Abstract
Genetic differentiation across populations that is maintained in the presence of gene flow is a hallmark of spatially varying selection. In Drosophila melanogaster, the latitudinal clines across the eastern coasts of Australia and North America appear to be examples of this type of selection, with recent studies showing that a substantial portion of the D. melanogaster genome exhibits allele frequency differentiation with respect to latitude on both continents. As of yet there has been no genome-wide examination of differentiated copy-number variants (CNVs) in these geographic regions, despite their potential importance for phenotypic variation in Drosophila and other taxa. Here, we present an analysis of geographic variation in CNVs in D. melanogaster. We also present the first genomic analysis of geographic variation for copy-number variation in the sister species, D. simulans, in order to investigate patterns of parallel evolution in these close relatives. In D. melanogaster we find hundreds of CNVs, many of which show parallel patterns of geographic variation on both continents, lending support to the idea that they are influenced by spatially varying selection. These findings support the idea that polymorphic CNVs contribute to local adaptation in D. melanogaster. In contrast, we find very few CNVs in D. simulans that are geographically differentiated in parallel on both continents, consistent with earlier work suggesting that clinal patterns are weaker in this species.
Keywords: population genetics, copy-number variation, natural selection.
Introduction
Latitudinal clines in Drosophila represent a classic system for investigating the role of spatially varying selection in the maintenance of genetic variation. The species that has received the most attention is Drosophila melanogaster. A large body of work amassed over several decades has revealed many genetically determined phenotypic clines, chromosome inversion clines, and allozyme clines (reviewed in De Jong and Bochdanovits 2003; Hoffmann and Weeks 2007; Adrion et al. 2015), whereas more recent work on spatial variation has focused on sequence variation (Sezgin et al. 2004), including genome-scale investigations (Turner et al. 2008; Kolaczkowski et al. 2011; Fabian et al. 2012; Reinhardt et al. 2014). Many traits and genetic variants exhibit patterns consistent with spatially varying selection on multiple continents, as expected under the simplest scenario of similar selection pressures correlated with latitude acting on ancestral variation. However, interesting differences between continents in patterns of spatial variation are also observed (Turner et al. 2008; Reinhardt et al. 2014).
The question of patterns of spatial variation in D. simulans, a sister species to D. melanogaster, has received relatively little attention. Both species are recent colonizers of Eurasia, the Americas, and Australia, and they are currently broadly sympatric (Capy and Gibert 2004). For these species, overall similarities and differences with respect to geographic differentiation could be properties of colonization histories, biology, or mechanisms by which selection acts. Most phenotypic work on clines in D. simulans has investigated variation in Australia (Arthur et al. 2008), and these studies suggest that clines in this species are less common or weaker than those observed in D. melanogaster. This observation supports earlier work suggesting that “clinality” is weaker in D. simulans than in D. melanogaster, even in North America (Singh and Long 1992; Gibert et al. 2004), though patterns of shared latitudinal gene expression differentiation in the two species (Zhao et al. 2015) have raised new questions about the influence of spatially varying selection in D. simulans.
Population genomic work on latitudinal differentiation in flies has largely focused on single-nucleotide polymorphisms (SNPs) (Turner et al. 2008; Kolaczkowski et al. 2011; Fabian et al. 2012; Reinhardt et al. 2014). However, because copy-number variants (CNVs) can be quite large and often contain genic sequence (Schrider and Hahn 2010), their phenotypic effects may typically be greater than those of SNPs, which suggests that such variants may play an important role in local adaptation. Copy-number variation is widespread in D. melanogaster (Huang et al. 2014) and D. simulans (Rogers et al. 2014), and population genetic evidence suggests that newly occurring CNVs in flies are often deleterious (Emerson et al. 2008; Cridland and Thornton 2010; Cardoso-Moreira et al. 2011; Langley et al. 2012; Schrider, Houle, et al. 2013), consistent with data from humans (McCarroll and Altshuler 2007; Stankiewicz and Lupski 2010; Girirajan et al. 2011). However, the extent to which segregating CNVs contribute to adaptive evolution in D. melanogaster and D. simulans is unknown. Here, we use genomic data to address this question and to determine the extent of parallelism in CNV differentiation in these closely related species.
Results
Spatially Varying Selection on Copy-Number Variation in D. Melanogaster
We sequenced pooled genomic DNA samples from four locations: Both the northern and southern ends of the East Cost of the United States (Maine and Florida), and the northern and southern ends of eastern Australia (northern Queensland and Tasmania; supplementary table S1, online). We then used read depth and paired-end information to identify CNVs differentiated in allele frequency between the northern and southern samples in the United States, and also read depth only on both continents (see Materials and Methods). This second approach was essential for the Australian samples for which only single-end reads were sequenced (see Materials and Methods). We found 190 CNVs >50 bp in length showing marked allele frequency differences in the United States (based on differences in read depth and numbers of discordant paired-ends from the two locations; see Materials and Methods). In Australia, we found 110 differentiated CNVs (supplementary table S2, Supplementary Material online), yielding a total of 203 distinct CNVs (64 duplications, and 139 deletions; see Materials and Methods) combined across both continents. Although the Maine sample had roughly 25% greater coverage than the Florida sample (supplementary table S1, Supplementary Material online), the set of CNVs found in the United States does not change dramatically after subsampling reads from Maine to match coverage in Florida (>86% of CNVs in the original set are recovered; see Materials and Methods). Thus, our results are not strongly affected by differences in sequencing depth. The smaller number of differentiated CNVs detected in Australia may be due to the lack of paired-end information—read depth information alone may have lower power to detect CNVs (Schrider, Begun, et al. 2013; see Materials and Methods).
