (fig. 2C and supplementary table S3, Supplementary Material online). This smaller set of traits still encompasses a broad range of mammalian anatomical and physiological characters related directly to human development and evolution. The largest fraction of the data consists of traits that characterize detailed bone structures in the inner and outer skull (27%), limbs (12.7% and 13.5% for the forelimb and hindlimb, respectively), ear (14.11%, including middle, inner, and outer ear), and axial skeleton (13.3%). Additional subsets include soft tissues and other organ-specific phenomic characters (e.g., digestive tract, urogenital tract, eye, oral cavity), vascular processes, brain morphology, reproduction, and development. Finally, we constructed individual phylogenetic trees (a “phenotree,” anchored at the 19 + 1 genotyped species) for each independently lost trait, where the species-specific phenotypic states (i.e., leaves marked with 1 or 0) are projected onto one-dimensional vectors. We later match these phenotrees against similar phylogenetic trees annotated with genomic conservation or loss of human CNEs, to predict functional links between human CNEs and these mammalian phenotypes (fig. 1B).
Assigning Independently Lost CNEs to Genes
We hypothesized that CNEs by and large regulate the expression of nearby genes (McLean et al. 2010). By performing a parameter search (described later), we converged on assigning CNEs to the two nearest transcription start sites (TSSs) up and downstream, up to 100 kb away. Furthermore, we focused on CNEs in regulatory domains for genes with annotations in either Human Phenotype ontology (Köhler et al. 2014) (HPO) or Mouse genome informatics (Eppig et al. 2012) (MGI), and were left with 69,614 CNEs near 6,197 annotated protein-coding genes. We further extracted a subset of 2,077 (fig. 3D and supplementary table S5, Supplementary Material online) independently lost CNEs (having at least two independent losses—see Materials and Methods) and projected the evolutionary profile of each onto one-dimensional vectors to match against each of the independently lost traits.
IL-CNEs Overlap with Functional Genomic Regions
Current technologies for identifying genomic regions that are likely to be functionally active in particular tissues and time points include ChIP-seq for histone modification patterns and TF binding, as well as DNase open chromatin (Thurman et al. 2012) experiments in large-scale projects like ENCODE (Ecker et al. 2012) and Roadmap Epigenomics project (REp) (Roadmap Epigenomics Consortium et al. 2015). To verify that our set of IL-CNEs significantly overlaps genomic regions likely to have cis-regulatory functions, we analyzed a set of H3K27ac and DNase open chromatin regions from REp (covering 30.5% and 23% of the human genome, respectively), and a set of ENCODE TF binding data (covering 5.3%, version3, available from the UCSC genome browser). We intersected these sets with our 2,077 human IL-CNEs, and found that 64.5%, 69.8%, and 17% of our IL-CNE set overlapped the H3K27ac, DNase, and ENCODE TF binding data sets, respectively. In all our four cases, our IL-CNE set exhibited a statistically significant enrichment for functionally active regions compared with the full genome background (P < 10−12, binomial test).
Reverse Genomics Associates Developmental and Evolutionary Functions with Human IL-CNEs
We identified putative functional genotype–phenotype links between coevolving human IL-CNEs and mammalian phenomic characters. Each of the 2,077 human IL-CNEs annotated for conservation or loss across 19 placental mammals and platypus were matched against each of the 496 traits scored across the species. We extracted trait-CNE pairs that highly match in their evolutionary profiles with at least two overlapping independent losses shared by both the trait and CNE. We required that at least an additional seven placental mammals preserve both the CNE and the trait, and eliminated any phenotype–genotype conflicts in platypus (i.e., phenotype 1/CNE 0, or vice versa). Our final set consisted of 2,759 unique trait-CNE pairs.
To assess the quality of our discovered trait-CNE associations and to perform a parameter search for maximizing phenotype–genotype association with putative biological significance (see below), we compared the anatomical definitions of the matched phenotypes with gene annotations contained in the mammalian phenotypic ontology from MGI (Eppig et al. 2012) as well as the human phenotypic abnormality ontology from HPO (Köhler et al. 2014). In lieu of HPO/MGI codes for the matched Morphobank traits, we created a mapping between each of the 496 traits from O’Leary et al. to one or more ontology terms from MGI and HPO using a free text–based mapping approach (see Materials and Methods). We hypothesized that a trait-CNE link predicted through reverse genomics is more likely to be functional if the trait matches via its textual definition to annotations of a gene to which the CNE is assigned. For example, an association that links a “phalanges” trait (digital bones in the hand and feet) with a CNE in the regulatory domain of a gene annotated with terms like “short phalanx of finger” yields a contextual “closed loop” we hypothesize to be putatively functional (fig. 4). We automatically detected closed loops within our set of independently lost CNEs and traits using our devised textual mapping. We validated a mapping accuracy of at least 85% between the Morphobank traits and ontology terms in the set of closed loop phenotype–genotype associations. For example, for all the “teeth”-related phenotype–genotype closed loop associations, we verified that 90.9% of the CNEs are indeed nearby teeth-related genes (e.g., annotated by teeth terms in either HPO or MGI ontology) (supplementary fig. S1, Supplementary Material online).
