Graphical abstract
Method names: QuEChERS (quick, easy, cheap, effective, rugged and Safe); Solvent extraction; SPE (solvent phase extraction)
Keywords: QuEChERS, solid phase extraction (SPE), solvent extraction, honey, honey bee, pesticide, LC–MS/MS
Abstract
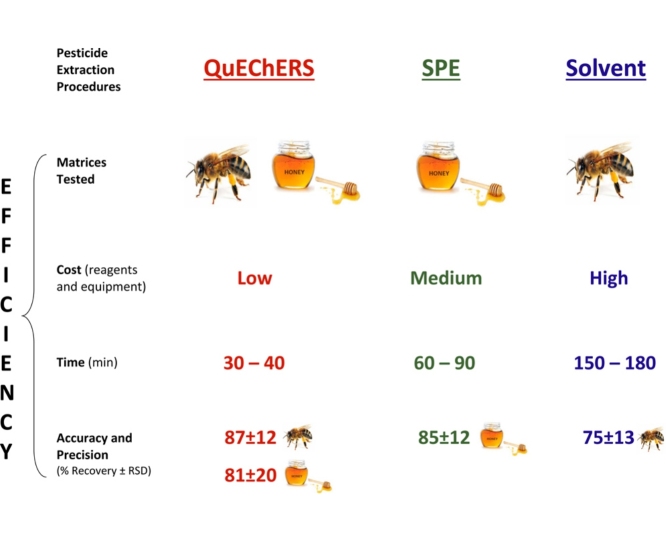
A comparison between QuEChERS and other pesticide extraction procedures for honey and honey bee matrices is discussed. Honey bee matrix was extracted by solvent based procedure whereas solid phase extraction was the protocol for the honey matrix. The citrate buffered QuEChERS method was used for both matrices. The methods were evaluated regarding cost (equipment and reagents), time, accuracy, precision, sensitivity and versatility. The results proved that the QuEChERS protocol was the most efficient method for the extraction of the selected pesticides in both matrices.
-
•
QuEChERS is the most economical and less time-consuming procedure.
-
•
SPE and solvent-based extraction procedures show equivalent recoveries to QuEChERS.
-
•
QuEChERS can be used to extract pesticide residues from both matrices.
Method details
QuEChERS approach for the extraction of pesticide residues in honey and honey bee matrices [1], [2], [3].
-
1)
Weigh 5 g of honey or honey bees into 50 mL centrifuge tubes and add 7.5 mL of water, 10 mL of acetonitrile, 6 g of MgSO4 and 1 g of NaCl. Homogenize the mixture immediately and then, centrifuge for 5 min at 300 rpm.
-
2)
Put 2 mL of the supernatant into another 15 mL centrifuge tube containing 50 mg C18, 50 mg PSA, and 150 mg MgSO4. Vortex the mix and centrifuge it for 5 min at 3000 rpm.
-
3)
Finally, filter the supernatant using a PTFE 13 mm × 0.22 μm into the autosampler vials for LC–MS analysis.
Solvent approach for the extraction of pesticide residues in honey bee matrix [4].
-
1)
Weigh 5 g of honey bees and pound thoroughly in a glass mortar. When homogenized place in a 250 mL flask and mix it vigorously for 10 min with 20 mL of acetone.
-
2)
Filter the mixture in a Kitassato flask through a Buchner funnel of 13 cm with a paper filter packed with a layer of Celite 545 (5–10 mm) and wash the filter cake with 20 mL of acetone.
-
3)
Prepare 100 mL, with 1% weight/volume (w/v) ammonium chloride and 2% volume/volume (v/v) ortophosphoric acid (85%) and add it to the filtrate. Allow it to stand for 30 min with occasional stirring and then filter with Celite 545.
-
4)
After filtration, dilute the sample with 200 mL of 2% aqueous sodium chloride (w/v) and extract twice with 100 mL of dichloromethane.
-
5)
Pass the resultant organic phase through a filter containing anhydrous sodium sulfate and evaporate it to dryness in a rotary evaporator at 35 °C.
-
6)
Dissolve the extract obtained from the honey bee samples in acetone, up to 2 mL, for GC analysis. For LC–MS determination, evaporate to dryness a 1-mL aliquot of the previous extract using a gentle stream of nitrogen and then dissolve it in the same volume of methanol.
Solid phase extraction (SPE) approach for the extraction of pesticide residues in honey matrix [5].
-
1)
Weigh honey (1.5 g) and mix it with 30 mL of hot water (<80 °C). Agitate by a stir bar for 10 min.