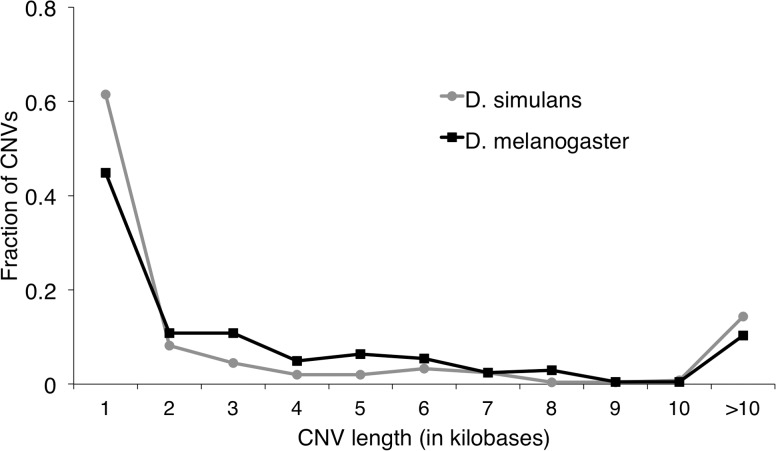
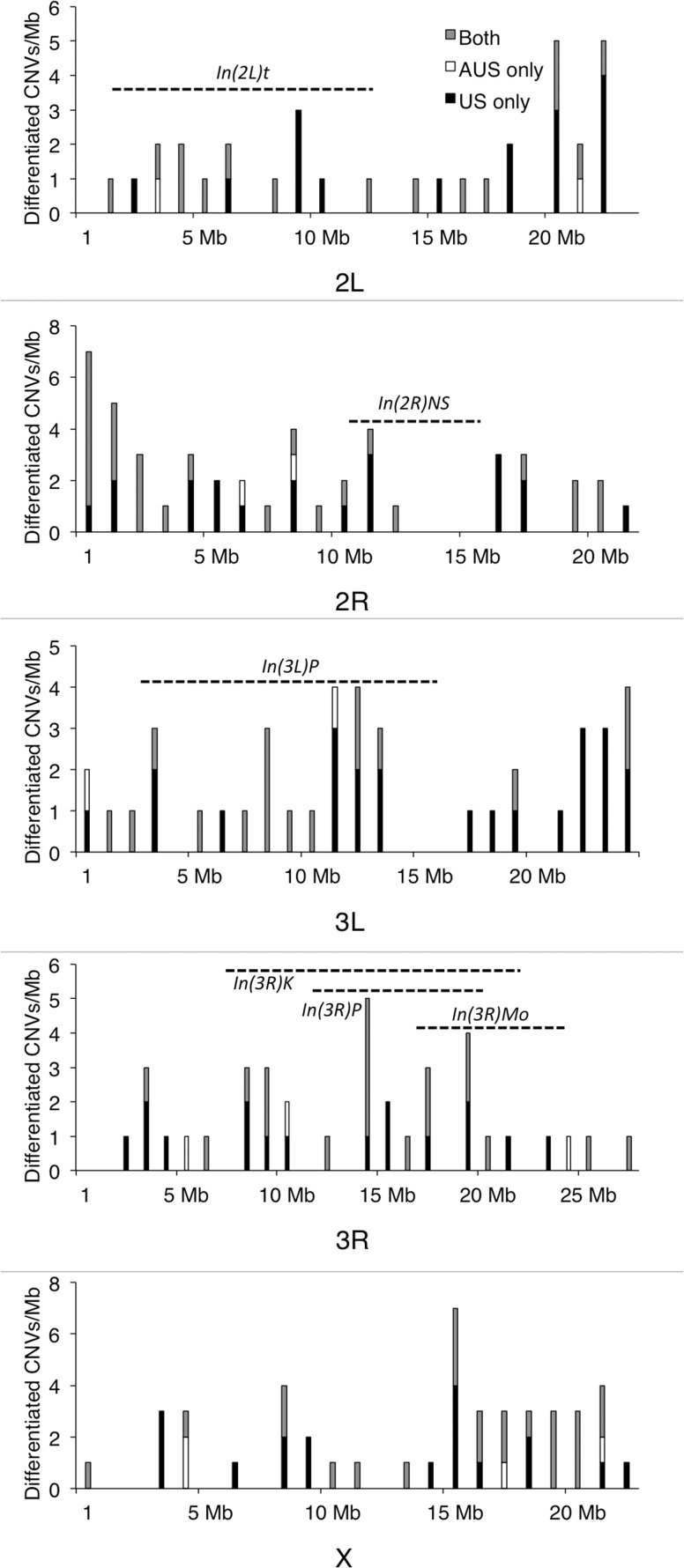
The differentiated CNVs have a median length of 1,500 bp; the length distribution is shown in figure 1. There is little evidence that these differentiated CNVs are heterogeneously distributed across chromosome arms (P = 0.096, χ2 test), and they are not enriched within large chromosomal inversions (fig. 2; P = 0.11, permutation test; see Materials and Methods). Differentiated CNV density is uncorrelated with recombination rate (P = 0.95, Spearman’s ρ; see Materials and Methods). Duplications are not significantly enriched for exonic sequence (1.25% of the expectation from permuted data sets; P = 0.0871 from one-sided permutation test; see Materials and Methods), complete genes (1.22% of the expectation; P = 0.1373), or intronic sequence (1.20% of the expectation; P = 0.3561). Deletions are depleted of complete genes (56.4% of the expectation from permuted data; P = 0.0185) and exonic sequence (60.1% of the P = 0.0094), but not significantly depleted of intronic sequence (72.6% of the expectation; P = 0.1986).
Fig. 1.
Histograms of lengths of all CNVs differentiated along one or both coastlines in Drosophila melanogaster (black) and D. simulans (gray).
Fig. 2.
Density of differentiated CNVs across the Drosophila melanogaster genome. The numbers of CNVs differentiated along the US cline (black), the Australian cline (white), or both (gray, whether in the same or opposite directions) are shown for each 1-Mb window on each chromosome arm. Approximate locations of large chromosomal inversions (from Corbett-Detig et al. 2012) are shown as dashed lines.
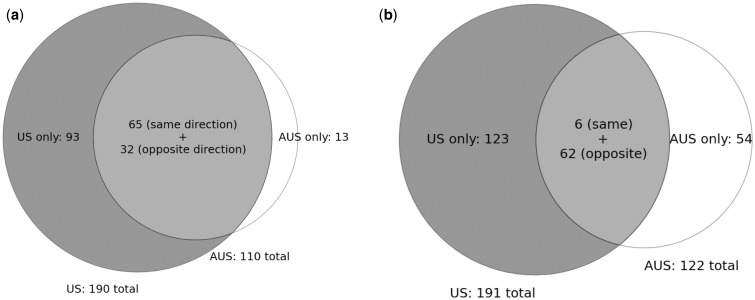
While genetic differentiation across heterogeneous environments despite extensive gene flow is a hallmark of natural selection, confidence in the biological relevance of such differentiation can be increased by asking whether differentiation occurs in parallel across different geographic regions exhibiting similar ecological gradients (e.g., Jones et al. 2012). Therefore, for each differentiated CNV detected on one continent, we asked whether it was also significantly differentiated on the other continent and in the same direction with respect to distance from the equator; that is, if allele A is at higher frequency in Maine, is this allele also at higher frequency in Tasmania? Of the 190 CNVs differentiated in the United States, 65 are also differentiated in the same direction with respect to latitude on both continents (fig. 3a; P < 2.2 × 10−16, binomial test), and 32 are differentiated but in the opposite direction on the two continents (P = 1.86 × 10−9). Importantly, we observe qualitatively similar results when examining CNVs detected from read depth alone: We detect 61 differentiated CNVs in the United States and 71 in Australia (supplementary table S3, Supplementary Material online; see Materials and Methods), with 39 of these differentiated along both clines in the same direction with respect to distance from the equator (P < 2.2 × 10−16), and 20 differentiated in opposite directions (P = 1.8 × 10−10). In our full set of CNVs, this excess of CNVs differentiated in the same direction holds for both duplications and deletions (P < 2.2 × 10−16 for each). However, the fraction of duplications that are differentiated in parallel across continents is greater than that of deletions (45% vs. 26%; P = 0.0094; Fisher’s exact test).