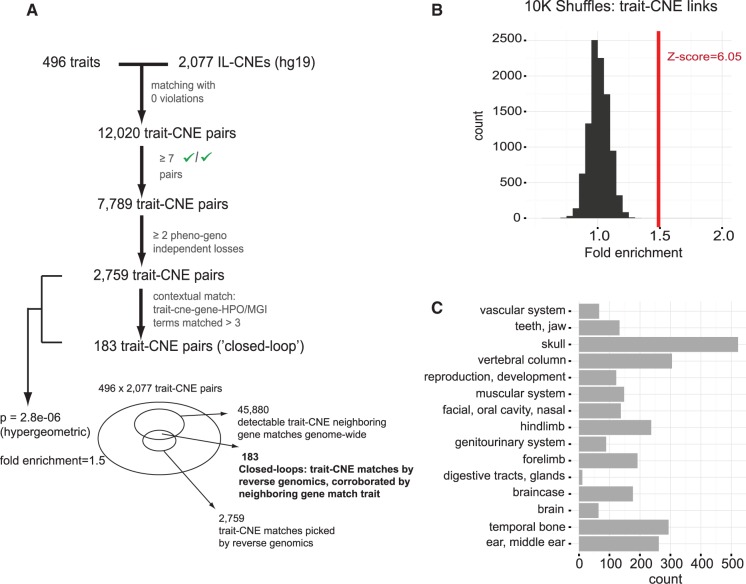
Fig. 4.
Matching human IL-CNEs to independently lost mammalian traits and identifying contextual closed loops via HPO/MGI gene annotations. Each of the IL-CNEs is matched by reverse genomics to hundreds of IL-traits from Morphobank, yielding an evolutionary matched phenotype–genotype (ph and g) pattern. An IL-CNE is associated with the closest gene (in either direction) if the distance from the TSS is less than 100 kb. A subset of predicted trait-CNE associations will generate closed loops (dashed blue) if the context of the matched phenotypic character matches the ontology terms annotating the nearby gene, which is presumed to be regulated by the IL-CNE (McLean et al. 2010). A predicted ph-g association is illustrated with a phenotypic character of the “phalanges” (bones of the hand and feet fingers). The matched IL-CNE resides in the regulatory domain of GPC3, a gene annotated with phalanges-related terms.
Our designed textual mapping between Morphobank traits and gene annotation terms from HPO/MGI allows us to run a parameter search for thresholds that maximize the proportion of closed loop associations in our predicted set of phenotype–genotype (i.e., the fold enrichment relative to expected number of closed loops). We scanned through thresholds of the minimal number of ontology terms in closed loop mappings (1–4 minimum terms), as well as the maximal distance between a CNE and nearest gene’s TSS (50 kb, 100 kb, 250 kb, 500 kb, 750 kb, 1 Mb). We converged on closed loop thresholds of at least 4 MGI or HPO ontology terms such that the CNE is within 100 kb of the TSS of the annotated gene and our textual mapping system maps the trait name to those ontology terms (fig. 4). Of all possible associations (1,030,192) between the 496 independently modified traits and 2,077 IL-CNEs, we counted 45,880 (4.45%) unique trait-CNE pairs that can yield a contextual closed loop between the matched trait and ontological terms annotating the gene next to the IL-CNE. Of these 45,880 closed loop trait-CNE pairs, 183 were in our set of 2,759 (6.63%) pairs extracted via reverse genomics, representing a significant enrichment (P = 2.83 × 10−6, hypergeometric test, Bonferroni corrected; fig. 5A). We additionally computed an empirical P value by performing 10,000 shuffles, where the fold enrichment was computed over a set of 2,759 unique pairs created at random between the input traits and IL-CNEs, and calculated a mean fold enrichment well above the value computed for the trait-CNE list from the screen (Z-score = 6; fig. 5B). The lists of trait-CNE associations are provided as supplementary tables with genomic coordinates (BED format, GRCh37/hg19) of the implicated CNEs and nearby genes (supplementary tables S6 and S7, Supplementary Material online).
