Abstract
E3 ligases represent an important class of enzymes, yet there are currently no chemical probes to profile their activity. We develop a new class of activity-based probe by reengineering of a ubiquitin-charged E2 conjugating enzyme and demonstrate their utility by profiling the transthiolation activity of the RING-in-between-RING (RBR) E3 ligase Parkin in vitro and in cellular extracts. Our study provides valuable insight into the roles, and cellular hierarchy, of distinct phosphorylation events in Parkin activation. We also profile Parkin patient disease-associated mutations and strikingly demonstrate that they largely mediate their effect by altering transthiolation activity. Furthermore, our probes enable direct and quantitative measurement of endogenous Parkin activity revealing that endogenous Parkin is activated in neuronal cell lines (≥75 %) in response to mitochondrial depolarization. This new technology also holds promise as a novel biomarker of PINK1-Parkin signalling as demonstrated by compatibility with Parkinson’s disease patient-derived samples.
Introduction
Ubiquitination of substrate proteins regulates most if not all aspects of eukaryotic biology and is frequently implicated in human disease1. Ubiquitination is carried out by an enzymatic cascade consisting of E1 activating enzymes (E1s), E2 conjugating enzymes (E2s) and E3 ligases (E3s)2. Initially, a catalytic cysteine residue in E1 is thioesterified with ubiquitin (Ub) forming a labile conjugate (E1~Ub). Next, E2 is recruited resulting in juxtaposition of its catalytic cysteine in E2 and the thioester linkage in E1~Ub. A transthiolation reaction ensues forming a thioester conjugate between E2 and Ub (E2~Ub). Finally, E3s catalyze Ub transfer to the ε-amino group of lysine residues in specific substrates3. In humans, there are >600 E3s with ~50 belonging to the RBR (RING-in-between-RING) and HECT (homologous to E6AP carboxy terminus) subfamilies3. These E3s possess a catalytic cysteine nucleophile that undergoes transthiolation reactions with E2~Ub forming a covalent intermediate that subsequently transfers Ub to substrate (Figure 1a).
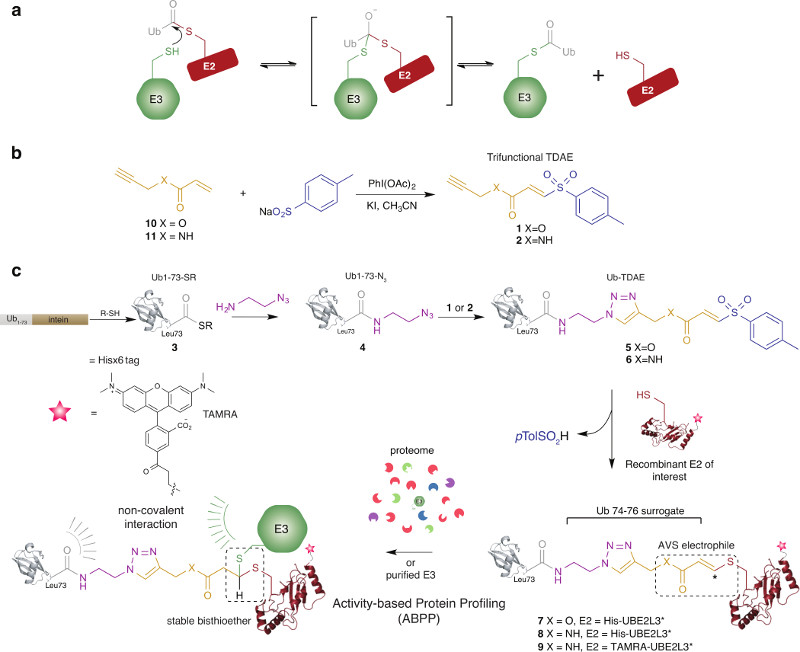
Figure 1. Synthesis of alkyne-functionalized TDAEs and their utility for assembling E2~Ub-based probes for profiling E3 transthiolation activity.
(a) Reaction scheme depicting transthiolation between E2~Ub and HECT/RBR E3 ligases. (b) Alkyne functionalized, electron deficient acrylate 10 and acrylamide 11 were used to prepare trifunctional TDAEs 1 and 2, respectively. This was achieved by PhI(OAc)2/KI-Mediated reaction of 10 and 11 with sodium arenesulfinate. (c) Probe construction involved production of Ub truncated to residues 1-73 bearing a C-terminal thioester (Ub1-73-SR). Ub1-73-SR was prepared by thiol (R-SH) treatment of a recombinant intein fusion protein. A truncated Ub species was chosen as molecular modelling, based on RING-E2~Ub and HECT-E2~Ub co-crystal structures, indicated that the AVS-triazole linker would satisfactorily mimic the Ub C-terminus and serve as a surrogate for residues 74-76 (Supplementary Figure 6). Preparation of Ub1-73-SR 3 was achieved by thiolysis of an intein fusion protein. Aminolysis of the Ub1-73-SR with azidoaminoethane afforded azide-functionalized Ub (Ub1-73-N3) 4. Copper-catalyzed Azide-Alkyne Cycloaddition (CuAAC) between 4 and TDAEs 1 and 2 yielded TDAE-functionalized Ub molecules (Ub-TDAEs) 5 and 6. Mild incubation with recombinant reporter tagged E2 generated E2~Ub-based probes bearing thioacrylate and thioacrylamide electrophiles poised for actvity-based labelling of catalytic cysteine nucleophiles in E3 ligases. Reporter tags were either a Hisx6 epitope or a 5-carboxytetramethylrhodamine (TAMRA) fluorophore. Ub in E2~Ub makes non-covalent contacts with E3 that are likely to be involved in coordinating a catalytically competent conformation.
The activity of human RBR/HECT E3s is often tightly regulated and aberrant activity is the hallmark of a number of diseases including Parkinson’s disease (PD), immune pathologies and cancer1. PD is the second most common neurodegenerative disorder and mutations in the RBR E3 ligase Parkin (encoded by PARK2) are causative of autosomal recessive early-onset forms4. The catalytic module of RBR E3s has a multi-domain architecture consisting of RING1 (Really Interesting New Gene 1), IBR (in-between-RING) and RING2 domains, the latter harboring the catalytic cysteine nucleophile5. Parkin also has N-terminal Ub-like (Ubl) and RING0 (also known as UPD) domains, and a repressor (REP) element. Collectively, these components mediate autoinhibition and PD patient mutations are found within all6. However, the mechanistic basis of these mutations is poorly defined.
The discovery that Parkin is autoinhibited in the resting state7–10, and is activated by phosphorylation of serine 65 (Ser65) of its Ubl domain and the paralogous Ser65 residue of Ub, by PTEN-induced kinase 1 (PINK1, encoded by PARK6), represented a breakthrough in our understanding of PD mechanisms11–15. However, the precise effects these phosphorylation events have on transthiolation activity and their cellular hierarchy in response to PINK1-Parkin signalling remain unclear.
A large body of evidence indicates that PD is associated with mitochondrial dysfunction6. PINK1 is located at the mitochondria and acts as a sensor of mitochondrial health. In response to mitochondrial depolarization, that can be induced by inhibition of oxidative phosphorylation with the protonophore carbonyl cyanide m-chlorophenyl hydrazone (CCCP), PINK1 is stabilized and activated15. Upon activation PINK1 phosphorylates Parkin and Ub 11,13–16. These signals trigger Parkin activation together with its recruitment and retention at the mitochondria by a feed-forward and feed-back amplification mechanism17–21. Parkin then ubiquitinates numerous outer mitochondrial membrane (MOM) proteins, thereby triggering clearance of the defective organelle by mitophagy6,22.
Biochemical and cellular analysis of E3 ligase activity is currently hampered by a lack of assays that directly monitor ligase activity. Moreover, transthiolation activity in particular is a pivotal step in RBR E3 ligase activity yet there are currently no tools that enable its direct measure. Therefore conventional assays provide limited mechanistic insight into E3 function. Furthermore, cellular Parkin activity is inferred indirectly by monitoring ubiquitination of known substrates, usually in combination with E3 overexpression23,24. Artefacts associated with this approach are potential pitfalls and these technical limitations have also prevented the direct demonstration of endogenous Parkin activation in response to mitochondrial depolarization.
Activity-based Protein Profiling (ABPP) is a powerful platform and an essential complement to transcriptomic and proteomic methodologies as it provides a direct readout of protein activity of a specific enzyme family25. ABPs that enable efficient profiling of the transthiolation activity of E1 activating enzymes have recently been reported26. This approach involves installing activated vinylsulfide (AVS) electrophiles onto E2 catalytic cysteines by labelling with small molecule tosyl-substituted doubly activated enes (TDAEs)26. However, these first-generation probes were ineffective at profiling HECT/RBR ligase activity. It was proposed that this is because they do not contain the Ub component in E2~Ub that is obligatory in conventional assays26. Ub-based probes, initially designed to profile the activity of deubiquitinating enzymes (DUBs)27, label the catalytic cysteine in certain E3s9,10,26,28. However, because E2~Ub is the native substrate of E3s, the biological relevance of Ub-based probes is unknown.
Here we report a scalable and modular strategy for the preparation of reengineered E2~Ub conjugates that harbor an electrophile that is positioned using structural and mechanistic knowledge. The conjugates serve as ABPs that profile the transthiolation activity of E3 ligases including Parkin. The ability to robustly quantify Parkin transthiolation activity, both in vitro and in cellular extracts, greatly expands our understanding of the mechanism and determinants of Parkin activation. Firstly, our experiments reveal that Ser65-phosphorylated ubiquitin (p-Ub) is required for sustained Ser65-phosphorylated parkin (p-Parkin) transthiolation activity. Our data also suggest that Parkin phosphorylation specifically, leads to displacement of the inhibitory REP component thereby contributing to relief of autoinhibition. Fluorescent derivatives of our probes enable the facile profiling of a panel of Parkin patient disease-associated mutations, spanning all Parkin domains, revealing that nearly all give rise to defects in transthiolation activity. We also demonstrate compatibility with cellular extracts verifying the requirements and shedding new insights into the hierarchy of Ub and Parkin phosphorylation in the context of cellular Parkin activation by PINK1. Our data strongly suggest that initial p-Ub binding is the primary cue for cellular Parkin activation. We also use our probes to demonstrate, and quantify, the direct activation (≥75 %) of endogenous Parkin, and its phosphorylation, in dopaminergic SH-SY5Y cells in response to mitochondrial depolarization. Finally, we demonstrate the clinical potential of our probes by profiling the general functionality of the PINK1-Parkin pathway in PD patient-derived cells harboring mutations in PARK6 or PARK2.