-
2)
Pre-condition an Oasis HLB cartridge [poly (divinylbenzene-co-N-pyrrolidone)] with 5 mL of methanol and 5 mL of Milli-Q water.
-
3)
Pass the mix through the cartridge at a flow rate of 10 mL min−1.
-
4)
Rinse the cartridge with 5 mL of Milli-Q water.
-
5)
Dry the cartridge under vacuum for 15 min.
-
6)
Elute the retained pesticides by passing 10 mL of methanol–dichloromethane (3:7).
-
7)
Evaporate the eluate to 0.5 mL using a gentle steam of nitrogen.
-
8)
Then, transfer it into 1-mL volumetric flask with methanol, obtaining a final extract in 100% methanol.
Liquid chromatography–mass spectrometry
Inject 5 μL of the extract in the LC–MS/MS according to the conditions already reported [1] and detailed below.
Ionization and fragmentation settings were optimized by direct injection of pesticide standard solutions. MS/MS was performed in the SRM mode using ESI in positive mode. For each compound, two characteristic product ions of the protonated molecule [M+H]+ were monitored, the first and most abundant one was used for quantification, while the second one was used as a qualifier. Collision energy and cone voltage were optimized for each pesticide (Table 1). Nitrogen was used as collision, nebulising and desolvation gas. The ESI conditions were: capillary voltage 4000 V, nebulizer 15 psi, source temperature 300 °C and gas flow 10 L min−1. In order to maximize sensitivity, dynamic MRM was used, with MS1 and MS2 at unit resolution and cell acceleration voltage of 7 eV for all the compounds.
Table 1.
Dynamic MRM conditions used for LC–MS/MS determination of pesticide residues.
| Target Pesticide | tRa (min) | Δ tRb | Precursor Ion | SRM1c | Fragd (V) | CEe (V) | SMR2f | Fragd (V) | CEe (V) | SMR2/SRM1 (%) (%RSD)g |
|---|---|---|---|---|---|---|---|---|---|---|
| Acetamiprid | 2.67 | 3.21 | 223 | 126 | 111 | 22 | 56 | 111 | 14 | 37.4 (12) |
| Acetochlor | 10.07 | 2 | 270 | 224 | 120 | 10 | 148 | 120 | 10 | 46.8 (22) |
| Alachlor | 10.07 | 2 | 270 | 238 | 80 | 15 | 162 | 80 | 10 | 50.4 (13) |
| Atrazine | 6.52 | 2.63 | 216 | 132 | 120 | 15 | 174 | 120 | 20 | 17.3 (14) |
| Atrazine-desethyl | 2.54 | 2.5 | 188 | 146 | 120 | 15 | 104 | 121 | 24 | 29.1 (15) |
| Atrazine-desisopropyl | 1.75 | 2.08 | 174 | 96 | 120 | 15 | 132 | 120 | 15 | 78.6 (13) |
| Azinphos-ethyl | 10.16 | 1.71 | 346 | 97 | 80 | 20 | 137 | 80 | 32 | 83.5 (12) |
| Azinphos-methyl | 8.17 | 1.24 | 318 | 125 | 80 | 8 | 132 | 80 | 12 | 85.4 (11) |
| Buprofezin | 14.5 | 1.1 | 306 | 201 | 120 | 10 | 116 | 120 | 15 | 64.6 (13) |
| Carbendazim | 4.54 | 4.74 | 192 | 160 | 95 | 17 | 132 | 95 | 25 | 11.4 (14) |
| Carbofuran | 4.37 | 2.91 | 222 | 123 | 120 | 10 | 165 | 70 | 15 | 98.0 (9.3) |
| Carbofuran-3-hydroxy | 1.85 | 2.48 | 255 | 163 | 70 | 5 | 220 | 70 | 15 | 90.8 (9) |
| Chlorfenvinphos | 11.74 | 1.61 | 359 | 155 | 120 | 10 | 127 | 120 | 15 | 63.8 (11) |
| Chlorpyriphos | 15.33 | 2.23 | 350 | 350 | 92 | 13 | 198 | 97 | 13 | 78.6 (14) |
| Coumpahos | 14.05 | 2.15 | 363 | 335 | 134 | 10 | 307 | 134 | 10 | 24.8 (10) |