Fig. 5.
(A) Matching 2,077 IL-CNEs against 496 independently modified traits. We extracted a total of 2,759 unique trait-CNE associations (P = 2.83 × 10−6, Bonferroni corrected, hypergeomtric test, fold enrichments = 1.5), among which we discovered 183 closed loop associations. (B) 10,000 shuffles where 2,077 IL-CNEs were randomly linked to 496 traits (fold computed over 2,759 random unique trait-CNE pairs). (C) A categorical classification of 2,759 trait-CNE associations by types of phenotypic characters.
IL-CNEs Are Associated with a Diverse Set of Mammalian Phenotypic Characters
Our set of 2,759 unique associations between IL-CNEs and traits spans a broad range of organs and tissue types with intriguing relations to morphological and physiological adaptations in mammals (fig. 5C and supplementary table S6, Supplementary Material online). Of the 15 categories of traits present in our data set, we found at least one representative from each category in our set of 183 closed loop trait-CNE associations, with the exception of “vascular system.” Manual inspection reveals appealing matches between vascular system traits and IL-CNEs near genes such as THBS1 and TLR2 that are implicated in vascular development and blood circulation; however, our automated textual mapping does not detect closed loops in such cases, due to the highly diverse nature of vascular traits and its distribution across many organs. Across our data set, we observed many compelling trait-CNE associations with matching annotations for nearby genes. A middle ear trait related to the morphology of the cochlea is associated with a region (IL-CNE605) about 20 kb upstream of BMP7, a gene that has been implicated in inner ear development (Mann et al. 2014). An association with a pelvis trait related to bone positioning in the iliac spine involves a CNE (IL-CNE107) in an intron of fibrillin 1 (FBN1), which has been associated with hip dysplasia and stature abnormalities in diseases like acromicric dysplasia (Klein et al. 2014) and Marfan syndrome (Haine et al. 2015). We also observe a midbrain morphology phenotype—exposure of the inferior colliculus in species from multiple lineages including rodents, microbats, and afrotherians—that is matched to a CNE (IL-CNE56) in a UBE4B intron. Interestingly, this CNE overlaps an H3K27ac histone modification mark in the substantia nigra, which is adjacent to the inferior colliculus in the midbrain (see supplementary table S7, Supplementary Material online, for detailed list of the 183 closed loop associations, with neighboring genes and inferred species).
To further motivate follow-up work on our set of 2,759 trait-CNE associations, we highlight an example of IL-CNE that has been lost in two independent aquatic mammalian lineages: Dolphins and manatees. The pectoral flipper of these aquatic mammals resembles the human arm in bone placement within the forelimb, having a ball and socket joint in the shoulder, and other homologous structures such as humerus, ulna, radius, carpals, and phalanges. The varying environment and functional tasks that accompanied the transition from land to water likely morphed forelimb bone structures in the flippers of these mammals (McGowen et al. 2014). Our screen identified a CNE lost in both dolphins and manatees that is located about 10 kb upstream of the EGR2 gene on human chromosome 10. This CNE was matched with a forelimb trait related to the skeletal structure of the elbow, as both aquatic species also have modified bone structures in their elbows that presumably prevent forelimb flexion and rotational motion (Lovejoy et al. 2009) (fig. 6A and B). We manually verified that the observed deletions in dolphin, manatee, as well as killer whale were not due to sequencing gaps or other artifacts, and that the region deleted was syntenic with EGR2 in dolphin, manatee, and killer whale. Notably, EGR2 is annotated in both HPO and MGI with functions related to forelimb bone morphology (fig. 6C). Taken together, this evidence suggests that this CNE influences forelimb structure by regulating EGR2.
Fig. 6.
An example of forelimb morphological trait associated with an IL-CNE (IL-CNE1686) near the EGR2 gene. (A) A bone trait involving the joint between the humerus, ulna, and radius (see arrows) exists in two states across placental mammals. (B) Left: a phenotree showing that an independent morphological modification pattern is observed in aquatic mammals and matches a CNE lost in those species. Right: A multiple alignment of the IL-CNEs with deletions in three delphinidae family members (belonging to the Odontoceti, toothed whale clade of cetacea): Dolphin, killer whale, and Yangtze River dolphin, as well as manatee from afrotheria. (C) The contextual logic linking the matched trait with the CNE near EGR2, through HPO/MGI terms related to the forelimb and the implicated bones.