Fig. 3.
Venn diagrams of differentiated CNVs detected in Drosophila melanogaster and D. simulans. (a) Drosophila melanogaster. Differentiated CNVs detected in the United States are represented by the gray circle, whereas those detected in Australia are in the white circle. The intersection represents all CNVs differentiated on both continents, whether in the same or opposite directions with respect to distance from the equator. The much lower number of CNVs differentiated only in Australia is likely due to the lack of paired-end reads from these samples. (b) Differentiated CNVs detected in D. simulans.
Although Kolaczkowski et al. (2011) report evidence of differentiated CNVs having on average elevated copy-numbers in Tasmania, we found no significant excess of CNVs with higher or lower read depth at the temperate cline endpoint on either continent (100 differentiated CNVs with higher read depth in Maine vs. 90 with elevated read depth in Florida; P = 0.51; 63 CNVs with higher depth in Tasmania vs. 47 in northern Queensland; P = 0.15; binomial tests). This may be a result of Kolaczkowski et al.’s treatment of 1-kb windows as independent observations, causing large CNVs to be counted multiple times and thereby inflating statistical significance.
To be considered differentiated on both continents, we required that a CNV detected on one continent exhibits a ratio of read depths in the 5% most extreme tail of the distribution of depth ratios on the other continent (see Materials and Methods). This requirement ensures that only a handful of CNVs would appear to be differentiated in the same direction in both clines by chance. Thus, the majority of CNVs showing evidence for geographic differentiation in both the United States and Australia (the intersection in fig. 3a) are likely to be influenced by spatially varying selection. Global hitchhiking events can also create patterns of differentiation in linked regions (Bierne 2010), but repeated spatially varying selection is a better explanation for the observed parallelism across clines. The hypothesis that the CNVs differentiated in parallel are the targets of this selection, rather than linked polymorphisms, is plausible for two reasons. First, the scale of linkage disequilibrium in D. melanogaster is generally small (Langley et al. 2012), meaning that it may be unlikely that a linked mutation is the one under selection. Second, the fact that a typical differentiated CNV includes a large number of base pairs suggests that a CNV may be more likely to be associated with a fitness effect compared with a SNP. Following this reasoning, we suggest below that several differentiated SNPs identified in early comparisons of pooled DNA sequences from ends of the US cline (Turner et al. 2008) are in fact associated with differentiated CNVs.
To investigate whether the CNVs differentiated on both continents are the same variants or independent mutational events, we estimated read depth around these CNVs and asked whether the ratio of tropical to temperate read depth returns to background levels at the same genomic location in the United States and Australia. Of the 34 CNVs detected with paired-ends and having a tropical:temperate or temperate:tropical ratio ≥1.25, 25 exhibited a ratio <1.25 within the 400-bp window flanking each putative breakpoint (estimated from the paired-end US data) of the CNV on each continent (supplementary table S2, Supplementary Material online). Thus, the breakpoints of differentiated CNVs appear to be similar for the Australian and North American data. Although we cannot rule out recurrent mutation causing CNVs with identical or very similar breakpoints for some of these pairs of highly overlapping CNVs, it is likely that much of the parallel differentiation on the two continents is the result of selection on ancestral, standing variation that was introduced independently to the two continents. Future studies that accurately resolve breakpoint sequences will be able to more precisely assess the relative contributions of previously standing variation and recurrent de novo mutations to shared clinal copy-number variation.
In contrast, a small number of the CNVs differentiated in parallel are consistent with different variants segregating on the two continents. For example, the previously described duplication of a > 100-kb region on chromosome arm 3R containing Ace (acetylcholine esterase)—which has been shown to influence insecticide resistance (Menozzi et al. 2004)—is differentiated in Australia (higher frequency in Queensland; Turner et al. 2008; Kolaczkowski et al. 2011) but does not appear to be differentiated in the United States. However, we did detect a > 10-kb region containing much of this gene with significantly higher copy-number in Florida than in Maine (also previously observed in Turner et al. 2008). Thus, it is plausible that Ace is the target of selection on both continents, but with independently arising variants spreading in a relatively short period of time. This result for CNVs parallels similar results suggesting that adaptive amino acid changes have also occurred independently multiple times in Ace (Karasov et al. 2010).
No Strong Correlation between Copy-Number Differentiation and Gene Expression Differentiation
To investigate whether differentiated CNVs affect gene expression, we examined transcript levels in genes wholly or partially contained within CNVs using RNA-seq data from flies originating from opposite ends of the North American cline and reared at either 21 or 29 °C (Zhao et al. 2015). Among genes overlapping CNVs differentiated in North America we found no significant correlation between the ratio of read depth in Florida to read depth in Maine and log2 fold-changes in expression between the two cline endpoints (Spearman’s ρ = 0.010 for flies reared at 21 °C, P = 0.93; Spearman’s ρ = 0.067 for flies reared at 29 °C, P = 0.59). Thus, these differentiated CNVs do not alter transcript abundance in a manner that is easily predicted from the observed copy-number change. However, this does not necessarily imply that these differentiated CNVs have no effect on gene expression: Although some CNVs predictably alter transcript levels of genes within their breakpoints, many CNVs have no detectable impact on transcript levels of constituent genes (Zhou et al. 2011), but some can impact expression of flanking or distant genes (Stranger et al. 2007).