| Diazinon | 11.77 | 1.89 | 305 | 169 | 128 | 17 | 153 | 128 | 21 | 66.3 (12) |
| Dichlofenthion | 14.68 | 2 | 315 | 259 | 120 | 10 | 287 | 120 | 5 | 44 (11) |
| Dimethoate | 2.06 | 2.59 | 230 | 199 | 80 | 10 | 171 | 80 | 5 | 45.3 (12) |
| Diuron | 7.5 | 1.25 | 233 | 72 | 120 | 20 | 160 | 120 | 20 | 3.2 (13) |
| DMF | 5.14 | 4.5 | 150 | 132 | 111 | 10 | 107 | 111 | 15 | 41.6 (16) |
| Ethion | 14.88 | 1.23 | 385 | 199 | 80 | 5 | 171 | 80 | 15 | 35.3 (11) |
| Fenitrothion | 10.03 | 1.18 | 278 | 125 | 140 | 15 | 109 | 121 | 12 | 95.5 (12) |
| Fenthion | 11.51 | 1.83 | 279 | 247 | 114 | 5 | 169 | 114 | 13 | 76.6 (10) |
| Fipronil | 13.33 | 2.85 | 437 | 368 | 150 | 15 | 290 | 150 | 25 | 21.8 (11) |
| Flumethrin | 18.53 | 1.85 | 527 | 267 | 50 | 10 | 239 | 50 | 10 | 48.3 (18) |
| Fluvalinate | 18.11 | 1.81 | 503 | 208 | 50 | 10 | 181 | 50 | 26 | 73.4 (10) |
| Hexythiazox | 15.11 | 1.15 | 353 | 228 | 120 | 20 | 168 | 120 | 10 | 67.4 (9) |
| Imazalil | 11.4 | 1.71 | 297 | 159 | 120 | 20 | 201 | 120 | 15 | 56 (14) |
| Imidacloprid | 1.61 | 1.96 | 256 | 209 | 80 | 10 | 175 | 80 | 10 | 75 (11) |
| Isoproturon | 6.83 | 2.37 | 207 | 72 | 120 | 20 | 165 | 120 | 10 | 16.8 (12) |
| Malathion | 9.36 | 1.96 | 331 | 99 | 80 | 10 | 127 | 80 | 5 | 98.5 (4) |
| Methiocarb | 8.64 | 1.93 | 226 | 121 | 80 | 5 | 169 | 80 | 10 | 66.6 (11) |
| Metholachlor | 10.49 | 2.04 | 284 | 252 | 120 | 15 | 176 | 120 | 10 | 10 (14) |
| Molinate | 9.41 | 1.98 | 188 | 126 | 80 | 20 | 55 | 80 | 10 | 61.7 (11) |
| Omethoate | 1.06 | 2.67 | 214 | 125 | 80 | 5 | 183 | 80 | 20 | 72.3 (12) |
| Parathion-ethyl | 11.11 | 1.91 | 292 | 236 | 88 | 4 | 264 | 88 | 8 | 45.5 (13) |
| Parathion-methyl | 8.17 | 1.5 | 264 | 125 | 120 | 20 | 232 | 110 | 5 | 34.5 (13) |
| Prochloraz | 12.08 | 1.91 | 376 | 308 | 80 | 10 | 266 | 80 | 10 | 14.3 (9) |
| Propanil | 8.6 | 2.01 | 218 | 162 | 120 | 20 | 127 | 120 | 15 | 92.4 (11) |
| Propazine | 8.74 | 2 | 230 | 146 | 120 | 15 | 188 | 120 | 20 | 93.3 (14) |
| Pyriproxyfen | 14.78 | 1.33 | 322 | 227 | 120 | 10 | 185 | 120 | 10 | 36.1 (12) |
| Simazine | 4.53 | 1.76 | 202 | 124 | 120 | 20 | 132 | 120 | 20 | 93.8 (12) |
| Tebuconazole | 13.82 | 2.87 | 308 | 125 | 95 | 25 | 70 | 95 | 21 | 6.6 (11) |
| Terbumeton | 10.98 | 2.89 | 226 | 170 | 95 | 17 | 114 | 95 | 25 | 13.8 (14) |
| Terbumeton-desethyl | 6.69 | 3.76 | 198 | 142 | 90 | 13 | 86 | 90 | 25 | 31.7 (12) |
| Terbuthylazine | 11.1 | 3.01 | 230 | 174 | 95 | 13 | 96 | 95 | 25 | 16.4 (13) |
| Terbuthylazine-2-hydroxy | 6.92 | 3.28 | 212 | 156 | 95 | 13 | 86 | 95 | 25 | 28 (13) |
| Terbuthylazine-desethyl | 6.98 | 2.81 | 202 | 146 | 95 | 13 | 79 | 95 | 25 | 13.2 (14) |
| Terbutryn | 10.63 | 1.2 | 242 | 186 | 120 | 20 | 71 | 120 | 15 | 4.6 (14) |
| Thiabendazole | 5.06 | 3.5 | 202 | 175 | 95 | 25 | 131 | 95 | 25 | 29.1 (18) |
| Thiamethoxam | 2 | 2.58 | 292 | 211 | 78 | 10 | 132 | 78 | 10 | 21.3 (11) |
| Tolclofos-methyl | 12.13 | 1.71 | 301 | 125 | 115 | 12 | 269 | 120 | 15 | 73.8 (19) |
tR = retention time.