Discussion
The repertoire of noncoding function in the human genome is far from fully understood. Epigenomic “active enhancer” measurements (e.g., H3K27ac Chip-seq marks) provide information about where and when an element is active, but cannot link enhancers to specific phenotypes or estimate how important the element is for the tissue (e.g., how knocking out the element will affect fitness). To address these challenges, we have developed an evolutionary-based reverse genomics approach to link human CNEs with phenomic characters on the basis of mutual independent phenotype–genotype modification patterns. Using a phenotypic matrix that scored trait states (i.e., trait presence or absence) across species, coupled with whole genome sequence alignments, we identified 2,759 functional trait-CNE associations. These associations are enriched for agreement with known gene annotations about mammalian phenotypes (Eppig et al. 2012) and human abnormalities (Köhler et al. 2014).
We link CNEs to traits related to a broad range of morphological and developmental traits. Among “two-state” traits sufficiently annotated across species with whole genome sequences, we have identified hundreds of phenotypic characters for which the ancestral state was modified independently at least twice during the evolution of placental mammals. In contrast to independent phenotype losses, we found that independent CNE losses are much less common: From an initial set of highly conserved human noncoding regions, we identified less than 5% as having multiple independent losses (the vast majority of which have independently lost exactly twice). It is possible that a higher than expected degree of pleiotropy of conserved cis-regulatory elements (i.e., driving expression in more than one anatomical structure) is a major factor constraining CNE losses (Hiller, Schaar, Bejerano, et al. 2012).
In carrying out a genomic screen for IL-CNEs, our work unifies, for the first time, large efforts from two disparate communities with convergent goals. We believe that genomicists could greatly benefit from the detailed curation efforts of morphologists (zoologists and paleontologists) (O’Leary and Kaufman 2011) in identifying novel links between genomic loci and function. Similarly, morphologists could use genomics to address the molecular basis for species anatomical and physiological diversity, or to infer ancestral traits from ancestral genomic states (Ma et al. 2006). There are concrete steps that members of both communities can take to facilitate this synergy. For example, genomicists could prioritize higher the whole genome sequencing of currently nonsequenced species with extensive morphological annotations (supplementary table S2, Supplementary Material online), thereby increasing the power of future screens similar to ours. In turn, morphologists could prioritize the development of a common vocabulary between free text trait descriptions in Morphobank and structured biological databases developed by the genomics community (e.g., by mapping scored trait terminology to the nearest ontology terms in MGI and HPO), thereby enabling both communities to combine each other’s data in their studies.
We believe that our proposed approach is able to identify a large number of important functional links between CNEs and morphological traits. However, no method is without limitations. First, associations involving pleiotropic CNEs may remain undetected, as the elements are constrained by their roles in additional traits. Second, our approach hinges on detecting ancestral traits that are encoded by ancestral genomic elements. The loss of similar phenotypic traits that are nonetheless encoded by distinct (nonorthologous) genomic regions will evade our current screen, as will be the case if an attempt is made to extend our approach to phenotypic gains that are mediated through nonorthologous genomic modifications. Looking forward, however, we expect our reverse genomics approach to become more powerful over time. Whole genome sequencing technology is rapidly marching forward, giving rise to novel sequence assemblies for many species (Lindblad-Toh et al. 2011) and significant improvements in the quality of existing assemblies. In parallel, our understanding of how different genes affect complex mammalian phenotypes is continuously expanding, making our gene annotations richer, and the proposed interpretation of nearby cis-regulatory elements via nearby genes more feasible. On the phenotypic side, structured phenotypic databases like Morphobank (O’Leary and Kaufman 2011) provide a rich and appealing resource for the investigation of genotype–phenotype links when intersected with comparative genomics analyses over multiple types of functional loci, including cis-regulatory elements, genes, and noncoding RNAs. Advances in automated text-mining approaches (Peters et al. 2014) have the potential to greatly increase the amount of available structured genomic and phenotypic information that can be easily processed computationally. Finally, experimental advances and the incorporation of accurate genome-editing tools like the CRISPR/Cas systems (Cong et al. 2013) will, over time, enhance the throughput of in vivo studies of trait-CNEs predictions and thus dramatically expand the landscape of opportunities for exploring cis-regulation in cross-species phenotypic diversity and in human biology.