Results
Design of an Activity-Based Probe Based on E2~Ub
We reasoned that an engineered E2~Ub conjugate that contained a mechanistically positioned electrophile would serve as an ABP for profiling the transthiolation activity of HECT/RBR E3 ligases. However, the labile thioester linkage in E2~Ub would need to be rendered stable by using an appropriate non-hydrolyzable mimetic. Additionally, an appropriately positioned and kinetically tuned electrophile would need to be installed between the Ub and E2 components. A further consideration is that E3s function with numerous E2s so modular production of probes built on recombinant E2s of choice would be desirable29. Furthermore, cross-reactivity with DUBs should be avoided30. Such probes would have utility as structural tools enabling the stabilization of otherwise transient enzyme intermediates31, therefore probe production in milligram quantities would also be advantageous. Although isopeptide analogues of diUb harboring an electrophile have been developed as DUB ABPs, these approaches are not applicable to recombinantly-derived analogues of E2~Ub32–35.
We reasoned that appending a TDAE26 to the C-terminus of Ub would generate a highly electrophilic, but thiol-specific, protein conjugate that could undergo addition-elimination with the catalytic cysteine residue of recombinant E2s allowing the production of a stable mimetic of E2~Ub harboring an electrophile. To achieve this the TDAE would need to be rendered tri-functional by the incorporation of an additional, yet orthogonal, reactive handle. We reasoned that an alkyne moiety would satisfy this requirement (Figure 1b). The Ub-TDAE conjugate could then be prepared by triazole formation between azide-functionalized Ub and alkyne-functionalized TDAEs using Copper-catalyzed Azide-Alkyne Cycloaddition (CuAAC) (Figure 1c).
Assembly of E2~Ub Conjugates with Internal Electrophiles
We designed and synthesized TDAEs 1 and 2 bearing alkyne functionality that would ultimately furnish E2~Ub conjugates harboring thioacrylate and thioacrylamide AVS electrophiles, respectively. Complementary electrophiles have been shown to improve enzyme family coverage with related ABPs36, and would also demonstrate generality of this strategy (Figure 1b, Supplementary Results and Supplementary Figures 1-4).
To generate azide-functionalized Ub we expressed truncated Ub as thioester 3 by thiolysis of an intein fusion (Figure 1c and Supplementary Figure 5 and 6). Azide functionality was subsequently installed by an aminolysis reaction between 3 and azidoethanamine furnishing 4 (Figure 1c and Supplementary Figure 5). Intermediate 4 was then conjugated to TDAEs 1 or 2 by CuAAC and purified providing Ub-TDAE conjugates 5 and 6, respectively (Figure 1c and Supplementary Figures 5 and 7). We next tested the reactivity of 5 and 6 towards an E2. To ensure mono addition we used a single cysteine mutant of UBE2L3 (UBE2L3*, also known as UbcH7)26 that was found to support Parkin activity comparable to wild type enzyme (Supplementary Figure 8). UBE2L3* underwent efficient formation of E2~Ub conjugates under native conditions when reacted with 5 or 6 (Supplementary Figure 9). Liquid Chromatography-Mass Spectrometry (LC-MS) analysis revealed that the anticipated elimination of p-toluene sulfinic acid (p-TolSO2H) was observed26, generating probes 7 and 8 (Figure 1b and c). This approach was also readily compatible with the E2, UBE2N (also known as Ubc13) (Supplementary Figure 10).
Construction of a TAMRA Conjugated E2~Ub Probe
To increase probe versatility we constructed derivatives bearing a bio-orthogonal alkyne handle. This would allow the incorporation fluorescent reporter tags enabling direct in-gel fluorescent scanning and grant access to other powerful ABPP methodologies reliant on probe fluorescence37. Affinity tags such as biotin could also be attached to allow enrichment of active enzymes and their identification/quantification by mass spectrometry37.
Alkyne functionality was genetically incorporated into UBE2L3* via the unnatural amino acid propargyloxycarbonyl-L-lysine (ProcK) positioned N-terminal to the Hisx6 purification/reporter tag in UBE2L3* (UBE2L3*-ProcK)38 (Supplementary Figure 11). Azide-functionalized 5-carboxytetramethylrhodamine (TAMRA) was then used to label UBE2L3*-ProcK by CuAAC yielding UBE2L3*-TAMRA (Supplementary Figure 12). UBE2L3*-TAMRA was then reacted with 6 and purified by size-exclusion chromatography yielding fluorescent probe 9 (Figure 1d and Supplementary Figure 13).
E2~Ub Probes Label Parkin in an Activity-dependent Manner
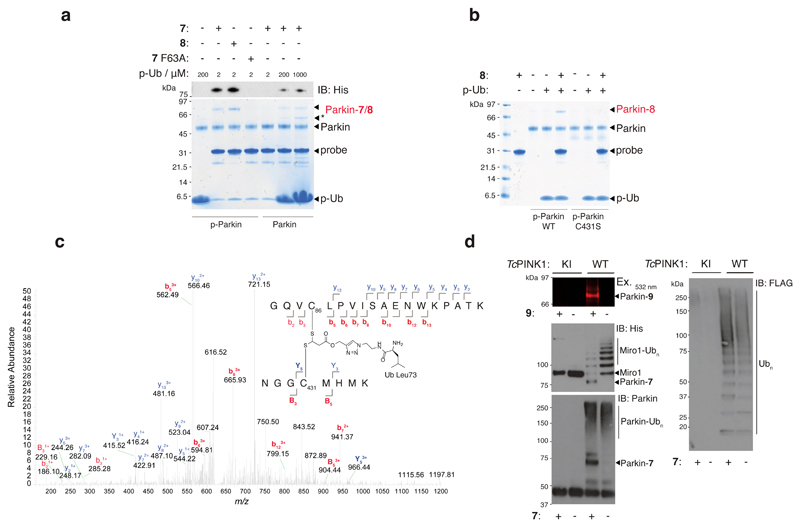
To validate our probes we profiled recombinant Parkin with probes 7 and 8. Since phosphorylation of Parkin and Ub have been demonstrated to maximally activate Parkin6,22, we initially carried out experiments using p-Parkin in the presence of p-Ub. Phosphoproteins p-Parkin and p-Ub were prepared by incubation of Parkin and Ub with PINK1 and ATP and were purified by size exclusion chromatography11. Phosphorylation of Ub was quantitative whilst phosphorylation of Parkin was ~60%, as determined by LC-MS and Phostag SDS-PAGE (Supplementary Figure 14). We tested the thioacrylate and thioacrylamide electrophiles by observing the labelling efficiency of 7 and 8 against p-Parkin/p-Ub. Probe-treated samples were resolved by SDS-PAGE and then visualized by coomassie staining and immunoblotting against the Hisx6 purification/reporter tag on 7 and 8. This revealed that Parkin was labelled by both probes with 8 being more efficient than 7 (Figure 2a). This was attributable to increased electrophilicity of the acrylamide electrophile, to it being a closer mimic of the Ub C-terminus, or a combination of both. To confirm that probes 7 and 8 were being recruited to Parkin via E2 engagement8, we built a mutant version of 7 (7 F63A) containing a Phe63Ala mutation in UBE2L3* (Supplementary Figure 15). Phe63 is required for efficient binding of E2 to RING, HECT and RBR E3 ligases39,40. Consistent with this, we found that 7 F63A failed to label p-Parkin/p-Ub (Figure 2a, all full gels and blots are shown in Supplementary Figure 22). We next assessed labelling of non-phosphorylated Parkin in the presence of p-Ub but did not observe labelling under initial test conditions (Figure 2a). As non-phosphorylated Parkin is activated with a molar excess of p-Ub11,18, as inferred by qualitative in vitro polyubiquitin assembly assays, we tested whether labelling of Parkin with 7 could be achieved with elevated levels of p-Ub. Indeed, we began to observe Parkin labelling at 0.2 and 1 mM p-Ub concentrations and labelling efficiency was concentration-dependent (Figure 2a).
Figure 2. E2~Ub-based probes label the RBR E3 ligase Parkin in an activity-dependant manner.
(a) Coomassie stained reducing SDS-PAGE and anti-His immunoblotting reveals that 7 and 8 (10 μM) form a covalent adduct with p-Parkin (2 μM) in the presence of p-Ub (2 μM) (lanes 2 and 3). Probe 7 F63A (predicted to abolish E3 binding) failed to label Parkin under the same conditions (lane 4). Non-phosphorylated Parkin failed to undergo labelling with probe 7 in the presence of p-Ub (2 μM) (lane 5). Labelling could be effected by the inclusion of molar excess levels of p-Ub (lanes 6 and 7). * Corresponds to contaminating band from p-Ub preparation. (b) Probe 8 does not label p-Parkin C431S in the presence of p-Ub (lane 8 vs. lane 5). All Parkin species and p-Ub were pre-phosphorylated by treatment with PhPINK1. (c) Annotated tryptic MS/MS spectrum for a tryptic 5+ charged precursor ion of the crosslinked peptide derived from labeling of Parkin with 7 (observed m/z = 625.7106; expected m/z = 625.7126) further confirms probe labeling of Parkin C431. (d) In situ probe labelling of reconstituted substrate ubiquitination assays. Parkin and FLAG-Ub in the reactions were phosphorylated by pre-incubation with TcPINK1. Parkin labelling with fluorescent probe 9 and probe 7 was strictly consistent with the Parkin activity readouts of His-SUMO-Miro1 substrate ubiquitination, Parkin autoubiquitination, and free polyubiquitin chain formation. In all cases, activity was strictly dependent on TcPINK1 activity. Consistent results were obtained over 3 replicate experiments.
We next confirmed that our probes were labelling the catalytic C431 residue in Parkin. We initially generated phosphorylated Cys431Ser (C431S) mutant Parkin (p-Parkin C431S) that was ~40 % phosphorylated (Supplementary Figure 14). Parallel profiling of p-Parkin WT and p-Parkin C431S, and tryptic MS/MS sequencing of the cross-linked peptide, confirmed probe labelling of C431 (Figure 2b and c). Taken together with the F63A experiment, these results indicate that our probes engage Parkin via a mechanism consistent with E2 binding and cysteine-cysteine juxtaposition of the catalytic C85 residue in UBE2L3 and C431 of Parkin.
To further confirm that labelling was consistent with Parkin E3 ligase activity we carried out post-probe labelling of Parkin ubiquitination assays that provided three independent readouts of Parkin activity (Figure 2d). In these experiments Parkin was activated by in situ phosphorylation with WT Tribolium castaneum PINK1 (TcPINK1 WT) in the presence of E1, UBE2L3, ATP and the substrate Miro1 that has been demonstrated to be a direct substrate of Parkin both in vitro and in cells12,41. As a negative control, parallel assays were performed with kinase inactive PINK1 (TcPINK1 KI). We also deployed fluorescent probe 9 which allowed rapid profiling by in-gel fluorescence and provided an orthogonal readout to anti-His immunoblotting, since the latter also detected Miro1 substrate ubiquitination. These assays confirmed that probe labelling of Parkin strictly correlated with the positive activity readouts of Parkin E3 ligase activity, namely autoubiquitination, multi-monoubiquitination of the Parkin substrate Miro1 and free polyUb chain formation.