Biological Characterization of Differentiated CNVs
There is an excess of cytochrome P450 genes contained either partially or completely within CNVs that are highly differentiated on one or both continents (P = 0.0002; based on 10,000 permutations of CNV coordinates). These include Cyp12d1-p and Cyp12d1-d, which are both completely encompassed by a deletion spanning approximately 10 kb on chromosome arm 2R; the presence allele is more common in the temperate population on both continents. Cyp12d1 confers improved insecticide resistance (Daborn et al. 2007). Cyp12a4, another cytochrome P450 gene associated with insecticide resistance when overexpressed, lies within a duplication that exhibits higher copy-number at low-latitudes; this CNV also partially duplicates Cyp12a5. Interestingly, the region containing Cyp12a4 was previously identified in a genome-wide scan for high-FST SNPs (Turner et al. 2008). In addition, we find that Cyp6g1 and part of Cyp6g2 and CG13175 lie within a duplication segregating at higher frequency in northern Queensland (low-latitude) than Tasmania—the region containing Cyp6g1 was also identified as having elevated FST at SNPs in Australia by Kolaczkowski et al. (2011). Some alleles at Cyp6g1 confer improved insecticide resistance when upregulated (Daborn et al. 2002), but we cannot be certain which Cyp6g1 allele(s) described by Schmidt et al. (2010) are differentiated in our data. Another duplicated cytochrome P450, Cyp12c1, has higher copy-number in Maine than in Florida. However, several other genes are included in this duplication event (Chmp1, CG34254, CG32202, and Sgf11), so it is unclear whether Cyp12c1 is a target of selection. Cyp28d2 lies entirely within a duplication with higher frequency in our tropical samples, whereas Cyp313a4 lies partially within a deletion with higher frequency in the tropics. Finally, a duplication that contains Cyp6a17 and parts of Cyp6a22 and Cyp6a23 (another high-FST region previously highlighted by Turner et al. 2008) exhibits higher copy-number in Florida (low-latitude) and Tasmania (high-latitude). Knockdown of Cyp6a17 has been shown to disrupt temperature preference (Kang et al. 2011). Interestingly, some cytochrome P450-associated CNVs have higher copy-number in temperate populations, whereas others have higher copy-number in the tropics. This suggests that the connection between gene dosage and fitness variation varies across different cytochromes. The observation that the CNV containing Cyp6a17 has higher copy-number in one tropical (Florida) and one temperate (Tasmania) population also implies that there may be spatially varying selection that is poorly correlated with latitude.
More generally, as noted above we find an excess of CNVs differentiated on both continents but in opposite directions with respect to distance from the equator. This surprising result could be due to CNVs that show different correlations between fitness variation and copy-number on the two continents, or could be the result of CNVs that show latitudinal differentiation as a result of selection at linked sites. The observation that many cytochrome P450-containing regions identified as containing an outlier FST SNP also contain differentiated CNVs implies that the targets of spatially varying selection in these regions could often be CNVs, rather than the SNP—some of these SNPs may actually be nucleotide differences between the two duplicate copies (when the CNV is a duplication), whereas others may be true SNPs in linkage disequilibrium with the selected CNV. On the other hand, the hypothesis that the CNVs are linked to the true targets of selection cannot be ruled out, in which case it is the linkage disequilibrium of these CNVs to other selected mutations that drives this antiparallel differentiation.
In order to gain broader insights into the types of genes targeted by spatially varying selection, we conducted a search for Gene Ontology (GO) terms enriched within the set of CNVs differentiated in the same direction on both continents using a permutation test (see Materials and Methods). Importantly, this search counted each term at most once per CNV and is therefore robust to the spatial clustering of functionally related genes (Pavlidis et al. 2012). Biological process terms overrepresented with a false discovery rate (FDR) less than 0.05 include several terms related to insecticide response (response to insecticide, response to carbamate, and response to organophosphorus), similar to terms found to be enriched by Turner et al. (2008). Additional terms include response to DNA damage checkpoint, acetylcholine catabolic process, and response to heat. All significant GO terms appearing in CNVs differentiated along both clines and their FDRs are listed in supplementary table S4, Supplementary Material online.
Little Evidence for Spatially Varying Selection on CNVs Repeated across Continents in D. simulans
Because D. simulans also migrated relatively recently to North America (David and Capy 1988) and Australia (Malloch 1923), and is broadly sympatric with D. melanogaster, we sought to compare geographic variation in copy-number polymorphism in the two species. Our D. simulans US population samples come from Maine and Florida, and our Australian samples come from Tasmania and Queensland (see Materials and Methods). We observed 191 differentiated CNVs in the United States and 122 differentiated CNVs in Australia, similar to the overall number of differentiated CNVs in D. melanogaster. Of these CNVs, however, only six are differentiated in the same direction on both continents (fig. 3b; supplementary table S5, Supplementary Material online); this does not represent a statistically significant excess of parallel differentiation (P > 0.3, using binomial tests for excess of US CNVs differentiated in the same direction Australia, and vice versa, with expected success rate of P = 0.05). Unexpectedly, we do observe a statistically significant excess of duplications and deletions differentiated along both clines but in opposite directions with respect to latitude (62 CNVs, P < 2.2 × 10−16 for both duplications and deletions). Although among these differentiated CNVs a larger fraction of duplications than deletions show this pattern of shared anti-parallel differentiation (33% vs. 21%), this difference is not significant (P = 0.063; Fisher’s exact test).
Overall, this result contrasts sharply with our findings in melanogaster where we find a highly significant excess of CNVs differentiated in the same direction on both continents. We did find three genes that showed evidence of copy-number differentiation in both species: Syx16, HERC2, and l(1)G0004, though this amount of overlap between our D. simulans and D. melanogaster sets is not in excess of that expected by chance (P > 0.3139; hypergeometric test). Thus, although our data do provide some support for spatially varying selection on CNVs in D. simulans, we find little evidence for parallel selection across the two continents.
Discussion
Uncovering the genetic basis for adaptation is a major goal of evolutionary genomics, but is often difficult to accomplish given the joint effects of demography and selection. The parallel environmental clines experienced by multiple species of Drosophila in the Northern and Southern hemispheres offer a promising system in which the effects of directional and stochastic changes may be partially disentangled. Here, we have taken advantage of these clines to compare cross-continent latitudinal differentiation within and between species. Our argument that genomic regions that are strongly differentiated on multiple continents are enriched for sites influenced by spatially varying selection hinges on the assumption that demographic processes are unlikely to generate substantial parallelism. Several recent studies have suggested that North American populations experienced recent African-European admixture (Caracristi and Schlötterer 2003; Duchen et al. 2013; Bergland et al. 2015; Kao et al. 2015), which may contribute to clinal variation. A recent study suggested a similar phenomenon in Australian populations (Bergland et al. 2015). However, given the very high levels of gene flow estimated between ends of the clines on both continents (Agis and Schlötterer 2001; Kennington et al. 2003; Schmidt et al. 2005), and the strong enrichment of clinal CNVs of genes involved in responses to insecticide, DNA damage, and heat, a purely demographic explanation seems unlikely. Moreover, these two explanations are not mutually exclusive: There may be CNVs previously differentiated between Africa and Europe due to local adaptation, leading to the establishment—and maintenance by selection—of clinal patterns following migration of both European and African flies to the Australia and North America.