Materials and Methods
Obtaining Genomic and Phenotypic Data
We obtained whole genome alignment data from the UCSC genome browser 46-way vertebrate multiz alignment (http://hgdownload.cse.ucsc.edu/goldenPath/hg19/multiz46way/, last accessed January 19, 2016). Morphological data were downloaded from Morphobank project 773 (http://morphobank.org/permalink/?P773, last accessed January 19, 2016)
Identifying Human CNEs from a 46-Way Vertebrate Alignment
We curated placental mammalian CNEs anchored to the human genome (assembly GRCh37/hg19). From an initial set of conserved regions in the human phastCons track (Siepel et al. 2005), generated from a 46-way vertebrate multiz alignment, we eliminated regions that overlap exons of known or predicted genes or functional RNA molecules. For that purpose, we generated a “might code” track from the union of regions annotated as exons by UCSC knownGene (Hsu et al. 2006), Ensembl (Flicek et al. 2013), the Mammalian Gene Collection (Temple et al. 2009), and RefSeq (Pruitt et al. 2007), including regions that matched RefSeq mRNAs found in other species. We also included regions predicted as exonic by Exoniphy (Siepel and Haussler 2004), and exons of pseudogenes annotated by VEGA (Ashurst et al. 2005) or the Yale Pseudogene Database (Karro et al. 2007). Finally, we included micro-RNAs from the miRNA Registry (Griffiths‐Jones 2004; Weber 2005), and small nucleolar RNA (snoRNA) and small cajal body-specific RNA (scaRNAs) from snoRNA-LBME-db (Lestrade and Weber 2006). We then removed conserved elements that had nonzero intersection with the “might code” track.
Next, we merged small conserved regions into larger ones, by grouping together all regions at most 20 bp apart, and selected only ones that were at least 50 bp in length. Finally, to exclude recent (e.g., primate specific) elements, we removed regions that were not conserved in at least 7 of the 19 sequenced and phenotyped placental mammals (fig. 2B and supplementary table S1, Supplementary Material online).
CNE Orthologous Chain Mapping
We analyzed the evolutionary history of each human (hg19) CNE, by mapping conserved regions in the human “reference” genome to other “query” species, through the UCSC liftOver chain pairwise alignments (Kent et al. 2003). A chain object depicts a pairwise alignment between orthologous genomic regions as a series of chain “links,” where each link corresponds to a gapless alignment, separated by either single-sided gaps (a deletion in one species or an insertion in the other) or double-sided gaps (representing multiple insertion or deletion events at the same locus).
Given a region in a reference species (GRCh37/hg19) and a query species, we find the orthologous coordinates in the query species, as defined by the liftOver chains. This procedure enables to identify deletions of the region in the query species, and distinguish them from false deletions created by assembly gaps. We first used the UCSC BigBed file format to store the reference genomic region spanned by the chain (i.e., storing each link with its chain identifier). We store the links in sorted order, as it is easier to get the links flanking the input region to either side. These are useful to eliminate errors due to assembly gaps.
Identification of CNE Loss Events
We extracted a pairwise alignment between CNEs in human and the orthologous locations in a query species from the liftOver chains. We compute two quantities: 1) percent identity (number of matches divided by the total alignment length) and 2) percent match (number of matches divided by the total number of bases in the reference). We mask away portions of the alignment where a deletion in the query species is flanked by chain links aligned to a region with an assembly gap. Similarly, we excluded insertions in the query species that contain assembly gaps.
We defined species-specific percent conservation cutoffs for a conservation and loss. By expectation, a neutrally evolving region should have a percent match of roughly 1−b, where b is the branch length between the two species (from UCSC multiple alignments, in units of substitutions per site). We therefore set the quantity 1−b as the loss cutoff. As this number can be quite high for species with small distance from humans (e.g., primates), we use a cutoff of 70% whenever 1−b is greater than 70%. For the conservation cutoff, we wished to estimate a parameter ρ (<1) such that the probability of a mutation at a given base within an evolutionarily conserved region is approximately ρb. Similar parameters were estimated by Siepel et al. (2005) when developing the phastCons model for identifying conserved elements. For various clades, they get estimates of ρ of approximately 1/3. We therefore set ρ = 1/3, and define the conservation cutoff for a species with a branch length b to be 1−ρb. We applied minor species-specific tuning (finalized independently of downstream calculations) to decrease this cutoff based on manual inspection of the percent identity histograms for each species.
The state of each CNE in a given query species is either conserved, lost, or unknown. An element is conserved if the entire interval lies above the conservation cutoff, and is lost if the entire interval lies below the loss cutoff (fig. 3). Because percent identity likely gives too high of a weight to insertions, and percent match ignores insertions, the interval these two values define can be interpreted as a “confidence interval” within which we expect the “true” conservation score to lie, if we knew exactly how deleterious each insertion was.