To gauge whether probes 7 and 8 had aberrant affinity for Parkin that could give rise to spurious labelling, we carried out pull-down assays against inactive p-Parkin C431S/p-Ub and compared this to native UBE2L3~Ub (Supplementary Figure 16). The probes only partially recapitulated the affinity of the native interaction and as such they may underestimate E3 activity in certain cases. However, we tested 7 and 8 with activated forms of the HECT E3 NEDD4L, the RBR E3 HOIP and the bacterial HECT-like E3 NleL42–44. Labelling was observed in all cases and efficiency was dependent on the catalytic cysteine nucleophiles (Supplementary Figure 17).
We next tested whether probes 7 and 8 also labelled DUBs or if they were susceptible to DUB-mediated hydrolytic degradation, both of which would restrict the utility of the probes. We found that that whilst the DUB ABP propargylated-Ub (Ub-Alk)45, efficiently labelled DUBs from 3 different subfamilies (USP, OTU and UCH), probes 7 and 8 exhibited no labelling, nor where they degraded by DUB isopeptidase activity (Supplementary Figure 18).
Requirements of Sustained Parkin Transthiolation Activity
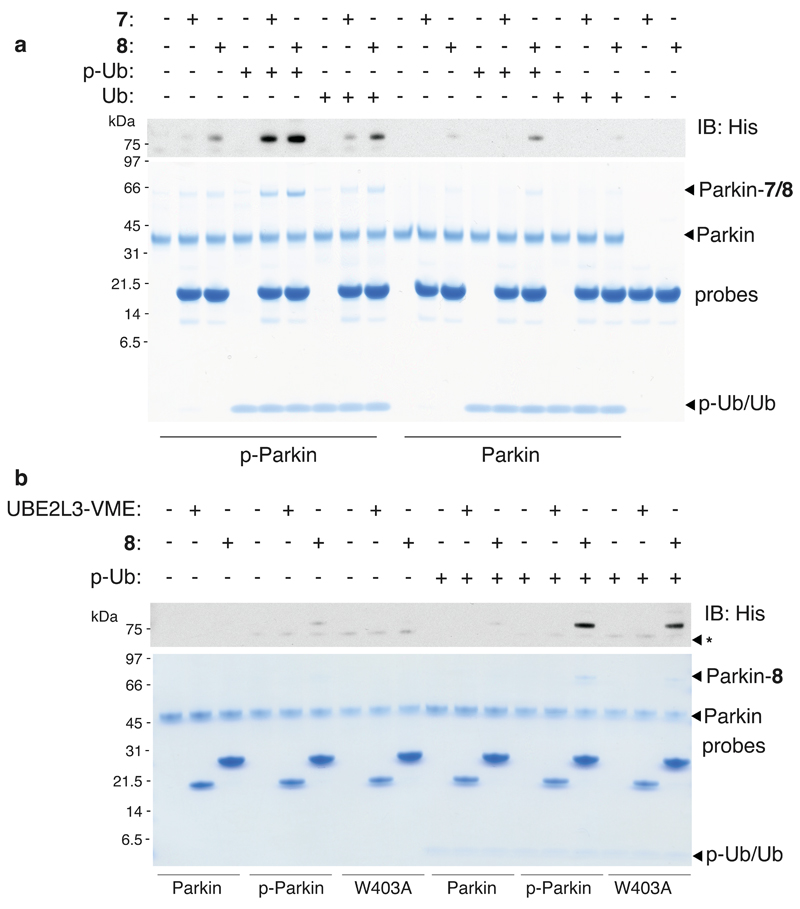
To gain a further understanding of the critical determinants of Parkin activity we next carried out a more comprehensive profiling experiment using probes 7 and 8 and combinations of Parkin and p-Parkin together with Ub and p-Ub. Interestingly, significant labelling was only observed with 7 and p-Parkin in the presence of p-Ub suggesting that both phosphorylation events are critical for optimal Parkin transthiolation activity (Figure 3a). Probe 8 was more sensitive, undergoing low levels of labelling when only a single phosphorylation site was present (Figure 3a)18. This result suggests that p-Ub is not redundant to Parkin activity after Parkin phosphorylation. PINK1-dependant phosphorylation of mitochondrial Ub is the primary mitophagy signal and Parkin serves to amplify this signal46. Therefore, maintaining p-Ub dependence on optimal Parkin activity could continuously ensure Parkin activity closely correlates with the levels of the p-Ub primary signal.
Figure 3. Sustained Parkin transthiolation activity requires phosphorylation of p-Parkin and p-Ub and Parkin Phosphorylation is Dispensible Upon Disruption of the REP Element.
(a) Pre-phosphorylated p-Parkin (3 μM) only underwent significant labelling with probe 7 in the presence of p-Ub (6 μM) (lane 5). Substantially reduced labeling was observed in the presence of Ub (lane 8). Parallel profiling with probe 8 demonstrated that 8 was more sensitive than 7. Labeling was consistently more efficient with 8, enabling detectable Parkin labelling in the presence of individual phosphorylation sites (lanes 9 and 15). (b) Probe 8 was used to determine whether a distinct phosphorylation event contributed to displacement of the autoinhibitory REP element. Non-phosphorylated Parkin W403A, in the presence of p-Ub, underwent robust labelling (lane 18). The first-generation E2-based probe UBE2L3*-VME, that bears a chemically analogous electrophile to 6 but lacks the Ub component, failed to label Parkin under any of the conditions tested. Consistent results were obtained over 2 replicate experiments.
Parkin Phosphorylation Mediates REP Element Displacement
The side chain of W403, located in the autoinhibitory REP element of Parkin, inserts into a pocket in RING1 and masks the predicted E2 docking site on Parkin8. Genetic disruption of this interaction with a W403A mutation results in increased Parkin autoubiquitination activity and accelerated Parkin recruitment to damaged mitochondria upon activation of PINK1 kinase activity8.
We explored whether relief of the autoinhibitory REP element was associated with phosphorylation of Parkin at Ser65. This was achieved by profiling the non-phosphorylated Parkin W403A mutant in parallel with Parkin and p-Parkin, in the presence and absence of p-Ub (Figure 3b). In the absence of p-Ub, no detectable activity of Parkin W403A was observed. However, Parkin W403A in the presence of p-Ub underwent probe labelling that was significantly greater than that of Parkin and p-Ub and a similar extent to p-Parkin in the presence of p-Ub (Figure 3b). However, upon consideration of the stoichiometric nature of the W403A mutation and the substoichiometry of Parkin phosphorylation (~60 %), parkin phosphorylation was only partially recapitulated by the W403A mutation. Control experiments confirmed that C431 was still being targeted in the context of the W403A mutation (Supplementary Figure 19). These data suggest that Parkin phosphorylation is directly associated with relieving the autoinhibitory constraints imposed by the REP element by an allosteric mechanism, thereby facilitating E2 binding, and REP displacement is independent of p-Ub binding. Interestingly, Parkin phosphorylation results in increased accessibility of C431S, as determined by labelling with the probe Ub-vinylsulfone (Ub-VS)17, suggesting that this event might also play a role in displacement of the RING0 domain that occludes C431 and account for the partial rescue of Parkin phosphorylation using the W403A mutation.
We also found that labelling was only observed with probe 8 and not the first generation UBE2L3-VME probe26, that bears a chemically similar thioacrylate electrophile, but does not contain the Ub component. As UBE2L3-VME did not undergo labelling of p-Parkin/p-Ub (Figure 3b), this result demonstrates that the Ub component in E2~Ub is required for adoption of a conformation where E2 and Parkin catalytic cysteines are juxtaposed40.
Rapid Profiling of Parkin Disease Mutants
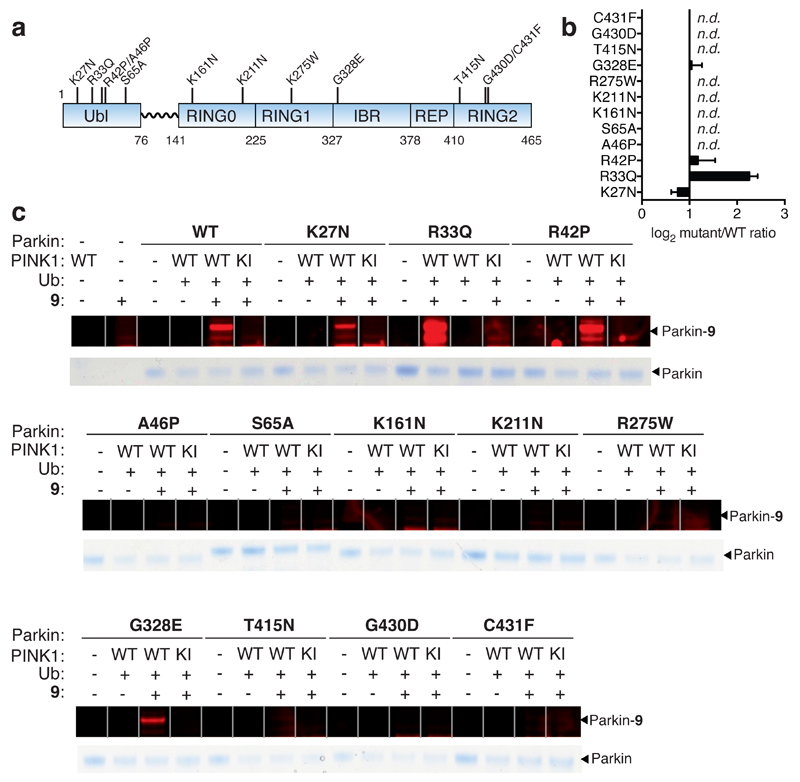
We next used fluorescent probe 9 to profile a panel of recombinant Parkin disease mutants (K27N, R33Q, R42P, A46P, K161N, K211N, R275W, G328E, T415N, G430D and C431F) in parallel, with Parkin WT and the phosphorylation defective S65A mutant, that had been treated with WT Pediculus humanus corporis PINK1 (PhPINK1) in the presence of Ub, to assess activity (Figure 4a - c). The ratio of mutant probe labelling relative to Parkin WT was determined revealing that 8 of the 12 mutants were inactive and K27N activity was reduced 0.7-fold. The R42P mutant was found to activate Parkin 2.2-fold (Figure 4b and c). These data illustrate that for 10 of the 12 mutants tested the observed affects on activity can be directly assigned to alterations in transthiolation activity12. However, we cannot formally exclude that the mutations may also affect the latter step of Ub transfer to substrate8,12. As these mutations spanned the full-length of the protein, this indicates that transthiolation activity is dependent on all of the domains within Parkin and leads towards a unifying pathogenic basis, despite their disparate distribution throughout the Parkin polypeptide.
Figure 4. Quantitative and direct activity-based Protein Profiling of transthiolation activity of Parkin patient mutations.