Three main patterns emerged from our characterization of geographic variation in CNVs on two continents in two species. First, in D. melanogaster, there is substantial sharing of differentiated CNVs in the United States and Australia, as has also been observed for differentiated SNPs along these clines (Reinhardt et al. 2014). These results support the idea that spatially varying selection is correlated on the two continents and that this adaptive differentiation is consistent with selection on ancestral variation (e.g., Aminetzach et al. 2005; Chan et al. 2010; Kolaczkowski et al. 2011; Domingues et al. 2012; Jones et al. 2012; Reinhardt et al. 2014). However, there are also several interesting exceptions that are candidates for independently evolved adaptations, such as the CNVs encompassing Ace. All else being equal, newer adaptive variants should be associated with larger-scale linkage disequilibrium. This prediction can be tested in the future for the candidate CNVs given appropriate genomic sequencing of the breakpoints and surrounding regions.
Second, we observe no connection between the extent of differentiation of CNVs and expression of genes residing within these CNVs. Thus, simple dosage changes of genes within differentiated CNVs do not appear to be the primary targets of spatially varying selection. Ohno (1970) referred to such changes as “Duplication for the sake of producing more of the same,” and they have been proposed to be a major force in duplicate gene retention (e.g., Sugino and Innan 2006), but they do not appear to account for strong patterns of clinal variation in gene expression in Drosophila (Zhao et al. 2015). However, there are also examples of CNVs that change overall levels of expression but that either encode proteins with different functions (e.g., Labbé et al. 2007) or have distinct expression domains from their parental copies (e.g., Schrider, Navarro, et al. 2013). In fact, an example of CNVs with different protein functions comes from the multiple copies of the Ace gene maintained in the mosquito, Culex pipiens: One copy is resistant to insecticide and one copy retains the ancestral function (Labbé et al. 2007). In this way, permanent heterozygosity is maintained in those populations that experience strong selection from insecticides, and again this mechanism has been proposed to act on many pairs of gene duplicates (reviewed in Hahn 2009). In addition, many of our CNVs only partially overlap genes; these CNVs may also affect expression through duplicating or deleting regulatory regions or by altering coding sequences. Our clinal CNVs may also alter expression levels of genes flanking or distant from the duplication or deletion (Stranger et al. 2007). Distinguishing between these multiple proposed mechanisms will require further molecular characterization of the differentiated CNVs that we have detected.
Finally, although our analyses support the idea that CNVs play an important role in adaptive differentiation within D. melanogaster, we observe less evidence for this in D. simulans: Though we do observe an equal number CNVs differentiated on both continents, very few of these are differentiated in the same direction with respect to distance from the equator. This result could imply that spatially varying selection is less pervasive in D. simulans than D. melanogaster, an interpretation that is consistent with previous studies suggesting that clinal variation is less prominent in D. simulans (Singh and Long 1992; Gibert et al. 2004; Arthur et al. 2008; Machado et al. 2015). Machado et al.’s (2015) recent analysis of SNP variation in D. melanogaster and D. simulans sampled along the American cline again found less differentiation in D. simulans. Nonetheless, Machado et al. (2015) did find more overlap among the sets of clinal genes in D. simulans and D. melanogaster than expected by chance. This suggests that spatially varying selection is contributing to clinal patterns of SNP variation in D. simulans as well as D. melanogaster, and that some of the same genes have been targeted in both species. These findings combined with our results may imply that the selective and stochastic forces affecting latitudinal differentiation have a greater effect on patterns of SNP than on copy-number variation in D. simulans. However, further study will be required to illuminate the biological significance of the intriguing excess of antiparallel differentiation in copy-number along the two coastlines in D. simulans. More generally, better data sets and additional analyses will be required to infer the relative importance of CNVs, SNPs, and other polymorphisms in latitudinal adaptation, and to explain the marked difference in the impact of geographically varying selection on genomic polymorphism in these species.
Materials and Methods
Flies, Sequence Data, and Read Mapping
Drosophila melanogaster isofemale lines from Maine (n = 16), Florida (n = 16), Tasmania (n = 15), and Queensland (n = 17) were previously described (Turner et al. 2008). North American D. simulans isofemale lines were from Fairfield, ME (n = 50) and Homestead, FL (n = 33) and were collected by Perot Saelao. Australian D. simulans samples were collected from Sorrell, Tas (n = 16) and Maryborough, Qld (n = 22) by Arthur et al (2008). For each population sample, a single female fly was randomly picked from each isofemale line. The flies from a population were then pooled and Illumina libraries were constructed—one per population (Kolaczkowski et al. 2011; Reinhardt et al. 2014). These libraries were then sequenced using the Illumina Genome Analyzer II (Reinhardt et al. 2014). Both of the Australian melanogaster sequencing libraries were single-end. The remaining sequencing libraries were all paired-end. The numbers of sequenced fragments, average insert sizes, and average depths-of-coverage after mapping for each of these eight pooled sequence runs are shown in supplementary table S1, Supplementary Material online. Using BWA version 0.5.9 (Li and Durbin 2009), we mapped D. melanogaster reads to release five of the melanogaster assembly with repetitive elements masked by RepeatMasker (http://www.repeatmasker.org), and D. simulans reads to an updated D. simulans assembly (Hu et al. 2013). The D. melanogaster data are from Reinhardt et al. (2014), and are available on the Short Read Archive (bioproject accession number PRJNA237820). The D. simulans data have also been uploaded to the Short Read Archive (bioproject number PRJNA308157 for Maine samples; PRJNA307610 for all others).
Detecting Differentiated CNVs from Pooled Paired-End Sequencing Data
We detected CNVs differentiated in allele frequency along each cline in each species using a combination of read depth and discordant paired-end mapping data as described in Schrider, Begun, et al. (2013). Briefly, from each pooled sample we clustered nearby pairs of reads mapped in an orientation indicative of a deletion (i.e., mapped further apart from one another than expected) or of a tandem duplication (in “everted” orientation; Cooper et al. 2008). We then counted the number of mapped read pairs (or single reads in single-end samples) supporting the CNV at the two endpoints of the cline (zero if the nonreference allele is completely undetected at that endpoint) and took the difference between the two. We considered CNVs for which this difference was in either the top or bottom 5% among all candidate CNVs to be potentially differentiated, but omitted all putative CNVs <50 bp in length.