Identifying IL-CNEs in Placental Mammal Phylogenies
We extracted a subset of IL-CNEs that are each conserved in at least 7 of the 19 placental mammals, as well as the human and armadillo lineages, to ensure that the element spans much of the placental mammalian phylogeny. We used a parsimony-based algorithm that, for a given rooted phylogenetic tree with leaves annotated with either 1 or 0 (representing CNE “presence” or “absence,” respectively), computes the minimum number of loss events that are needed to explain the particular evolutionary pattern of ones and zeros, restricting 0 to 1 transitions (Gould 1970). We start at the leaves of the tree and work our way back toward the root to infer the state of each ancestral node. The number of independent losses in the tree equals the number of unique state 1 internal tree nodes with at least one child node of state 0 and another of state 1.
For each CNE, we required at least two independent loss events in the eutherian tree, and that independent loss events have outgroups with intact CNEs (i.e., bracketed by leaves annotated with 1). We applied the same method to trait phylogenetic trees to identify independently lost traits, as well as the matched trait-CNE phenotrees.
Mapping Morphobank Traits to MGI and HPO Ontologies Using Free Text Matching
To facilitate the automated bulk assessment of our candidate trait-CNE pairs, we mapped Morphobank phenotypic characters (O’Leary et al. 2013) to gene ontology terms related to mammalian anatomy and physiology. Specifically, we wanted to map the free text descriptions (O’Leary et al. 2013) of the 496 independently lost traits to one or more terms from the MGI and HPO ontologies, thus creating a unified vocabulary that links our phenotypic and genomic data. We started by preprocessing the textual description of each Morphobank trait. First, we removed two and three letter words (excluding C1–C7, which represents column vertebrae, and meaningful keywords like eye, pad, and ear). Next, using a table of English word frequency, we removed an additional set of general terms. Finally, we lemmatized each remaining word (using the Python stemming package stemming.porter2), ending up with at least two informative keywords for each phenotype. We then mapped each Morphobank phenotype to MGI and HPO ontology terms whose descriptions contained at least one of the keywords for the phenotype. To assess and refine the accuracy of the mapping, keywords were ranked according to how often they were used to link traits to ontology terms, and the top 100 keywords were inspected manually to ensure that they yielded correct phenotype-term mappings at least 90% of the time. Thirty-five of these 100 keywords were either removed from the table for reducing the accuracy (e.g., neck, which mapped “neck” related traits to “small neck of uterus”), required to match as a standalone keyword (e.g., “ear” should not match “heart”), or replaced with synonyms that better match the MGI/HPO vocabulary. The final accuracy of the resultant mapping from Morphobank traits to MGI and HPO is estimated to be higher than 85% when analyzing closed loop traits, IL-CNEs, and genes over particular types of traits (supplementary fig. S1, Supplementary Material online).
Supplementary Material
Supplementary tables S1–S7 and figure S1 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We thank Terence D. Capellini (Harvard) for discussions of the phenotypic characters and input about forelimb bone morphology in aquatic mammals, J. Gray Camp for manuscript advice, and Bejerano laboratory members for data analysis advice. This work was supported by a Stanford CEHG postdoctoral fellowship (A.M.), a Stanford School of Medicine postdoctoral Dean’s fellowship (A.M.), a National Science Foundation Graduate Research Fellowship under grant no. DGE-114747 (R.J.), the Packard Foundation (G.B.), a Microsoft Faculty Fellowship (G.B.), and an NIH U01MH105949 grant (G.B.).