(a) Amino acid boundaries of the multi-domain architecture of Parkin. (b) Fold-change of mutant Parkin transthiolation activity relative to WT represented on a Log2 scale (n.d. means no detectable labeling). Data represent mean values and error bars correspond to ±s.d, n = 3 . (c) Recombinant Parkin mutants were incubated with PhPINK1 in presence of Ub and ATP. Mutations resided throughout the multi-domain architecture of Parkin. Incubations were then directly profiled for Parkin transthiolation activity with fluorescent probe 9 Mutations mildy perturbed, activated or abolished transthiolation activity as determined by fluorescence of labeled Parkin.
Determinants of Parkin Activation in Cells
We next profiled the activity of Parkin using probes 7 and 8 in cells by analysing untagged Parkin WT, Parkin S65A and Parkin H302A stably expressing HeLa cells, that do not express endogenous Parkin46. The H302A mutation significantly impairs p-Ub binding20,21,47,48 therefore in combination with S65A, the effects of Ub and Parkin phosphorylation in the context of PINK1-Parkin signalling could be assessed by probe profiling of cell extracts. We treated HeLa cells expressing the various forms of Parkin with CCCP for 3 h, in parallel with untreated cells, and then extracted cellular proteomes by mild lysis via sonication. Extracts where then profiled with probes 7 and 8 (5 μM) for 4 h, resolved by SDS-PAGE, and immunoblotted against total Parkin and p-Ser65-Parkin specific antibodies20 (Figure 5a). As expected, no phosphorylation of Parkin or probe labelling was observed in the absence of CCCP treatment for Parkin WT or either of the Parkin mutants. However, after CCCP treatment, total Parkin immunoblotting revealed a pool of activated Parkin WT as judged by the presence of new bands corresponding to Parkin WT labelled with probes 7 or 8. Probe labelling was also observed by p-Parkin immunoblotting and labelling efficiency of the phosphorylated pool was significantly greater than for the total pool. In contrast to Parkin WT, no labelling was observed with the Parkin S65A mutant indicating that phosphorylation at Ser65 of Parkin in cells plays a fundamental role in its activation15,16. Furthermore, results from additional experiments were in further support of a model whereby Parkin phosphorylation allosterically mediates C431 accession in addition to displacement of the REP element (Supplementary Figure 20).
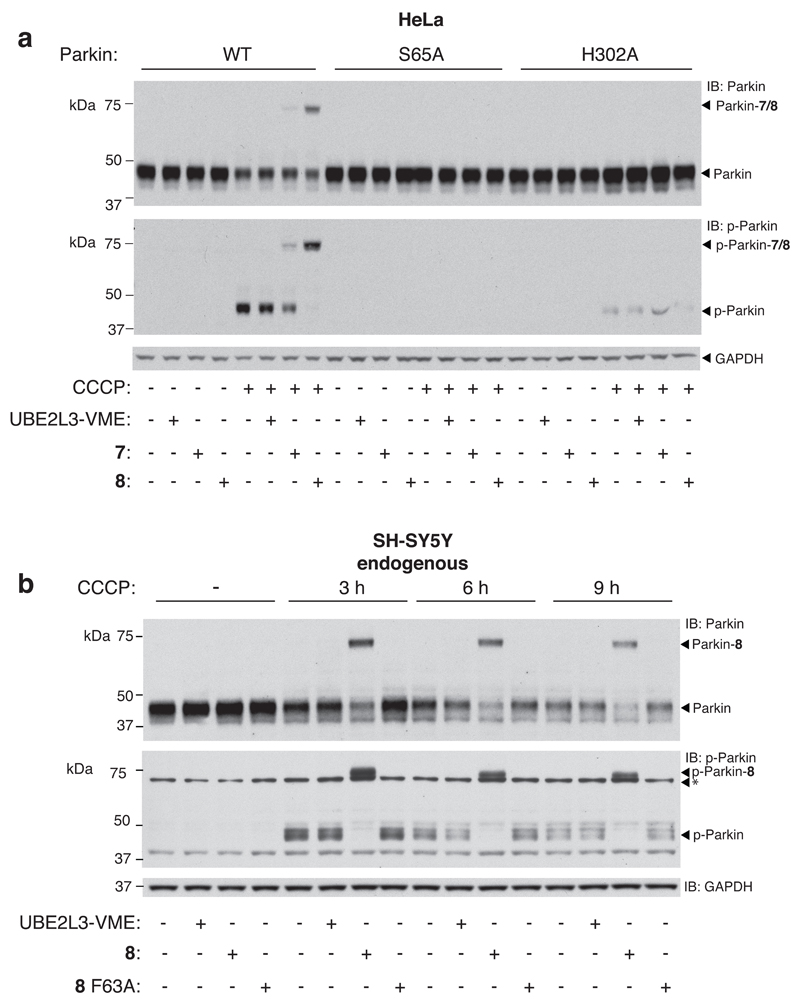
Figure 5. Activity-based profiling of cellular Parkin provides insights into the hierachy of Parkin and Ub phosphorylation in the context of PINK1-Parkin signalling, and reveals quantifiable, protonophore responsive activation of endogenous Parkin in SH-SY5Y dopaminergic cells.
(a) HeLa cells stably expressing Parkin WT, Parkin S65A and Parkin H302A were untreated or treated with the CCCP (10 μM) for 3 h to promote mitochondrial depolarization. Extracts were were profiled in parallel with probes UBE2L3*-VME, 7 and 8 (5 μM). Labelling of Parkin WT was only observed with probes 7 and 8 in response to CCCP treatment. Probe labeling correlated with Parkin phosphorylation and labeling of the phosphorylated pool was more efficient than that of the total Parkin pool. Non-phosphorylatable Parkin S65A did not undergo labeling with probe 7 or 8. Parkin H302A with significantly impaired p-Ub binding ability exhibited no detectable labeling with 7 or 8. Parkin phosphorylation was also significantly compromised. (b) Activity-based profiling of endogenous Parkin in SH-SY5Y cells directly reveals Parkin phosphorylation and activation of transthiolation in response to CCCP treatment. Labelling of the total Parkin pool is more efficient than with overexpressed Parkin. Labelling efficiency of the p-Parkin pool approached 100 %. Labeling was not observed with the control probes UBE2L3*-VME or 8 F63A. Over the time course, probe labeling efficiency of Parkin remains unchanged whilst Parkin levels reduce suggestive of Parkin clearance in the activated state. Asterisk corresponds to a cross-reactive band. Consistent results were obtained over 3 replicate experiments.
Strikingly, Parkin H302A exhibited reduced phosphorylation and probe labelling was undetectable, suggesting that phosphorylation of Ub by endogenous PINK1 contributes to Parkin activation (Figure 5a). This result supports a model where the primary mechanism of Parkin activation in response to mitochondrial depolarization involves initial p-Ub binding to Parkin that primes it for PINK1 phosphorylation and optimal activation20,21,48. p-Parkin has enhanced affinity for p-Ub17,47,48, and our in vitro data indicate that both p-Ub and p-Parkin are required for optimal activity. This supports a new modality of the Parkin feed-forward mechanism ensuring Parkin is not only recruited to mitochondria via binding to p-Ub and phosphorylated by PINK1, but p-Ub abundance is continuously sensed, post phosphorylation of Parkin, to drive sustained activation as well as mitochondrial retention. UBE2L3-VME failed to label under these conditions further suggestive of the role of the Ub component in E2~Ub for cysteine-cysteine juxtaposition (Figure 5a).
Profiling Endogenous Parkin in Dopaminergic SH-SY5Y Cells
To date the majority of studies in cells examining Parkin activation in response to mitochondrial depolarisation have employed conditions in which Parkin is overexpressed24,49 (often with N-terminal tagging that can aberrantly activate Parkin7) and studies on endogenous Parkin have been limited. We therefore used probe 8 to determine the activation status of endogenous Parkin in response to mitochondrial uncoupling in dopaminergic neuroblastoma SH-SY5Y cells. Cells were untreated or treated with CCCP (10 μM) for 3, 6 and 9 h and then lysed. Profiling was then carried out with the inclusion of inactive probe 8 F63A as a further control (Supplementary Figure 21). Probe labelling and phosphorylation was undetectable in untreated cells (Figure 5b). However, probe labelling and Parkin phosphorylation was detected in CCCP treated cells and peaked at 3 h. As expected, 8 F63A failed to label under any conditions. Labelling efficiency of the total Parkin and p-Parkin pools were ≥75 % and >95 %, respectively, higher than that observed with Parkin overexpression in Hela cells (likely attributable to stoichiometric imbalance where factors such as PINK1, p-Ub and mitochondrial abundance were limiting). Overall, our probe analysis has provided the first physiological and quantitative assessment of the stoichiometry of endogenous Parkin activation in response to CCCP treatment. The finding ≥75 % highlights the rapid activation induced by PINK1-dependent phosphorylation of Parkin and Ub. Furthermore, the finding that probe labelling of the phosphorylated pool approached 100 % permits estimation of the stoichiometry of endogenous Parkin phosphorylation upon CCCP treatment (≥75 %) underscoring the significance of the phosphorylation of endogenous Parkin for its activation. Interestingly, over the time course degradation of Parkin was observed whilst probe labelling efficiency remained unchanged (Figure 5b). This suggests that the Parkin activation signal is attenuated by degradation of Parkin in the activated state rather than its deactivation.
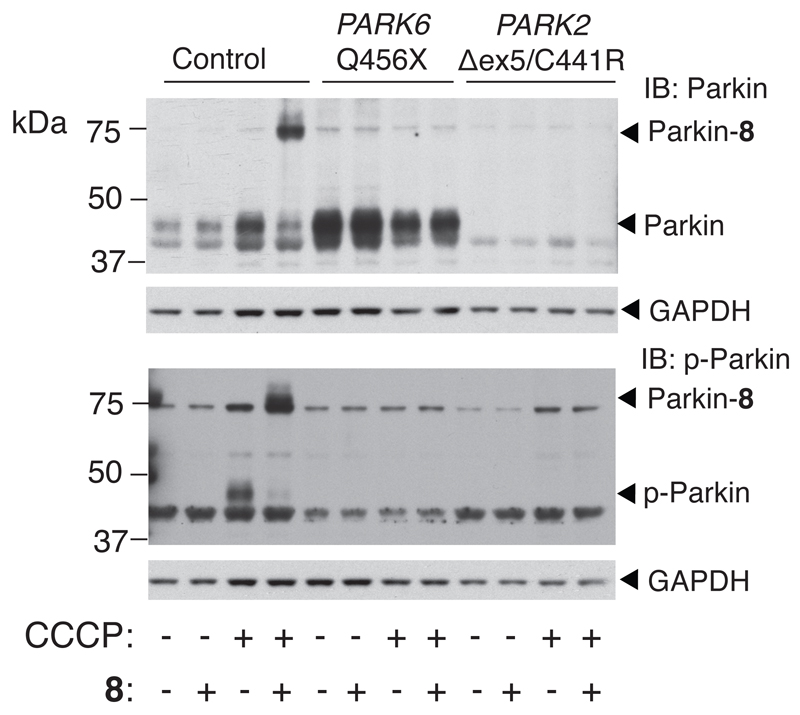
Profiling of Primary Fibroblasts Derived from PD Patients
We next explored the potential of our probes for clinical diagnostic purposes. We took primary fibroblasts derived from a skin biopsy from PD patients carrying a homozygous Q456X mutation in the PARK6 gene and an exon 5 deletion/C441R (∆ex5/C441R) compound heterozygous mutation in the PARK2 gene. A sample was also derived from a healthy individual. The Q456X mutation abolishes PINK1 activity50, therefore Parkin should be refractory to activation in response to mitochondrial depolarization whereas the PARK2 compound heterozygous mutation would be predicted to result in aberrant Parkin expression and/or stability50. Parkin from healthy control cells was activated upon CCCP treatment, similarly to that observed in SH-SY5Y cells, whereas Parkin activation was abolished in the PARK6 Q456X patient-derived cells indicating an absolute requirement for PINK1 activation (Figure 6). Parkin was undetectable in the PARK2 ∆ex5/C441R derived cells and as expected, no probe labelling signals were observed. These data provide the first quantitative assessment of Parkin activity in patient-derived cells and indicate that our probes can be deployed to determine the general functionality of the PINK1-Parkin signalling pathway in PD patients.