We then used read depth information to confirm or reject each potentially differentiated CNV by asking whether the ratio of read depths between the two cline ends (again, ignoring repetitive DNA) departed significantly from 1:1 in the direction predicted by the difference in the number of read pairs supporting the CNV. The significance cutoffs were determined empirically by randomly selecting genomic regions of a given length and measuring the ratio of read depths counted from the two ends of the cline, and selecting the top and bottom 5% cutoffs from the resulting distribution. CNVs with both a significant difference in supporting read pairs and a concordantly extreme read depth ratio were considered to be differentiated between the cline endpoints. Rather than compute these read depth ratio cutoffs for every CNV length, we computed cutoffs for various lengths and for a given CNV we used the cutoffs for the closest length of lesser or equal value. These lengths were: 50, 100, 150, 200, 250, 300, 350, 400, 450, 500, 600, 700, 800, 900 bp, 1, 1.25, 1.5 kb, 1.75 bp, 2, 2.5, 3, 3.5, 4, 4.5, 5, 6, 7, 8, 9, 10, 20, 30, 400, 50, and 100 kb. Although the number of sampled flies can impact estimates of allele frequency within pooled samples and therefore differentiation between populations (Kolaczkowski et al. 2011; Zhu et al. 2018), our empirical outlier-based approach should be unaffected, as we constrain the probability of a differentiated CNV detected on one continent appearing to be differentiated on the other by chance alone to be 0.05.
In D. simulans, we detected a large number of CNVs on the X chromosome between positions 7000000 and 8300000 that overlapped one another, and a similar region on chromosome arm 2R between positions 331000 and 722000. Although these may represent distinct CNVs rather than artifacts of poor assembly or repetitive sequence in this region, from these two regions we conservatively removed all overlapping CNVs of the same type (i.e., duplication or deletion) except for the largest one to prevent overcounting from affecting tests of overlap between continents and GO enrichment. When testing for significance of the overlap between CNVs differentiated in North America and Australia in D. simulans, we used a binomial test for an unusual fraction of North American differentiated CNVs also differentiated in Australia (expected fraction = 0.05). We then tested the fraction of Australian CNVs for overlap with North American CNVs in the same manner. We conservatively took the greater of the two P values resulting from these two tests.
Subsampling Reads to Assess the Impact of Differences in Coverage
In order to assess the effect of the greater coverage in Maine than Florida on our CNV calls, we subsampled the set of mapped fragments from the Maine data set to match the number of fragments in Florida. This resulted in a fraction (roughly 23%) of read pairs supporting the presence of CNVs in Maine being thrown out. We then asked what fraction of the differentiated CNVs in our full data set are also classified as differentiated (according to the cutoffs described above) after this subsampling, and repeated this process 1,000 times. On average, 86.5% of the CNVs in our original North American were recovered as differentiated in the subsampled set.
Detecting Differentiated CNVs from Read Depth Alone and Merging CNV Calls
Because the Australian D. melanogaster sequences were single-end only, we used a hidden Markov model (HMM) to detect differentiated CNVs from read depth along both clines following Schrider, Begun, et al. (2013). Transition and emission probabilities were estimated from CNVs differentiated along both clines. For the North American HMM, the ratios of read depths from the North American data in these CNVs were used to estimate the parameters. For the Australian HMM, the ratios of read depths from the Australian data in these CNVs were used. We then used the Viterbi algorithm to segment the genome into three states: Higher copy-number in the temperate cline endpoint, higher copy-number in the tropical cline endpoint, and no difference in copy-number. To determine which CNVs uncovered by this HMM were also differentiated along the other continent, we counted read pairs mapping within the CNV from each cline endpoint and asked whether the ratio was in either 5% tail of the empirical distribution as described above. We also counted CNVs detected on both clines (by any method) as a single event if each overlapped the other across at least 50% of its sequence; in such cases, the CNV was assumed to span the entire region encompassed by either of these two CNVs. CNVs detected from read depth only are listed in supplementary table S3, Supplementary Material online.
For each CNV detected by the HMM, we examined the average read depth in each cline endpoint, and asked which deviated more from the genome-wide average. If read depth in this pooled sample was greater than the genome-wide average, we inferred that the CNV is a duplication relative to the reference genome, and otherwise inferred the CNV to be a deletion relative to the reference.
Comparing Expression Differences with Copy-Number Differentiation
We took the log2 fold-differences in expression between the northern and southern endpoints of the North American cline from Zhao et al. (2015) for each gene at least partially residing in a CNV differentiated in North America (or both continents). For each of these genes, we then took the ratio of Florida:Maine read depths as a measure of copy-number differentiation between the two cline endpoints. We then asked whether across genes differentiated in copy-number there was a correlation between fold difference in expression and read depth ratio. We performed this analysis twice: Once using expression data from flies reared at 21 °C, and once using 29 °C. If a gene was present in multiple differentiated CNVs, we randomly selected the read depth ratio from only one of these CNVs for inclusion before computing Spearman’s rank correlation coefficient. Similarly, if a CNV contained multiple genes, we randomly selected the expression value from one of these genes for inclusion in the analysis.
Identifying CNVs Differentiated on Both Continents
We used two approaches to identify CNVs differentiated on both continents within a given species. First, if we found two CNVs, one from each continent, with at least 50% of the length of each CNV overlapping the other, we treated them as the same CNV. Second, for any CNV detected in one continent, we counted the CNV as differentiated on the other continent if the read depth ratio in that continent was found within either 5% tail of the distribution of read depths found in randomly selected regions of a similar (but no greater) size than the CNV.
Testing for a Correlation between Recombination Rates and CNV Densities
We used Comeron et al.’s (2012) recombination rate estimates (downloaded from http://www.recombinome.com/), and counted the number of differentiated CNVs in D. melanogaster whose beginning (leftmost) position were found within each 100-kb recombination rate window. One CNV was located very close to the proximal telomere of chr3L where no recombination rate estimate was available, and was therefore omitted from the analysis. We then asked whether there was a significant correlation between the number of CNVs found in each window and its estimated recombination rate.
Testing for Enrichment of Annotation Categories
We tested for statistical enrichment of complete genes, exonic base pairs, and intronic base pairs, within all differentiated CNVs found in a species, as well as the number of such CNVs overlapping large inversions. We also tested for enrichment of GO terms associated with genes overlapping CNVs differentiated on both clines in the same direction with respect to distance from the equator. For enrichment testing in D. melanogaster, we used gene locations and GO annotations from FlyBase release 33 (Tweedie et al. 2009). For D. simulans, we used the locations of orthologs to FlyBase release 33 genes found by Hu et al. (2013) and again used the FlyBase GO annotations. We then calculated P values for each GO term or annotation category by permuting the set of CNV coordinates 10,000 times and comparing the number of occurrences of each annotation feature in these permuted sets to the true set. For observed GO terms, we calculated FDR (q values) using the approach of Storey (2002), treating the three GO namespaces (biological process, molecular function, and cellular component) separately. To avoid overcounting of GO terms due to spatial clustering of functionally related genes, we counted each GO term encountered by a given CNV only once even if that term appeared in multiple genes overlapping the CNV.