Δ tR = delta retention time, that is the centered retention time window.
SRM1 = selected product ion for quantification.
Frag = Fragmentor.
CE = Collision energy.
SRM2 = selected product ion for qualification.
(%RSD) = relative standard deviation of the ratio SRM2/SRM1, calculated from mean values obtained from the matrix-matched calibration curves.
Quality assurance/quality control (QA/QC)
In order to compare QuEChERS to other routine procedures, methods were validated according to the European Union Guideliness [6]. Furthermore, the main elements of uncertainty as the amount of sample used for a determination, the recovery value of the analytical procedure and the repeatability of determinations for a true sample [7], were considered through the validation process (for detailed information of the validation parameters, see Supplementary material Table S1 and S2).
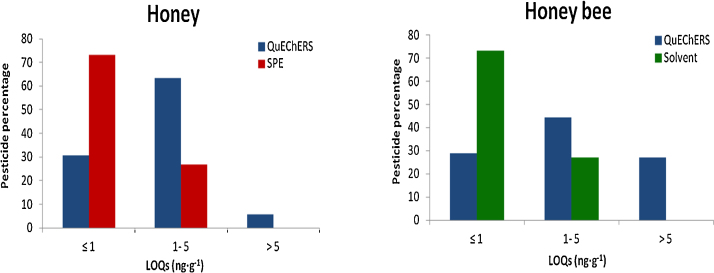
The sensitivity of the method was estimated by establishing the limits of quantification (LOQs) (Fig. 1). The LOQs were determined in pure solvent and in spiked honey and honey bees samples. LOQs were calculated as the lowest concentration or mass of the analyte that has been validated with acceptable accuracy by applying the complete analytical method. LOQs were from 0.2 to 10 ng g−1 and from 0.03 to 10 ng g−1 for honey and honey bee matrices respectively. Solvent and SPE methods were slightly more sensitive than QuEChERS approach.
Fig. 1.
Limits of quantitation (LOQs) of QuEChERS, SPE and solvent methods in honey and honey bee matrices.
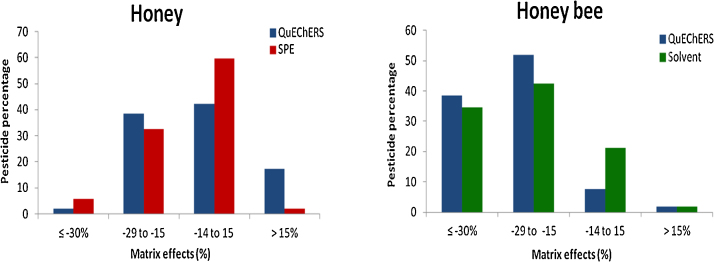
Matrix effects were evaluated by comparing the slope of the previous calibration curve and the slope of that prepared in the extract of honey or honey bee matrix with six concentration levels of standard solutions (Fig. 2). Matrix effects were mostly suppressive in both matrices and ranged from −60 to 50 and from −60 to 35% in honey and honey bee matrices, respectively.
Fig. 2.
Matrix effects of QuEChERS, SPE and solvent methods in honey and honey bee matrices.