Figure 6. Activity-based profiling of PD patient-derived fibroblasts revals biomarker potential.
Low passage primary fibroblasts derived from skin biopsies from a healthy age-matched family member, a PD patient harbouring a Q456X homozygous mutation in the PARK6 gene, and a PD patient harboring a ∆ex5/C441R compound heterozygous mutation in the PARK2 gene were profiled with probe 8. Control cells undergo robust Parkin phosphorylation and activation (lane 4). In contrast, Parkin in either PARK6 or PARK2 mutant PD patient cells cannot be activated in response to CCCP treatment (lanes 8 and 12).
Discussion
Herein we have reported the construction of a new class of activity-based probe (ABP) that can be used to profile the activity of E3 ligases including the PD-linked enzyme Parkin. In vitro profiling experiments of recombinant Parkin demonstrates that phosphorylation of Parkin and Ub both contribute to sustained transthiolation activity. We also provide evidence that Parkin phosphorylation plays a prominent role in displacement of the inhibitory REP element that occludes E2 binding. Several models have been proposed to define the phosphorylation hierarchy; some in favour of initial Parkin phosphorylation17,47, contrasted with those favoring initial Ub phosphorylation20,21,48. Our data are consistent with the latter model that initial binding of p-Ub to Parkin then renders Parkin a more effective PINK1 substrate in cells and both phosphorylation events are then absolutely required for sustained Parkin activity, as judged by transthiolation. As PINK1 is the only Ub kinase that has thus far been identified, and activated PINK1 is associated with the MOM, we speculate that basal levels of Ub are present at the MOM that become phosphorylated upon PINK1 activation and this cues Parkin activation.
We also described the construction of fluorescent probe 9 that provides a rapid platform for profiling ligase activity. Strikingly, this revealed that 10 out of 12 tested mutations can exert their pathogenicity by altering transthiolation activity. We have also demonstrated that our probes can be used to profile the activation status of endogenous Parkin in neuronal SH-SY5Y cells. These data clearly demonstrated that ≥75 % of endogenous Parkin is phosphorylated and activated in response to protonophore treatment. To our knowledge our analysis also represents the first report that Parkin can be phosphorylated at Ser65 under endogenous conditions providing physiological evidence for this posttranslational modification. Furthermore, time course profiling demonstrated that Parkin levels reduce (whilst probe labelling efficiency remains unchanged) upon prolonged activation, suggestive of Parkin clearance in the activated state.
As there are currently no methods for early diagnosis of PD and the disease is heterogeneous in nature, novel biomarker approaches are urgently required. In addition, genetic testing for mutations in autosomal recessive forms of PD, such as PARK2 and PARK6 mutations, is not trivial. The ability of our probe to profile aberrations of the PINK1-Parkin signalling pathway in general, using fibroblasts derived from PD patients, could provide a much needed clinical tool and future studies will be required to validate its utility in the diagnosis of familial forms of PD, and for patient stratification in idiopathic cases.
A large body of powerful experimental platforms have been developed that are based on fluorescent ABPs. These include in-gel based multiplexed profiling of endogenous enzyme activities in cellular proteomes, inhibitor screening and inhibitor selectivity profiling37. Fluorescent probe 9 should be valuable tool for discovering small molecules that activate the transthiolation activity of Parkin. In principle, high affinity enrichment tags such as azide-functionalized biotin could also be conjugated to our probes allowing multiplexed profiling in combination with E3 ligase activity quantification and identification. As E3 ligase activities are stringently regulated and deregulated in pathophysiology1, we envisage that such ABPP platforms based on our probes will provide great insight into the roles of other E3 ligases in disease. This could lead to novel biomarkers, therapeutic targets and as drivers for further biological investigation.
Another potential application of our probes is as structural tools. There is a long-standing interest in the conformational changes and mechanistic aspects of Ub transfer throughout the Ub conjugation cascade. Key intermediates, such as transthiolation intermediates between E1-E2 and E2-E3, are inherently unstable yet define the mechanistic features of these important enzymes and our probes could be used to facilitate their structural characterization.
Online Methods
General Materials
All DNA constructs were verified by DNA sequencing, (School of Life Sciences, University of Dundee). DNA for bacterial protein expression was transformed into E. coli BL21-DE3 (Merck), BL21 DE3 RIL (codon plus) cells (Stratagene) or ER2566 (NEB). With the exception of pCDF-PylST, all cDNA plasmids and antibodies generated for this study are available to request through our reagents website (https://mrcppureagents.dundee.ac.uk/).
LC-MS and Semi-preparative peptide HPLC was carried out as previously described26. All solvents and reagents were purchased from Sigma Aldrich or VWR unless otherwise stated.
Synthesis of alkyne-functionalized TDAE 1
PhI(OAc)2 (966 mg, 2 mmol) was added to a suspension of propargyl acrylate 10 (2 mmol), sodium arenesulfinate (8.0 mmol), and KI (328 mg, 2.0 mmol) in acetonitrile (MeCN) (8 mL)51. The reaction mixture was stirred vigorously at room temperature for 1 h under an inert atmosphere. The reaction mixture was then quenched by the addition of saturated Na2S2O3 aq. (20 mL) followed by a saturated aqueous solution of NaHCO3 aq. (20 mL). Further stirring was followed by extraction with ethyl acetate (EtOAc) (3 x 50 mL). The combined organic phases were washed with saturated NaCl aq. (75 mL) and dried over Na2SO4. The organic solvent was removed in vacuo and the crude product was directly purified by flash chromatography using a Reveleris® X2 flash chromatography system (Grace) (Reveleris® 12g silica column; EA:Hexane; 15% to 25% to 100% EA gradient elution) to yield 1.
1H NMR (500 Mhz, CDCl3) δ 7.79 (2H, d, 8.4 Hz), 7.35-7.40 (3H, m), 6.81 (1H, d, 15.2 Hz), 4.78 (2H, d, 2.5 Hz), 2.51 (1H, t, 2.5 Hz), 2.46 (3H, s); 13C NMR (126 MHz, CDCl3) δ 162.75, 145.83, 144.60, 135.21, 130.32, 129.19, 128.48, 76.50, 75.87, 53.23, 21.74; HR-MS: observed [M-H]- 263.0361 (calculated 263.0373).
Synthesis of alkyne-functionalized TDAE 2
Starting material N-propargylacrylamide, 11 was first prepared as previously described52. Purification was carried out by flash chromatography using a Reveleris® X2 flash chromatography system (Grace) (Reveleris® 12g silica column; EA:Hexane; 20% to 35% EA gradient elution). Analytical data corresponded to literature values. PhI(OAc)2 (966 mg, 2 mmol) was added to a suspension of 11 (2 mmol), sodium arenesulfinate (8.0 mmol), and KI (328 mg, 2.0 mmol) in CH3CN (8 mL). Compound 2 was then prepared as described for 1. The organic solvent was removed in vacuo and the crude product was directly purified by flash chromatography using a Reveleris® X2 flash chromatography system (Grace) (Reveleris® 12 g silica column; Toluene:EA; 0% to 1% to 10% EA gradient elution).
1H NMR (500 Mhz, CDCl3) δ 7.78 (2H, d, 8.3 Hz), 7.37 (2H, d, 8.5 Hz), 7.34 (1H, d, 14.9 Hz), 6.98 (1H, d, 14.8 Hz), 6.60 (1H, t, 5.1 Hz), 4.10-4.12 (2H, dd, 5.3, 2.6 Hz), 2.45 (3H, s), 2.23 (1H, t, 2.6 Hz); 13C NMR (126 MHz, CDCl3) δ 161.70, 145.60, 141.00, 135.58, 132.38, 130.25, 128.23, 78.28, 72.39, 29.77, 21.72; HR-MS: observed [M-H]- 262.0537 (calculated 262.0543).
Preparation of UBE2L3* (C17S, C137S), UBE2D3* (C21S, 107S, C111S) and UBE2N
Full-length E2s were cloned into the pET expression vectors downstream of an N-terminal Hisx6 tag and Precision protease site. E. coli strain BL21(DE3) transformed with E2 clones were cultured at 37 °C in LB medium supplemented with 100 μg mL-1 of ampicillin. When the OD600 reached 0.6, the culture was induced by addition of 0.4 mM isopropyl thio-β-D-galactoside (IPTG) and incubated at 37 °C for 5.0 hours. The cells were harvested by centrifugation and stored at -80 °C. Cells were resuspended in ice cold lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 25 mM imidazole, 0.5 mg/mL-1 lysozyme, 50 μg/mL-1 DnaseI, Complete, Mini, EDTA-free protease inhibitor cocktail, (one tablet per 50 mL of buffer, Roche)) and incubated on ice for 30 minutes followed by sonication. The clarified lysate was then subjected to Ni-NTA affinity chromatography (Qiagen) and washed with wash buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 25 mM imidazole). Protein was eluted with elution buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 300 mM imidazole) and dialyzed into storage buffer (20 mM Tris, 150 mM NaCl, 1 mM DTT). The protein was then further purified by size exclusion chromatography using a HiLoad 16/600 Superdex 75 pg column (GE Life Sciences) coupled to an ÄKTA Purifier FPLC system (1.0 mL min-1, running buffer: 20 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM DTT). Fractions were collected, concentrated and flash frozen for storage at -80 °C.
General Method for ∆Ub-SR Preparation
A plasmid composed of full-length Ub cloned into the pTXB1 vector53, was taken and the bases coding for the C-terminal Ub residues 74-76 were deleted by Quikchange mutagenesis. To suppress cellular intein activity54, a T3C mutation in the GyrA intein was also introduced yielding plasmid pTXB1-Ub∆74-76-T3C. Expression and purification was carried out as previously described for full-length Ub53. After HPLC purification fractions containing ∆Ub-SR were pooled and lyophilized yielding ~30 mg.