Supplementary Material
Supplementary tables S1–S5 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
This study was supported by the National Institute of Health under Ruth L. Kirschstein National Research Service Award F32 GM105231 (to D.R.S.), National Institute of Health grant GM084056 (to D.J.B.), and National Science Foundation grant DBI-0845494 (to M.W.H.).
References
- Adrion JR, Hahn MW, Cooper BS. 2015. Revisiting classic clines in Drosophila melanogaster in the age of genomics. Trends Genet. 31:434–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agis M, Schlötterer C. 2001. Microsatellite variation in natural Drosophila melanogaster populations from New South Wales (Australia) and Tasmania. Mol Ecol. 10:1197–1205. [DOI] [PubMed] [Google Scholar]
- Aminetzach YT, Macpherson JM, Petrov DA. 2005. Pesticide resistance via transposition-mediated adaptive gene truncation in Drosophila. Science 309:764–767. [DOI] [PubMed] [Google Scholar]
- Arthur A, Weeks A, Sgro C. 2008. Investigating latitudinal clines for life history and stress resistance traits in Drosophila simulans from eastern Australia. J Evol Biol. 21:1470–1479. [DOI] [PubMed] [Google Scholar]
- Bergland AO, Tobler R, González J, Schmidt P, Petrov D. 2015. Secondary contact and local adaptation contribute to genome-wide patterns of clinal variation in Drosophila melanogaster. Mol Ecol. doi: 10.1111/mec.13455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne N. 2010. The distinctive footprints of local hitchhiking in a varied environment and global hitchhiking in a subdivided population. Evolution 64:3254–3272. [DOI] [PubMed] [Google Scholar]
- Capy P, Gibert P. 2004. Drosophila melanogaster, Drosophila simulans: so similar yet so different. Genetica 120:5–16. [DOI] [PubMed] [Google Scholar]
- Caracristi G, Schlötterer C. 2003. Genetic differentiation between American and European Drosophila melanogaster populations could be attributed to admixture of African alleles. Mol Biol Evol. 20:792–799. [DOI] [PubMed] [Google Scholar]
- Cardoso-Moreira M, Emerson J, Clark AG, Long M. 2011. Drosophila duplication hotspots are associated with late-replicating regions of the genome. PLoS Genet. 7:e1002340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YF, Marks ME, Jones FC, Villarreal G Jr, Shapiro MD, Brady SD, Southwick AM, Absher DM, Grimwood J, Schmutz J, et al. 2010. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science 327:302–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron JM, Ratnappan R, Bailin S. 2012. The many landscapes of recombination in Drosophila melanogaster. PLoS Genet. 8:e1002905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GM, Zerr T, Kidd JM, Eichler EE, Nickerson DA. 2008. Systematic assessment of copy number variant detection via genome-wide SNP genotyping. Nat Genet. 40:1199–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett-Detig RB, Cardeno C, Langley CH. 2012. Sequence-based detection and breakpoint assembly of polymorphic inversions. Genetics 192:131–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cridland JM, Thornton KR. 2010. Validation of rearrangement break points identified by paired-end sequencing in natural populations of Drosophila melanogaster. Genome Biol Evol. 2:83–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daborn P, Yen J, Bogwitz M, et al. 2002. A single P450 allele associated with insecticide resistance in Drosophila. Science 297:2253–2256. [DOI] [PubMed] [Google Scholar]
- Daborn PJ, Lumb C, Boey A, Wong W, Batterham P. 2007. Evaluating the insecticide resistance potential of eight Drosophila melanogaster cytochrome P450 genes by transgenic over-expression. Insect Biochem Mol Biol. 37:512–519. [DOI] [PubMed] [Google Scholar]
- David JR, Capy P. 1988. Genetic variation of Drosophila melanogaster natural populations. Trends Genet. 4:106–111. [DOI] [PubMed] [Google Scholar]
- De Jong G, Bochdanovits Z. 2003. Latitudinal clines in Drosophila melanogaster: body size, allozyme frequencies, inversion frequencies, and the insulin-signalling pathway. J Genet. 82:207–223. [DOI] [PubMed] [Google Scholar]
- Domingues VS, Poh YP, Peterson BK, Pennings PS, Jensen JD, Hoekstra HE. 2012. Evidence of adaptation from ancestral variation in young populations of beach mice. Evolution 66:3209–3223. [DOI] [PubMed] [Google Scholar]
- Duchen P, Živković D, Hutter S, Stephan W, Laurent S. 2013. `Demographic inference reveals African and European admixture in the North American Drosophila melanogaster population. Genetics 193:291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson J, Cardoso-Moreira M, Borevitz JO, Long M. 2008. Natural selection shapes genome-wide patterns of copy-number polymorphism in Drosophila melanogaster. Science 320:1629–1631. [DOI] [PubMed] [Google Scholar]
- Fabian DK, Kapun M, Nolte V, Kofler R, Schmidt PS, Schlötterer C, Flatt T. 2012. Genome‐wide patterns of latitudinal differentiation among populations of Drosophila melanogaster from North America. Mol Ecol. 21:4748–4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibert P, Capy P, Imasheva A, Moreteau B, Morin J, Pétavy G, David J. 2004. Comparative analysis of morphological traits among Drosophila melanogaster and D. simulans: genetic variability, clines and phenotypic plasticity. Genetica 120:165–179. [DOI] [PubMed] [Google Scholar]
- Girirajan S, Campbell CD, Eichler EE. 2011. Human copy number variation and complex genetic disease. Annu Rev Genet. 45:203–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW. 2009. Distinguishing among evolutionary models for the maintenance of gene duplicates. J Hered. 100:605–617. [DOI] [PubMed] [Google Scholar]
- Hoffmann AA, Weeks AR. 2007. Climatic selection on genes and traits after a 100 year-old invasion: a critical look at the temperate-tropical clines in Drosophila melanogaster from eastern Australia. Genetica 129:133–147. [DOI] [PubMed] [Google Scholar]
- Hu TT, Eisen MB, Thornton KR, Andolfatto P. 2013. A second-generation assembly of the Drosophila simulans genome provides new insights into patterns of lineage-specific divergence. Genome Res. 23:89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Massouras A, Inoue Y, et al. 2014. Natural variation in genome architecture among 205 Drosophila melanogaster Genetic Reference Panel lines. Genome Res. 24:1193–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones FC, Grabherr MG, Chan YF, et al. 2012. The genomic basis of adaptive evolution in threespine sticklebacks. Nature 484:55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Kim J, Choi K-W. 2011. Novel cytochrome P450, Cyp6a17, is required for temperature preference behavior in Drosophila. PLoS One 6:e29800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao JY, Zubair A, Salomon MP, Nuzhdin SV, Campo D. 2015. Population genomic analysis uncovers African and European admixture in Drosophila melanogaster populations from the south-eastern United States and Caribbean Islands. Mol Ecol. 24:1499–1509. [DOI] [PubMed] [Google Scholar]