References
- Ashurst JL, Chen CK, Gilbert JGR, Jekosch K, Keenan S, Meidl P, Searle SM, Stalker J, Storey R, Trevanion S, et al. 2005. The Vertebrate Genome Annotation (Vega) database. Nucleic Acids Res. 33:D459–D465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D. 2004. Ultraconserved elements in the human genome. Science 304:1321–1325. [DOI] [PubMed] [Google Scholar]
- Bradley RK, Li XY, Trapnell C, Davidson S, Pachter L, Chu HC, Tonkin LA, Biggin MD, Eisen MB. 2010. Binding site turnover produces pervasive quantitative changes in transcription factor binding between closely related Drosophila species. PLoS Biol. 8:e1000343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. 2005. Evolution at two levels: on genes and form. PLoS Biol. 3:e245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. 2008. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134:25–36. [DOI] [PubMed] [Google Scholar]
- Chan YF, Marks ME, Jones FC, Villarreal G, Shapiro MD, Brady SD, Southwick AM, Absher DM, Grimwood J, Schmutz J, et al. 2010. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science 327:302–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker JR, Bickmore WA, Barroso I, Pritchard JK, Gilad Y, Segal E. 2012. Genomics: ENCODE explained. Nature 489:52–55. [DOI] [PubMed] [Google Scholar]
- Eppig JT, Blake JA, Bult CJ, Kadin JA, Richardson JE, Mouse Genome Database Group. 2012. The Mouse Genome Database (MGD): comprehensive resource for genetics and genomics of the laboratory mouse. Nucleic Acids Res. 40:D881–D886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Ahmed I, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, et al. 2013. Ensembl 2013. Nucleic Acids Res. 41:D48–D55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ. 1970. Dollo on Dollo’s law: Irreversibility and the status of evolutionary laws. J Hist Biol. 3:189–212. [DOI] [PubMed] [Google Scholar]
- Griffiths‐Jones S. 2004. The microRNA Registry. Nucleic Acids Res. 32:D109–D111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guturu H, Doxey AC, Wenger AM, Bejerano G. 2013. Structure-aided prediction of mammalian transcription factor complexes in conserved non-coding elements. Philos Trans R Soc Lond B Biol Sci. 368:20130029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haine E, Salles JP, Khau Van Kien P, Conte-Auriol F, Gennero I, Plancke A, Julia S, Dulac Y, Tauber M, Edouard T. 2015. Muscle and bone impairment in children with Marfan syndrome: correlation with age and FBN1 genotype. J Bone Miner Res. 30:1369–1376. [DOI] [PubMed] [Google Scholar]
- Hiller M, Schaar BT, Bejerano G. 2012. Hundreds of conserved non-coding genomic regions are independently lost in mammals. Nucleic Acids Res. 40:11463–11476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller M, Schaar BT, Indjeian VB, Kingsley DM, Hagey LR, Bejerano G. 2012. A “forward genomics” approach links genotype to phenotype using independent phenotypic losses among related species. Cell Rep. 2:817–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. 2009. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 106:9362–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu F, Kent WJ, Clawson H, Kuhn RM, Diekhans M, Haussler D. 2006. The UCSC Known Genes. Bioinformatics 22:1036–1046. [DOI] [PubMed] [Google Scholar]
- Johnson MB, Wang PP, Atabay KD, Murphy EA, Doan RN, Hecht JL, Walsh CA. 2015. Single-cell analysis reveals transcriptional heterogeneity of neural progenitors in human cortex. Nat Neurosci. 18:637–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karro JE, Yan Y, Zheng D, Zhang Z, Carriero N, Cayting P, Harrrison P, Gerstein M. 2007. Pseudogene.org: a comprehensive database and comparison platform for pseudogene annotation. Nucleic Acids Res. 35:D55–D60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Baertsch R, Hinrichs A, Miller W, Haussler D. 2003. Evolution’s cauldron: duplication, deletion, and rearrangement in the mouse and human genomes. Proc Natl Acad Sci U S A. 100:11484–11489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein C, Le Goff C, Topouchian V, Odent S, Violas P, Glorion C, Cormier-Daire V. 2014. Orthopedics management of acromicric dysplasia: follow up of nine patients. Am J Med Genet A. 164A:331–337. [DOI] [PubMed] [Google Scholar]
- Köhler S, Doelken SC, Mungall CJ, Bauer S, Firth HV, Bailleul-Forestier I, Black GCM, Brown DL, Brudno M, Campbell J, et al. 2014. The Human Phenotype Ontology project: linking molecular biology and disease through phenotype data. Nucleic Acids Res. 42:D966–D974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lestrade L, Weber MJ. 2006. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res. 34:D158–D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh K, Garber M, Zuk O, Lin MF, Parker BJ, Washietl S, Kheradpour P, Ernst J, Jordan G, Mauceli E, et al. 2011. A high-resolution map of human evolutionary constraint using 29 mammals. Nature 478:476–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovejoy CO, Simpson SW, White TD, Asfaw B, Suwa G. 2009. Careful climbing in the Miocene: the forelimbs of Ardipithecus ramidus and humans are primitive. Science 326:70e1–70e8. [PubMed] [Google Scholar]