∆Ub-N3 Aminolysis Reaction
∆Ub-SR (25.8 mg) was reconstituted55, by the addition of DMSO (200 μL). On complete dissolution of ∆Ub-SR in DMSO, H2O (800 μL) was added to give a final DMSO concentration of 20% (v/v). 500 μL of 2-azidoethanamine (Enamine) in 50% (v/v) aqueous DMSO/MQ (8 M) was then added to the ∆Ub-SR solution. 60.0 μl of triethylamine (TEA) was then added raising the solution pH to 9 and the mixture was then briefly vortexed. The solution incubated at 30 °C for 16 hours and monitored by LC-MS. The protein (∆Ub-N3) was then further purified by semi-preparative RP-HPLC (Column: BioBasic-4; Part number: 72305-259270). A gradient of 20 % buffer A to 50 % buffer B was applied at a flow rate of 10 mL min-1 over 60 min (buffer A=0.1% TFA in H2O, buffer B=0.1% TFA in acetonitrile). Fractions containing ∆Ub-N3 were pooled and lyophilized (Yield: 80%).
Conjugation of ∆Ub-N3 with TDAE by CuAAC click reaction
∆Ub-N3 (3.5 mg) was reconstituted by dissolution into 100 μl DMSO. H2O (900 μL) was added to give a final DMSO concentration of 10% (v/v) and a final ∆Ub-N3 concentration of 418.6 μM. DMSO stock solutions of TDAEs 1 or 2 were prepared (10 mM). TDAEs 1 or 2 (200 μL) were then mixed with a pre-prepared DMSO/MQ solution (456.2 μL, 20% (v/v)). Phosphate buffer (50.0 μL, 100 mM Na2HPO4 pH 7.5, 150 mM NaCl) and ∆Ub-N3 stock solution (238.6 μL, 418.6 μM) were then subsequently added. Tris(3-hydroxypropyltriazolylmethyl)amine (THPTA)56 in H2O (25 μL, 12.5 eq., 50 mM stock solution) was pre-mixed with a freshly prepared solution of CuSO4 (aq.) (5 μL, 2.5 eq., 50 mM stock solution). The THPTA/CuSO4 (aq.) solution (30 μL) was then added to the previously prepared TDAE/∆Ub-N3 solution. L-ascorbic acid in H2O (25 μL, 100 mM stock solution) was then added to the previously mixed components. The reaction was incubated at 23 °C with shaking (1400 rpm) for 15 minutes. The reaction was then quenched by the addition of EDTA (2.5 mM final concentration) and the product (5 or 6) was purified by semi-preparative RP-HPLC (Column: Thermo Scientific BioBasic-4; Part number: 72305-259270) using the Dionex system. A gradient of 20 % buffer A to 50 % buffer B was applied at a flow rate of 10 mL min-1 over 60 minutes (buffer A=0.1% TFA in H2O, buffer B=0.1% TFA in acetonitrile). Fractions containing ∆Ub-TDAE were pooled and lyophilized (Yield: 60%).
Synthesis E2~Ub Probes 7, 8, 9, 7 F63A and 8 F63A
∆Ub-TDAE (5 7.0 mg) was reconstituted by the addition of 50 μl DMSO. On complete dissolution of ∆Ub-TDAE, H2O (450 μL) was added to give a final DMSO concentration of 10% (v/v) and a final ∆Ub-TDAE concentration of 1.6 mM. ∆Ub-TDAE was then incubated with 0.5 eq. of the desired E2 in 0.8 mL phosphate buffer (100 mM Na2HPO4 pH 7.5, 150 mM NaCl, reducing agent free) at 23 °C for two hours and monitored by LC-MS. To demonstrate compatibility with other E2s, a probe based on UBE2N was constructed by analogous reaction of UBE2N with 5. For thioacrylamide probes 8, 8 F63A and 9, the above procedure was carried out with ∆Ub-TDAE 6 and 30 °C incubation for 12 h was required to consistently achieve quantitative elimination to the unsaturated AVS species. The products were purified by size exclusion chromatography (0.5 mL min-1, 20 mM Tris-HCl pH 7.5, 150 mM NaCl as mobile buffer, HiLoad 16/600 Superdex 75pg).
Molecular Modelling of RING-, HECT-, and RBR-probe Complexes
The RING-E2~Ub model was generated by importing the PDB coordinates 4AP457 into the BioLuminate biologics modelling suite (Schrödinger). The AVS-triazole linker present between E2 and Ub in probe 8 was manually built and geometry optimized. Hydrogen atoms were then added using the Protein Preparation tool. The E2 residue Cys85, the Ub residue Leu73, and the linkage between them were selected along with all protein residues within 5 Å. This substructure was then energy minimized using the OPLS2005 forcefield. Minimized coordinates were then exported as mol2 files and imported into Pymol (Schrödinger) for figure generation. The same procedure was repeated for the HECT-E2~Ub and RBR-E2~Ub models using PDB 3JW042 and PDB 5EDV40, respectively.
Parkin Activity Assay with UBE2L3*
Parkin (3 μg, 760 nM) was incubated with ubiquitylation assay components in a final volume of 50 μl (50 mM Tris-HCl pH 7.5, 5 mM MgCl2, 240 nM UBA1, 2 μM UBE2L3 variant, 2 mM ATP, 58 μM Ub and 380 nM of GST-PhPINK1 WT. The reaction was incubated at 30 °C for 1 h. The reaction was quenched by the addition of reducing 4X LDS loading buffer, resolved by SDS-PAGE, and visualized by coomassie staining.
Preparation of UBE2L3-ProcK
An amber stop codon was introduced at position 3 of the open reading frame in pET156-UBE2L3* by Quikchange site-directed mutagenesis. This yielded the plasmid pET156-UBE2L3*-TAG3 (* corresponds to a C17S and C137S mutant). BL21(DE3) cells (Merck Biosciences) were transformed with plasmids pET156-UBE2L3*-TAG3 and pCDF-PylST (plasmid harbouring constitutive copies of the MbPylRS/tRNACUA pair, a kind gift from Dr. Jason Chin) and used to inoculate 200 mL LB media containing ampicillin (100 μg mL-1) and spectinomycin (50 μg mL-1). After overnight incubation at 37 C, 6 × 1 L of LB medium containing ampicillin (100 μg mL-1) and spectinomycin (25 μg mL-1) were each inoculated with 30 mL overnight culture and incubated at 37 °C, 200 rpm, until cell density reached OD600 = 0.8-0.9. Propargyloxycarbonyl-L-lysine (ProcK; Iris Biotech GmbH, #HAA2095) (300 mM stock solution in H2O and pH adjusted to ~7) was then added to a final concentration of 3 mM. Cultures were then incubated for a further 20 min at 37 °C and then induced with IPTG (500 μM). After 4 h cells were harvested and cell pellets were resuspended in 200 mL lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 25 mM imidazole, 0.5 mg mL-1 lysozyme, 50 μg mL-1 DnaseI, 4 tablets of Complete, EDTA-free protease inhibitor cocktail (Roche) and incubated on ice for 30 min. The suspension was then sonicated before clarification at 18,000 rpm, 30 min at 4 °C. Ni-NTA resin (600 μl of settled resin, Qiagen) was then added to the lysates and rotated at 4 °C for 1 h. Resin was then washed with wash buffer (50 mM Tris pH 7.5, 150 mM NaCl, 25 mM imidazole) and then eluted with elution buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 300 mM Imidazole). The eluent was then concentrated to 2.5 mL and further purified by size exclusion chromatography using a HiLoad 16/600 Superdex 75 pg column (GE Life Sciences) coupled to an ÄKTA Purifier FPLC system (running buffer: 100 mM Na2HPO4 pH 8, 150 mM NaCl, 0.1 mM TCEP). Fractions were collected and concentrated (2.4 mg mL-1, ~118 μM).
TAMRA Labelling of UBE2L3-ProcK by Copper-catalyzed Azide-Alkyne Cycloaddition (CuAAC)
UBE2L3-ProcK was diluted in buffer (100 mM Na2HPO4 pH 8, 150 mM NaCl, 0.1 mM TCEP) to a final concentration of 50 μM. CuSO4 and Tris(3-hydroxypropyltriazolylmethyl)amine (THPTA)56, stock solutions were freshly prepared in H2O and then premixed before being added to the protein at final concentrations of 0.1 mM and 1.0 mM, respectively. TAMRA-azide (Jena Bioscience, #CLK-FA008, M.W. = 512.56 g mol-1) was then added to a final concentration of 150 μM. Ascorbic acid was added to a final concentration of 4 mM. The reaction was incubated at 23 °C for 1-2 h (or until starting material had been consumed as determined by LC-MS). The reaction was then quenched with 5 mM EDTA for 15 min at 23 °C. Small molecule components were then removed by dialysis against 2 × 1L buffer (100 mM Na2HPO4 pH 8.0, 150 mM NaCl, 5 mM EDTA, 0.1 mM TCEP) at 4 °C over 48 h. Protein was snap frozen in liquid nitrogen and stored at -80 °C.
In vitro probe-labeling assay
The indicated E3 ligases were added into Tris buffer (20 mM Tris-HCl pH 7.5, 150 mM NaCl). 5.0 e.q (unless otherwise indicated) of E2~Ub (probe 7 or 8) was incubated with E3 ligase at 30 °C for the indicated time. Reactions were quenched by the addition of 4X LDS loading buffer (supplemented with ~680 mM 2-mercaptoethanol) and samples were analyzed by SDS-PAGE (4-12% NuPage gel) followed by coomassie staining or immunoblotting.
Ubiquitination assay and in situ profiling
Parkin (1.3 μM) was incubated with ubiquitylation assay components in a final volume of 30 μl (50 mM Tris-HCl pH 7.5, 5 mM MgCl2, 0.12 μM UBA1, 2 μM UBE2L3, 0.83 μM His-SUMO-Miro1, 2 mM ATP, 88 μM Flag-ubiquitin and 0.31 μM TcPINK1 (WT/KD). Ubiquitylation reactions were incubated at 30 °C and 1050 rpm for 60 min. Ubiquitylation reactions were stopped by the addition of 5 U mL-1 Apyrase (New England BioLabs) and incubated for 10 min at 30 °C and agitated at 1050 rpm. Probe profiling was carried out by the addition of 7 or 9 (8 μM) and incubated for 10 h at 30 °C and 650 rpm. The reactions were terminated by the addition of LDS sample buffer containing 4 % 2-mercaptoethanol. Reaction mixtures were resolved by SDS-PAGE and immunoblotted with the following antibodies: anti-FLAG (Sigma, 1:10000), anti-Parkin (Santa-Cruz, 1:500) or anti-His (1:5000). For Fluorescence detection, 20 μl of sample were loaded on a 4-12 % gel and subjected to SDS-PAGE. Gels were washed with water for 30 minutes and then visualized by scanning with a FLA-5100 imaging system (Filter: LPG, Laser: 532 nm, Voltage: 400, FUJIFILM Life Science). The gel was subsequently coomassie stained.