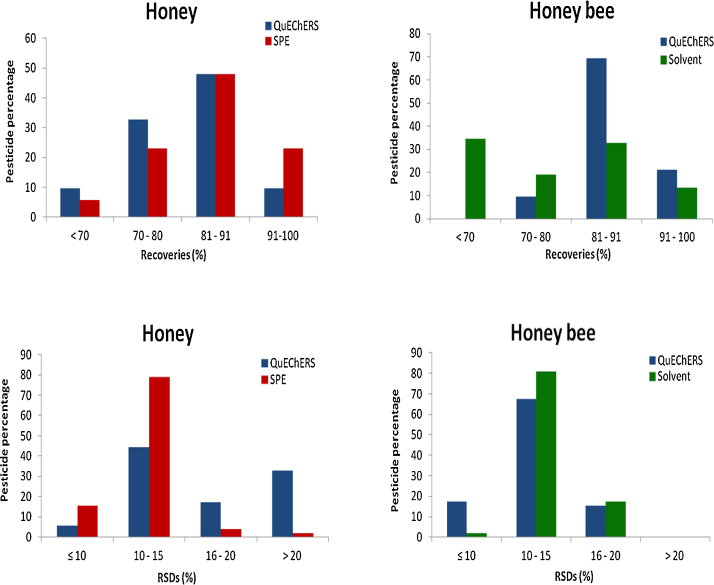
Mean recovery (as accuracy) and relative standard deviation (as precision) were evaluated by spiking the samples at the LOQ and 10 x LOQ, with a minimum of 5 replicates (Fig. 3). Recovery values of honey bee matrix were from 34 to 96%, whereas RSDs were in all cases <20%. Honey matrix showed recoveries that ranged from 30 to 96% and RDS were <20% except for 17 compounds that were from 21 to 42%. QuEChERS approach showed better results than solvent method in the honey bee matrix while SPE was slightly better both in accuracy and precision than QuEChERS extraction procedure for honey.
Fig. 3.
Accuracy (Recoveries) and precision (RSDs) validation parameters of QuEChERS, SPE and solvent methods in honey and honey bee matrices.
Additional information
The use of pesticides in agricultural cropping systems is often discussed as a factor influencing honey bee health [1]. Furthermore, honey, which is considered a healthy natural product, can be contaminated during its production from both agricultural and beekeeping practices [8], [5]. The development of extraction procedures able to process samples in an economic way is crucial.
This paper presents some of the currently applied sample preparation methods for the separation and pre-concentration of pesticides in honey and honey bee samples. The composition of honey and honey bees is very different but both are complex matrices. In order to achieve an accurate and reliable analytical result, an efficient pre-concentration/separation step is usually required prior to determination, even when such a sensitive detection method as LC–MS/MS is used.
From an analytical point of view, honey can be considered as a highly concentrated sugar solution (mostly fructose). Then, after water dilution it can be extracted using protocols similar to those applied to water as SPE. The protocol described here requires a medium cost in reagent and equipment because the SPE sorbents involve a high cost. The extraction of a sample requires between 60 and 90 min, being evaporation the step that takes more time. The performance of the method provides the best sensitivity and lower matrix effects.
On the contrary, honey bees are rich in lipids and proteins, requiring most sophisticated and extensive sample preparation methods. Traditional methods as the solvent approach are long, tedious and require high amounts of expensive organic solvents [4]. Considering the use of reagents and equipment this method has high cost, requires between 150 and 180 min to process a sample and provides recoveries slightly lower for more polar pesticides
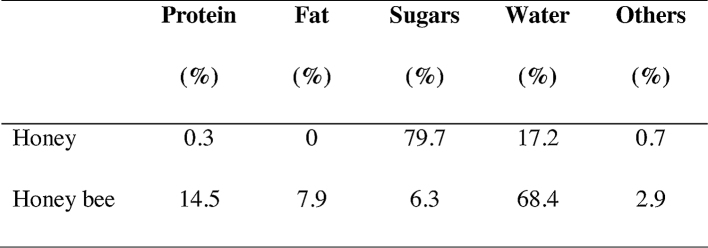
The results pointed out that QuEChERS approach is used in many different matrices as hive products (beeswax, pollen, honey, honey bee) [9], [3], [10]. Honey and honey bee composition (Fig. 4) evidence the versatility of the QuEChERS method compared to other extraction procedures as those used in the present work. Appropriate results in terms of specificity, selectivity, accuracy and sensitivity, low cost and quickness make QuEChERS a suitable procedure for determining pesticides in less studied hive matrices as royal jelly and propolis. Furthermore, QuEChERS approach meets important components of green analytical chemistry [11] due to its small amounts of solvent needed compared to the traditional methods.
Fig. 4.
Acknowledgements
This work has been supported by the agreement No. OTR2013-11072ASESO between the Agrupación de Defensa Sanitaria Apicola (apiADS) and the Environmental and Food Safety Research Group (SAMA-UV), Department of Medicine Preventive, Faculty of Pharmacy, University of Valencia (Spain).