- Lowe CB, Kellis M, Siepel A, Raney BJ, Clamp M, Salama SR, Kingsley DM, Lindblad-Toh K, Haussler D. 2011. Three periods of regulatory innovation during vertebrate evolution. Science 333:1019–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Zhang L, Suh BB, Raney BJ, Burhans RC, Kent WJ, Blanchette M, Haussler D, Miller W. 2006. Reconstructing contiguous regions of an ancestral genome. Genome Res. 16:1557–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann ZF, Thiede BR, Chang W, Shin JB, May-Simera HL, Lovett M, Corwin JT, Kelley MW. 2014. A gradient of Bmp7 specifies the tonotopic axis in the developing inner ear. Nat Commun. 5:3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowen MR, Gatesy J, Wildman DE. 2014. Molecular evolution tracks macroevolutionary transitions in Cetacea. Trends Ecol Evol. 29:336–346. [DOI] [PubMed] [Google Scholar]
- McLean C, Bejerano G. 2008. Dispensability of mammalian DNA. Genome Res. 18:1743–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. 2010. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 28:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Reno PL, Pollen AA, Bassan AI, Capellini TD, Guenther C, Indjeian VB, Lim X, Menke DB, Schaar BT, et al. 2011. Human-specific loss of regulatory DNA and the evolution of human-specific traits. Nature 471:216–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary MA, Bloch JI, Flynn JJ, Gaudin TJ, Giallombardo A, Giannini NP, Goldberg SL, Kraatz BP, Luo ZX, Meng J, et al. 2013. The placental mammal ancestor and the post–K-Pg radiation of placentals. Science 339:662–667. [DOI] [PubMed] [Google Scholar]
- O’Leary MA, Kaufman S. 2011. MorphoBank: phylophenomics in the “cloud.” Cladistics 27:529–537. [DOI] [PubMed] [Google Scholar]
- Pennacchio LA, Bickmore W, Dean A, Nobrega MA, Bejerano G. 2013. Enhancers: five essential questions. Nat Rev Genet. 14:288–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters SE, Zhang C, Livny M, Ré C. 2014. A machine reading system for assembling synthetic paleontological databases. PLoS One 9:e113523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR. 2007. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 35:D61–D65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragvin A, Moro E, Fredman D, Navratilova P, Drivenes Ø, Engström PG, Alonso ME, Mustienes E, de la C, Skarmeta JLG, Tavares MJ, et al. 2010. Long-range gene regulation links genomic type 2 diabetes and obesity risk regions to HHEX, SOX4, and IRX3. Proc Natl Acad Sci U S A. 107:775–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, et al. 2015. Integrative analysis of 111 reference human epigenomes. Nature 518:317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Wilson MD, Ballester B, Schwalie PC, Brown GD, Marshall A, Kutter C, Watt S, Martinez-Jimenez CP, Mackay S, et al. 2010. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science 328:1036–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15:1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Haussler D. 2004. Computational identification of evolutionarily conserved exons. In: Gusfield D, Bourne P, Istrail S, Pevzner P, Waterman M, editors. Proceedings of the Eighth Annual International Conference on Research in Computational Molecular Biology. RECOMB ’04. New York: ACM. p. 177–186. Available from: http://doi.acm.org/10.1145/974614.974638.
- Smith RP, Taher L, Patwardhan RP, Kim MJ, Inoue F, Shendure J, Ovcharenko I, Ahituv N. 2013. Massively parallel decoding of mammalian regulatory sequences supports a flexible organizational model. Nat Genet. 45:1021–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temple G, Gerhard DS, Rasooly R, Feingold EA, Good PJ, Robinson C, Mandich A, Derge JG, Lewis J, Shoaf D, et al. 2009. The completion of the Mammalian Gene Collection (MGC). Genome Res. 19:2324–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, et al. 2012. The accessible chromatin landscape of the human genome. Nature 489:75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuch BB, Galgoczy DJ, Hernday AD, Li H, Johnson AD. 2008. The evolution of combinatorial gene regulation in fungi. PLoS Biol. 6:e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman NF, Aneas I, Nobrega MA. 2010. An 8q24 gene desert variant associated with prostate cancer risk confers differential in vivo activity to a MYC enhancer. Genome Res. 20:1191–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber MJ. 2005. New human and mouse microRNA genes found by homology search. FEBS J. 272:59–73. [DOI] [PubMed] [Google Scholar]
- Woolfe A, Goodson M, Goode DK, Snell P, McEwen GK, Vavouri T, Smith SF, North P, Callaway H, Kelly K, et al. 2005. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 3:e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray GA. 2007. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 8:206–216 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.