Expression and Purification of NEDD4L, HOIP and NleL
GST-NEDD4L, GST-NEDD4L C922A, HOIP, HOIP C885S, GST-NleL and GST-NleL C753A, were expressed and purified as previously described42–44.
DUB Resistance Assay
His-USP2259–605, His-USP21196-565, OTUB2 and GST-UCHL3 were expressed in E. coli and purified. The DUB-reactive ubiquitin-based probe, Ub-Alk45,58, was used as a positive control and tested in parallel with probes 7 and 8. Probes were diluted to 40 μM and DUBs were diluted to 10 μM in buffer (20 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM TCEP). Probe (10 μL) and DUB (10 μL) solutions were then mixed and incubated for 2 h at 30 °C. Proteins were then resolved by SDS-PAGE and visualized by coomassie staining.
Tryptic MS/MS sequencing of crosslinked peptides from Parkin and 7
The coomassie stained SDS-PAGE band corresponding to the Parkin labeled with probe 7 was excised, dehydrated and resuspended using standard procedures. LC-MS/MS analysis was performed on an LTQ Orbitrap Velos instrument (Thermo Scientific) coupled to an Ultimate nanoflow HPLC system (Dionex). A gradient running from 3 % solvent A to 60 % solvent B over 45 min was applied (solvent A = 0.1 % formic acid in H2O; solvent B = 0.08 % formic acid in 80 % MeCN). Fragment ions were generated by CID and 1+ and 2+ precursor ions were excluded. Thermo .raw data was converted to .mgf format using the MSConvert software (ProteoWizard). Raw data was searched using the pLink software against UBE2L3* and Parkin sequences with trypsin specificity (up to 3 missed cleavages)59. A crosslinker monoisotopic mass of 307.1644 was manually added which accounted for the theoretical mass difference associated with formation of a bisthioether between 2 Cys residues together with the acrylate AVS, the triazole linkage and the tryptic Leu73 remnant from the Ub C-terminus.
Probe Pull-Down Assay
UBE2L3* was thioesterified with Ub by incubation with E1, ATP Mg2+ for 3.5 h at 30 °C. UBE2L3*-Ub was then purified by size exclusion chromatography at 4 °C (running buffer: 20 mM Na2HPO4 pH 6, 150 mM NaCl). 50 μL of probes 7, 8 and UBE2L3*~Ub (40 μM) were mixed with 10 μL of a 1:1 mixture of p-Parkin C431S/pUb (20 μM) and incubated with 5 μL Ni-NTA resin (Qiagen) on ice for 2 min. Resin was washed twice with buffer (10 mM Tris-HCl pH 8, 100 mM NaCl, 15 mM imidazole, 0.1 mM TCEP). Buffer was aspirated and protein eluted with elution buffer (10 mM Tris-HCl pH 8, 100 mM NaCl, 300 mM imidazole, 0.1 mM TCEP). Eluents were resolved by SDS-PAGE and visualized by coomassie staining and anti-Parkin immunoblotting.
Cell culture and lysis Protocol
Flp-In T-Rex HeLa (Life Technologies) stable cell lines (HeLa cells stably expressing untagged Parkin WT, S65A, H302A or IBR-RING2) were cultured (37 °C, 10 % CO2) in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% (v/v) Fetal Bovine Serum (FBS), 2.0 mM L-glutamine, 1X minimum essential medium (MEM, Life Technologies), 1X non-essential amino acids (NEAA, Life Technologies), 1.0 mM of sodium pyruvate (Life Technologies) and antibiotics (100 units mL-1 penicillin, 0.1 mg mL-1 streptomycin, 15 μg mL-1 blasticidin, 100 μg mL-1 hygromycin). At 80 % confluency cells were either untreated or treated with 10 μM CCCP60, and incubated for a further 3 h. Cells were rinsed with ice-cold PBS and extracted in lysis buffer (50 mM Tris-HCl pH 7.5, 5 μM EDTA, 0.27 M sucrose, 10 mM sodium 2-glycerophosphate, 0.2 mM phenylmethane sulfonyl fluoride (PMSF), 1.0 mM benzamidine, 1.0 mM sodium ortho-vanadate, 50 mM sodium fluoride, 5.0 mM sodium pyrophosphate and 10 μM TCEP). Lysis was carried out by sonication (Sonic & Materials INC, VC 100, Jencons Scientific LTD, CT, USA, 55% amplitude, 12 times (1 sec on, 1 sec off)) and then clarified by centrifugation at 4 °C for 30 min at 14,800 rpm. Supernatants were collected (total cell extracts) and protein concentration determined by Bradford assay.
SH-SY5Y (PHE Culture Collection) cells were grown in DMEM/F-12 media supplemented with 15% (v/v) FBS, 2.0 mM L-glutamine and antibiotics (100 units mL-1 penicillin, 100 μg mL-1 streptomycin). To depolarize mitochondria, cells were treated with 10 μM Carbonyl cyanide m-chlorophenyl hydrazine (CCCP), dissolved in DMSO, for the indicated time. Cell extracts were prepared as described for HeLa cells. All cells were cultured at 37 °C in a 5 % CO2 humidified atmosphere.
Testing of cell stocks for mycoplasma contamination was routinely carried out in accordance with departmental protocols.
ABPP of Total Cell Extracts
E2~Ub probes were diluted with buffer (20 mM Tris-HCl pH 7.5, 150 mM NaCl) and added to total cell extracts at a final concentration of 5.0 μM (final extract concentration 1.6 mg mL-1). Reactions were then incubated at 30 °C for the indicated time. Reactions were quenched by the addition of 4X LDS loading buffer (supplemented with betamercaptoethanol) and samples were analyzed by SDS-PAGE (4-12% NuPage gel) followed by immunoblotting.
Immunoblotting
Samples were resolved by SDS–PAGE (4–12% NuPage gel, Invitrogen) with MOPS or MES running buffer (without boiling) and transferred on to 0.45 μm nitrocellulose membranes (GE Healthcare Life Science). Membranes were blocked with PBS-T buffer (PBS + 0.1% Tween-20) containing 5% (w/v) non-fat dried skimmed milk powder (PBS-TM) at room temperature for 1 h. Membranes were subsequently probed with the indicated antibodies in PBS-T containing 5 % (w/v) Bovine Serum Albumin (BSA) overnight at 4 °C. Detection was performed using HRP-conjugated secondary antibodies in PBS-TM for 1 h at 23 °C. ECL Prime substrate (GE Life Sciences) was used for visualization in accordance with the manufacturers protocol.
Antibodies
His-tagged species were probed with 1:10000 anti-His primary antibody (Clontech, cat number: 631212). Anti-Parkin mouse monoclonal was obtained from Santa Cruz (sc-32282) at 1:2000 dilution (HeLa and SH-SY5Y samples); anti-Parkin phospho-serine 65 rabbit monoclonal antibody was obtained as previously described20, and used at 1:2500 dilution. GAPDH (14C10) rabbit mAb (HRP Conjugate, Cell Signaling Technology) was used at 1:5000 dilution.
Parkin Disease Mutant in vitro Profiling
The different recombinant Parkin mutants12, (1.0 μM, 1.535 μg) were incubated with Pediculus humanus corporis PINK1 (PhPINK1) or kinase dead PhPINK1 (0.77 μg), TCEP (1.0 mM), ATP (1.0 mM) and Ub (75 μM) in 1X Tris buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 5.0 mM MgCl2) at 30 °C for 1.0 h. After 1.0 hour incubation, fluorescent probe 9 was added (5.0 μM) and incubated for another 4 or 8 hours in the dark at 30 °C. Reactions were quenched by the addition of 4X LDS loading buffer (supplemented with betamercaptoethanol) for the indicated time point and samples were resolved by SDS-PAGE (4-12% NuPage gel). Gels were washed with water for 30 minutes and then visualized by scanning with a FLA-5100 imaging system (Filter: LPG, Laser: 532 nm, Voltage: 400, FUJIFILM Life Science). Experiments were carried out in triplicate and quantification was carried out using the Fiji software variant of ImageJ. For each replicate the ratio of the band size relative to the WT signal was taken. The mean of these values were plotted on a Log2 scale using Prism 6 (GraphPad Software).
Profiling of Patient-derived Fibroblasts
Low passage fibroblasts from a PD patient carrying a Q456X mutation in the PARK6 gene and from a healthy age-matched family member, were cultured as previously described50. Fibroblasts were obtained from skin biopsies following routine clinical procedures, underwritten informed consent and approval by a local ethics committee (Comité de Protection des Personnes “Ile de France”). Cell lysis, probe profiling with 7 and immunoblotting was carried out as described for SH-SY5Y extracts.
Supplementary Material
Acknowledgments
We are grateful to the MRC-PPU Proteomics Facility and the SLS DNA Sequencing Facility. We are also grateful to I. Gilbert for support with chemistry instrumentation and access to modelling software. We also thank J. W. Chin for critical reading of the manuscript. This work was funded by the Scottish Funding Council, the Medical Research Council and pharmaceutical companies supporting the Division of Signal Transduction Therapy (AstraZeneca, Boehringer-Ingelheim, GlaxoSmith- Kline, Merck KGaA, Janssen Pharmaceutica and Pfizer). K.B. is funded by an A J. Macdonald Menzies Charitable Trust Prize Studentship. M.M.K.M. is funded by a Wellcome Trust Senior Research Fellowship in Clinical Science (101022/Z/13/Z).