MethodsX thanks the reviewers of this article for taking the time to provide valuable feedback.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.mex.2016.05.005.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.Calatayud-Vernich P., Calatayud F., Simó E., Suarez-Varela M.M., Picó Y. Influence of pesticide use in fruit orchards during blooming on honeybee mortality in 4 experimental apiaries. Sci. Total Environ. 2016;541:33–41. doi: 10.1016/j.scitotenv.2015.08.131. [DOI] [PubMed] [Google Scholar]
- 2.Wiest L., Bulete A., Giroud B., Fratta C., Amic S., Lambert O. Multi-residue analysis of 80 environmental contaminants in honeys, honeybees and pollens by one extraction procedure followed by liquid and gas chromatography coupled with mass spectrometric detection. J. Chromatogr. A. 2011;1218:5743–5756. doi: 10.1016/j.chroma.2011.06.079. [DOI] [PubMed] [Google Scholar]
- 3.Niell S., Jesus F., Perez C., Mendoza Y., Diaz R., Franco J. QuEChERS adaptability for the analysis of pesticide residues in beehive products seeking the development of an agroecosystem sustainability monitor. J. Agric. Food. Chem. 2015;63:4484–4492. doi: 10.1021/acs.jafc.5b00795. [DOI] [PubMed] [Google Scholar]
- 4.Ghini S., Fernandez M., Pico Y., Marin R., Fini F., Manes J. Occurrence and distribution of pesticides in the province of Bologna, Italy, using honeybees as bioindicators. Arch. Environ. Contam. Toxicol. 2004;47:479–488. doi: 10.1007/s00244-003-3219-y. [DOI] [PubMed] [Google Scholar]
- 5.Blasco C., Vazquez-Roig P., Onghena M., Masia A., Pico Y. Analysis of insecticides in honey by liquid chromatography–ion trap-mass spectrometry: comparison of different extraction procedures. J. Chromatogr. A. 2011;1218:4892–4901. doi: 10.1016/j.chroma.2011.02.045. [DOI] [PubMed] [Google Scholar]
- 6.SANCO/12571 . European Commission; Brussels, Belgium: 2013. Guidance Document on Analytical Quality Control and Validation Procedures for Pesticide Residues Analysis in Food and Feed, Supersedes SANCO/12495/2011, Implemented by 01/01/2014. [Google Scholar]
- 7.Konieczka P., Namieśnik J. Estimating uncertainty in analytical procedures based on chromatographic techniques. J. Chromatogr. A. 2010;1217:882–891. doi: 10.1016/j.chroma.2009.03.078. [DOI] [PubMed] [Google Scholar]
- 8.Barganska Z., Slebioda M., Namiesnik J. Honey bees and their products: bioindicators of environmental contamination. Crit. Rev. Environ. Sci. Technol. 2016;46:235–248. [Google Scholar]
- 9.Barganska Z., Slebioda M., Namiesnik J. Determination of pesticide residues in honeybees using modified QUEChERS sample work-up and liquid chromatography–tandem mass spectrometry. Molecules. 2014;19:2911–2924. doi: 10.3390/molecules19032911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niell S., Cesio V., Hepperle J., Doerk D., Kirsch L., Kolberg D. QuEChERS-Based method for the multiresidue analysis of pesticides in beeswax by LC–MS/MS and GCxGC-TOF. J. Agric. Food. Chem. 2014;62:3675–3683. doi: 10.1021/jf405771t. [DOI] [PubMed] [Google Scholar]
- 11.Gałuszka A., Migaszewski Z., Namieśnik J. The 12 principles of green analytical chemistry and the significance mnemonic of green analytical practices. TrAC Trends Anal. Chem. 2013;50:78–84. [Google Scholar]
- 12.Bogdanov S., Jurendic T., Sieber R., Gallmann P. Honey for nutrition and health: a review. J. Am. Coll. Nutr. 2008;27:677–689. doi: 10.1080/07315724.2008.10719745. [DOI] [PubMed] [Google Scholar]
- 13.Rumpold B.A., Schlueter O.K. Nutritional composition and safety aspects of edible insects. Mol. Nutr. Food Res. 2013;57:802–823. doi: 10.1002/mnfr.201200735. [DOI] [PubMed] [Google Scholar]
- 14.Studier E.H., Sevick S.H. Live mass, water-content, nitrogen and mineral levels in some insects from South-Central Lower Michigan. Comp. Biochem. Physiol. A: Physiol. 1992;103:579–595. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.