- Karasov T, Messer PW, Petrov DA. 2010. Evidence that adaptation in Drosophila is not limited by mutation at single sites. PLoS Genet. 6:e1000924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennington WJ, Gockel J, Partridge L. 2003. Testing for asymmetrical gene flow in a Drosophila melanogaster body-size cline. Genetics 165:667–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolaczkowski B, Kern AD, Holloway AK, Begun DJ. 2011. Genomic differentiation between temperate and tropical Australian populations of Drosophila melanogaster. Genetics 187:245–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbé P, Berthomieu A, Berticat C, Alout H, Raymond M, Lenormand T, Weill M. 2007. Independent duplications of the acetylcholinesterase gene conferring insecticide resistance in the mosquito Culex pipiens. Mol Biol Evol. 24:1056–1067. [DOI] [PubMed] [Google Scholar]
- Langley CH, Stevens K, Cardeno C, et al. 2012. Genomic variation in natural populations of Drosophila melanogaster. Genetics 192:533–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado HE, Bergland AO, O’Brien KR, Behrman EL, Schmidt PS, Petrov DA. 2016. Comparative population genomics of latitudinal variation in D. simulans and D. melanogaster. Mol Ecol. 25(3): 723–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malloch JR. 1923. Notes on Australian Diptera with descriptions. Proc Linn Soc N S W. 48:601–622. [Google Scholar]
- McCarroll SA, Altshuler DM. 2007. Copy-number variation and association studies of human disease. Nat Genet. 39:S37–S42. [DOI] [PubMed] [Google Scholar]
- Menozzi P, Shi MA, Lougarre A, Tang ZH, Fournier D. 2004. Mutations of acetylcholinesterase which confer insecticide resistance in Drosophila melanogaster populations. BMC Evol Biol. 4:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. 1970. Evolution by gene duplication: London: George Alien & Unwin Ltd; /Berlin, Heidelberg, and New York: Springer-Verlag. [Google Scholar]
- Pavlidis P, Jensen JD, Stephan W, Stamatakis A. 2012. A critical assessment of storytelling: gene ontology categories and the importance of validating genomic scans. Mol Biol Evol. 29:3237–3248. [DOI] [PubMed] [Google Scholar]
- Reinhardt JA, Kolaczkowski B, Jones CD, Begun DJ, Kern AD. 2014. Parallel geographic variation in Drosophila melanogaster. Genetics 197:361–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers RL, Cridland JM, Shao L, Hu TT, Andolfatto P, Thornton KR. 2014. Landscape of standing variation for tandem duplications in Drosophila yakuba and Drosophila simulans. Mol Biol Evol. 31:1750–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt JM, Good RT, Appleton B, et al. 2010. Copy number variation and transposable elements feature in recent, ongoing adaptation at the Cyp6g1 locus. PLoS Genet. 6:e1000998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt PS, Matzkin L, Ippolito M, Eanes WF. 2005. Geographic variation in diapause incidence, life-history traits, and climatic adaptation in Drosophila melanogaster. Evolution 59:1721–1732. [PubMed] [Google Scholar]
- Schrider DR, Begun DJ, Hahn MW. 2013. Detecting highly differentiated copy-number variants from pooled population sequencing. Pac Symp Biocomput. 18:344–355. [PMC free article] [PubMed] [Google Scholar]
- Schrider DR, Hahn MW. 2010. Gene copy-number polymorphism in nature. Proc R Soc Lond B Biol Sci. 277:3213–3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrider DR, Houle D, Lynch M, Hahn MW. 2013. Rates and genomic consequences of spontaneous mutational events in Drosophila melanogaster. Genetics 194:937–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrider DR, Navarro FC, Galante PA, Parmigiani RB, Camargo AA, Hahn MW, de Souza SJ. 2013. Gene copy-number polymorphism caused by retrotransposition in humans. PLoS Genet. 9:e1003242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sezgin E, Duvernell DD, Matzkin LM, Duan Y, Zhu C-T, Verrelli BC, Eanes WF. 2004. Single-locus latitudinal clines and their relationship to temperate adaptation in metabolic genes and derived alleles in Drosophila melanogaster. Genetics 168:923–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RS, Long AD. 1992. Geographic variation in Drosophila: from molecules to morphology and back. Trends Ecol Evol. 7:340–345. [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR. 2010. Structural variation in the human genome and its role in disease. Annu Rev Med. 61:437–455. [DOI] [PubMed] [Google Scholar]
- Storey JD. 2002. A direct approach to false discovery rates. J R Stat Soc Series B Stat Methodol. 64:479–498. [Google Scholar]
- Stranger BE, Forrest MS, Dunning M, et al. 2007. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science 315:848–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugino RP, Innan H. 2006. Selection for more of the same product as a force to enhance concerted evolution of duplicated genes. Trends Genet. 22:642–644. [DOI] [PubMed] [Google Scholar]
- Turner TL, Levine MT, Eckert ML, Begun DJ. 2008. Genomic analysis of adaptive differentiation in Drosophila melanogaster. Genetics 179:455–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tweedie S, Ashburner M, Falls K, et al. 2009. FlyBase: enhancing Drosophila gene ontology annotations. Nucleic Acids Res. 37:D555–D559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Wit J, Svetec N, Begun DJ. 2015. Parallel gene expression differences between low and high latitude populations of Drosophila melanogaster and D. simulans. PLoS Genet. 11:e1005184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Lemos B, Dopman EB, Hartl DL. 2011. Copy-number variation: the balance between gene dosage and expression in Drosophila melanogaster. Genome Biol Evol. 3:1014–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Bergland AO, González J, Petrov DA. 2012. Empirical validation of pooled whole genome population re-sequencing in Drosophila melanogaster. PLoS One 7:e41901. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.