Footnotes
Author Contributions
S.V. conceived the project. Experiments were designed by S.V., M.M.K.M., K-C.P. K-C.P. carried out experiments with assistance from M.S., C.H., Y-C.L, P.M., K.B. and S.V. M.S. also carried out small molecule synthesis. N.T.W. performed cloning. J-C.C. and O.C. generated and provided patient fibroblasts. S.V. and M.M.K.M. wrote the manuscript with input from other authors.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Popovic D, Vucic D, Dikic I. Ubiquitination in disease pathogenesis and treatment. Nat Med. 2014;20:1242–1253. doi: 10.1038/nm.3739. [DOI] [PubMed] [Google Scholar]
- 2.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 3.Berndsen CE, Wolberger C. New insights into ubiquitin E3 ligase mechanism. Nat Struct Mol Biol. 2014;21:301–307. doi: 10.1038/nsmb.2780. [DOI] [PubMed] [Google Scholar]
- 4.Kitada T, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 5.Wenzel DM, Lissounov A, Brzovic PS, Klevit RE. UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature. 2011;474:105–108. doi: 10.1038/nature09966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pickrell AM, Youle RJ. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson’s disease. Neuron. 2015;85:257–273. doi: 10.1016/j.neuron.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaugule VK, et al. Autoregulation of Parkin activity through its ubiquitin-like domain. EMBO J. 2011;30:2853–2867. doi: 10.1038/emboj.2011.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trempe JF, et al. Structure of parkin reveals mechanisms for ubiquitin ligase activation. Science. 2013;340:1451–1455. doi: 10.1126/science.1237908. [DOI] [PubMed] [Google Scholar]
- 9.Riley BE, et al. Structure and function of Parkin E3 ubiquitin ligase reveals aspects of RING and HECT ligases. Nat Commun. 2013;4:1982. doi: 10.1038/ncomms2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wauer T, Komander D. Structure of the human Parkin ligase domain in an autoinhibited state. EMBO J. 2013;32:2099–2112. doi: 10.1038/emboj.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kazlauskaite A, et al. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem J. 2014;460:127–139. doi: 10.1042/BJ20140334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kazlauskaite A, et al. Phosphorylation of Parkin at Serine65 is essential for activation: elaboration of a Miro1 substrate-based assay of Parkin E3 ligase activity. Open Biol. 2014;4:130213. doi: 10.1098/rsob.130213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kane LA, et al. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol. 2014;205:143–153. doi: 10.1083/jcb.201402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koyano F, et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510:162–166. doi: 10.1038/nature13392. [DOI] [PubMed] [Google Scholar]
- 15.Kondapalli C, et al. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol. 2012;2:120080. doi: 10.1098/rsob.120080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shiba-Fukushima K, et al. PINK1-mediated phosphorylation of the Parkin ubiquitin-like domain primes mitochondrial translocation of Parkin and regulates mitophagy. Sci Rep. 2012;2:1002. doi: 10.1038/srep01002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ordureau A, et al. Quantitative Proteomics Reveal a Feedforward Mechanism for Mitochondrial PARKIN Translocation and Ubiquitin Chain Synthesis. Mol Cell. 2014;56:360–375. doi: 10.1016/j.molcel.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ordureau A, et al. Defining roles of PARKIN and ubiquitin phosphorylation by PINK1 in mitochondrial quality control using a ubiquitin replacement strategy. Proc Natl Acad Sci U S A. 2015;112:6637–6642. doi: 10.1073/pnas.1506593112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okatsu K, et al. Phosphorylated ubiquitin chain is the genuine Parkin receptor. J Cell Biol. 2015;209:111–128. doi: 10.1083/jcb.201410050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kazlauskaite A, et al. Binding to serine 65-phosphorylated ubiquitin primes Parkin for optimal PINK1-dependent phosphorylation and activation. EMBO Rep. 2015;16:939–954. doi: 10.15252/embr.201540352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wauer T, Simicek M, Schubert A, Komander D. Mechanism of phospho-ubiquitin-induced PARKIN activation. Nature. 2015;524:370–374. doi: 10.1038/nature14879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kazlauskaite A, Muqit MM. PINK1 and Parkin - mitochondrial interplay between phosphorylation and ubiquitylation in Parkinson’s disease. FEBS J. 2015;282:215–223. doi: 10.1111/febs.13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lazarou M, et al. PINK1 drives Parkin self-association and HECT-like E3 activity upstream of mitochondrial binding. J Cell Biol. 2013;200:163–172. doi: 10.1083/jcb.201210111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuda N, et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol. 2010;189:211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cravatt BF, Wright AT, Kozarich JW. Activity-based protein profiling: from enzyme chemistry to proteomic chemistry. Annu Rev Biochem. 2008;77:383–414. doi: 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- 26.Stanley M, et al. Orthogonal Thiol Functionalization at a Single Atomic Center for Profiling Transthiolation Activity of E1 Activating Enzymes. ACS Chem Biol. 2015;10:1542–1554. doi: 10.1021/acschembio.5b00118. [DOI] [PubMed] [Google Scholar]
- 27.Borodovsky A, et al. A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. EMBO J. 2001;20:5187–5196. doi: 10.1093/emboj/20.18.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Love KR, Pandya RK, Spooner E, Ploegh HL. Ubiquitin C-terminal electrophiles are activity-based probes for identification and mechanistic study of ubiquitin conjugating machinery. ACS Chem Biol. 2009;4:275–287. doi: 10.1021/cb9000348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smit JJ, Sixma TK. RBR E3-ligases at work. EMBO Rep. 2014;15:142–154. doi: 10.1002/embr.201338166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Komander D, Clague MJ, Urbé S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 31.Olsen SK, Capili AD, Lu X, Tan DS, Lima CD. Active site remodelling accompanies thioester bond formation in the SUMO E1. Nature. 2010;463:906–912. doi: 10.1038/nature08765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGouran JF, Gaertner SR, Altun M, Kramer HB, Kessler BM. Deubiquitinating enzyme specificity for ubiquitin chain topology profiled by di-ubiquitin activity probes. Chem Biol. 2013;20:1447–1455. doi: 10.1016/j.chembiol.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mulder MP, El Oualid F, ter Beek J, Ovaa H. A native chemical ligation handle that enables the synthesis of advanced activity-based probes: diubiquitin as a case study. Chembiochem. 2014;15:946–949. doi: 10.1002/cbic.201402012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haj-Yahya N, et al. Dehydroalanine-based diubiquitin activity probes. Org Lett. 2014;16:540–543. doi: 10.1021/ol403416w. [DOI] [PubMed] [Google Scholar]
- 35.Li G, Liang Q, Gong P, Tencer AH, Zhuang Z. Activity-based diubiquitin probes for elucidating the linkage specificity of deubiquitinating enzymes. Chem Commun (Camb) 2014;50:216–218. doi: 10.1039/c3cc47382a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borodovsky A, et al. Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chem Biol. 2002;9:1149–1159. doi: 10.1016/s1074-5521(02)00248-x. [DOI] [PubMed] [Google Scholar]
- 37.Niphakis MJ, Cravatt BF. Enzyme inhibitor discovery by activity-based protein profiling. Annu Rev Biochem. 2014;83:341–377. doi: 10.1146/annurev-biochem-060713-035708. [DOI] [PubMed] [Google Scholar]
- 38.Nguyen DP, et al. Genetic encoding and labeling of aliphatic azides and alkynes in recombinant proteins via a pyrrolysyl-tRNA Synthetase/tRNA(CUA) pair and click chemistry. J Am Chem Soc. 2009;131:8720–8721. doi: 10.1021/ja900553w. [DOI] [PubMed] [Google Scholar]
- 39.Weissman AM. Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol. 2001;2:169–178. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- 40.Lechtenberg BC, et al. Structure of a HOIP/E2~ubiquitin complex reveals RBR E3 ligase mechanism and regulation. Nature. 2016 doi: 10.1038/nature16511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sarraf SA, et al. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature. 2013;496:372–376. doi: 10.1038/nature12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kamadurai HB, et al. Insights into ubiquitin transfer cascades from a structure of a UbcH5B~ubiquitin-HECT(NEDD4L) complex. Mol Cell. 2009;36:1095–1102. doi: 10.1016/j.molcel.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stieglitz B, Morris-Davies AC, Koliopoulos MG, Christodoulou E, Rittinger K. LUBAC synthesizes linear ubiquitin chains via a thioester intermediate. EMBO Rep. 2012;13:840–846. doi: 10.1038/embor.2012.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin DY, Diao J, Zhou D, Chen J. Biochemical and structural studies of a HECT-like ubiquitin ligase from Escherichia coli O157:H7. J Biol Chem. 2011;286:441–449. doi: 10.1074/jbc.M110.167643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ekkebus R, et al. On terminal alkynes that can react with active-site cysteine nucleophiles in proteases. J Am Chem Soc. 2013;135:2867–2870. doi: 10.1021/ja309802n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lazarou M, et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kumar A, et al. Disruption of the autoinhibited state primes the E3 ligase parkin for activation and catalysis. EMBO J. 2015;34:2506–2521. doi: 10.15252/embj.201592337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sauve V, et al. A Ubl/ubiquitin switch in the activation of Parkin. EMBO J. 2015;34:2492–2505. doi: 10.15252/embj.201592237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Okatsu K, et al. A dimeric PINK1-containing complex on depolarized mitochondria stimulates Parkin recruitment. J Biol Chem. 2013;288:36372–36384. doi: 10.1074/jbc.M113.509653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lai YC, et al. Phosphoproteomic screening identifies Rab GTPases as novel downstream targets of PINK1. EMBO J. 2015;34:2840–2861. doi: 10.15252/embj.201591593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Katrun P, Chiampanichayakul S. PhI(OAc)2/KI-Mediated Reaction of Aryl Sulfinates with Alkenes, Alkynes, and α,β-Unsaturated Carbonyl Compounds: Synthesis of Vinyl Sulfones and β-Iodovinyl Sulfones. Eur J Org Chem. 2010;29:5633–5641. [Google Scholar]
- 52.Dadová J, et al. Vinylsulfonamide and acrylamide modification of DNA for cross-linking with proteins. Angew Chem Int Ed Engl. 2013;52:10515–10518. doi: 10.1002/anie.201303577. [DOI] [PubMed] [Google Scholar]
- 53.Virdee S, Ye Y, Nguyen DP, Komander D, Chin JW. Engineered diubiquitin synthesis reveals Lys29-isopeptide specificity of an OTU deubiquitinase. Nat Chem Biol. 2010;6:750–757. doi: 10.1038/nchembio.426. [DOI] [PubMed] [Google Scholar]
- 54.Cui C, Zhao W, Chen J, Wang J, Li Q. Elimination of in vivo cleavage between target protein and intein in the intein-mediated protein purification systems. Protein Express Purif. 2006;50:74–81. doi: 10.1016/j.pep.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 55.El Oualid F, et al. Chemical synthesis of ubiquitin, ubiquitin-based probes, and diubiquitin. Angew Chem Int Ed Engl. 2010;49:10149–10153. doi: 10.1002/anie.201005995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hong V, Presolski SI, Ma C, Finn MG. Analysis and optimization of copper-catalyzed azide-alkyne cycloaddition for bioconjugation. Angew Chem Int Ed Engl. 2009;48:9879–9883. doi: 10.1002/anie.200905087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Plechanovova A, Jaffray EG, Tatham MH, Naismith JH, Hay RT. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature. 2012;489:115–120. doi: 10.1038/nature11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sommer S, Weikart ND, Linne U, Mootz HD. Covalent inhibition of SUMO and ubiquitin-specific cysteine proteases by an in situ thiol-alkyne addition. Bioorg Med Chem. 2013;21:2511–2517. doi: 10.1016/j.bmc.2013.02.039. [DOI] [PubMed] [Google Scholar]
- 59.Yang B, et al. Identification of cross-linked peptides from complex samples. Nat Methods. 2012;9:904–906. doi: 10.1038/nmeth.2099. [DOI] [PubMed] [Google Scholar]
- 60.Narendra DP, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.